Abstract
Human epidermal growth factor receptor 2 (HER2) is overexpressed in 20–25% of breast cancers. Increased HER2 expression is an adverse prognostic factor and correlates with decreased patient survival. HER2-positive (HER2(+)) breast cancer is treated with trastuzumab. Unfortunately, some patients are intrinsically refractory to therapy, and many who do respond initially become resistant within 1 year. Understanding the molecular mechanisms underlying HER2 signaling and trastuzumab resistance is essential to reduce breast cancer mortality. IQGAP1 is a ubiquitously expressed scaffold protein that contains multiple protein interaction domains. By regulating its binding partners IQGAP1 integrates signaling pathways, several of which contribute to breast tumorigenesis. We show here that IQGAP1 is overexpressed in HER2(+) breast cancer tissue and binds directly to HER2. Knockdown of IQGAP1 decreases HER2 expression, phosphorylation, signaling, and HER2-stimulated cell proliferation, effects that are all reversed by reconstituting cells with IQGAP1. Reducing IQGAP1 up-regulates p27, and blocking this increase attenuates the growth inhibitory effects of IQGAP1 knockdown. Importantly, IQGAP1 is overexpressed in trastuzumab-resistant breast epithelial cells, and reducing IQGAP1 both augments the inhibitory effects of trastuzumab and restores trastuzumab sensitivity to trastuzumab-resistant SkBR3 cells. These data suggest that inhibiting IQGAP1 function may represent a rational strategy for treating HER2(+) breast carcinoma.
Keywords: Breast Cancer, Calmodulin, Cancer Therapy, Signal Transduction, Tumor Marker, IQGAP1, Trastuzumab
Introduction
The ErbB family of transmembrane receptors has four members, namely epidermal growth factor receptor (EGFR)2/HER1/ErbB1, HER2/ErbB2, HER3/ErbB3, and HER4/ErbB4 (1). Each member is a typical receptor-tyrosine kinase and comprises an extracellular ligand binding region, a single membrane-spanning region, and a cytoplasmic C-terminal region that contains the tyrosine kinase domain. By controlling intracellular signaling pathways that govern essential cellular processes, including proliferation, migration, metabolism and survival, ErbB receptors have fundamental roles during development and in adult physiology (2). In addition, EGFR and HER2 in particular have been implicated in the pathogenesis of several types of human cancer (3). For example, HER2 is overexpressed in 20–25% of breast cancers (4), and increased HER2 expression correlates strongly with a shorter time to relapse and a decrease in overall survival (5).
Unlike other members of the ErbB family, HER2 does not bind a specific ligand directly. Instead, it exists in a constitutively phosphorylated state (6, 7). Phosphorylated (active) HER2 signals primarily through the phosphatidylinositol 3-kinase (PI3K)/AKT and mitogen-activated protein kinase (MAPK) pathways (8), both of which participate in HER2-stimulated tumorigenesis. HER2 signaling induces degradation of p27, a cyclin-dependent kinase (CDK) inhibitor (9–12). p27 inhibits CDK2 activity, resulting in G1 arrest and apoptosis (13). Activation of HER2, therefore increases cell proliferation, transformation, and oncogenesis (8).
The importance of HER2 to the pathogenesis of human breast cancer has led to an enormous effort to develop HER2-targeted therapeutics. The first agent licensed by the United States Food and Drug Administration for the treatment of HER2(+) breast cancer was trastuzumab (herceptin). Trastuzumab is a recombinant humanized monoclonal antibody directed against domain IV of the HER2 extracellular region (14). The mechanisms by which trastuzumab induces regression of HER2(+) tumors are not completely defined, but several hypotheses have been proposed. These are based on data from preclinical models and include internalization and degradation of HER2 (15, 16), inhibition of signaling pathways downstream of HER2 (17), stimulation of cell cycle arrest through the induction of p27 (18, 19), and antibody-dependent cellular cytotoxicity (17). The initial clinical trials of trastuzumab evaluated its use as a single agent for the treatment of metastatic breast cancer and demonstrated responses ranging from 12 to 34% with a median response duration of 9 months (20–22). When combined with adjuvant chemotherapy, the disease-free and overall survival rate for patients with early stage breast cancer is significantly improved (23). Nevertheless, ∼15% of women in this category still progress to metastatic disease. Moreover, the majority of patients with metastatic breast cancer who initially respond to trastuzumab-based treatment develop resistance within 1 year (24). Several mechanisms have been proposed to contribute to trastuzumab resistance, including accumulation of truncated HER2 (p95HER2) (25), which lacks an extracellular domain, constitutive activation of the PI3K/AKT pathway as a result of phosphatase and tensin homolog (PTEN) deficiency (26) or PIK3CA mutation (27), or the overexpression of other receptor-tyrosine kinases (such as insulin-like growth factor-1 receptor (28, 29)). Understanding the molecular mechanisms underlying trastuzumab resistance is critical to improving the survival of patients with HER2(+) tumors.
IQGAP1 is a ubiquitously expressed 189-kDa scaffold protein that contains multiple protein interaction domains (30). Moving from the N to the C terminus, these include a calponin homology domain, a polyproline binding domain, four IQ (calmodulin binding) motifs, and a RasGAP-related region. IQGAP1 binds multiple proteins thereby integrating diverse signaling pathways. Proteins that are known to bind IQGAP1 include Rac1/Cdc42 (but not RhoA or H-Ras), actin, calmodulin, E-cadherin, β-catenin, components of the MAPK pathway, adenomatous polyposis coli, vascular endothelial growth factor receptor 2 (VEGFR2) (30) and EGFR (31). By interacting with these proteins, IQGAP1 regulates multiple cellular activities, such as cytoskeletal organization, cell-cell adhesion, cell migration, gene transcription, and signal transduction. For example, binding of IQGAP1 to β-catenin both disrupts the E-cadherin-catenin complex, inhibiting epithelial cell-cell adhesion (32), and increases β-catenin-mediated transcriptional activation (33). Furthermore, IQGAP1-VEGFR2 interaction modulates reactive oxygen species-dependent signaling by vascular endothelial growth factor (34).
Accumulating evidence strongly supports a role for IQGAP1 in tumorigenesis (35, 36). More than 50% of the identified IQGAP1 binding partners have defined roles in neoplastic transformation and/or tumor progression, and many cellular functions regulated by IQGAP1 are important in cancer biology (35, 36). Genomic and proteomic studies of primary tumors also provide compelling data. For example, the Iqgap1 gene is up-regulated in oligodendroglioma (37) and colorectal (38) and lung (39) carcinomas. Moreover, IQGAP1 protein is overexpressed in squamous cell (40) and hepatocellular (41) carcinomas, astrocytoma (42), and aggressive forms of gastric cancer (43). The relevance to tumor biology of many of the known IQGAP1 binding partners coupled with the existing evidence for its role in neoplasia discussed above strongly suggests that IQGAP1 is an oncogene. Consistent with this hypothesis, overexpression of IQGAP1 stimulates tumorigenesis of human breast epithelial cells (44). We report here that IQGAP1 regulates HER2 expression, phosphorylation, and signaling. Furthermore, we show that IQGAP1 is overexpressed in trastuzumab-resistant human breast epithelial cells and that specific knockdown of IQGAP1 both enhances the inhibitory effects of trastuzumab in vitro and abrogates trastuzumab resistance.
EXPERIMENTAL PROCEDURES
Materials
SkBR3 cells were obtained from American Type Culture Collection (Manassas, VA). All tissue culture reagents were obtained from Invitrogen. Trastuzumab was generously provided by Ian Krop (Dana-Farber Cancer Institute, Boston, MA). Anti-pHER2 (Tyr1221/Tyr1222), anti-HER2, anti-pAKT (Ser473), anti-AKT, anti-pERK (Thr202/Tyr204), anti-ERK, anti-p27, and anti-β-tubulin antibodies and a glutathione S-transferase (GST) fusion construct encoding the entire HER2 intracellular domain (designated GST-HER2; residues 676–1255 (numbering corresponds to the immature sequence)) were obtained from Cell Signaling Technology (Danvers, MA). The anti-IQGAP1 polyclonal antibodies have been previously characterized (45). Secondary antibodies for enhanced chemiluminescence detection were obtained from GE Healthcare. GST-Cdc42V12 was generously provided by Ann Ridley (Kings College London). siRNA oligomers (with dTdT overhangs at each 3′ terminus) against IQGAP1 were obtained from Sigma (designated siIQ12) and Dharmacon (Lafayette, CO; designated siIQ14). The siRNA oligomer (with dTdT overhangs at each 3′ terminus) against p27 (designated sip27) was obtained from Sigma, and that against renilla luciferase (designated siRen) was synthesized by Invitrogen. The siRNA sequences are as follows: siIQ12, 5′-GGCAAUUUAAAUGACCCAA-3′ (sense (S)), 5′-UUGGGUCAUUUAAAUUGCC-3′ (antisense (AS)); siIQ14, 5′-GGCAUAUCAAGAUCGGUUA-3′ (S), 5′-UAACCGAUCUUGAUAUGCC-3′ (AS); sip27, 5′-GAAUGGACAUCCUGUAUAA-3′ (S), 5′-UUAUACAGGAUGUCCAUUC-3′ (AS); siRen, 5′-AAACAUGCAGAAAAUGCUG-3′ (S), 5′-CAGCAUUUUCUGCAUGUUU-3′ (AS). siIQ12-resistant IQGAP1 was made by introducing 3 silent mutations (A4511G, T4514C and C4517T) into wild-type IQGAP1 using the QuikChange II site-directed mutagenesis system (Agilent Technologies, Santa Clara, CA). Unless otherwise stated, all other reagents were of standard analytical grade.
Patient Tissue
A total of 42 routinely processed formalin-fixed paraffin-embedded surgical resection specimens (male/female patient ratio, 0/39; median patient age, 58 years) were selected from the tissue archives of the Department of Pathology, Brigham and Women's Hospital, between the years 1998 and 2002. These included 19 HER2(+) (3+ by immunohistochemistry and/or amplified by fluorescence in situ hybridization (FISH)) invasive breast carcinomas, 20 HER2(−) invasive breast carcinomas (18 hormone receptor-positive and 2 triple-negative tumors), and 3 normal breast specimens (supplemental Table S1). Hematoxylin and eosin-stained sections were reviewed by a board-certified pathologist (D. A. Dillon, Brigham and Women's Hospital) for confirmation of the diagnoses. Patient age and hormone receptor status were extracted from the corresponding surgical pathology reports (supplemental Table S1). This study was approved by the Institutional Review Board of Brigham and Women's Hospital.
Immunohistochemistry
Formalin-fixed paraffin-embedded blocks were cut into 5-μm-thick tissue sections, and slides were prepared using standard techniques. Mounted tissue sections were baked for 20 min at 60 °C, deparaffinized in xylene, and rehydrated through graded alcohols. Antigens were retrieved by heating in 1 μm sodium citrate (pH 6.0) in a pressure cooker for 30 s at 125 °C. Nonspecific staining was blocked using Dako Protein Block (Dako, Carpinteria, CA) according to the manufacturer's instructions. Rabbit polyclonal anti-IQGAP1 (dilution 1:2,000) and rabbit polyclonal anti-HER2 (dilution 1:100) antibodies were diluted in Dako antibody diluent and incubated with tissue sections for 1 h at room temperature. Staining was visualized using Dako Envision and developed with a DAB Chromogen substrate. Immediately after visualization, sections were dipped in DAB Enhancer, counterstained with hematoxylin, dehydrated through graded alcohols and xylene, and mounted. Appropriate positive and negative controls were used throughout all staining and interpretation.
Immunostaining Interpretation
Immunohistochemical staining was evaluated by a board-certified pathologist (D. A. Dillon). IQGAP1 immunostaining was scored as follows: 0 (no immunoreactivity), 1+ (weak immunoreactivity), 2+ (moderate immunoreactivity in greater than 10% of tumor cells), or 3+ (strong immunoreactivity in greater than 30% of tumor cells). A score of 0 or 1+ was considered negative, whereas 2+ or 3+ was considered positive.
Preparation of Fusion Proteins
GST-IQGAP1, GST-HER2, and GST-Cdc42V12 were expressed in Escherichia coli and isolated using glutathione-Sepharose essentially as previously described (45). Where indicated, the GST tag was cleaved from GST-IQGAP1 using tobacco etch virus protease as previously described (46).
TNT Product Production
[35S]Methionine-labeled TNT (transcription and translation) products were produced with the TNT Quick Coupled Transcription/Translation system (Promega, Madison, WI) according to the manufacturer's instructions. Briefly, 1–2 μg of pcDNA3-IQGAP1, -IQGAP1-N, -IQGAP1-IQ, -IQGAP1-C, or -IQGAP1ΔIQ was incubated with 40 μl of TNT Quick Master Mix and 20 μCi of [35S]methionine (PerkinElmer Life Sciences) for 1 h at 30 °C. TNT product production was confirmed by SDS-PAGE and autoradiography.
In Vitro Binding Assays
For in vitro binding experiments using pure proteins, purified untagged IQGAP1 (>90% pure) was incubated with GST alone, GST-HER2, or calmodulin-Sepharose in 500 μl of buffer A (50 mm Tris (pH 7.4), 150 mm NaCl, 1 mm EGTA, and 1% (v/v) Triton X-100) for 3 h at 4 °C. GST- or GST-HER2-bound complexes were isolated using glutathione-Sepharose beads, washed 6 times in buffer A, resolved by SDS-PAGE, and processed by Western blotting. Calmodulin-Sepharose-bound complexes were isolated by centrifugation and washed and processed in the same way. For in vitro binding experiments using the [35S]methionine-labeled TNT products, the IQGAP1 constructs described in the previous paragraph were incubated with GST-HER2, GST-Cdc42V12, or GST alone in 500 μl of buffer A for 3 h at 4 °C. Complexes were isolated using glutathione-Sepharose, washed as above, resolved by SDS-PAGE, and processed by autoradiography.
Cell Culture and Transfection
SkBR3 cells, a malignant human breast epithelial cell line that overexpresses HER2, were maintained in McCoy's 5A medium supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin/streptomycin. All cultures were regularly confirmed to be free of mycoplasma contamination. Trastuzumab-resistant SkBR3 cells (termed SkBR3TR) were derived by culturing SkBR3 cells in the presence of 21 μg/ml trastuzumab for 8 months essentially as previously described (47). SkBR3TR cells were maintained in McCoy's 5A medium supplemented with 10% (v/v) fetal bovine serum, 1% (v/v) penicillin/streptomycin, and 21 μg/ml trastuzumab. siRNA transfections were performed using Lipofectamine 2000 (Invitrogen) according the manufacturer's instructions. Briefly, cells were plated in 6-well plates at a density of 2 × 105 cells/well and allowed to attach overnight. The following day each well was transfected with 250 pm siRNA in a complex volume of 2.5 ml (yielding a final siRNA concentration of 100 nm). Untransfected cells were included in all experiments and compared with cells transfected with control siRNA (siRen). No significant differences were observed (data not shown). Transfection of siRNA-resistant IQGAP1 was performed using X-tremeGENE HP (Roche Applied Science) according to the manufacturer's instructions. Untransfected cells were included in all experiments and compared with cells transfected with vector. No significant differences were observed (data not shown).
Immunoprecipitation and GST Pulldown Assays
SkBR3 cells were plated in 100-mm dishes at a density of 5 × 106 cells/dish and allowed to attach overnight. The following day cells were washed twice with ice-cold phosphate-buffered saline (PBS) and lysed in lysis buffer (50 mm Tris (pH 8.0), 100 mm NaF, 30 mm Na4P2O7, 2 mm Na2MoO4, 5 mm EDTA, and 2 mm Na3VO4) supplemented with 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mm phenylmethylsulfonyl fluoride (together designated buffer B). Immunoprecipitation was performed essentially as previously described (48, 49). Briefly, clarified cell lysates were equalized for protein concentration using the modified Bradford Assay (Bio-Rad), and equal amounts of protein were incubated with non-immune rabbit serum or anti-IQGAP1 or anti-HER2 polyclonal antibodies for 3 h at 4 °C. Immune complexes were isolated using protein A-Sepharose beads (GE Healthcare), washed six times in buffer B, resolved by SDS-PAGE, and processed by Western blotting. GST pulldown assays were performed essentially as previously described (50). Briefly, equal amounts of protein from SkBR3 cell lysates were incubated with GST alone, GST-HER2, or calmodulin-Sepharose in 500 μl of buffer B for 3 h at 4 °C. GST- or GST-HER2-bound complexes were isolated using glutathione-Sepharose beads, washed six times in buffer B, resolved by SDS-PAGE, and processed by Western blotting. Calmodulin-Sepharose-bound complexes were isolated by centrifugation, then washed and processed in the same way.
Measurement of HER2 Phosphorylation and Signaling
36 h after transfection, cells were serum-starved in the presence of vehicle (double distilled H2O) or 21 μg/ml trastuzumab for 48 h. Cell monolayers were then placed on ice, washed twice with ice-cold PBS, and lysed in 50 μl/well buffer B. Clarified cell lysates were equalized for protein concentration using the modified Bradford assay, resolved by SDS-PAGE, and processed by Western blotting. p27 expression was measured by probing blots with anti-p27 antibody (dilution 1:2000). HER2 phosphorylation, AKT phosphorylation, and extracellular signal-regulated kinase (ERK) phosphorylation were measured by probing blots with phospho-specific HER2 (dilution 1:1000), AKT (dilution 1:1000), and ERK (dilution 1:1000) antibodies, respectively. In all experiments blots were stripped by incubating with stripping buffer (62.5 mm Tris (pH 6.8), 2% (w/v) SDS, and 0.7% (v/v) β-mercaptoethanol) for 30 min at 50 °C then reprobed with antibodies against HER2 (dilution 1:1000), AKT (dilution 1:1000), and ERK (dilution 1:1000). All blots were also probed with anti-IQGAP1 antibodies (dilution 1:1000; to verify IQGAP1 expression) and anti-β-tubulin antibody (dilution 1:2000; to verify protein loading).
Measurement of HER2-stimulated Cell Growth
HER2-stimulated cell growth was measured using sulforhodamine B staining essentially as previously described (51). Briefly, 36 h after transfection cells were serum-starved as described above for 48 h. Cell monolayers were then placed on ice, and 1 ml of ice-cold 25% (w/v) trichloroacetic acid was added directly to the culture medium. Cells were left for 1 h at 4 °C before being stained with 0.4% (w/v) sulforhodamine B (in 1% (v/v) acetic acid). Protein-bound dye was dissolved in 10 mm Tris (pH 10.5), and absorbencies were read at 510 nm.
Quantitative RT-PCR
RT-PCR was performed on an iCycler IQ real time PCR detection system (Bio-Rad). 36 h after siRNA transfection, total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer's instructions. After ethanol precipitation and DNase treatment, 2 μg of RNA was reverse-transcribed to cDNA using the iScript cDNA Synthesis kit (Bio-Rad) according to the manufacturer's instructions. RT-PCR reactions were performed using 1× iQ SYBR Green Supermix (Bio-Rad) and 1 μm forward and reverse primers essentially as previously described (52). The primer sequences are as follows: HER2, 5′-ACAGTGGCATCTGTGAGCTG-3′ (forward (F)), 5′-CCCACGTCCGTAGAAAGGTA-3′ (reverse (R)); GAPDH, 5′-GGCCTCCAAGGAGTAAGACC-3′ (F), 5′-AGGGGTCTACATGGCAACTG-3′ (R). RT-PCR reactions were initiated for 3 min at 95 °C and cycled 40 times (10 s at 95 °C, 30 s at 57 °C). All samples and standards were run in triplicate, and GAPDH was used as an internal control. Every RT-PCR plate also contained RT and H2O controls to verify the specificity of PCR amplification. Results were analyzed using the standard curve method. HER2 gene expression was quantified relative to that of GAPDH.
Measurement of HER2 Half-life
Immediately after siRNA transfection, cells were treated with 100 μg/ml cyclohexamide (Sigma). 0, 4, 16, 24, and 40 h later, cell monolayers were placed on ice, washed twice with ice-cold PBS, and lysed in 50 μl/well buffer B. Clarified cell lysates were equalized for protein concentration using the modified Bradford Assay, resolved by SDS-PAGE, and processed by Western blotting. HER2 and β-tubulin expression was measured as described above.
Statistical Analysis
All experiments were repeated independently at least three times. Statistical significance was set at p < 0.05, and analyses were performed using Student's t test. In Figs. 5–7, 9, and 10, the * denotes statistical significance from control cells transfected with siRen and treated with vehicle, θ denotes statistical significance from cells treated with vehicle, and # denotes statistical significance from cells transfected with siIQ12. In Fig. 8, the * denotes statistical significance from trastuzumab-sensitive SkBR3 cells.
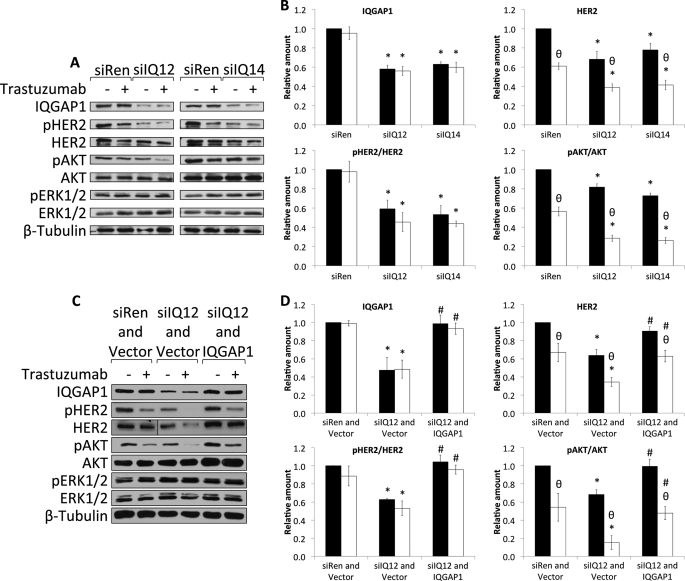
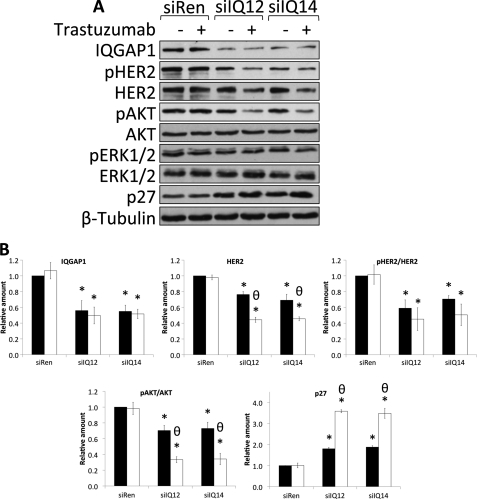
FIGURE 5.
Knockdown of IQGAP1 reduces HER2 expression, phosphorylation, and signaling and enhances the inhibitory effects of trastuzumab. A, SkBR3 cells were transiently transfected with siRNA against renilla luciferase (siRen) or siRNAs against IQGAP1 (siIQ12 and siIQ14). 36 h after siRNA transfection, cells were serum-starved in the presence of vehicle (−) or 21 μg/ml trastuzumab (+) for 48 h. Equal amounts of protein were resolved by SDS-PAGE, transferred to PVDF membranes, and probed with anti-IQGAP1, anti-pHER2, anti-HER2, anti-pAKT, anti-AKT, anti-pERK, anti-ERK, and anti-β-tubulin antibodies. The data are representative of five independent experiments. B, the amounts of IQGAP1 and HER2 were quantified by densitometry and corrected for the amount of β-tubulin in the corresponding lysate. The amounts of pHER2 and pAKT were quantified by densitometry and corrected for the amounts of total HER2 and total AKT, respectively, in the corresponding lysate. Samples that were treated with vehicle are depicted by black bars, and those treated with trastuzumab are depicted by white bars. The data, expressed relative to the amount of each protein in control cells transfected with siRen and treated with vehicle represent the mean ± S.E. (n = 5). *, significantly different from control cells transfected with siRen and treated with vehicle (p < 0.05); θ, significantly different from cells treated with vehicle (p < 0.05). C, SkBR3 cells were transiently transfected with siRNA against renilla luciferase and vector (siRen and Vector), siRNA against IQGAP1 and vector (siIQ12 and Vector), or siRNA against IQGAP1 and siRNA-resistant IQGAP1 (siIQ12 and IQGAP1). 36 h after transfection, cells were serum-starved in the presence of vehicle (−) or 21 μg/ml trastuzumab (+) for 48 h. Equal amounts of protein were resolved by SDS-PAGE, transferred to PVDF membranes, and probed with anti-IQGAP1, anti-pHER2, anti-HER2, anti-pAKT, anti-AKT, anti-pERK, anti-ERK, and anti-β-tubulin antibodies. The data are representative of three independent experiments. D, the amounts of IQGAP1 and HER2 were quantified by densitometry and corrected for the amount of β-tubulin in the corresponding lysate. The amounts of pHER2 and pAKT were quantified by densitometry and corrected for the amounts of total HER2 and total AKT, respectively, in the corresponding lysate. Samples that were treated with vehicle are depicted by black bars, and those treated with trastuzumab are depicted by white bars. The data, expressed relative to the amount of each protein in control cells transfected with siRen and vector and treated with vehicle, represent the mean ± S.E. (n = 3). *, significantly different from control cells transfected with siRen and vector and treated with vehicle (p < 0.05); θ, significantly different from cells treated with vehicle (p < 0.05); #, significantly different from cells transfected with siIQ12 and vector (p < 0.05).
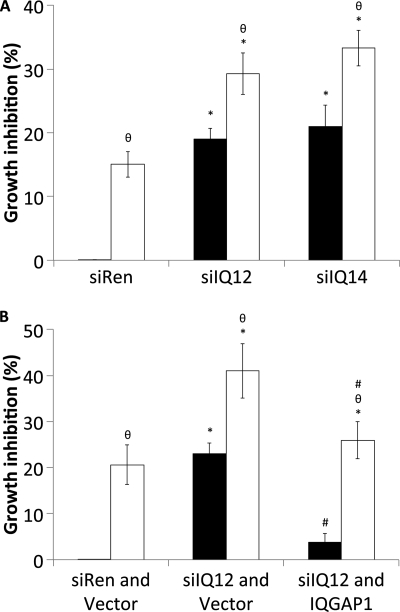
FIGURE 6.
Knockdown of IQGAP1 inhibits SkBR3 cell growth and increases the sensitivity of SkBR3 cells to trastuzumab. A, SkBR3 cells were transiently transfected with siRNA against renilla luciferase (siRen) or siRNAs against IQGAP1 (siIQ12 and siIQ14). 36 h after siRNA transfection cells were serum-starved in the presence of vehicle (black bars) or 21 μg/ml trastuzumab (white bars) for 48 h. Cell growth was measured using sulforhodamine B staining. The data, expressed relative to the number of cells in wells transfected with siRen and treated with vehicle, represent the mean ± S.E. (n = 5). *, significantly different from control cells transfected with siRen and treated with vehicle (p < 0.05); θ, significantly different from cells treated with vehicle (p < 0.05). B, SkBR3 cells were transiently transfected with siRNA against renilla luciferase and vector (siRen and vector), siRNA against IQGAP1 and vector (siIQ12 and vector), or siRNA against IQGAP1 and siRNA-resistant IQGAP1 (siIQ12 and IQGAP1). 36 h after transfection, cells were serum-starved in the presence of vehicle (black bars) or 21 μg/ml trastuzumab (white bars) for 48 h. Cell growth was measured using sulforhodamine B staining. The data, expressed relative to the number of cells in wells transfected with siRen and vector and treated with vehicle, represent the mean ± S.E. (n = 3). *, significantly different from control cells transfected with siRen and vector and treated with vehicle (p < 0.05); θ, significantly different from cells treated with vehicle (p < 0.05); #, significantly different from cells transfected with siIQ12 and vector (p < 0.05).
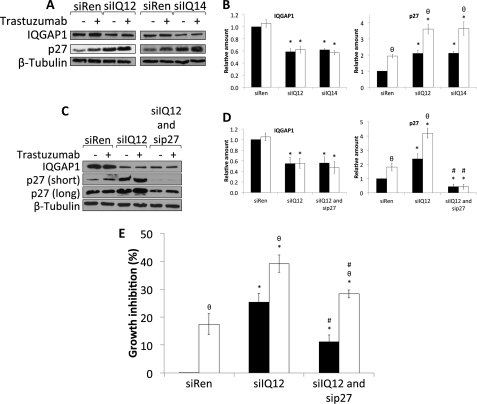
FIGURE 7.
Knockdown of IQGAP1 increases p27 expression and augments trastuzumab-induced p27 up-regulation. A, SkBR3 cells were transiently transfected with siRNA against renilla luciferase (siRen) or siRNAs against IQGAP1 (siIQ12 and siIQ14). 36 h after siRNA transfection, cells were serum-starved in the presence of vehicle (−) or 21 μg/ml trastuzumab (+) for 48 h. Equal amounts of protein were resolved by SDS-PAGE, transferred to PVDF membranes, and probed with anti-IQGAP1, anti-p27, and anti-β-tubulin antibodies. The data are representative of five independent experiments. B, the amounts of IQGAP1 and p27 were quantified by densitometry and corrected for the amount of β-tubulin in the corresponding lysate. Samples that were treated with vehicle are depicted by black bars, and those treated with trastuzumab are depicted by white bars. The data, expressed relative to the amount of each protein in control cells transfected with siRen and treated with vehicle, represent the mean ± S.E. (n = 5). *, significantly different from control cells transfected with siRen and treated with vehicle (p < 0.05); θ, significantly different from cells treated with vehicle (p < 0.05). C, SkBR3 cells were transiently transfected with siRNA against renilla luciferase (siRen), siRNA against IQGAP1 (siIQ12), or siRNAs against IQGAP1 and p27 (siIQ12 and sip27). 36 h after siRNA transfection, cells were serum-starved in the presence of vehicle (−) or 21 μg/ml trastuzumab (+) for 48 h. Equal amounts of protein were resolved by SDS-PAGE, transferred to PVDF membranes, and probed with anti-IQGAP1, anti-p27, and anti-β-tubulin antibodies. The data are representative of three independent experiments. D, the amounts of IQGAP1 and p27 were quantified by densitometry and corrected for the amount of β-tubulin in the corresponding lysate. Samples that were treated with vehicle are depicted by black bars, and those treated with trastuzumab are depicted by white bars. The data, expressed relative to the amount of each protein in control cells transfected with siRen and treated with vehicle, represent the mean ± S.E. (n = 3). *, significantly different from control cells transfected with siRen and treated with vehicle (p < 0.05); θ, significantly different from cells treated with vehicle (p < 0.05); #, significantly different from cells transfected with siIQ12 (p < 0.05). E, SkBR3 cells were transiently transfected with siRNA against renilla luciferase (siRen), siRNA against IQGAP1 (siIQ12), or siRNAs against IQGAP1 and p27 (siIQ12 and sip27). 36 h after siRNA transfection, cells were serum-starved in the presence of vehicle (black bars) or 21 μg/ml trastuzumab (white bars) for 48 h. Cell growth was measured using sulforhodamine B staining. The data, expressed relative to the number of cells in wells transfected with siRen and treated with vehicle, represent the mean ± S.E. (n = 3). *, significantly different from control cells transfected with siRen and treated with vehicle (p < 0.05); θ, significantly different from cells treated with vehicle (p < 0.05); #, significantly different from cells transfected with siIQ12 (p < 0.05).
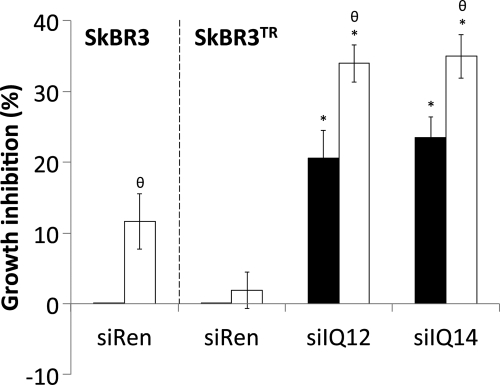
FIGURE 9.
Knockdown of IQGAP1 abrogates resistance to the growth inhibitory effects of trastuzumab. Trastuzumab-sensitive (SkBR3) or trastuzumab-resistant (SkBR3TR) SkBR3 cells were transiently transfected with siRNA against renilla luciferase (siRen) or siRNAs against IQGAP1 (siIQ12 and siIQ14). 36 h after siRNA transfection, cells were serum-starved in the presence of vehicle (black bars) or 21 μg/ml trastuzumab (white bars) for 48 h. Cell growth was measured using sulforhodamine B staining. The data, expressed relative to the number of control cells in wells transfected with siRen and treated with vehicle, represent the mean ± S.E. (n = 5). *, significantly different from control cells transfected with siRen and treated with vehicle (p < 0.05); θ, significantly different from cells treated with vehicle (p < 0.05).
FIGURE 10.
Knockdown of IQGAP1 reduces HER2 expression, phosphorylation, and signaling and restores trastuzumab sensitivity to trastuzumab-resistant SkBR3 cells. A, trastuzumab-resistant SkBR3 cells were transiently transfected with siRNA against renilla luciferase (siRen) or siRNAs against IQGAP1 (siIQ12 and siIQ14). 36 h after siRNA transfection, cells were serum-starved in the presence of vehicle (−) or 21 μg/ml trastuzumab (+) for 48 h. Equal amounts of protein were resolved by SDS-PAGE, transferred to PVDF membranes, and probed with anti-IQGAP1, anti-pHER2, anti-HER2, anti-pAKT, anti-AKT, anti-pERK, anti-ERK, anti-p27, and anti-β-tubulin antibodies. The data are representative of five independent experiments. B, the amounts of IQGAP1, HER2, and p27 were quantified by densitometry and corrected for the amount of β-tubulin in the corresponding lysate. The amounts of pHER2 and pAKT were quantified by densitometry and corrected for the amounts of total HER2 and total AKT, respectively, in the corresponding lysate. Samples that were treated with vehicle are depicted by black bars, and those treated with trastuzumab are depicted by white bars. The data, expressed relative to the amount of each protein in control cells transfected with siRen and treated with vehicle, represent the mean ± S.E. (n = 5). *, significantly different from control cells transfected with siRen and treated with vehicle (p < 0.05); θ, significantly different from cells treated with vehicle (p < 0.05).
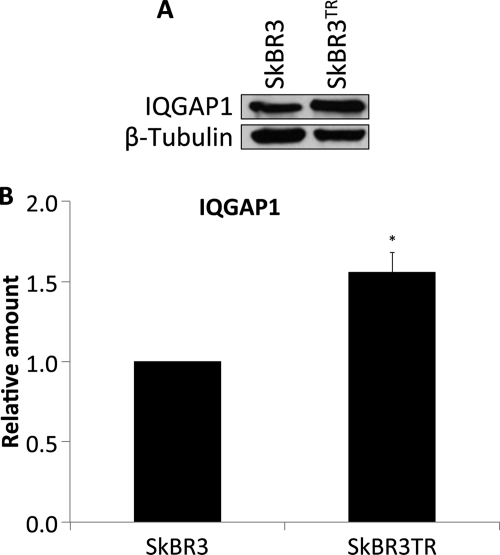
FIGURE 8.
IQGAP1 is overexpressed in trastuzumab-resistant SkBR3 cells. A, equal amounts of protein from trastuzumab-sensitive (SkBR3) and trastuzumab-resistant (SkBR3TR) SkBR3 cells were resolved by SDS-PAGE, transferred to PVDF membranes, and probed with anti-IQGAP1 and anti-β-tubulin antibodies. The data are representative of three independent experiments. B, the amount of IQGAP1 was quantified by densitometry and corrected for the amount of β-tubulin in the corresponding lysate. The data, expressed relative to the amount of IQGAP1 in SkBR3 cells, represent the mean ± S.E. (n = 3). *, significantly different from SkBR3 cells (p < 0.05).
RESULTS
IQGAP1 Is Overexpressed in HER2(+) Breast Cancer Tissue
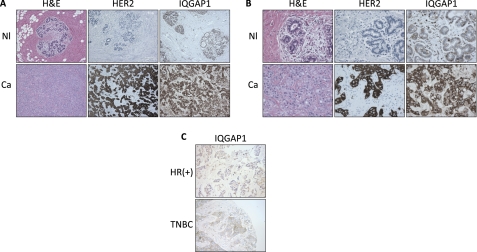
IQGAP1 expression was analyzed in three normal breast specimens as well as 19 HER2(+) (3+ by immunohistochemistry and/or amplified by FISH) and 20 HER2(−) invasive breast carcinomas. Immunostaining was reviewed by a board-certified pathologist (D. A. Dillon), and IQGAP1 expression was scored as described under “Experimental Procedures.” Minimal IQGAP1 immunoreactivity was apparent in the luminal epithelial cells of any (3/3) normal cases and 16/20 HER2(−) tumors (Fig. 1; supplemental Table S1). In contrast, 16/19 HER2(+) carcinomas showed at least moderate IQGAP1 immunoreactivity in at least 10% of tumor cells (Fig. 1; supplemental Table S1). Of note, 8/19 HER2(+) tumors displayed strong IQGAP1 immunoreactivity in at least 30% of tumor cells (3+) compared with 0/20 HER2(−) neoplasms (supplemental Table S1).
FIGURE 1.
IQGAP1 is overexpressed in HER2(+) breast cancer. A and B, IQGAP1 protein expression was evaluated by immunohistochemistry in normal breast specimens (Nl, upper panels) and HER2(+) (3+ by immunohistochemistry and/or amplified by FISH) invasive breast carcinomas (Ca, lower panels). Representative images are shown. The final magnification is 100× (A) and 400× (B). H&E, hematoxylin and eosin. C, IQGAP1 protein expression was evaluated by immunohistochemistry in hormone receptor-positive (HR(+); upper panel) and triple-negative (TNBC; lower panel) invasive breast carcinomas. Representative images are shown. The final magnification is 100×.
IQGAP1 Binds HER2
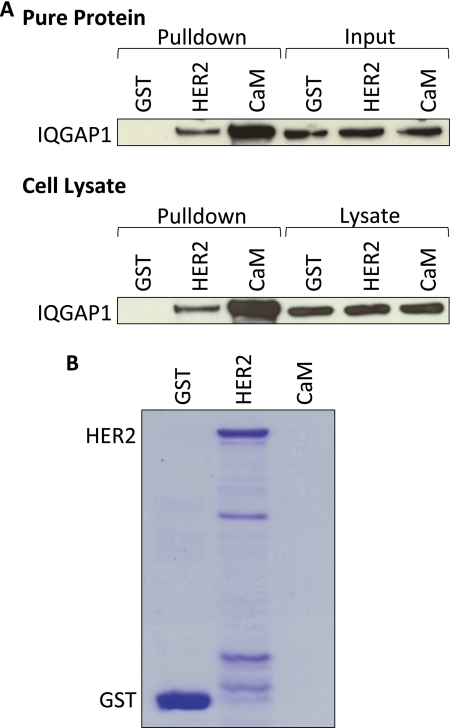
In vitro analysis with pure proteins was used to examine a possible interaction between IQGAP1 and HER2. GST alone or GST-HER2 was incubated with purified IQGAP1, and complexes were isolated with glutathione-Sepharose. Analysis by Western blotting reveals that IQGAP1 binds HER2 (Fig. 2A, upper panel). The binding is specific, as no HER2 is present in samples incubated with GST alone. Calmodulin-Sepharose was used as a positive control (Fig. 2A, upper panel). Analysis of inputs reveals equivalent amounts of IQGAP1 in all samples, and Coomassie staining confirms similar amounts of GST and GST-HER2 in each pulldown assay (Fig. 2B) (note that calmodulin does not come off the Sepharose beads under denaturing conditions and, therefore, cannot be seen on the gel).
FIGURE 2.
IQGAP1 binds HER2. A, GST alone, GST-HER2 (HER2), or calmodulin-Sepharose (CaM) was incubated with equal amounts of purified IQGAP1 (upper panel) or equal amounts of protein from SkBR3 cell lysates (lower panel). Complexes were isolated and washed as described under “Experimental Procedures.” The samples were resolved by SDS-PAGE, transferred to polyvinylidene fluoride (PVDF) membranes, and probed with anti-IQGAP1 antibodies. An aliquot of each sample (equivalent to 2% of the amount in each pulldown) was also processed by Western blotting (Input, top panel; Lysate, bottom panel). The data are representative of five independent experiments. B, equivalent amounts of GST alone, GST-HER2 (HER2), or calmodulin-Sepharose (CaM) to those used in the pulldown assays in A were resolved by SDS-PAGE. The gel was stained with Coomassie Blue. The data are representative of five independent experiments.
We next evaluated whether IQGAP1 interacts with HER2 in a normal cell milieu. SkBR3 cells were lysed and incubated with GST alone or GST-HER2. Endogenous IQGAP1 in cell lysates binds GST-HER2 (Fig. 2A, lower panel). The specificity of the binding is validated by the absence of IQGAP1 from samples that were incubated with GST alone. Calmodulin-Sepharose was used as a positive control (Fig. 2A, lower panel). The amount of IQGAP1 in all lysates was equivalent, as was the amount of bait in each pulldown (Fig. 2B). Collectively, these data reveal a direct interaction between IQGAP1 and HER2.
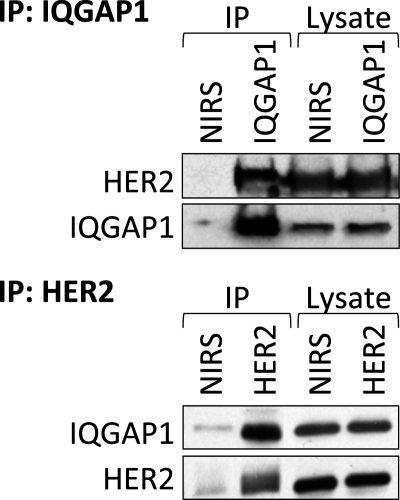
IQGAP1 and HER2 Co-immunoprecipitate
Immunoprecipitation of IQGAP1 demonstrated that endogenous HER2 binds endogenous IQGAP1 in SkBR3 cells (Fig. 3, upper panel). The specificity of the interaction is confirmed by the absence of HER2 from samples that were precipitated with non-immune rabbit serum. Binding was also demonstrated by immunoprecipitating HER2. IQGAP1 co-immunoprecipitates with HER2 (Fig. 3, lower panel), whereas minimal IQGAP1 and HER2 are present in samples precipitated with non-immune rabbit serum. Probing the lysates for HER2 and IQGAP1 reveals equal loading among samples (Fig. 3). These data indicate that IQGAP1 and HER2 associate in SkBR3 human breast epithelial cells.
FIGURE 3.
IQGAP1 and HER2 co-immunoprecipitate. Equal amounts of protein from SkBR3 cell lysates were immunoprecipitated (IP) with anti-IQGAP1 antibodies (upper panel) or anti-HER2 antibody (lower panel). Non-immune rabbit serum (NIRS) was used as a negative control. Both unfractionated lysates (Lysate) and complexes (IP) were resolved by SDS-PAGE, transferred to PVDF membranes, and probed with anti-IQGAP1 and anti-HER2 antibodies. The data are representative of five independent experiments.
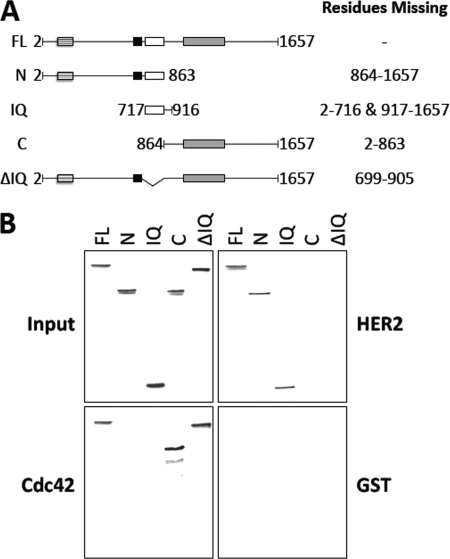
Identification of the HER2 Binding Domain on IQGAP1
The region of IQGAP1 to which HER2 binds was investigated using GST pulldown assays and [35S]methionine-labeled TNT products of selected IQGAP1 fragments and mutants (Fig. 4A). IQGAP1 constructs were incubated with GST-HER2, GST-Cdc42V12, or GST alone. Complexes were isolated with glutathione-Sepharose, resolved by SDS-PAGE, and identified by autoradiography. Analogous to the isolation of IQGAP1 by GST-HER2 (Fig. 2A), [35S]methionine-labeled full-length IQGAP1 (amino acids 2–1657) binds GST-HER2 (Fig. 4B, top right panel). Examination of the 2 halves of IQGAP1 reveals that only the N-terminal half (amino acids 2–863) binds GST-HER2; no interaction between the C-terminal half of IQGAP1 (amino acids 864–1657) and HER2 was detected (Fig. 4B, top right panel). GST-Cdc42V12, which binds the C-terminal half of IQGAP1 (45, 53), was used as a positive control (Fig. 4B, bottom left panel). No IQGAP1 binds GST alone (Fig. 4B, bottom right panel). The amount of [35S]methionine-labeled TNT product in each sample was essentially identical (Fig. 4B, top left panel). These data reveal that the region of IQGAP1 containing amino acids 2–863 is necessary for HER2 binding.
FIGURE 4.
The IQ region of IQGAP1 is both necessary and sufficient for HER2 binding. A, shown is a schematic representation of IQGAP1 constructs depicting full-length IQGAP1, truncated IQGAP1 fragments, and a deletion mutant. The specific amino acids absent from each construct are indicated. FL, full-length IQGAP1 (amino acids 2–1657); N, N-terminal half of IQGAP1 (amino acids 2–863); IQ, IQ region of IQGAP1 (amino acids 717–916); C, C-terminal half of IQGAP1 (amino acids 864–1657); ΔIQ, full length IQGAP1 from which the IQ region has been deleted. B, [35S]methionine-labeled full length IQGAP1 (FL), IQGAP1-N (N), IQGAP1-IQ (IQ), IQGAP1-C (C), and IQGAP1ΔIQ (ΔIQ) were incubated with equal amounts of GST-HER2 (HER2, top right panel), GST-Cdc42V12 (Cdc42, bottom left panel), or GST alone (GST, bottom right panel). Complexes were isolated and washed as described under “Experimental Procedures.” The samples were resolved by SDS-PAGE, and the gels were dried and processed by autoradiography. An aliquot of each [35S]methionine-labeled TNT product (equivalent to 2% of the amount in each pulldown) was also resolved by SDS-PAGE and processed by autoradiography (Input, top left panel). The data are representative of five independent experiments.
A [35S]methionine-labeled IQGAP1 construct from which the IQ region (amino acids 699–905) has been deleted (designated ΔIQ) was used to narrow the HER2 binding site. Deletion of the IQ region abrogates the interaction of IQGAP1 with HER2 (Fig. 4B, top right panel), whereas interaction with GST-Cdc42V12 indicates that ΔIQ can associate with a binding partner (Fig. 4B, bottom left panel). Moreover, an IQGAP1 fragment comprising only the IQ region (amino acids 717–916) (designated IQ) binds GST-HER2 (Fig. 4B, top right panel) but not GST alone (Fig. 4B, bottom right panel). These data strongly suggest that the IQ region of IQGAP1 is both necessary and sufficient for HER2 binding.
IQGAP1 Regulates HER2 Expression, Phosphorylation, and Signaling
We employed a siRNA-based loss of function strategy to ascertain the biological relevance of the interaction between IQGAP1 and HER2. Before beginning, a BLAST (blast.ncbi.nlm.nih.gov) search of the entire human RefSeq data base was performed to exclude the possibility of siRNA-induced off-target effects. Two independent siRNAs (from two different suppliers) directed against different regions of IQGAP1 were transfected into SkBR3 cells. HER2 expression, phosphorylation, and signaling were evaluated by Western blotting. Transfection of siRNAs against IQGAP1 (termed siIQ12 and siIQ14) into SkBR3 cells reduces IQGAP1 by 40–50% (Fig. 5, A and B, top left panel). No significant decrease in IQGAP1 was observed when untransfected cells were compared with cells transfected with control siRNA (against renilla luciferase; designated siRen) (data not shown). Moreover, IQGAP1 knockdown does not significantly alter the levels of AKT, ERK, or β-tubulin (Fig. 5A). Reducing endogenous IQGAP1 decreases the expression of HER2 protein by 20–30% (Fig. 5, A and B, top right panel). Interestingly, no decrease in HER2 mRNA was observed after IQGAP1 knockdown (supplemental Fig. S1A), whereas the half-life of HER2 protein was significantly reduced (supplemental Fig. S1, B and C). Because reducing IQGAP1 decreases HER2 expression, the extent of HER2 phosphorylation was corrected for total HER2 in the corresponding sample. Transfection of siIQ12 or siIQ14 significantly attenuates HER2 phosphorylation by 40–50% (Fig. 5, A and B, bottom left panel). To evaluate a possible role for IQGAP1 in HER2 signaling, the activation status of the PI3K/AKT and MAPK pathways was determined. Knockdown of IQGAP1 significantly decreases pAKT (Fig. 5, A and B, bottom right panel). In contrast, no significant change in the levels of pERK was observed (Fig. 5A; supplemental Fig. S2A).
To validate the effects of the siRNA, we rescued IQGAP1 knockdown. Transfection of a siRNA-resistant IQGAP1 plasmid into SkBR3 cells with reduced IQGAP1 restores IQGAP1 levels to those in control cells (i.e. without knockdown) (Fig. 5, C and D, top left panel). Replacing IQGAP1 completely restores HER2 expression (Fig. 5, C and D, top right panel). Similarly, the amounts of both phosphorylated HER2 (Fig. 5, C and D, bottom left panel) and pAKT (Fig. 5, C and D, bottom right panel) revert to basal levels when IQGAP1 expression is returned to normal. Consistent with our previous data (Fig. 5A; supplemental Fig. S2A), no significant difference in ERK activity was observed after reconstitution of SkBR3 cells with siRNA-resistant IQGAP1 (Fig. 5C; supplemental Fig. S2B). These data identify a role for IQGAP1 in the regulation of HER2 expression, phosphorylation, and signaling through the PI3K/AKT cascade.
Knockdown of IQGAP1 Enhances the Inhibitory Effects of Trastuzumab
Patients with HER2(+) breast cancer are treated with trastuzumab (4, 17). Therefore, we determined whether knockdown of IQGAP1 might alter the response of SkBR3 cells to trastuzumab. Incubation with trastuzumab for 48 h had no significant effect on the levels of IQGAP1 (Fig. 5, A and B, top left panel), AKT, or β-tubulin (Fig. 5A). Trastuzumab reduces HER2 expression by ∼40% in cells transfected with control siRNA (Fig. 5, A and B, top right panel) and by ∼60% in cells in which IQGAP1 has been knocked down. Trastuzumab does not significantly alter HER2 phosphorylation in either control cells or in cells with reduced IQGAP1 (Fig. 5, A and B, bottom left panel). In contrast, trastuzumab reduces pAKT by ∼40 and ∼70% in cells transfected with control siRNA and IQGAP1 siRNA, respectively (Fig. 5, A and B, bottom right panel). Although trastuzumab slightly increased the expression of pERK and ERK (Fig. 5A), the change was not statistically significant (supplemental Fig. S2A).
The effects of restoring IQGAP1 on trastuzumab function in SkBR3 cells were examined. Cells with reduced IQGAP1 were transfected with siRNA-resistant IQGAP1 before being incubated with trastuzumab. Reconstitution of IQGAP1 abrogates the additional inhibitory effects of trastuzumab on both HER2 expression (Fig. 5, C and D, top right panel) and AKT phosphorylation (Fig. 5, C and D, bottom right panel) induced by IQGAP1 knockdown. These data reveal that lowering IQGAP1 expression augments the inhibitory effects of trastuzumab on HER2 expression and signaling through the PI3K/AKT cascade.
Knockdown of IQGAP1 Inhibits HER2-stimulated SkBR3 Cell Growth and Increases the Sensitivity of SkBR3 Cells to Trastuzumab
To evaluate whether reducing IQGAP1 expression alters HER2 function, we measured HER2-stimulated cell proliferation. Knockdown of IQGAP1 significantly attenuates HER2-stimulated cell growth by ∼20% (Fig. 6A). Consistent with prior publications (54, 55), trastuzumab reduces cell growth by ∼15%. Importantly, reducing IQGAP1 expression augments the growth inhibitory effects of trastuzumab by ∼2-fold (Fig. 6A). Replacing IQGAP1 in the knockdown cells (with siRNA-resistant IQGAP1) eliminates the growth inhibitory effects produced by reducing endogenous IQGAP1 (Fig. 6B). Similarly, reconstitution of IQGAP1 reduces the growth inhibition elicited by trastuzumab from ∼40 to ∼25%. These data indicate that knockdown of IQGAP1 attenuates HER2-stimulated cell growth and enhances the growth inhibition produced by trastuzumab.
Knockdown of IQGAP1 Increases p27 Expression and Augments Trastuzumab-induced p27 Up-regulation
Trastuzumab inhibits cell proliferation, at least in part, by inducing the expression of the CDK inhibitor p27 (18, 19, 56). We investigated whether IQGAP1 knockdown impairs cell proliferation by modulating the levels of p27. Reducing IQGAP1 augments p27 expression by 2-fold (Fig. 7, A and B, right panel). The magnitude of this increase was virtually identical to that elicited by trastuzumab. Interestingly, knockdown of IQGAP1 significantly enhances the effects of trastuzumab on p27 levels (Fig. 7, A and B, right panel). These data indicate that reducing IQGAP1 increases both p27 expression and the up-regulation of p27 elicited by trastuzumab.
To validate the role of p27 in the effects on cell growth produced by reducing IQGAP1, we simultaneously knocked down both proteins. Transfection of siRNAs against IQGAP1 and p27 reduces the levels of both proteins in SkBR3 cells (Fig. 7, C and D). The p27 siRNA abrogates the increased expression of p27 elicited by IQGAP1 knockdown, although residual p27 expression remains. Reducing p27 in cells with decreased IQGAP1 significantly (but not completely) impairs the inhibition of growth we observed (Fig. 7E). The enhanced inhibition of growth effected by trastuzumab in cells with reduced IQGAP1 is significantly attenuated by concomitant reduction of p27. Collectively, these data suggest that the inhibition of growth caused by IQGAP1 knockdown is, at least in part, due to an increase in p27 levels.
IQGAP1 Is Overexpressed in Trastuzumab-resistant SkBR3 Cells
Resistance to trastuzumab severely limits the successful treatment of HER2(+) breast cancer (17, 24). To determine whether the levels of IQGAP1 are altered in trastuzumab-resistant cells, we developed a trastuzumab-resistant SkBR3 cell line (termed SkBR3TR). IQGAP1 expression in SkBR3TR cells was increased by ∼1.6-fold compared with SkBR3 cells (Fig. 8). Similar results were obtained using the JIMT-1 cell line, which was established from a pleural metastasis of a trastuzumab-resistant breast cancer patient (57) (data not shown). These data reveal that IQGAP1 is up-regulated in trastuzumab-resistant breast cancer.
Knockdown of IQGAP1 Abrogates Resistance to the Growth Inhibitory Effects of Trastuzumab
To examine whether manipulating IQGAP1 expression affects trastuzumab resistance, we transfected siRNAs against IQGAP1 into SkBR3TR cells and measured HER2-stimulated cell proliferation. Trastuzumab did not significantly reduce SkBR3TR cell growth, thereby confirming that these cells are trastuzumab-resistant (Fig. 9). In contrast, IQGAP1 knockdown impairs growth of SkBR3TR cells by ∼20–25% (Fig. 9), approximately the same extent as that produced by reducing IQGAP1 in trastuzumab-sensitive SkBR3 cells (Fig. 6). Knockdown of IQGAP1 in SkBR3TR cells restores their sensitivity to trastuzumab (Fig. 9). Trastuzumab inhibits growth of both SkBR3 and SkBR3TR cells by ∼35% when IQGAP1 levels are reduced. These data strongly suggest that lowering IQGAP1 expression abrogates resistance to the growth inhibitory effects of trastuzumab.
Knockdown of IQGAP1 Reduces HER2 Expression, Phosphorylation, and Signaling and Restores Trastuzumab Sensitivity to Trastuzumab-resistant SkBR3 Cells
To elucidate the molecular mechanism underlying the role of IQGAP1 in trastuzumab resistance, we analyzed HER2 levels and signaling in SkBR3TR cells. siIQ12 or siIQ14 reduced IQGAP1 by ∼40–50% in SkBR3TR cells (Fig. 10, A and B, top left panel). IQGAP1 knockdown does not significantly alter the levels of AKT, ERK, or β-tubulin (Fig. 10A). Reducing endogenous IQGAP1 in SkBR3TR cells significantly decreases HER2 expression by ∼20–30% (Fig. 10, A and B, top middle panel) and HER2 phosphorylation by ∼30–40% (Fig. 10, A and B, top right panel). IQGAP1 knockdown also decreases pAKT (Fig. 10, A and B, bottom left panel) and increases the levels of p27 (Fig. 10, A and B, bottom right panel) but does not significantly change pERK (Fig. 10A; supplemental Fig. S2C). Note that the magnitude of the alterations in HER2 signaling and p27 expression produced by reducing IQGAP1 in SkBR3TR cells (Fig. 10) are essentially the same as those that IQGAP1 knockdown elicits in SkBR3 cells (Figs. 5 and 7).
In SkBR3TR cells, trastuzumab alone has no significant effect on the levels of IQGAP1, HER2, pHER2, pAKT, AKT, pERK, ERK, or p27 (Fig. 10; supplemental Fig. S2C). By contrast, when IQGAP1 was decreased, the ability of trastuzumab to reduce HER2 expression and signaling was restored (Fig. 10). These data strongly suggest that reducing IQGAP1 abrogates trastuzumab resistance.
DISCUSSION
Over the past four decades, considerable advances have been made in our comprehension of the pathogenesis of human breast cancer. Analyses of the presence of selected hormone receptors, such as those for estrogen and progesterone, are performed routinely on patient tissue and influence therapy and prognosis (58). More recent evidence reveals that increased expression of HER2, a member of the ErbB family of receptor-tyrosine kinases, correlates strongly with a shorter time to relapse and a decrease in overall survival (5). HER2(+) breast cancer is treated with trastuzumab, but ∼15% of women in the early stages of disease still develop metastases (17). Moreover, in the metastatic setting, resistance to trastuzumab develops within 1 year in the majority of patients (24). Several mechanisms have been proposed to underlie trastuzumab resistance, including accumulation of p95HER2 (25), constitutive activation of the PI3K/AKT pathway (26, 27), or the overexpression of other receptor-tyrosine kinases (28, 29). Here, we provide strong evidence to suggest that IQGAP1 is a potent regulator of HER2 expression, phosphorylation, and signaling. Moreover, although the number of samples we examined is relatively small, our data suggest that HER2(+) breast tumors have more IQGAP1 than either HER2(−) breast neoplasms or normal breast tissue. Importantly, IQGAP1 is overexpressed in trastuzumab-resistant human breast epithelial cells, and reducing IQGAP1 expression both augments the growth inhibitory effects of trastuzumab in vitro and abrogates trastuzumab resistance.
IQGAP1 is a scaffold protein that integrates several signaling pathways (30), and a solid body of evidence supports its involvement in neoplasia. For example, many IQGAP1 binding partners have established roles in tumorigenesis (35, 36). Moreover, increased IQGAP1 gene and protein expression have been documented in several human neoplasms (37–44). Importantly, we previously showed that modulating IQGAP1 expression levels in malignant human breast epithelial cells significantly alters their tumorigenicity (44, 59). Overexpression of IQGAP1 enhances in vitro motility and invasion of both MCF-7 and MDA-MB-231 cells (44, 59). Conversely, siRNA-mediated knockdown of IQGAP1 reduces MCF-7 anchorage-independent growth, motility, and invasion in vitro as well as growth and invasion in an in vivo tumor model system (44). Collectively, these data suggest that IQGAP1 overexpression is not merely a consequence of neoplastic transformation but instead that it contributes to tumorigenesis of human breast epithelium. These findings support the concept that IQGAP1 is an oncogene (35, 36).
IQGAP1 binds to and modulates the function of several growth factor receptors, including VEGFR2 (34, 60), fibroblast growth factor receptor 1 (61), nerve growth factor receptor (62), and EGFR (31). Here we demonstrate that IQGAP1 also binds directly to HER2, an interaction that elicits several effects. IQGAP1 binding appears to stabilize HER2 protein expression. Knockdown of IQGAP1 decreases the half-life of HER2 in SkBR3 cells, thereby reducing HER2 protein without significantly altering HER2 mRNA. Although considerable attention has been directed toward endocytosis and trafficking of EGFR (63), comparatively little is known about the regulation of HER2 turnover. HER2 appears to be degraded primarily by endocytic down-regulation followed by targeting to the lysosome (64). These processes are less efficient for HER2 than for EGFR, and many HER2 receptors are recycled back to the plasma membrane (63). The molecular mechanism by which IQGAP1 stabilizes HER2 remains to be determined. IQGAP1 is an important component of trafficking. For example, overexpression of IQGAP1 enhances targeting of β-catenin to the plasma membrane (65), and IQGAP1 is required for plasma membrane insertion of caveolae (66). It is tempting to speculate that increased IQGAP1 in HER2(+) tumors promotes recycling of HER2 to the plasma membrane, augmenting HER2 signaling and HER2-stimulated cell proliferation. In this model, knockdown of IQGAP1 would induce internalization and degradation of HER2. This is exactly what we observed.
IQGAP1 is also necessary for maximum HER2 phosphorylation. Reducing IQGAP1 expression significantly attenuates HER2 phosphorylation. Phosphorylation of HER2 is an absolute requirement for HER2 signaling, and several studies have demonstrated that reducing HER2 phosphorylation inhibits activation of the PI3K/AKT and MAPK pathways (54, 67, 68). Conventional models of receptor-tyrosine kinase activation suggest that ligand binding results in receptor dimerization (69, 70). Although HER2 does not bind a specific ligand directly, it is the preferred dimerization partner of all ErbB family members (71). Ligand binding to EGFR, HER3, or HER4 induces the formation of HER2-containing heterodimers, whereas HER2 self-association gives rise to homodimeric complexes. After dimerization, intermolecular contacts between the membrane-spanning and C-terminal regions facilitate receptor phosphorylation in trans (72–74). IQGAP1 binding may be necessary for proper orientation of the HER2 intracellular domain, thereby facilitating catalytic activity and/or enabling the tyrosine kinase to access the tyrosine residues for phosphate attachment. Solving the structure of HER2 bound to IQGAP1 is necessary to obtain additional insight.
IQGAP1 has been implicated in both PI3K/AKT and MAPK signaling (34, 48, 49, 75). For example, knockdown of IQGAP1 impairs AKT activation by vascular endothelial growth factor (34). Our observations are analogous to this finding, as reducing IQGAP1 impairs AKT activation by HER2. HER2-induced AKT phosphorylation stimulates cell proliferation by down-regulating p27 (76), a CDK inhibitor that reduces CDK2 activity resulting in G1 arrest and apoptosis (13). Consistent with these data, we showed that IQGAP1 knockdown increases p27 expression and reduces HER2-stimulated cell proliferation. Moreover, blocking the increase in p27 induced by IQGAP1 knockdown significantly attenuates the inhibition of SkBR3 cell growth that occurs in cells transfected with IQGAP1 siRNA. We propose that residual p27 is responsible for the remaining grown inhibition we observed after simultaneously reducing both IQGAP1 and p27. Consistent with this suggestion, under our experimental conditions knocking down p27 does not completely prevent the inhibition of cell growth induced by trastuzumab. Other investigators have demonstrated complete reduction of trastuzumab-induced cell growth inhibition by apparently more pronounced p27 silencing (18), suggesting that residual p27 contributes to the remaining inhibition of SkBR3 growth that we observe. Regardless, these data support the hypothesis that reducing IQGAP1 inhibits growth of SkBR3 cells at least in part by increasing the levels of p27.
Reducing endogenous IQGAP1 expression does not significantly alter MAPK signaling in SkBR3 cells, as ERK phosphorylation was unchanged. This observation was surprising as IQGAP1 is a scaffold in the MAPK cascade. IQGAP1 binds directly to B-Raf (75), MAPK kinase (49), and ERK (48), and IQGAP1 knockdown in MCF-7 cells attenuates ERK activation by both epidermal growth factor and insulin-like growth factor 1 (48). The reasons for these differences are unknown, but there are several possible explanations. For example, variation between cell types and/or intracellular signaling components may influence MAPK activity (77). This is exemplified by HER2-stimulated MAPK signaling. Inhibition of HER2 signaling in BT474 cells frequently reduces activation of both AKT and ERK (54). In contrast, changes in ERK phosphorylation are not always observed in SkBR3 cells when HER2 signaling is modulated (54), an observation that is consistent with our data. It is also possible that MAPK signaling downstream of HER2 may differ from that of EGFR or VEGFR2.
Initial studies suggested that trastuzumab inhibits HER2(+) tumor growth by stimulating endocytosis and degradation of HER2, with subsequent impairment of downstream signaling through the PI3K/AKT and MAPK cascades (15). Later analyses reported that reduced PI3K/AKT and MAPK signaling induces the expression of p27 (10, 11, 15, 16). Our data reveal that IQGAP1 modulates trastuzumab function. Reducing endogenous IQGAP1 significantly augments the ability of trastuzumab to decrease HER2 expression and HER2-stimulated activation of the PI3K/AKT cascade. Moreover, trastuzumab-mediated inhibition of HER2-stimulated cell growth and up-regulation of p27 were enhanced in cells in which IQGAP1 had been knocked down. Either reconstitution of IQGAP1 knockdown cells with IQGAP1 or preventing the increase in p27 blunted the inhibitory effects of trastuzumab on cell growth. Conceptually, it is unlikely that IQGAP1 alters the interaction of trastuzumab with HER2. Trastuzumab binds to the HER2 extracellular region (14), and IQGAP1 is located exclusively within the cell. Moreover, reducing IQGAP1 expression and trastuzumab treatment had additive effects. Therefore, it appears that IQGAP1 regulates trastuzumab function solely by modulating HER2.
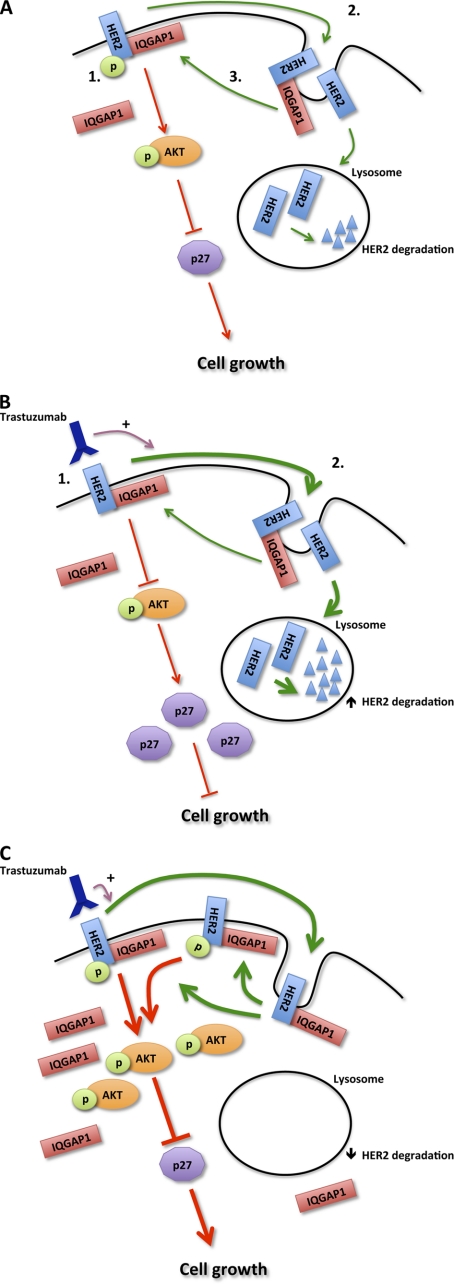
As outlined previously, resistance to trastuzumab is a major concern. We observed that IQGAP1 is overexpressed in trastuzumab-resistant SkBR3 cells, and knockdown of IQGAP1 abrogates trastuzumab resistance. When IQGAP1 levels are reduced in trastuzumab-resistant cells, sensitivity to trastuzumab is restored. The effects of trastuzumab on HER2 expression, signaling, and HER2 function in SkBR3TR (trastuzumab-resistant) cells after IQGAP1 knockdown are indistinguishable from those in the parental SkBR3 cell line. These findings imply that IQGAP1 overexpression contributes at least in part to trastuzumab resistance. Based on these data, we propose a model of the molecular mechanism for the participation of IQGAP1 in HER2 function and trastuzumab resistance (Fig. 11). IQGAP1 binds HER2, both augmenting HER2 signaling and reducing HER2 degradation. The latter increases the number of HER2 receptors. In trastuzumab-resistant cells, the increased IQGAP1 levels counteract the growth inhibitory effects of trastuzumab by stabilizing HER2 protein expression, thereby facilitating HER2 signaling (Fig. 11C). It is also possible that IQGAP1 may regulate trastuzumab resistance independently of the canonical HER2 pathway. For example, hyperactivation of Rac1 in trastuzumab-resistant SkBR3 cells inhibits trastuzumab-induced HER2 endocytosis (78). IQGAP1 is an important regulator of Rac1/Cdc42 function and stabilizes these small GTPases in their active GTP-bound form (79). Overexpression of IQGAP1 in SkBR3TR cells may, therefore, induce trastuzumab resistance by increasing the levels of active Rac1. Importantly, the two mechanisms are not mutually exclusive, and both may operate. Our data strongly suggest that IQGAP1 overexpression is a component of HER2 function and trastuzumab resistance.
FIGURE 11.
Model of IQGAP1 involvement in HER2 function. A1, HER2 (blue), which is constitutively phosphorylated, signals (red arrows) primarily through the AKT (orange ovals) and MAPK (not shown) pathways. HER2 signaling degrades p27 (light purple decagon), a CDK inhibitor, and thereby stimulates cell growth. A2, HER2 is internalized (green arrows) followed by degradation in the lysosome. A3, some receptors, particularly those bound to IQGAP1 (red), escape degradation and are recycled back to the plasma membrane where they are biologically active. B, trastuzumab-sensitive cells are shown. B1, trastuzumab (dark purple) inhibits signaling by HER2. The reduced activation of AKT results in accumulation of p27, which arrests the cell cycle, reducing growth. B2, trastuzumab also stimulates (purple arrow) HER2 internalization and degradation, reducing the number of HER2 receptors in the cell. C, trastuzumab-resistant cells are shown. These cells contain increased amounts of IQGAP1. The excess IQGAP1 stabilizes HER2 protein expression by reducing HER2 degradation. The resultant increase in the number of HER2 receptors enhances HER2 signaling, inducing trastuzumab resistance and promoting cell growth. p, phosphate.
Other than members of the ErbB family and selected intracellular signaling molecules, very few proteins are known to bind HER2 directly (8). We demonstrate that IQGAP1 interacts with HER2 and modulates HER2 expression and function. Combined with our data which show that IQGAP1 contributes to trastuzumab resistance, these findings imply that IQGAP1 is a potential target for the development of additional therapeutic strategies for patients with HER2(+) breast neoplasms.
Supplementary Material
Acknowledgments
We thank the staff of the Specialized Histopathology Core at Brigham and Women's Hospital for tissue slide preparation and immunostaining, Adrian Marino-Enriquez from the Department of Pathology, Brigham and Women's Hospital, for help with immunostaining interpretation, Simin Arad from the Department of Pathology, Brigham and Women's Hospital, for assistance with microscopy, and all members of the Sacks laboratory, past and present, for insightful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA075205-09 (to D. B. S.). This work was also funded by the Department of Defense Breast Cancer Research Program (BC087504; to C. D. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- EGFR
- epidermal growth factor receptor
- CDK
- cyclin-dependent kinase
- HER2
- human epidermal growth factor receptor 2
- VEGFR2
- VEGF receptor 2.
REFERENCES
- 1. Hynes N. E., Lane H. A. (2005) Nat. Rev. Cancer 5, 341–354 [DOI] [PubMed] [Google Scholar]
- 2. Yarden Y., Sliwkowski M. X. (2001) Nat. Rev. Mol. Cell Biol. 2, 127–137 [DOI] [PubMed] [Google Scholar]
- 3. Hynes N. E., MacDonald G. (2009) Curr. Opin. Cell Biol. 21, 177–184 [DOI] [PubMed] [Google Scholar]
- 4. Nahta R., Esteva F. J. (2006) Cancer Lett. 232, 123–138 [DOI] [PubMed] [Google Scholar]
- 5. Slamon D. J., Clark G. M., Wong S. G., Levin W. J., Ullrich A., McGuire W. L. (1987) Science 235, 177–182 [DOI] [PubMed] [Google Scholar]
- 6. Cho H. S., Mason K., Ramyar K. X., Stanley A. M., Gabelli S. B., Denney D. W., Jr., Leahy D. J. (2003) Nature 421, 756–760 [DOI] [PubMed] [Google Scholar]
- 7. Garrett T. P., McKern N. M., Lou M., Elleman T. C., Adams T. E., Lovrecz G. O., Kofler M., Jorissen R. N., Nice E. C., Burgess A. W., Ward C. W. (2003) Mol. Cell 11, 495–505 [DOI] [PubMed] [Google Scholar]
- 8. Moasser M. M. (2007) Oncogene 26, 6469–6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang H. Y., Zhou B. P., Hung M. C., Lee M. H. (2000) J. Biol. Chem. 275, 24735–24739 [DOI] [PubMed] [Google Scholar]
- 10. Lane H. A., Beuvink I., Motoyama A. B., Daly J. M., Neve R. M., Hynes N. E. (2000) Mol. Cell. Biol. 20, 3210–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neve R. M., Sutterlüty H., Pullen N., Lane H. A., Daly J. M., Krek W., Hynes N. E. (2000) Oncogene 19, 1647–1656 [DOI] [PubMed] [Google Scholar]
- 12. Yang H. Y., Shao R., Hung M. C., Lee M. H. (2001) Oncogene 20, 3695–3702 [DOI] [PubMed] [Google Scholar]
- 13. Lloyd R. V., Erickson L. A., Jin L., Kulig E., Qian X., Cheville J. C., Scheithauer B. W. (1999) Am. J. Pathol. 154, 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carter P., Presta L., Gorman C. M., Ridgway J. B., Henner D., Wong W. L., Rowland A. M., Kotts C., Carver M. E., Shepard H. M. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4285–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sliwkowski M. X., Lofgren J. A., Lewis G. D., Hotaling T. E., Fendly B. M., Fox J. A. (1999) Semin. Oncol. 26, 60–70 [PubMed] [Google Scholar]
- 16. Baselga J., Albanell J., Molina M. A., Arribas J. (2001) Semin. Oncol. 28, 4–11 [DOI] [PubMed] [Google Scholar]
- 17. Nahta R., Esteva F. J. (2007) Oncogene 26, 3637–3643 [DOI] [PubMed] [Google Scholar]
- 18. Le X. F., Claret F. X., Lammayot A., Tian L., Deshpande D., LaPushin R., Tari A. M., Bast R. C., Jr. (2003) J. Biol. Chem. 278, 23441–23450 [DOI] [PubMed] [Google Scholar]
- 19. Nahta R., Takahashi T., Ueno N. T., Hung M. C., Esteva F. J. (2004) Cancer Res. 64, 3981–3986 [DOI] [PubMed] [Google Scholar]
- 20. Baselga J., Tripathy D., Mendelsohn J., Baughman S., Benz C. C., Dantis L., Sklarin N. T., Seidman A. D., Hudis C. A., Moore J., Rosen P. P., Twaddell T., Henderson I. C., Norton L. (1996) J. Clin. Oncol. 14, 737–744 [DOI] [PubMed] [Google Scholar]
- 21. Cobleigh M. A., Vogel C. L., Tripathy D., Robert N. J., Scholl S., Fehrenbacher L., Wolter J. M., Paton V., Shak S., Lieberman G., Slamon D. J. (1999) J. Clin. Oncol. 17, 2639–2648 [DOI] [PubMed] [Google Scholar]
- 22. Vogel C. L., Cobleigh M. A., Tripathy D., Gutheil J. C., Harris L. N., Fehrenbacher L., Slamon D. J., Murphy M., Novotny W. F., Burchmore M., Shak S., Stewart S. J., Press M. (2002) J. Clin. Oncol. 20, 719–726 [DOI] [PubMed] [Google Scholar]
- 23. Piccart-Gebhart M. J., Procter M., Leyland-Jones B., Goldhirsch A., Untch M., Smith I., Gianni L., Baselga J., Bell R., Jackisch C., Cameron D., Dowsett M., Barrios C. H., Steger G., Huang C. S., Andersson M., Inbar M., Lichinitser M., Láng I., Nitz U., Iwata H., Thomssen C., Lohrisch C., Suter T. M., Rüschoff J., Suto T., Greatorex V., Ward C., Straehle C., McFadden E., Dolci M. S., Gelber R. D. (2005) N. Engl. J. Med. 353, 1659–1672 [DOI] [PubMed] [Google Scholar]
- 24. Nahta R., Esteva F. J. (2006) Breast Cancer Res. 8, 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Scaltriti M., Rojo F., Ocaña A., Anido J., Guzman M., Cortes J., Di Cosimo S., Matias-Guiu X., Ramon y Cajal S., Arribas J., Baselga J. (2007) J. Natl. Cancer Inst. 99, 628–638 [DOI] [PubMed] [Google Scholar]
- 26. Nagata Y., Lan K. H., Zhou X., Tan M., Esteva F. J., Sahin A. A., Klos K. S., Li P., Monia B. P., Nguyen N. T., Hortobagyi G. N., Hung M. C., Yu D. (2004) Cancer Cell 6, 117–127 [DOI] [PubMed] [Google Scholar]
- 27. Berns K., Horlings H. M., Hennessy B. T., Madiredjo M., Hijmans E. M., Beelen K., Linn S. C., Gonzalez-Angulo A. M., Stemke-Hale K., Hauptmann M., Beijersbergen R. L., Mills G. B., van de Vijver M. J., Bernards R. (2007) Cancer Cell 12, 395–402 [DOI] [PubMed] [Google Scholar]
- 28. Lu Y., Zi X., Zhao Y., Mascarenhas D., Pollak M. (2001) J. Natl. Cancer Inst. 93, 1852–1857 [DOI] [PubMed] [Google Scholar]
- 29. Nahta R., Yuan L. X., Zhang B., Kobayashi R., Esteva F. J. (2005) Cancer Res. 65, 11118–11128 [DOI] [PubMed] [Google Scholar]
- 30. Brown M. D., Sacks D. B. (2006) Trends Cell Biol. 16, 242–249 [DOI] [PubMed] [Google Scholar]
- 31. McNulty D. E., Li Z., White C. D., Sacks D. B., Annan R. S. (2011) J. Biol. Chem. 286, 15010–15021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuroda S., Fukata M., Nakagawa M., Fujii K., Nakamura T., Ookubo T., Izawa I., Nagase T., Nomura N., Tani H., Shoji I., Matsuura Y., Yonehara S., Kaibuchi K. (1998) Science 281, 832–835 [DOI] [PubMed] [Google Scholar]
- 33. Briggs M. W., Li Z., Sacks D. B. (2002) J. Biol. Chem. 277, 7453–7465 [DOI] [PubMed] [Google Scholar]
- 34. Yamaoka-Tojo M., Ushio-Fukai M., Hilenski L., Dikalov S. I., Chen Y. E., Tojo T., Fukai T., Fujimoto M., Patrushev N. A., Wang N., Kontos C. D., Bloom G. S., Alexander R. W. (2004) Circ. Res. 95, 276–283 [DOI] [PubMed] [Google Scholar]
- 35. Johnson M., Sharma M., Henderson B. R. (2009) Cell. Signal. 21, 1471–1478 [DOI] [PubMed] [Google Scholar]
- 36. White C. D., Brown M. D., Sacks D. B. (2009) FEBS Lett. 583, 1817–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. French P. J., Swagemakers S. M., Nagel J. H., Kouwenhoven M. C., Brouwer E., van der Spek P., Luider T. M., Kros J. M., van den Bent M. J., Sillevis Smitt P. A. (2005) Cancer Res. 65, 11335–11344 [DOI] [PubMed] [Google Scholar]
- 38. Bertucci F., Salas S., Eysteries S., Nasser V., Finetti P., Ginestier C., Charafe-Jauffret E., Loriod B., Bachelart L., Montfort J., Victorero G., Viret F., Ollendorff V., Fert V., Giovaninni M., Delpero J. R., Nguyen C., Viens P., Monges G., Birnbaum D., Houlgatte R. (2004) Oncogene 23, 1377–1391 [DOI] [PubMed] [Google Scholar]
- 39. Sun W., Zhang K., Zhang X., Lei W., Xiao T., Ma J., Guo S., Shao S., Zhang H., Liu Y., Yuan J., Hu Z., Ma Y., Feng X., Hu S., Zhou J., Cheng S., Gao Y. (2004) Cancer Lett. 212, 83–93 [DOI] [PubMed] [Google Scholar]
- 40. Patel V., Hood B. L., Molinolo A. A., Lee N. H., Conrads T. P., Braisted J. C., Krizman D. B., Veenstra T. D., Gutkind J. S. (2008) Clin. Cancer Res. 14, 1002–1014 [DOI] [PubMed] [Google Scholar]
- 41. White C. D., Khurana H., Gnatenko D. V., Li Z., Odze R. D., Sacks D. B., Schmidt V. A. (2010) BMC Gastroenterol. 10, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou R., Skalli O. (2000) Differentiation 66, 165–172 [DOI] [PubMed] [Google Scholar]
- 43. Walch A., Seidl S., Hermannstädter C., Rauser S., Deplazes J., Langer R., von Weyhern C. H., Sarbia M., Busch R., Feith M., Gillen S., Höfler H., Luber B. (2008) Mod. Pathol. 21, 544–552 [DOI] [PubMed] [Google Scholar]
- 44. Jadeski L., Mataraza J. M., Jeong H. W., Li Z., Sacks D. B. (2008) J. Biol. Chem. 283, 1008–1017 [DOI] [PubMed] [Google Scholar]
- 45. Ho Y. D., Joyal J. L., Li Z., Sacks D. B. (1999) J. Biol. Chem. 274, 464–470 [DOI] [PubMed] [Google Scholar]
- 46. Ren J. G., Li Z., Crimmins D. L., Sacks D. B. (2005) J. Biol. Chem. 280, 34548–34557 [DOI] [PubMed] [Google Scholar]
- 47. Nahta R., Esteva F. J. (2004) Cancer Chemother. Pharmacol. 53, 186–190 [DOI] [PubMed] [Google Scholar]
- 48. Roy M., Li Z., Sacks D. B. (2004) J. Biol. Chem. 279, 17329–17337 [DOI] [PubMed] [Google Scholar]
- 49. Roy M., Li Z., Sacks D. B. (2005) Mol. Cell. Biol. 25, 7940–7952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li Z., Sacks D. B. (2003) J. Biol. Chem. 278, 4347–4352 [DOI] [PubMed] [Google Scholar]
- 51. White C. D., Coetsee M., Morgan K., Flanagan C. A., Millar R. P., Lu Z. L. (2008) Mol. Endocrinol. 22, 2520–2530 [DOI] [PubMed] [Google Scholar]
- 52. Li L., Li Z., Sacks D. B. (2005) J. Biol. Chem. 280, 13097–13104 [DOI] [PubMed] [Google Scholar]
- 53. Mataraza J. M., Briggs M. W., Li Z., Frank R., Sacks D. B. (2003) Biochem. Biophys. Res. Commun. 305, 315–321 [DOI] [PubMed] [Google Scholar]
- 54. Shattuck D. L., Miller J. K., Carraway K. L., 3rd, Sweeney C. (2008) Cancer Res. 68, 1471–1477 [DOI] [PubMed] [Google Scholar]
- 55. Wu K., Wang C., D'Amico M., Lee R. J., Albanese C., Pestell R. G., Mani S. (2002) Mol. Cancer Ther. 1, 695–706 [PubMed] [Google Scholar]
- 56. Le X. F., Pruefer F., Bast R. C., Jr. (2005) Cell Cycle 4, 87–95 [DOI] [PubMed] [Google Scholar]
- 57. Tanner M., Kapanen A. I., Junttila T., Raheem O., Grenman S., Elo J., Elenius K., Isola J. (2004) Mol. Cancer Ther. 3, 1585–1592 [PubMed] [Google Scholar]
- 58. Subramaniam D. S., Isaacs C. (2005) Curr. Treat. Options Oncol. 6, 147–159 [DOI] [PubMed] [Google Scholar]
- 59. Mataraza J. M., Briggs M. W., Li Z., Entwistle A., Ridley A. J., Sacks D. B. (2003) J. Biol. Chem. 278, 41237–41245 [DOI] [PubMed] [Google Scholar]
- 60. Meyer R. D., Sacks D. B., Rahimi N. (2008) PLoS ONE 3, e3848–e3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Benseñor L. B., Kan H. M., Wang N., Wallrabe H., Davidson L. A., Cai Y., Schafer D. A., Bloom G. S. (2007) J. Cell Sci. 120, 658–669 [DOI] [PubMed] [Google Scholar]
- 62. Com E., Lagadec C., Page A., El Yazidi-Belkoura I., Slomianny C., Spencer A., Hammache D., Rudkin B. B., Hondermarck H. (2007) Mol. Cell. Proteomics 6, 1842–1854 [DOI] [PubMed] [Google Scholar]
- 63. Sorkin A., Goh L. K. (2009) Exp. Cell Res. 315, 683–696 [DOI] [PubMed] [Google Scholar]
- 64. Citri A., Kochupurakkal B. S., Yarden Y. (2004) Cell Cycle 3, 51–60 [PubMed] [Google Scholar]
- 65. Sharma M., Henderson B. R. (2007) J. Biol. Chem. 282, 8545–8556 [DOI] [PubMed] [Google Scholar]
- 66. Wickström S. A., Lange A., Hess M. W., Polleux J., Spatz J. P., Krüger M., Pfaller K., Lambacher A., Bloch W., Mann M., Huber L. A., Fässler R. (2010) Dev. Cell 19, 574–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li H., Sánchez-Torres J., Del Carpio A., Salas V., Villalobo A. (2004) Biochem. J. 381, 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nahta R., Hung M. C., Esteva F. J. (2004) Cancer Res. 64, 2343–2346 [DOI] [PubMed] [Google Scholar]
- 69. Landgraf R. (2007) Breast Cancer Res. 9, 202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Warren C. M., Landgraf R. (2006) Cell. Signal. 18, 923–933 [DOI] [PubMed] [Google Scholar]
- 71. Graus-Porta D., Beerli R. R., Daly J. M., Hynes N. E. (1997) EMBO J. 16, 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Di Fiore P. P., Pierce J. H., Kraus M. H., Segatto O., King C. R., Aaronson S. A. (1987) Science 237, 178–182 [DOI] [PubMed] [Google Scholar]
- 73. Bargmann C. I., Weinberg R. A. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 5394–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schaefer G., Akita R. W., Sliwkowski M. X. (1999) J. Biol. Chem. 274, 859–866 [DOI] [PubMed] [Google Scholar]
- 75. Ren J. G., Li Z., Sacks D. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10465–10469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Baselga J., Swain S. M. (2009) Nat. Rev. Cancer 9, 463–475 [DOI] [PubMed] [Google Scholar]
- 77. Sacks D. B. (2006) Biochem. Soc. Trans. 34, 833–836 [DOI] [PubMed] [Google Scholar]
- 78. Dokmanovic M., Hirsch D. S., Shen Y., Wu W. J. (2009) Mol. Cancer Ther 8, 1557–1569 [DOI] [PubMed] [Google Scholar]
- 79. Swart-Mataraza J. M., Li Z., Sacks D. B. (2002) J. Biol. Chem. 277, 24753–24763 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.