Abstract
An early step in the intracellular processing of vitamin B12 involves CblC, which exhibits dual reactivity, catalyzing the reductive decyanation of cyanocobalamin (vitamin B12), and the dealkylation of alkylcobalamins (e.g. methylcobalamin; MeCbl). Insights into how the CblC scaffold supports this chemical dichotomy have been unavailable despite it being the most common locus of patient mutations associated with inherited cobalamin disorders that manifest in both severe homocystinuria and methylmalonic aciduria. Herein, we report structures of human CblC, with and without bound MeCbl, which provide novel biochemical insights into its mechanism of action. Our results reveal that CblC is the most divergent member of the NADPH-dependent flavin reductase family and can use FMN or FAD as a prosthetic group to catalyze reductive decyanation. Furthermore, CblC is the first example of an enzyme with glutathione transferase activity that has a sequence and structure unrelated to the GST superfamily. CblC thus represents an example of evolutionary adaptation of a common structural platform to perform diverse chemistries. The CblC structure allows us to rationalize the biochemical basis of a number of pathological mutations associated with severe clinical phenotypes.
Keywords: Adenosylcobalamin, Flavin, Homocysteine, Protein Structure, Vitamins and Cofactors, Glutathione Transferase
Introduction
A network of trafficking proteins tailor and escort B12, a reactive and rare cofactor, from its point of entry into the cell to its two client enzymes in mammals, methionine synthase, and methylmalonyl-CoA mutase (1). An early step in this processing pathway involves accepting the B12 cargo as it exits the lysosome and converting the varied incoming derivatives into a common intermediate that can be partitioned for synthesis into the two biologically active forms, methylcobalamin (MeCbl)2 and 5′-deoxyadenosylcobalamin (AdoCbl). This step is catalyzed by the CblC protein (2, 3), the product of the cblC locus, a hotbed of mutations in the group of inherited cobalamin disorders (4). CblC is unique among B12 proteins in being able to manipulate the cobalt-carbon bond in B12 in both a homolytic and heterolytic fashion. It catalyzes the reductive decyanation of cyanocobalamin (CNCbl or vitamin B12) in the presence of a flavoprotein oxidoreductase (3). Remarkably, when presented with an alkylcobalamin, CblC also catalyzes the nucleophilic displacement of the alkyl group by glutathione (2) (see Fig. 1a). The lack of obvious sequence similarity between CblC and any protein of known structure has limited mechanistic insights into the structural basis of its remarkable chemical versatility. In this study, we report the crystal structures of human CblC in the apo (2.0 Å) and B12-bound (1.95 Å) forms, which reveal the presence of a large cavity for housing B12 and an N-terminal flavodoxin nitroreductase domain. The latter suggested that CblC might be a flavin-dependent B12 processing enzyme, which was confirmed by the demonstration that CblC catalyzes decyanation of CNCbl in the presence of FMNH2. The structures provide a framework for understanding the biochemical penalties associated with patient mutations leading to methylmalonic aciduria and homocystinuria.
FIGURE 1.
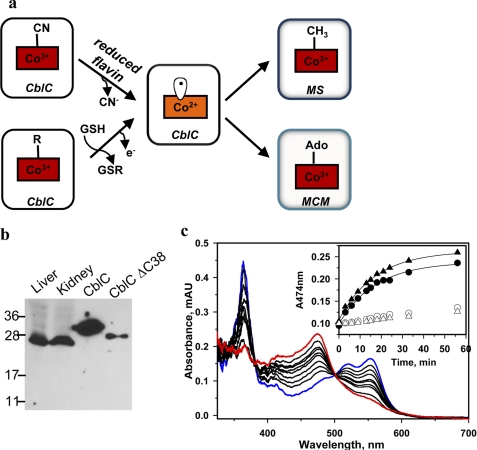
The predominance of t-CblC in tissues and the reactions it catalyzes. a, decyanation and dealkylation reactions catalyzed by CblC. b, the predominant form of CblC in murine liver and kidney extracts (∼26 kDa) migrates at a molecular mass consistent with its truncation (28.9 kDa with His tag versus 32.8 kDa with His tag for full-length CblC). c, t-CblC (50 μm) catalyzes conversion of 35 μm CNCbl (blue) to cob(II)alamin (red) in the presence of 40 μm FMNH2. The inset shows the kinetics of decyanation in the presence of t-CblC/FMNH2 (●), t-CblC/methionine synthase reductase (▴) or in the absence of the enzyme with FMNH2 (○) or methionine synthase reductase (△). MS and MCM denote methionine synthase and methylmalonyl-CoA mutase, respectively.
EXPERIMENTAL PROCEDURES
Expression and Purification of CblC Constructs
Truncated CblC(1–244)ΔC38 (construct 1) was cloned into the pET28b(+) vector (Novagen) between the NcoI and XhoI restriction sites using full-length CblC as a template and the following primers: 5′-CGTCTCCCATGGAGCCGAAAGTCGCAGAGCTGAAGCAG-3′ (forward) and 5′-ATGTGACTCGAGACTAGGCTTCTCTGAGGGCTGAG-3′ (reverse). To aid with phasing, the quadruple methionine mutant L48M/V83M/V107M/I164M CblC(1–244)ΔC38 (construct 2) was generated using the QuikChange site-directed mutagenesis kit (Stratagene) using the following sense mutagenic primers: L48M (5′-CTGCCAGGACCTACCATGGCCTTCCT-3′), V83M (5′-CCAGTGGACCAGTGTATGGCCTACCA-3′), V107M (5′-ATTGCTGACTACGAGATGCACCCCAA-3′), and V164M (5′-GCCATCCGAGGGGTAATGCTGCTGC-3′).
CblC(1–238)ΔC44 (construct 3) was generated by LIC cloning into pMCSG7 (5). t-CblC(1–244) (construct 1) was expressed and purified as described previously for full-length CblC (2) with the following modifications. Cells were resuspended in 50 mm potassium phosphate, pH 7.8, 300 mm KCl, 20 mm imidazole buffer containing 0.2 mg/ml lysozyme, and one tablet of EDTA-free complete protease inhibitor (Roche Applied Science). Following sonication, the cell-free extract was subjected to nickel-nitrilotriacetic acid chromatography, followed by size exclusion chromatography on a Superdex 200 column. Purification of CblC(1–237)ΔC45, construct 3, containing a cleavable N-terminal His6 tag, included an additional step of tag removal by tobacco etch virus protease prior to size exclusion chromatography. For crystallization, purified recombinant proteins were exchanged in 50 mm Tris, pH 8.0, and 1 mm tris (2-carboxyethyl) phosphine and were concentrated to ∼10–15 mg/ml. For enzymatic studies, CblC proteins were exchanged into 100 mm Hepes, pH 8, 150 mm KCl, 10% glycerol or 50 mm Tris, pH 8. Selenomethionine-labeled (SeMet) CblC (construct 2) was expressed in an identical manner to constructs 1 and 3 with the exception that the cells were grown in minimal media supplemented with 50 mg/liter of SeMet.
CblC Expression in Murine Tissues and Protein Stability Studies
Western blot analysis using CblC antibody generated in-house was performed to determine the predominant form of the CblC protein in murine tissues. The tissues were homogenized in liquid nitrogen, resuspended in an equal volume of lysis buffer (0.1 m sodium phosphate (pH 7.4) containing 0.1% Triton X-100, 10 μl/ml protease inhibitor mixture (Sigma), including 25 μg/ml tosyllysine chloromethyl ketone, 25 μg/ml phenylmethylsulfonyl fluoride, 27 μg/ml aprotinin, and 10 μg/ml leupeptin) and stored at −80 °C until further use. The lysate was centrifuged at 4 °C for 30 min at ≈14,000 × g, and the protein concentration in the supernatant was determined by the Bradford assay (Bio-Rad). Alternatively, the pulverized tissue homogenate powder was boiled directly in loading buffer. Protein (200 μg/lane) samples were separated by electrophoresis on a 10% polyacrylamide gel followed by transfer to a PVDF membrane, and the expression of CblC was probed by using the respective primary and secondary antibodies and detected by using the chemiluminescent horseradish peroxidase system (Pierce).
The stability of full-length and t-CblC was monitored in a turbidity assay where the change in absorbance at 600 nm was recorded for each protein (0.5 mg/ml) incubated at temperatures ranging from 25 to 75 °C. Tm was determined using Equation 1.
CblC Dealkylation and Decyanation Activities
Dealkylation and decyanation were monitored as published previously (2, 3). In addition, decyanation of CNCbl by t-CblC was examined in the absence of methionine synthase reductase/NADPH using either FMNH2 (30–40 μm) or FMN/FAD (30–40 μm)/NADPH (200 μm) as source of reducing equivalents. Reduced flavins were generated prior to use by photochemical reduction in the presence of 10–20 mm sodium oxalate (6).
Binding of FMN and FAD to t-CblC by Protein Fluorescence
Flavin binding to apo-t-CblC was determined from quenching of protein fluorescence monitored at 340 nm following excitation at 280 nm. Fluorimetric titrations were carried out at room temperature (20 °C) in 1-ml quartz cuvettes on a Shimadzu RF-5301 PC spectrofluorometer, and the emission spectra at 300–500 nm were monitored with excitation and emission slit widths of 3 and 5 mm, respectively. Aliquots of 0–50 μm flavin were titrated in 0.1 μm protein in 100 mm Hepes, pH 8, 150 mm KCl, and 10% glycerol. The change in protein fluorescence was recorded after allowing 5 min for equilibration. The binding constant was determined using Equation 2.
Crystallization and Cryoprotection
Three constructs were used for crystallization. Construct 1, t-CblC(1–244), was a C-terminal His-tagged CblCΔC38 that crystallized in the apo-form even in the presence of various Cbl forms. Construct 2, t-CblC(1–244), is a variant of construct 1 with four methionine substitutions at positions Leu-48, Val-83, Val-107, and Val-164. The three Met residues in wild-type CblC did not provide a sufficient anomalous signal for phasing with data from crystals of the SeMet variant. Thus, we engineered the extra methionines in construct 2, which afforded diffraction quality crystals with a sufficient anomalous signal for phasing (see details below). Construct 3, t-CblC(1–238), ΔC44, is a “tag-less” construct derived from an N-terminal His-tagged construct after tobacco etch virus protease cleavage. After the removal of the His tag, the CblC protein was concentrated to 25–30 mg/ml, and MeCbl was added to a final concentration of 0.75–1.0 mm. The mixture was then passed over a size exclusion (S200) column to remove any free cofactor. The final concentration of MeCbl·CblC for crystallography was 10–15 mg/ml. All of the steps described above were performed in the dark to avoid photolysis of the cofactor. Construct 3 was engineered to address the problem of crystallization of a MeCbl·CblC complex using construct 1 with its C-terminal His tag.
All protein samples were concentrated to ∼10 mg/ml in 50 mm Tris, pH 8.0 and 1 mm TCEP before crystallization at 4 °C by the vapor diffusion method from 1:1 mixtures of protein solution with reservoir solution in sitting drop plates (Corning and Intel). The reservoir solution for construct 1 was 0.9 m sodium malonate, 5% Jeffamine 600, pH 7.0, 0.1 m Hepes, pH 6.7, and the solution for construct 2 was 0.96 m sodium malonate, 10% Jeffamine 600, pH 7.0, 0.1 m Hepes, pH 6.7. The resulting crystals of apo-t-CblC and the SeMet-apo-t-CblC were cryoprotected for a few minutes by soaking in 0.8 m sodium malonate, 5% Jeffamine 600, pH 7.0, 0.1 m Hepes, pH 6.7, and 20% (v/v) glycerol prior to flash cooling in liquid N2. Apo-t-CblCΔC38 (construct 1) crystals were of space group P6222 (a = b = 112.5, c = 112.1) with one molecule in the asymmetric unit (Matthews' coefficient VM = 3.2 Å3/Da for one molecules per asymmetric unit, 61.3% solvent content), whereas crystals of apo-SeMet-t-CblCΔC38 (construct 2) were of space group P65 (a = b = 69.3, c = 201.8) with two molecules in the asymmetric unit (Matthews' coefficient VM = 2.4 Å3/Da for two molecules per asymmetric unit, 49.0% solvent content).
Co-crystallization of t-CblCΔC44 (construct 3) with MeCbl was achieved using a reservoir solution of 2.0 m ammonium sulfate and 0.1 m Tris-HCl, pH 8.5. The resulting colored crystals of MeCbl·t-CblC were cryoprotected by a 1-h soak in 1.6 m ammonium sulfate, 0.1 m Tris-HCl, pH 8.5, and 25% (v/v) glycerol prior to flash cooling in liquid N2. MeCbl·t-CblCΔC44 (construct 3) crystals were of space group P6222 (a = b = 105.6, c = 84.2, α = β = 90.0, γ = 120.0) with one molecule in the asymmetric unit (Matthews' coefficient VM = 3.5 Å3/Da for one molecules per ASU, 65.3% solvent content).
Data Collection and Structure Determination
Diffraction data were collected at 100 K for all t-CblC crystals on beamline GM/CA-CAT 23-ID-D at the Advanced Photon Source, Argonne National Laboratory (Argonne, IL). Data were recorded on a Mar300 detector and processed with HKL200 (7). Phenix (8) AutoSol was used to identify the selenium sites and calculate density-modified 2.9-Å experimental maps based on a single-wavelength SAD data set from one crystal of SeMet-“enriched” protein. Specifically, 13 of 14 selenium sites were located and used for SAD phasing, using phenix.hyss. Subsequently, Phaser (9) was used to calculated experimental phases, followed by density modification by RESOLVE (10), including 2-fold averaging (figure of merit 0.35 before and 0.65 after density modification). The experimental density map showed clear features of the protein backbone and well defined side chains. Modeling of electron density was done manually using COOT (11). Restrained refinement with isotropic individual B-factors was performed using REFMAC5 (12) of the CCP4 suite (13). The final SeMet-CblC model is missing residues 135–145 and 230–244 in chain A and 1–4 and 233–244 in chain B. The final SeMet-CblC model was used to solve the other structures by molecular replacement with Phaser using the 2.0 Å apo-t-CblC data set and the 1.95 Å MeCbl·t-CblC data set. Both molecular replacement solutions were refined with PHENIX, including rigid body refinement of the individual domains, followed by simulated annealing in torsional space, individual atomic refinement, and restrained individual B-factor adjustment with maximum likelihood targets. Initial density allowed for the MeCbl cofactor to be modeled. The geometric quality of the models was assessed with MolProbity (14). PyMOL (15) was used to create molecular images.
RESULTS AND DISCUSSION
Truncated CblC Is Predominant Cellular Form of Protein
Human CblC is 282 amino acids in length; however, the last ∼40 residues are predicted to be highly unstructured and are absent in nonmammalian CblCs (supplemental Fig. S1). The cblC gene is predicted to be alternatively spliced by ECgene (16) to generate products that are either 245 or 282 amino acids in length and, despite the relatively large number of patient mutations (>700 alleles) mapped to CblC (4, 17), no disease-causing mutations have been confirmed beyond residue 245.3 These data beg the question as to whether the truncated form of CblC predominates intracellularly. Western blot analysis of murine tissue reveals the presence of a single band with a molecular mass of ∼26 kDa, corresponding to the truncated human protein, t-CblC (28.9 kDa for the His-tagged recombinant form), and in excellent agreement with the calculated size for murine t-CblC (27 kDa) (Fig. 1b). This result indicates that t-CblC is the predominant, if not only, form of CblC. The decyanation activity of t-CblC(1–244) is similar to full-length CblC, whereas the dealkylation activity is ∼2-fold higher (2, 3). The full-length protein (Tm = 41.5 + 0.5 °C) is slightly less stable than t-CblC (Tm = 46.6 + 1.5 °C).
Crystallization of CblC
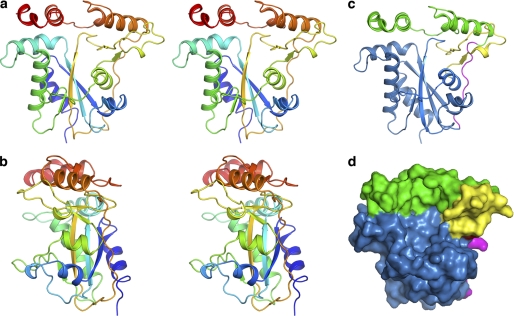
The apo- and Cbl-bound forms of human t-CblC were crystallized, and phases were obtained through Se anomalous diffraction for apo-CblC and by molecular replacement for the Cbl-bound form (Table 1). CblC is comprised of two modules (N-terminal 1–172; C-terminal 183–244) connected by a long linker (residues 173–182) (Fig. 2). The N-terminal core module exhibits a fold characteristic of the NADP oxidase/flavin reductase family and contains a four-stranded antiparallel β-sheet flanked by α-helices and a short antiparallel two-stranded β-sheet (supplemental Fig. S2). The C-terminal module (cap) is composed of four helices that cap the core N-terminal module (Fig. 2a). The fourth helix is ordered in apo-CblC but disordered in the MeCbl·t-CblC complex. A large cavity is formed at the interface of the cap and core of CblC (Fig. 2). CblC was predicted to house a C-terminal TonB-like domain (residues 185–282) (4) found in bacterial two-protein transporter systems. However, the C-terminal cap domain does not exhibit a TonB-like fold.
TABLE 1.
Data Collection and Refinement Statistics (SAD Phasing)
NA, not available.
| Protein | Apo-CblC | SeMet CblC (peak) | MeCbl CblC |
|---|---|---|---|
| Data collection | |||
| Space group | P6222 | P65 | P6222 |
| Cell dimensions a, b, c (Å) | 112.4, 112.4, 112.1 | 69.3, 69.3, 201.8 | 105.6, 105.6, 84.2 |
| Wavelength (Å) | |||
| Resolution (Å) | 50-2.00 (2.07-2.00) | 50-2.70 (2.80-2.70) | 50-1.95 (2.02-1.95) |
| Rsym (%) | 6.6 (50.2) | 13.3 (47.3) | 7.5 (49.5) |
| I/σI | 21.5 (5.8) | 16.1 (5.8) | 15.9 (2.9) |
| Completeness (%) | 99.9 (100.0) | 100.0 (100.0) | 99.9 (99.8) |
| Redundancy | 11.4 (10.9) | 7.7 (7.5) | 6.6 (4.1) |
| Refinement | |||
| Resolution (Å) | 50-2.00 | 50-2.70 | 50-1.95 |
| No. of reflections | 27786 | 14911 | 20022 |
| Rwork/Rfree (%) | 0.189/0.230 | 0.228/0.290 | 0.186/0.244 |
| No. of atoms | |||
| Protein | 1885 | 3614 | 1915 |
| Ligands | 27 | NA | NA |
| MeCbl | NA | NA | 92 |
| Water | 142 | 30 | 179 |
| B-factors (Å2) | |||
| Protein | 43.5 | 42.7 | 30.9 |
| Ligands | 49.2 | NA | NA |
| MeCbl | NA | NA | 25.1 |
| Water | 48.1 | 42.4 | 37.8 |
| Root mean square deviations | |||
| Bond lengths (Å) | 0.008 | 0.007 | 0.012 |
| Bond angles | 1.192° | 1.052° | 1.552° |
| Ramachandran plot (%) | |||
| Favored/allowed/outliers | 98.7/0.9/0.4 | 96.1/3.7/0.2 | 97.0/2.6/0.4 |
| Protein Data Bank code | 3SBZ | 3SBY | 3SC0 |
FIGURE 2.
Structure of apo-CblC. a and b, stereoview of apo-t-CblC in two orientations, related by a 90° rotation. Ribbon (c) and space-filling (d) representations of the arrangement of the apo-t-CblC modules. The core module is shown in blue, the cap is displayed in green, the linker is in pink, and the two β-sheet antiparallel insertion of the core domain is shown in yellow.
CblC Has a Flavin Reductase Fold
Although CblC has the fold of a flavin reductase, it has very low sequence identity (<12%) and poor structure alignment with other members of the flavin reductase family. Two of the closest structural homologues of CblC are also examples of noncanonical flavin reductases, BluB (18) (root mean square deviation = 4.2 Å based on 117 residues, 9% identity) and iodotyrosine deiodinase (19) (root mean square deviation = 3.0 Å based on 102 residues, 7% identity) (supplemental Fig. S3a). Both enzymes are members of a subfamily with novel catalytic activities atypical of flavin reductases; BluB cannibalizes flavin to synthesize the cobalamin tail and iodotyrosine deiodinase uses mono- and di-iodotyrosine to salvage iodine in the thyroid. Although CblC possesses the overall flavin reductase fold, it is the most divergent member of the flavin reductase family and is the first member of a new subfamily. CblC catalyzes the dealkylation of alkylcobalamins, a reaction that is flavin-independent. Decyanation of CNCbl requires reducing equivalents, and the diflavin oxidoreductases, methionine synthase reductase, and novel reductase 1, can couple to CblC (3). The structural similarity of CblC to members of the flavin reductase family suggests that it has intrinsic flavin oxidoreductase activity, consistent with a report of reduced NADH-dependent cobalamin reductase activity in cblC cell lines (20) and the reported decyanation activity in the presence of NADPH and FAD (21).
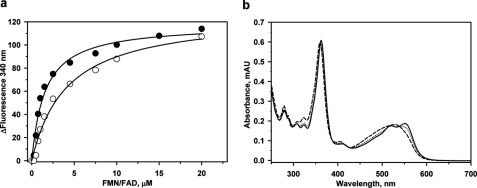
We characterized the reductive decyanation of CNCbl in the presence of FMN/FAD and NADPH (Fig. 1c). The observed decyanation rate (k = 0.07 ± 0.01 min−1), is comparable with those observed in the presence of methionine synthase reductase (0.10 ± 0.004 min−1) and novel reductase 1 (0.068 ± 0.007 min−1) (3). Intrinsic protein fluorescence quenching was used to monitor binding of FMN (KD = 4.4 ± 1.1 μm) and FAD (1.4 ± 0.3 μm) to apo-t-CblC (Fig. 4).
FIGURE 4.
Binding of flavin and CNCbl to CblC. a, FMN (○) and FAD (●) to apo-t-CblC binding to CblC. A solution of t-CblC was titrated with increasing concentrations of FMN or FAD, respectively as described under “Experimental Procedures.” Upon excitation at 280 nm, changes in the emission spectra were recorded between 300 and 450 nm. The binding constant for each flavin was determined by plotting the changes at 340 nm as a function of ligand concentration as described under ”Experimental Procedures.” b, time-dependent spectral change from base-on to base-off CNCbl in the presence of t-CblC. t-CblC (50 μm) was incubated with 30 μm CNCbl and changes in the absorption spectra were recorded over a period of time. The initial spectra of CNCbl-bound to t-CblC (dashed line) changed over time to an intermediate spectrum at 3 min (dotted line) and finally to the spectrum of base-off CNCbl-bound t-CblC after 20 min (dashed line). No further changes were observed after this time point. The authenticity of the base-off CNCbl-bound CblC was confirmed by comparing the spectra to that of CNCbl in 7% HCl (not shown). mAu, milli-arbitrary units.
Unlike other flavin reductases, which bind flavin at a dimer interface, CblC is a monomer in solution and in crystals. The inability of CblC to dimerize can be explained structurally. Other flavin reductases form intersubunit β-sheets in which each subunit contributes a β-strand to the central sheet of the partner subunit. Helix HD and the following loop (LDE) block formation of this contact in CblC (supplemental Figs. S2 and S3). Attempts to soak either FMN or FAD into apo- and MeCbl-bound t-CblC crystals were unsuccessful. However, based on structural alignments with other flavin reductases, we can model a probable flavin-binding site (supplemental Fig. S3). In the apo-t-CblC structure, the predicted flavin pocket is blocked by loop L3F (supplemental Fig. S3a). The putative flavin site becomes accessible upon Cbl binding, which induces structural changes as described below.
B12 Binds to CblC in “Base-off” Conformation
The structure of t-CblC in the presence of MeCbl reveals that Cbl binds in the large cavity between core and cap in a base-off form, i.e. where the endogenous lower axial base, dimethylbenzimidazole, is not a ligand to the cobalt as predicted by absorption spectroscopy (3) (Fig. 3). On the other hand, CblC has been reported to bind CNCbl in both the “base-on” (3) and base-off (21) state. This discrepancy is explained by the observation that the conversion from the base-on to CblC-bound base-off state is slow (Fig. 4b). The pKa for protonation of dimethylbenzimidazole in CNCbl is 0.1 and governs the base-on/base-off equilibrium in solution (22). By binding the cofactor in the base-off form, the protein enhances the reactivity of cobalt for reductive decyanation. By removing the electron-releasing nitrogen ligand, the base-off state facilitates cobalt reduction (23), providing a chemical rationale for binding the substrate in this conformation.
FIGURE 3.
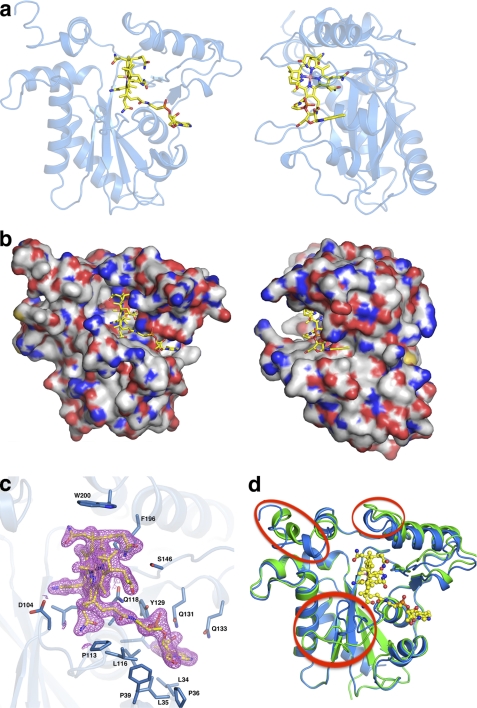
Structure of holo-CblC. MeCbl·t-CblC shown in two orientations and displayed in schematic (a) and space-filling (b) representations. c, 2Fo − Fc electron density contoured at 1.5σ (magenta) and stick model (yellow) of MeCbl is shown with its immediate protein environment. Residues in close proximity to MeCbl are shown as sticks (blue). d, superposition of the apo- (green) and MeCbl- (blue) t-CblC structures in which the three major differences are encircled in red.
The cobalamin tail resides in a crevice on the surface of CblC (Fig. 3, a and b), which is formed between helix HF and the two-stranded antiparallel β-sheet (supplemental Fig. S3). Contacts in the crevice are largely hydrophobic with only two hydrogen bonds between the cofactor tail and residues Tyr-129 and Gln-131 (Fig. 3c and supplemental Table S1). The corrin ring binds in the large cavity with its nonalkylated, “lower ligand” side against the two-stranded anti-parallel β-sheet. This is in striking contrast to the mammalian B12-dependent enzymes, which bind cobalamin in a base-off/His on conformation, wherein a histidine residue in a conserved DXHXXGX41SXLX26–28GG motif serves as a lower axial ligand to the cobalt. CblC lacks the His motif and does not provide a lower ligand. The cofactor is secured at the cavity wall by ten polar interactions between the side chains of the corrin ring and backbone and side chain atoms from both the core and cap of CblC (Fig. 3 and supplemental Table S1). The “upper ligand” face of the corrin ring is oriented toward the unfilled portion of the active site cavity. The cobalamin “cage” is further defined on the top by part of the cap module (helix I and loop LIJ) and on the floor by loop L3F. Electron density is clear for the upper axial methyl group of MeCbl. The large cavity is only partially filled by MeCbl. The cavity size explains the latitude of CblC to accommodate a structural diversity of Cbl derivatives ranging from a methyl to a 5′-deoxyadenosyl group (24). The cavity volume is large enough to also accommodate glutathione adjacent to the cobalt alkyl ligand (supplemental Fig. S4), consistent with the glutathione-dependent dealkylation activity of CblC.
Conformational Changes Induced by B12 Binding
MeCbl binding results in three conformational changes in CblC (Fig. 3d). The most significant is movement of loop L3F toward Cbl, creating the floor of the binding pocket. In the apo-t-CblC structure, the L3F loop blocks the putative flavin binding site, and the conformational change suggests that Cbl binding induces formation of or stabilizes the flavin pocket. This is consistent with the higher affinity of CblC for FMN in the presence (1.5 ± 0.2 μm) versus absence (0.5 ± 0.1 μm) of CNCbl. The affinity of CblC for flavin might be further modulated by interaction between CblC and the next protein in the B12-trafficking pathway, e.g. CblD.
A second difference is the repositioning of the LIJ loop in the cap to form part of the Cbl-binding pocket. Finally, helix HL in the C-terminal cap is disordered in the MeCbl·t-CblC structure. Although our attempts to build into electron density for this region were unsuccessful, the cap region is clearly in a different and more flexible conformation in the MeCbl·t-CblC structure. Glutathione was modeled into the MeCbl·t-CblC structure by placing the sulfur near the methyl ligand of MeCbl (supplemental Fig. S4). We speculate that glutathione binding may order the flexible C terminus of the cap. Although CblC exhibits GST activity, it lacks both the conserved G-site for glutathione binding and the thioredoxin domain with distinct βαβαββα topology, which are hallmarks of the GST superfamily (25, 26).
Structural Insights into Pathogenic Mutations in CblC
Our data allow us to rationalize the potential biochemical penalties associated with a number of patient mutations (supplemental Fig. S1). Most of the pathological mutations map to residues (supplemental Fig. S1 and Fig. 5) that can be grouped into two major categories: positions important for cobalamin binding and for structural integrity. The significance of another category of residues that are mutated in a small subset of patients is not obvious. Most mutations within this group are benign with the notable exception of R161G/R161Q. Arg-161 is proximal to but does not contact the upper axial ligand of cobalamin. Arg-161 may be important for glutathione binding and/or the dealkylation activity. Interestingly, none of the patient mutations map to the putative FMN binding site. We speculate that mutations in the FMN binding site may be “silent” because CblC can use alternative reducing partners such as methionine synthase reductase and novel reductase 1.
FIGURE 5.
Patient mutations mapped onto the MeCbl·t-CblC structure. Residues are shown in ball-and-stick representation with those colored red representing residues involved in Cbl binding or structural integrity. The functions of residues in black are not obvious, and we speculate that Arg-73, due to its surface exposure, might be involved in protein-protein interactions, whereas Arg-161 might be involved in GSH binding due to its vicinity to the putative GSH binding site.
Summary
Our study provides structural insights into how catalytic versatility is embedded within the CblC scaffold enabling catalysis of radically distinct chemical transformations. The CblC structure reveals that it is the most divergent member of the NADPH-dependent flavin reductase family and suggests that the flavin site is formed on demand, i.e. when Cbl is bound. The cavernous Cbl-binding site explains the relaxed specificity for the Cbl upper axial ligand (24), an advantage for a processing enzyme that accepts varied B12 derivatives entering the cytoplasm. The CblC structure illustrates that its GST activity is housed in a scaffold that is unrelated to the GST superfamily. Hence, CblC represents a novel architectural solution for both the decyanation and dealkylation activities.
Supplementary Material
Acknowledgments
We thank Rowena G. Matthews for critical discussions and for input on the manuscript. We acknowledge the General Medicine and Cancer Collaborative Access Team, supported by the National Institute of General Medical Sciences (Y1-GM-1104) and the National Cancer Institute (Y1-CO-1020), for beam time and assistance at the Advanced Photon Source. We thank W. C. Brown and J. Delproposto (Life Sciences Institute, University of Michigan, Ann Arbor, MI) for cloning CblC in one of the expression plasmids (construct 3).
This work was supported, in whole or in part, by National Institutes of Health Grants DK45776 (to R. B.) and GM024908 (to Rowena G. Matthews).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
The atomic coordinates and structure factors (codes 3SBY, 3SBZ, and 3SC0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Of the three mutations described in Ref. 6 beyond residue 245, two (R267Q and S271G) are benign and one (a nine-amino acid deletion between Ser-264–Pro-272) is downstream of a frameshift-terminating mutation on the same allele.
- MeCbl
- methylcobalamin
- AdoCbl
- 5′-deoxyadenosylcobalamin
- SeMet
- selenomethionine
- CNCbl
- cyanocobalamin or vitamin B12
- t-CblC
- truncated CblC.
REFERENCES
- 1. Banerjee R., Gherasim C., Padovani D. (2009) Curr. Opin. Chem. Biol. 13, 484–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim J., Hannibal L., Gherasim C., Jacobsen D. W., Banerjee R. (2009) J. Biol. Chem. 284, 33418–33424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim J., Gherasim C., Banerjee R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14551–14554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lerner-Ellis J. P., Tirone J. C., Pawelek P. D., Doré C., Atkinson J. L., Watkins D., Morel C. F., Fujiwara T. M., Moras E., Hosack A. R., Dunbar G. V., Antonicka H., Forgetta V., Dobson C. M., Leclerc D., Gravel R. A., Shoubridge E. A., Coulton J. W., Lepage P., Rommens J. M., Morgan K., Rosenblatt D. S. (2006) Nat. Genet. 38, 93–100 [DOI] [PubMed] [Google Scholar]
- 5. Stols L., Gu M., Dieckman L., Raffen R., Collart F. R., Donnelly M. I. (2002) Protein Expr. Purif. 25, 8–15 [DOI] [PubMed] [Google Scholar]
- 6. Massey V., Hemmerich P. (1977) J. Biol. Chem. 252, 5612–5614 [PubMed] [Google Scholar]
- 7. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 8. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Terwilliger T. C. (2000) Acta Crystallogr. D. Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 12. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 13. (1994) Acta Crystallogr. D. Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 14. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Schrodinger, LLC, Portland, OR [Google Scholar]
- 16. Kim P., Kim N., Lee Y., Kim B., Shin Y., Lee S. (2005) Nucleic Acids Res. 33, D75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lerner-Ellis J. P., Anastasio N., Liu J., Coelho D., Suormala T., Stucki M., Loewy A. D., Gurd S., Grundberg E., Morel C. F., Watkins D., Baumgartner M. R., Pastinen T., Rosenblatt D. S., Fowler B. (2009) Hum. Mutat. 30, 1072–1081 [DOI] [PubMed] [Google Scholar]
- 18. Taga M. E., Larsen N. A., Howard-Jones A. R., Walsh C. T., Walker G. C. (2007) Nature 446, 449–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thomas S. R., McTamney P. M., Adler J. M., Laronde-Leblanc N., Rokita S. E. (2009) J. Biol. Chem. 284, 19659–19667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pezacka E. H. (1993) Biochem. Biophys. Acta. 1157, 167–177 [DOI] [PubMed] [Google Scholar]
- 21. Froese D. S., Zhang J., Healy S., Gravel R. A. (2009) Mol. Genet. Metab. 98, 338–343 [DOI] [PubMed] [Google Scholar]
- 22. Brown K. L., Hakimi J. M., Jacobsen D. W. (1984) J. Am. Chem. Soc. 106, 7894–7899 [Google Scholar]
- 23. Lexa D., Saveant J. M., Jacobsen D. W. (1983) Acc. Chem. Res. 16, 235–243 [Google Scholar]
- 24. Hannibal L., Kim J., Brasch N. E., Wang S., Rosenblatt D. S., Banerjee R., Jacobsen D. W. (2009) Mol. Genet. Metab. 97, 260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Allocati N., Federici L., Masulli M., Di Ilio C. (2009) FEBS J. 276, 58–75 [DOI] [PubMed] [Google Scholar]
- 26. Oakley A. J. (2005) Curr. Opin. Struct. Biol. 15, 716–723 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.