Abstract
ATP synthase uses a unique rotational mechanism to convert chemical energy into mechanical energy and back into chemical energy. The helix-turn-helix structure in the C-terminal domain of the β subunit containing the conserved DELSEED motif, termed “DELSEED-loop,” was suggested to be involved in coupling between catalysis and rotation. If this is indeed the role of the loop, it must have a critical length, the minimum length required to sustain its function. Here, the critical length of the DELSEED-loop was determined by functional analysis of mutants of Bacillus PS3 ATP synthase that had 7–14 amino acids within the loop deleted. A 10 residue deletion lost the ability to catalyze ATP synthesis, but was still an active ATPase. Deletion of 14 residues abolished any enzymatic activity. Modeling indicated that in both deletion mutants the DELSEED-loop was shortened by ∼10 Å; fluorescence resonance energy transfer experiments confirmed the modeling results. This appears to define the minimum length for DELSEED-loop required for coupling of catalysis and rotation. In addition, we could demonstrate that the loss of high-affinity binding to the catalytic site(s) that had been observed previously in two deletion mutants with 3–4 residues removed was not due to the loss of negative charged residues of the DELSEED motif in these mutants. An AALSAAA mutant in which all negative charges of the DELSEED motif were removed showed a normal pattern for MgATP binding to the catalytic sites, with a clearly present high-affinity site.
Keywords: ATP Synthase, ATPases, Enzyme Catalysis, Enzyme Mechanisms, Molecular Motors, Rotational Catalysis
Introduction
F1Fo-ATP synthase catalyzes the final step of oxidative phosphorylation and photophosphorylation, the synthesis of ATP from ADP and inorganic phosphate. F1Fo-ATP synthase consists of the membrane-embedded Fo subcomplex with, in most bacteria, a subunit composition of ab2c10, and the peripheral F1 subcomplex, with a subunit composition of α3β3γδϵ. The energy necessary for ATP synthesis is derived from an electrochemical transmembrane proton (or, in some organisms, sodium ion) gradient. Proton flow, down the gradient, through Fo is coupled to ATP synthesis on F1 by a unique rotary mechanism. The protons flow through (half) channels at the interface of a and c subunits, which drives rotation of the ring of c subunits. The c10 ring, together with F1 subunits γ and ϵ, forms the rotor. Rotation of γ leads to conformational changes in the catalytic nucleotide binding sites on the β subunits, where ADP and Pi are bound. The conformational changes result in formation and release of ATP. Thus, ATP synthase converts electrochemical energy, the proton gradient, into mechanical energy in form of subunit rotation, and back into chemical energy as ATP. In bacteria, under certain physiological conditions, the process runs in reverse. ATP is hydrolyzed to generate a transmembrane proton gradient which the bacterium requires for such functions as nutrient import and locomotion (for reviews, see Refs. 1–6).
F1 (or “F1-ATPase”) has three catalytic nucleotide binding sites, located on the β subunits, at the interface to the adjacent α subunit. The catalytic sites have pronounced differences in their nucleotide binding affinity. During rotational catalysis, the sites switch their affinities in a synchronized manner; the position of γ determines which catalytic site is the high-affinity site (Kd1 in the nanomolar range), which site is the medium-affinity site (Kd2 ≈ 1 μm), and which site is the low-affinity site (Kd3 ≈ 30–100 μm; see Refs. 7, 8). In the original crystal structure of bovine mitochondrial F1 (9), one of the three catalytic sites was filled with the ATP analog AMPPNP,2 a second one with ADP (plus azide; see Ref. 10), and the third site was empty. Hence, the β subunits are referred to βTP, βDP, and βE. The occupied β subunits, βTP and βDP, were in a closed conformation, the empty βE subunit was in an open conformation. The main difference between these two conformations is found in the C-terminal domain. Here, the “DELSEED-loop,” a helix-turn-helix structure containing the conserved DELSEED motif, is in an “up” position when the catalytic site on the respective β subunit is filled with nucleotide, and in a “down” position when the site is empty. When all three catalytic sites are occupied by nucleotide, the previously open βE subunit assumes an intermediate, half-closed (“βHC”) conformation. It cannot close completely because of steric clashes with γ (11).
The DELSEED-loop of each of the three β subunits makes contact with the γ subunit. These interactions, the movement of the DELSEED-loop during catalysis, the conservation not only of the DELSEED motif, but of the whole loop (12), and a number of mutagenesis experiments (12–14) led to the assumption that the DELSEED-loop might play an essential role in coupling between catalysis and rotation of γ. According to a model favored by several authors (Refs. 5, 15, 16; see also Refs. 17–19), binding of ATP (or, more precisely, MgATP) to the low-affinity catalytic site on βE and the subsequent closure of this site, accompanied by its conversion into the high-affinity site, are responsible for driving the large (∼80°) rotation substep during ATP hydrolysis, with the DELSEED-loop acting as a “pushrod” on γ. The finding that an AALSAAA mutant was able to drive rotation of γ upon ATP hydrolysis with the same torque as the wild-type enzyme (14) showed that the negatively charged amino acid residues of the DELSEED motif are not essential. In agreement with this result, there is increasing evidence that it is the bulk of the DELSEED-loop that makes γ rotate, more so than individual interactions. A molecular dynamics (20) study implicated mainly the region around several hydrophobic residues upstream of the DELSEED motif (specifically βI386 and βL387)3 as being responsible for making contact with γ during the large rotation substep. We could recently show (12) that no individual residue at the β/γ interface is essential, using deletion mutants of Bacillus PS3 ATP synthase that had their DELSEED-loops shortened by 3–7 residues. Every residue between βL380 and βR402 was included in at least one of the deletion mutants. According to the crystal structures, residues between βD382 and βD394 are involved in β/γ contacts. The deletion mutants covering this latter stretch were all active ATPases and ATP synthases, albeit most of them were less well coupled than the wild-type (12).
If the DELSEED-loop drives rotation of γ during ATP hydrolysis, it should not be possible to reduce its length beyond a certain limit. In the present study, our main goal was to define the critical length of the DELSEED-loop, the minimal length necessary to sustain catalysis. For this purpose, we generated deletion mutants where longer stretches of between 7 and 14 residues in the DELSEED-loop of PS3 ATP synthase were removed. A deletion of 10 residues essentially abolished ATP synthesis; a deletion of 14 residues resulted in the additional loss of ATPase activity. According to modeling data, in these mutants the DELSEED-loop is shortened by 10 Å or more. This was confirmed by fluorescence resonance energy transfer (FRET). A reduction by 10 Å appears to define the critical length of the loop; beyond this point the capability to catalyze ATP synthesis and hydrolysis ceases. In the previous study (12), it was demonstrated that two deletions encompassing residues of the DELSEED motif itself resulted in “loss” of the high-affinity catalytic nucleotide binding site. Here we show that this loss is not due to the associated removal of the negative charges. The quintuple point mutation, AALSAAA, exhibited normal binding behavior, with the high-affinity catalytic site clearly present. The functional role of the DELSEED-loop will be discussed in light of the new information.
MATERIALS AND METHODS
Bacterial Strains and Plasmids
Plasmid pTR19-ASDS, which carries the genes for the F1Fo-ATP synthase from thermophilic Bacillus PS3 (21), was used to generate deletion mutants. All deletion mutations investigated here were based on combinations of mutants from a previous study. In addition, we generated the quintuple point mutant, β390AALSAAA396. The mutagenic oligonucleotides were designed in such a way that, in addition to the desired mutation, a restriction site would be eliminated or generated, to facilitate screening. Deletions were introduced by polymerase chain reaction using the QuikChange II XL mutagenesis kit (Stratagene). Wild-type and mutated plasmids were transformed into Escherichia coli strain DK8 which does not express E. coli ATP synthase (22).
The AALSAAA mutation was also introduced in plasmid pNM2. Plasmid pNM2 is a derivative of plasmid pKAGB1 (23). pKAGB1 is used to express a Cys- and Trp-less form of the α3β3γ subcomplex of PS3. pNM1 contains an additional mutation to generate a His10 tag at the N terminus of the β subunits, to facilitate protein purification. pNM2 is pNM1 with a βY341W mutation, allowing to monitor nucleotide binding to the three catalytic sites. For expression, pNM2 and the derived AALSAAA mutant were transformed into E. coli strain JM103 Δ(uncB-uncD).
For construction of a plasmid that only expresses the β subunit, a TAA stop codon and an NheI restriction site were inserted immediately downstream of the ATG start codon for the α subunit gene. The presence of a natural NheI site at the end of the gene encoding γ subunit allowed us to remove the genes for α and γ by removing the NheI- NheI fragment. The resultant plasmid was named pNM1A6. The same construct was made in plasmid encoding the Δ381QDIIAIL387+392LSD394 (“Δ10”) deletion mutant. Additional point mutations (βY364W, βM389C, and βL398C) were introduced into the wt and Δ10 pNM1A6 plasmids to enable the fluorescence energy transfer measurements in isolated β subunits. All mutations were confirmed by DNA sequencing. For expression, the plasmids were transformed into strain DK8.
Isolation of Inverted Membrane Vesicles, Determination of F1Fo Content in E. coli Membranes
E. coli strain DK8 harboring wild-type or mutated pTR19-ASDS plasmids was aerobically cultivated at 37 °C for 18 h in 2× YT medium containing 100 μg/ml ampicillin. Inverted membrane vesicles from E. coli cells expressing thermophilic F1Fo were prepared as described (21, 24). The amount of wild-type F1Fo in E. coli membrane preparations was determined by SDS-PAGE, visualized by staining with Coomassie Brilliant Blue (21). The relative amount of mutant F1Fo the membranes was estimated via Western blots, using an anti-β antibody (Agrisera, Vännäs, Sweden). The staining intensity was quantified using a Photodyne imaging system and Image J acquisition software (NIH).
Preparation of α3β3γ Subcomplex and of Isolated β Subunit
The purification method of α3β3γ subcomplex is modified from a previously described procedure (25). Cells were grown aerobically at 37 °C in terrific broth medium containing 100 μg/ml ampicillin. After cell lysis by French Press the cell debris was removed by centrifugation at 35,000 rpm for 30 min. The supernatant containing complex was applied to a Ni2+-NTA column (Qiagen) equilibrated with 20 mm imidazole and 100 mm NaCl, pH 7.0. The column was washed with 50 mm imidazole and 100 mm NaCl, pH 7.0, and the enzyme was eluted with 500 mm imidazole and 100 mm NaCl, pH 7.0. The preparation of the isolated β subunit followed the same protocol. The proteins were stored as precipitate in 70% saturated ammonium sulfate at 4 °C.
Functional Analysis of Mutant Strains and Enzymes
Growth of strains expressing wild-type or mutant PS3 ATP synthase in limiting glucose, ATPase activities (including temperature dependence), ATP synthase activities, NADH- and ATP-driven H+-pumping, and nucleotide binding to the catalytic sites were determined as described previously (12, 26).
Fluorescence Resonance Energy Transfer and Anisotropy Measurements
FRET experiments were performed to measure the distance between a Trp in position β364 and an IAEDANS-labeled Cys in positions β389 and β398 in an isolated β subunit. (To simplify the nomenclature, we will refer to the extrinsic probe as IAEDANS even after its reaction with Cys, which removes the “I” standing for “iodo”.) A stock solution of IAEDANS (Invitrogen, Carlsbad, CA) was prepared in dimethyl sulfoxide. Isolated β subunit was passed through a 1 ml Sephadex G-50 centrifuge column, equilibrated with 50 mm HEPES/KOH, pH 7.0. The protein was labeled by incubation with 1 mm IAEDANS for 1 h at 23 °C in the dark. Τhe reaction was stopped by passing the protein through two Sephadex G-50 centrifuge columns, equilibrated with 50 mm Tris/H2SO4, pH 8.0. Unlabeled β subunit was passed through two Sephadex G-50 centrifuge columns, equilibrated with the same buffer.
Fluorescence measurements were carried out at 23 °C in a buffer containing 50 mm Tris/H2SO4, pH 8.0, using a spectrofluorometer type Fluorolog 3 (HORIBA Jovin Yvon, Edison, NY). In the FRET experiments, the fluorescence of the donor βW364 was measured in absence and in presence of the acceptor, the IAEDANS-labeled βC389 or βC398. From the decrease in fluorescence intensity (λexc = 295 nm; λem = 340 nm) the energy transfer efficiency was calculated. For determination of the critical transfer distance, R0, where the transfer efficiency is 50%, the quantum yield of βW364 was determined to be 0.10, using N-acetyl-tryptophanamide as standard (Φ = 0.14; Ref. 27). Assuming values of 2/3 and 1.4 for the orientation factor, κ2, and the refractive index, n, respectively, the R0 value for the donor/acceptor pair Trp/IAEDANS was calculated to be 20 Å. This is close to the value of 22 Å frequently cited in the literature (28). Obviously, the value will vary for different Trp donors, based on their quantum yield and wavelength position (which affects the overlap integral). From the transfer efficiency and R0, the distance between donor and acceptor was calculated (Ref. 29; see also supplemental Fig. S1).
For anisotropy measurements, the instrument was equipped with Glan-Thompson polarizers. For determination of the anisotropy of βW364, λexc was 300 nm and λem was 340 nm; for determination of the anisotropy of β-bound IAEDANS, λexc was 360 nm and λem was 480 nm. Anisotropies were measured as described in Ref. 29, using the L-format method. For estimation of the rotational mobility of the protein-bound fluorophors, the values used were as follows. Trp: fundamental anisotropy, r0, 0.31; fluorescence lifetime, τ, 3 ns (29). IAEDANS: r0, 0.33 (30); τ, 15 ns (31). The rotational correlation time of the isolated β subunit was assumed to be 30 ns (29).
Modeling
Homology modeling including energy minimization refinement was performed using the program PRIME (Schroedinger Inc.). The template was the structure of bovine mitochondrial enzyme, Protein Data Bank file 1h8e (11).
RESULTS
Deletion Mutants in the DELSEED-loop: an Overview
In a previous study (12), we had removed stretches of 3–4 contiguous residues from the helix-turn-helix structure known as “DELSEED-loop” in the C-terminal domain of the β subunit of ATP synthase: Δ380LQDI383, Δ384IAIL387, Δ388GMDE391, Δ392LSD394, Δ395EDKL398, and Δ399VVHR402 (it should be noted that in Bacillus PS3 the DELSEED motif is actually 390DELSDED396). In addition, we could obtain one longer deletion mutant with 7 residues removed, Δ381QDIIAIL387. Modeling of these deletion mutants suggested that in all cases, including the 7 residue deletion, Δ381QDIIAIL387, the overall length of the DELSEED-loop was hardly affected (reduced by ≤ 3 Å). Thus, it was obvious that longer deletion were necessary to shorten the loop significantly. In the present study, we combined several of the previous deletions, Δ380LQDI383+392LSD394 (termed “Δ7” in the following), Δ381QDIIAIL387+392LSD394 (“Δ10”), and Δ381QDIIAILGMDELSD394 (“Δ14”). We also included the quintuple point mutant β390AALSAAA396 in the investigation. It had been shown that an α3β3γ subcomplex containing AALSAAA was able to rotate γ with the same torque as the wild-type (14).
Oxidative Phosphorylation in Vivo
In growth yield assays in limiting glucose strain pTR19-ASDS/DK8, expressing wild-type PS3 F1Fo in E. coli, grew to a turbidity (measured as absorbance at 590 nm) of 62% of that of the control strain pBWU13.4/DK8, expressing “native” wild-type E. coli ATP synthase. The negative (unc−) control, strain pUC118/DK8, reached 38% of the value for strain pBWU13.4/DK8 and 62% of the value for strain pTR19-ASDS/DK8. One deletion mutant, Δ7, showed growth yields significantly higher than the negative control, but clearly below the value for the wild-type PS3 enzyme (Table 1). One of the other deletion mutants, Δ10, exhibited growth yields similar to those of the negative control, indicating loss of the capability of oxidative phosphorylation. Δ14 grew less well than the negative control, which might be the result of severe uncoupling and/or of proton leaks through the membrane, possibly due to an incorrectly assembled enzyme. In contrast, the AALSAAA mutant was only slightly impaired in its ability to catalyze ATP synthesis in vivo.
TABLE 1.
Oxidative phosphorylation in vivo and ATP synthesis activities in vitro
Growth yields in limiting glucose were measured via the turbidity (A590) and are expressed as percentage of the value for the positive control. ATP synthesis activities of membrane preparations were measured at 42 °C and pH 7.5 (12). The given values represent the average of at least four independent measurements ± S.D. The positive control was strain pTR19-ASDS/DK8, which expresses wild-type Bacillus PS3 ATP synthase in E. coli; this strain served as background for the deletions. The negative control was strain pUC118/DK8, which expresses neither PS3 ATP synthase nor the endogenous E. coli enzyme. The amount of F1Fo in the membrane preparations was measured by quantitative immunoblot analysis, as described under “Materials and Methods,” it should be noted that all individual readings of the staining intensity, even for Δ14, were higher than that of the respective negative control. Turnover rates were calculated using a molecular mass of 531 kDa for the holoenzyme, taking into account the differing amounts of ATP synthase in the individual membrane preparations.
| Strain/mutation | Growth yields in limiting glucose | Amount of F1Fo on membranes | NADH-driven ATP synthesis activity | Turnover rate kcat |
|---|---|---|---|---|
| % wild-type | % total protein | mU/mg | s−1 | |
| Wild-type | 100 ± 3 | 20 ± 5 | 49 ± 12 | 2.2 |
| pUC118/DK8 (unc−) | 62 ± 3 | 0 | 0.2 ± 2.3 | |
| Δ7 (Δ380LQDI383+392LSD394) | 76 ± 3 | 13 ± 3 | 14 ± 4 | 1.0 |
| Δ10 (Δ381QDIIAIL387+392LSD394) | 60 ± 3 | 20 ± 4 | 0.4 ± 2.4 | < 0.1 |
| Δ14 (Δ381QDIIAILGMDELSD394) | 47 ± 3 | 0.9 ± 0.8 | −0.6 ± 3.0 | 0a |
| 390AALSAAA396 | 89 ± 4 | 22 ± 3 | 91 ± 11 | 3.7 |
a See text.
ATP Synthesis Activity of Membrane Preparations
In addition to the in vivo growth assay to monitor oxidative phosphorylation, we measured NADH-driven ATP synthesis in vitro. At 42 °C, the wild-type PS3 enzyme in E. coli membrane vesicles showed an ATP synthesis activity of 49 mU per mg membrane protein. As in the in vivo assay, the deletion mutant Δ7 exhibited some ATP synthesis activity, while Δ10 and Δ14 had no significantly higher activity than the negative control lacking ATP synthase (Table 1). Besides being a direct consequence of a mutation, lack of activity can be due to lack of expression or to oligomeric instability of the enzyme; thus, it was necessary to quantify the amount of enzyme on the membranes. The amount of wild-type PS3 F1Fo in E. coli membrane preparations was found to be ∼20% of the total membrane protein (this study, and Ref. 21). Membrane preparations containing PS3 ATP synthase with the deletions Δ7 and Δ10 had at least 65% of the amount of the wild-type enzyme (Table 1), demonstrating that the lack of ATP synthesis activity observed for Δ10 is indeed a characteristic of the enzyme and not due to expression or stability problems. The Δ14 deletion mutant exhibited only a small amount of ATP synthase on the membranes. Whereas the mutant appears to have lost its capability to catalyze ATP synthesis, it should be noted that even wild-type-like activity of the remaining ATP synthesis molecules would not have resulted in activity values significantly higher than the standard deviation. The AALSAAA mutant synthesized ATP at a higher rate than the wild-type enzyme.
ATPase Activity of Membrane Preparations
As can be seen in Table 2, 2 deletion mutants, Δ7 and Δ10, showed ATPase activity roughly similar to wild-type. Δ14 had no ATPase activity beyond the background, even when the assay was performed using a high concentration of membranes (0.5 mg protein per ml instead of 0.05–0.15 mg/ml) and an extended incubation time (20 min instead of 1–5 min). We estimated that under these conditions we would have been able to detect an activity of ∼5 mU/mg over background. Even taking the low amount of ATP synthase in the membranes into account, this means that the Δ14 mutant has a turnover rate of less than 5 s−1, i.e. less than 5% of the activity of the wild-type. The AALSAAA mutant reached Vmax turnover rates similar to those of the wild-type enzyme (Table 2). When the ATPase assay was performed at lower MgATP concentrations (20 μm MgSO4 and 50 μm ATP instead of 4 mm MgSO4 and 10 mm ATP), the activity of the AALSAAA mutant was significantly higher than that of the wild-type enzyme (3.8 s−1 for AALSAAA, 1.7 s−1 for wild-type). It should be noted that a mutant where the C-terminal domain of the ϵ subunit was removed, ϵI88stop, behaved similar to the AALSAAA mutant: increased ATP synthesis activity compared with wild-type, no effect on ATPase activity at saturating MgATP concentrations (26), and increased ATPase at lower MgATP concentrations (2.7 s−1 at 20 μm MgSO4 and 50 μm ATP).
TABLE 2.
ATPase activities of membrane vesicles of deletion mutants
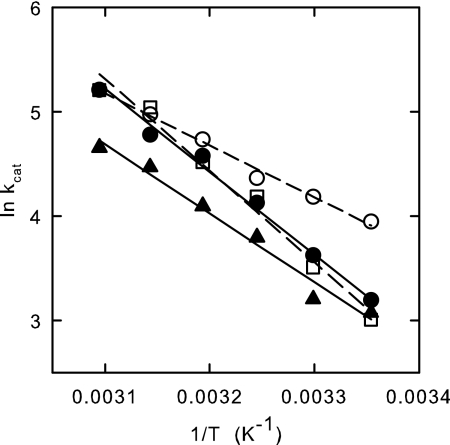
ATPase activities of membrane preparations of mutants and controls (column 2) were measured at 42 °C and pH 8.0 (12). The given values represent the average of at least four independent measurements ± S.D. The positive control was strain pTR19-ASDS/DK8, which expresses Bacillus PS3 ATP synthase in E. coli; this strain served as background for the deletions. The negative control was strain pUC118/DK8, which expresses neither PS3 ATP synthase nor the endogenous E. coli enzyme. Turnover rates were calculated as described in the legend to Table I. The ATPase activity found for the negative control was subtracted in calculation of turnover rates. To determine the temperature dependence, ATPase activities were measured at temperatures between 25 and 50 °C as described (12). From the resulting Arrhenius plots (ln kcat versus 1/T; see Fig. 1), the activation energy, Ea, was calculated.
| Strain/mutation | Membrane ATPase activity | Turnover rate kcat | Activation energy Ea |
|---|---|---|---|
| unit/mg | s−1 | kJ/mol | |
| Wild-type | 2.6 ± 0.4 | 114 | 70.1 |
| pUC118/DK8 (unc−) | 0.018 ± 0.014 | ||
| Δ7 (Δ380LQDI383+392LSD394) | 2.0 ± 0.2 | 136 | 73.0 |
| Δ10 (Δ381QDIIAIL387+392LSD394) | 1.6 ± 0.2 | 71 | 54.9 |
| Δ14 (Δ381QDIIAILGMDELSD394) | 0.016 ± 0.022 | 0a | |
| 390AALSAAA396 | 3.2 ± 0.4 | 129 | 41.4 |
a See text.
Arrhenius Analysis of ATPase Activities
Turnover rates at saturating ATP concentrations were measured as a function of temperature. Because of its lack of membrane ATPase activity, the Δ14 deletion mutant could not be included in this analysis. The results for the other deletion mutants, the AALSAAA mutant, and the wild-type enzyme are shown in Fig. 1 in form of Arrhenius plots (ln kcat versus 1/T). From the slope of the regression lines the activation energy, Ea, was calculated. In the preceding study with deletions of 3–7 residues (12), the Ea values for the deletion mutants were dependent on the length of the deletion, about 70% of the wild-type value for deletions of 3–4 amino acids, and about 50% of the wild-type value for the deletion of 7 amino acids. The results of the present study show some deviations from this pattern. The Δ7 mutant is the first of the investigated deletion mutants that has an activation energy close to or even slightly higher than wild-type, indicating that the number and strength of interactions that have to be broken or formed in the rate-limiting step of the overall hydrolysis reaction is similar in the mutant and in the wild-type enzyme. In Δ10, Ea was about 80% of wild-type, and in AALSAAA about 60%. The latter result indicates that (some of) the negative charges of the DELSEED motif itself contribute to the energetically relevant interactions in the transition state.
FIGURE 1.
Arrhenius analysis of the ATPase activity of the deletion mutants. ATPase activities were measured at different temperatures (25 to 50 °C) and were calculated as turnover rates (kcat, in s−1). From the negative slope of the regression lines, the activation energy, Ea, of the rate-limiting step of the overall reaction can be determined. Filled circles, wild-type PS3 ATP synthase; open squares, Δ7 deletion mutant; filled triangles, Δ10; open circles, AALSAAA mutant.
NADH- and ATP-induced Proton Pumping
NADH-induced ACMA quenching was not reduced in membrane vesicles containing any of the deletion mutants or the AALSAAA mutant. The quenching reached values around 80–85%, just as with wild-type PS3 enzyme (data not shown), indicating that the mutations do not prevent the build-up of a considerable proton gradient by the electron transport chain. Thus, the deletions did not cause stability problems, which increased the “leakiness” of the membranes. It should be noted, however, that reduced stability does not always manifest itself in enhanced proton leak rates.
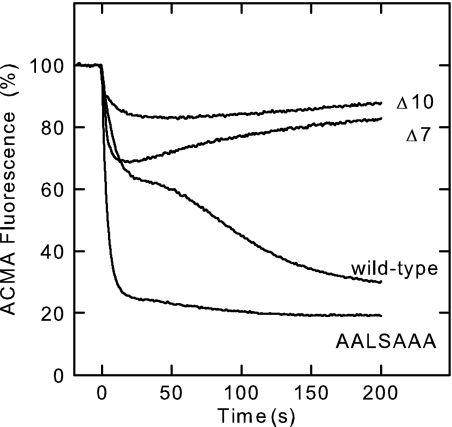
Upon ATP hydrolysis, wild-type PS3 ATP synthase in E. coli membrane vesicles forms a proton gradient which results in 70–75% quenching of the ACMA fluorescence. The reaction is biphasic (Fig. 2); in the second, slow phase the majority of the enzyme molecules appear to be in an MgADP-inhibited state (12). Of the deletion mutants, Δ14 did not produce measurable ATP-induced ACMA quenching, in agreement with the absence of ATPase activity described above. In contrast, the two other deletion mutants could use ATP hydrolysis to form a proton gradient, although a rather small one. With the Δ7 and Δ10 deletion mutants the initial fast phase resulted in quenching of 25 and 15%, respectively; in the slower phase, ATP-driven H+-pumping appears reduced to such an extent that it can no longer compensate for the natural leak rate of the membranes. The AALSAAA mutant reached 85% ACMA quenching in a single, fast phase, similar to the Δ392LSD394 deletion mutant in the previous study (12) and a mutant where the C-terminal domain of ϵ was removed (32). It should be noted that in all cases after addition of 1 μm CCCP, to dissipate the proton gradient, the ACMA fluorescence returned to within 5% of its original value.
FIGURE 2.
ATP-driven H+ pumping. The quenching of ACMA fluorescence upon addition of ATP (at t = 0 s) is shown. “Wild-type” refers to membrane preparations of strain pTR19-ASDS/DK8, which expresses wild-type ATP synthase from the thermophilic Bacillus PS3 in E. coli membranes. This strain served as the background of the deletion mutants. In all cases, after addition of 1 μm CCCP to dissipate the proton gradient the ACMA fluorescence returned to within 5% of its original value.
The Structure of the Loop in the Deletion Mutants: Modeling and Experimental Verification
Modeling of the deletion mutants in the previous study (12) had suggested that in all cases, including the 7 residue deletion Δ381QDIIAIL387, the overall length of the DELSEED-loop was hardly affected (reduced by ≤ 3 Å). Instead, in most cases the deletions were accommodated by straightening the protein backbone, thereby “unraveling” the helices flanking the deletions or the single helical turn at the tip of the loop. Here, for the Δ7 deletion a similar pattern was observed. As can be seen in Fig. 3, while the length of the loop is not affected, its tip is narrower than in the wild-type enzyme. In contrast, in the Δ10 and Δ14 deletion mutants the DELSEED-loop appears to be significantly shortened (Fig. 3B). This decreases the contact area between the DELSEED-loop and γ significantly. In the conformation represented by the crystal structure(s), the number of possible β/γ hydrogen bonds involving the DELSEED-loop in the wild-type enzyme is ∼7. Besides interactions of β with the N- and C-terminal helices of γ, (Fig. 3C, top) this number includes hydrogen bonds to a loop-helix segment in the globular portion of γ, residues γ82–100 (not shown in Fig. 3C; see Fig. 6A in Ref. 12). In Δ7, the number of β/γ hydrogen bonds is reduced to ∼ 5, in Δ10 and Δ14 to one; in both latter cases, the hydrogen bond involves βK397 and γS90. Bulk contacts show a similar decrease (for Δ10, see Fig. 3C, bottom).
FIGURE 3.
The βDELSEED-loop and its interaction with the γ subunit. A, overview of the interaction of the βE and βTP subunits with γ. β Subunits are in yellow, γ is in blue; the DELSEED-loop is colored orange, with the DELSEED motif itself in green. The transparent part shows a spacefill representation of the DELSEED-loops and γ. A nucleotide molecule (shown in stick representation) occupies the catalytic site of βTP, and the subunit is in the closed conformation. The catalytic site on βE is empty, and the subunit is in the open conformation. The DELSEED-loop of βE makes contact with the convex portion of γ, formed mainly by the N-terminal helix. βE cannot close completely because of steric clashes with γ. Based on Protein Data Bank file 1e79 (43). B, modeling the βDELSEED-loop of the deletion mutants. The main chain of the models of deletion mutants Δ7, Δ10, and Δ14 is shown as the thicker line in cartoon representation; the thinner line is the main chain of the wild-type enzyme (from 1h8e; see Ref. 11). The orientation of the loop is approximately the same as in the βTP subunit in A. Side chains (in stick representation) are only given for the deletion mutants. Segments of 3–4 residues are color-coded: 380LQDI383, pink; 384IAIL387, green; 388GMDE391, yellow; 392LSD394, cyan; 395EDKL398, orange; 399VVHR402, blue (12). N and C terminus of the loop are indicated in the panel of deletion mutant Δ7. The Cα atom of residue βY364 is marked as a sphere; a Trp in this position is used as the donor in the FRET experiments. C, contacts between the DELSEED-loop and γ in wild-type (top) and Δ10 (bottom). βDELSEED-loop, yellow; γ, blue. For clarity, only portions of the N- and C-terminal helices of γ are shown. Residues forming β/γ hydrogen bonds in the wild-type enzyme are shown in stick representation and labeled (in the top panel). Further contacts are made between the DELSEED-loop and a segment of the globular portion of γ (see text). Residues involved in bulk interactions with γ in wild-type (βI383, βI386, βL387) are shown in stick representation and highlighted by dots. Transparent portions show spacefill representations of γ (both panels) and the βDELSEED-loop (bottom panel), illustrating the reduction in contacts between the loop and γ in the Δ10 mutant. D, distances between residues in the DELSEED-loop tip and βY364. Distances were measured between the Cα atoms in wild-type (1h8e) and in the models of the deletion mutants. Red line, wild-type; green line, Δ7; blue line, Δ10; purple line, Δ14; black line, Δ381QDIIAIL387 (12). Dotted/dashed portion of the lines indicate missing residues. E, comparison of results from modeling and FRET experiments. The DELSEED-loops of wild-type (thin line) and Δ10 (thick line) are shown in a different perspective than in B. Gray sphere, Cα of βY364; yellow spheres, Cα of βM389; orange sphere, Cα of βL398. Red arrow, distance in wild-type; blue arrow, distance in Δ10; red/blue arrow, distance unchanged by deletion. Numbers without brackets are distances (in Å) from FRET experiments; distances in brackets are from the model. A, B, C, and E were generated using the program PyMOL (Schroedinger Inc.).
Fig. 3D shows a numerical evaluation of the models, including the one for Δ381QDIIAIL387 (12). The distance between the Cα atom of residue βY364 (located in the N-terminal helix of the DELSEED-loop; see Fig. 3E) and Cα of each of the amino acids in the tip of the loop was measured. As can be seen, the 7 residue deletions, Δ7 and Δ381QDIIAIL387, appear to shorten the loop by only <4 and ∼2 Å, respectively. In contrast, the Δ10 and Δ14 deletions seem to reduce the maximum length of the loop substantially, by ∼10 Å. For the individual residues βG388 to βE391 in Δ10 (which are absent in Δ14), the models indicate that the distance to βY364 is shortened by 7–10 Å compared with the wild-type. Therefore, these residues are ideally suitable for experimental assessment of the accuracy of the models.
We used FRET to determine the distance between residues β364 and β389 and between residues β364 and β398 in wild-type and in Δ10. According to the modeling results, in the deletion mutant the distance between β364 and β389 is expected to be reduced by 7 Å or 23%, from 31 Å to 24 Å. In contrast, serving as control for the experimental approach and for the structural integrity of the Δ10 mutant, the distance between residues β364 and β398 should not be affected by the deletion (22–23 Å in both cases). As donor for the FRET experiment, a Trp residue was inserted in position β364, in form of a βY364W mutation. The FRET acceptor was IAEDANS attached to a Cys in the mutants βM389C and βL398C. The results of the FRET experiments (supplemental Fig. S1) confirmed the model. The distance between β364 and β389 was reduced by 6 Å or 23%, from 26 ± 1 Å in wild-type to 20 ± 1 Å in Δ10. In contrast, the deletion did not affect the distance between β364 and β398, which was found to be 25 ± 1 Å in wild-type and 26 ± 2 Å in Δ10 (Fig. 3E). As can be seen, the absolute distances vary somewhat between the model and the FRET measurements, due to the fact that the model data use the coordinates of the Cα atoms, whereas the FRET results are based on fluorophors formed by or attached to the side chain of the respective amino acid. It should be noted that in order to avoid complications due to presence of multiple donors and acceptors the FRET experiments were performed using isolated β subunits. NMR studies (33–35) showed that the open conformation of the isolated β-subunit, which is assumed in absence of nucleotide, is very similar to that in the assembled F1 subcomplex. The closed conformation of β, obtained in presence of nucleotide, displays differences between the isolated and enzyme-integrated forms; however, the intra-domain structure of the C-terminal domain with the DELSEED-loop is not affected. Thus, the distances determined here by FRET should also apply to β in the assembled enzyme.
Anisotropy measurements gave values of 0.19 for βW364, 0.14 for βC389-IAEDANS and 0.15 for βC398-IAEDANS, for wild-type as well as for Δ10. In all cases, this is well below the fundamental anisotropy for a totally immobilized fluorophor, which would be 0.31 for Trp and 0.33 for IAEDANS. Depolarization due to rotational diffusion of the entire β subunit during the fluorescence lifetime would reduce these values to 0.28 and 0.22, respectively. The remaining depolarization can be ascribed to segmental motion within the protein, including motion of the side chain. The data show that both fluorophors have some degree of rotational mobility, which justifies the use of κ2 = 2/3 in calculation of the critical transfer distance, R0 (see “Materials and Methods”). Furthermore, the results indicate that the Δ10 deletion does not affect the mobility of the tip of the DELSEED-loop.
MgATP Binding to the Catalytic Sites of the α3β3γ Subcomplex
In the preceding study using deletion mutants (12) we could show that those mutants that we were able to express as α3β3γ subcomplexes, Δ388GMDE391 and Δ392LSD394, had “lost” their high-affinity binding site. The Kd1 value for binding of MgATP to the high-affinity site went from ∼10 nm in the wild-type enzyme to 1–3 μm in the deletion mutants. Here, we investigated MgATP binding to the AALSAAA mutant, using the fluorescence of the inserted Trp βW341 as signal. The results are shown in Fig. 4. From the titration curves, we determined affinities of Kd1 ≈ 25 nm, Kd2 = 13 μm, and Kd3 = 13 μm for the AALSAAA mutant. The values for the wild-type enzyme are Kd1 ≈ 10 nm, Kd2 = 5 μm, Kd3 = 38 μm. While the numbers indicate differences in corresponding Kd values of up to a factor of three between wild-type and mutant enzymes, from Fig. 4 it is obvious that the actual binding pattern for both enzymes is quite similar. Specifically, the high-affinity binding site is preserved.
FIGURE 4.
MgATP binding to the catalytic sites. MgATP binding to the three catalytic sites of the α3β3γ subcomplex of PS3 ATP synthase was measured in a buffer containing 50 mm Tris/H2SO4, 2.5 mm MgSO4, pH 8.0, at 23 °C (12). α3(βY341W)3γ, filled circles; 390AALSAAA396 in α3(βY341W)3γ, open circles. The lines are fitted binding curves based on the Kd values given in the text. α3(βY341W)3γ, solid line; 390AALSAAA396 in α3(βY341W)3γ, dashed line.
DISCUSSION
The central goal of the present study was to determine the critical length of the β390DELSEED396-loop of ATP synthase, the minimum length necessary to sustain its function of coupling of catalysis and subunit rotation. Shortening of the loop has to be achieved by deleting amino acid residues. Previously, we had analyzed the functional consequences of deleting stretches of 3–7 residues (12). In that study, all tested deletion mutants showed some ATPase activity, and all mutants that eliminated residues at the β/γ interface were still capable of ATP synthesis. Two mutants that had residues deleted downstream of the β/γ contact region, Δ395EDKL398 and Δ399VVHR402, had lost their ATP synthesis activity, most likely due to interference with β/α interactions combined with the low amount of enzyme present on the membranes (12). Modeling suggested that none of the mutations had a pronounced effect on the overall length of the DELSEED-loop (for the 7 residue deletion, Δ381QDIIAIL387, see Fig. 3D). To actually shortening the loop, the logical approach seemed to be to delete more than 3–7 residues. In the present study, we deleted stretches of 7, 10, and 14 residues and analyzed the effects on the function of the enzyme. Again, modeling was used to determine the structure of the shortened loop. In the most important case of Δ10, FRET was applied to confirm the accuracy of the model as far as the length of the loop is concerned. The results of the present study demonstrate that the applied approach is indeed able to determine the critical length of the DELSEED-loop.
In many aspects, the Δ7 deletion included in the present report, Δ380LQDI383+392LSD394, behaved similar to the 3–7 residue deletions at the β/γ interface in the previous study (12). Δ7 was capable of ATP hydrolysis with similar rates as the wild-type enzyme, although it did not reach the extraordinarily rapid ATPase rates (2.5 to 5 times as fast as wild-type) of Δ384IAIL387, Δ392LSD394, and Δ381QDIIAIL387. In contrast to the hydrolysis rates, the ATP synthesis rate of Δ7 was below that for the wild-type enzyme. Reduced ATP synthesis rates combined with normal or higher-than-normal hydrolysis rates can be an indication of uncoupling. This conclusion is supported further by the finding of reduced proton gradient formation upon ATP hydrolysis. Upon analysis of the temperature-dependence of the hydrolysis reaction, we obtained an activation energy, Ea, similar to wild-type and higher than in any other deletion mutant previously investigated. This means that in Δ7 the number and strength of bonds that have to be rearranged during the rate-limiting step of the overall hydrolysis reaction is similar than in wild-type. It should be noted that it is not known exactly which bonds are affected in the rate-limiting step. Both individual mutants that are combined in Δ7, Δ380LQDI383+392LSD394, had shown a reduced activation energy, illustrating that the effects of the deletions are not necessarily additive. A possible reason for such behavior is the fact that combinations of deletions in close proximity, as present here, might affect the folding of the protein backbone in a different way and therefore might have different functional consequences than the individual deletions. In contrast, the combination of point mutations is more likely to show additive effects. Several other examples of “non-additive” behavior of the deletion mutants can be found in the results of this study.
One example is the wild-type-like amount of ATP synthase found on membranes containing the Δ10 (Δ381QDIIAIL387+392LSD394) mutation. While one of the deletions combined here, Δ392LSD394, had also shown similar amounts of enzyme as the wild-type, the Δ381QDIIAIL387 mutation found to reduce the amount to 12% of wild-type (12). Obviously, the combination of both deletions in Δ10 can overcome the negative effect of the Δ381QDIIAIL387 deletion on expression and/or stability, quite likely for the reasons outlined above.
Δ10 appears to represent a borderline case with regard to the length of the DELSEED-loop that is required to sustain catalytic activity. With the loop reduced by ∼10 Å, the mutant lost the capability to synthesize ATP; however, it is still an active ATPase. It is interesting to note that Δ10 combines two deletions which individually resulted in very high ATPase rates (12). Clearly, Δ10 fails to reach the hydrolysis rates of either precursor. To exclude the possibility that the deletions might affect the structural integrity of the loop, we demonstrated for the Δ10 mutant that the distance between β364 and β398 is similar as in wild-type, making it highly likely that the helix connecting both residues is preserved. In addition, we could show that the Δ10 deletion did not affect the segmental mobility of protein in the loop region. The Δ381QDIIAILGMDELSD394 (“Δ14”) mutant, with a DELSEED-loop tip that is narrower and on one side slightly shorter than in Δ10, had lost in addition the ability to hydrolyze ATP. Considering the low amount of Δ14 enzyme found on the membranes, we cannot exclude that in this case the β subunit is not correctly folded, resulting in lack of properly assembled ATP synthase.
The finding that reducing the size of the DELSEED-loop abolishes catalytic activity is one of the strongest arguments so far for a direct involvement of the loop in mechanochemical coupling of catalysis and rotation. In rotational catalysis by F1, each 120° rotation step is divided into an 80° and a 40° substep (17, 36). As described in the introduction, during ATP hydrolysis MgATP binding and the subsequent closure of the β subunit is believed to drive the 80° rotation substep (5, 15–17, 36). If the DELSEED-loop is shortened sufficiently, the β subunit will be able to close upon binding of MgATP without driving rotation of γ. Similarly, in ATP synthesis rotation of γ will be no longer able to force the high-affinity site open to release the newly-formed ATP. The results presented here show that the DELSEED-loop reaches its critical length when shortened by ∼10 Å. The finding that the Δ10 deletion mutant just abolishes ATP synthesis, but not hydrolysis, suggests that both pathways are not the exact reversal of each other (see also Refs. 26, 37, 38). The molecular basis for the differences between forward and backward reaction is unknown. However, it is possible that the ϵ subunit is involved (see below).
As described in the preceding study (12), the 3–4 residue deletion mutants Δ388GMDE391 and Δ392LSD394 had lost their ability to bind MgATP with high affinity. These deletions encompassed three of the negatively charged residues of the DELSEED motif itself, βD390, βE391, and βD394. A comparable decrease in affinity of the high-affinity site had been described for a βD390C point mutation (39). We could show here that the 390AALSAAA396 mutant, where all the negative charges of the DELSEED motif are removed, could still bind MgATP with high affinity. It appears possible that not only the deletions Δ388GMDE391 and Δ392LSD394, but also βD390C point mutation cause larger scale conformational rearrangements, which affect nucleotide binding to the high-affinity catalytic site.
Whereas the ATPase activity of the 390AALSAAA396 mutant was close to wild-type, its ATP synthesis activity in vitro was significantly higher than that of the wild-type control. This difference could be due to perturbation of the interaction with the ϵ subunit in the AALSAAA mutant. ϵ, or more precisely, the C-terminal domain of ϵ, exists in different conformations, which can be broadly categorized as “up” (two different up conformations have been demonstrated, a third one was postulated; see refs. 40–42) and “down” (43). In the “up” conformation it acts as intrinsic inhibitor of ATPase activity; more recently it was also found to inhibit ATP synthesis (26, 44). In the wild-type enzyme, the “up” conformation of ϵ interacts with the βDELSEED-loop (35, 45, 46). Specifically, contacts with the DELSEED motif itself have been described (38, 47, 48). If the relevant interactions are reduced in the AALSAAA mutant, this might shift the equilibrium to the non-inhibitory “down” conformation during ATP synthesis. As far as the absence of an effect of the mutation on ATP hydrolysis at Vmax is concerned, it should be noted that, at least in the PS3 enzyme, ϵ inhibition of the ATPase activity is strongly dependent on the ATP concentration and virtually absent at high ATP concentrations (49). At lower substrate concentrations the ATPase activity of the AALSAAA mutant is indeed higher than that of the wild-type enzyme. Strong support for the notion of involvement of ϵ in the effects of the AALSAAA mutation on the enzymatic activities comes from experiments with mutants of PS3 ATP synthase in which the C-terminal domain of ϵ was removed. These mutants, just like AALSAAA, had increased ATP synthesis activity, normal ATPase activity at Vmax (26), increased ATPase at lower ATP concentrations (this study), and rapid, monophasic proton gradient formation upon ATP hydrolysis (38).
Taken together, the data obtained previously (14) and here for the AALSAAA mutant show that the negative charges of the DELSEED motif are not directly involved in the catalytic mechanism. Instead, the reason for the conservation of these charges appears to be their involvement in the regulation of the enzymatic activity via interaction with the C terminus of the ϵ subunit (this work and Ref. 45).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM071462 (including ARRA Administrative Supplement) (to J. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Bacillus PS3 numbering is used throughout.
- AMPPNP
- 5′-adenylyl-β,γ-imidodiphosphate
- ACMA
- 9-amino-6-chloro-2-methoxyacridine
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- FRET
- fluorescence resonance energy transfer
- IAEDANS
- 5-((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1-sulfonic acid.
REFERENCES
- 1. Noji H., Yoshida M. (2001) J. Biol. Chem. 276, 1665–1668 [DOI] [PubMed] [Google Scholar]
- 2. Weber J., Senior A. E. (2003) FEBS Lett. 545, 61–70 [DOI] [PubMed] [Google Scholar]
- 3. Wilkens S. (2005) Adv. Protein Chem. 71, 345–382 [DOI] [PubMed] [Google Scholar]
- 4. Weber J. (2006) Biochim. Biophys. Acta 1757, 1162–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakamoto R. K., Baylis Scanlon J. A., Al-Shawi M. K. (2008) Arch. Biochem. Biophys. 476, 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Junge W., Sielaff H., Engelbrecht S. (2009) Nature 459, 364–370 [DOI] [PubMed] [Google Scholar]
- 7. Weber J., Wilke-Mounts S., Lee R. S., Grell E., Senior A. E. (1993) J. Biol. Chem. 268, 20126–20133 [PubMed] [Google Scholar]
- 8. Weber J., Senior A. E. (2004) Methods Enzymol. 380, 132–152 [DOI] [PubMed] [Google Scholar]
- 9. Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. (1994) Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 10. Bowler M. W., Montgomery M. G., Leslie A. G., Walker J. E. (2007) J. Biol. Chem. 282, 14238–14242 [DOI] [PubMed] [Google Scholar]
- 11. Menz R. I., Walker J. E., Leslie A. G. W. (2001) Cell 106, 331–341 [DOI] [PubMed] [Google Scholar]
- 12. Mnatsakanyan N., Krishnakumar A. M., Suzuki T., Weber J. (2009) J. Biol. Chem. 284, 11336–11345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ketchum C. J., Al-Shawi M. K., Nakamoto R. K. (1998) Biochem. J. 330, 707–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hara K. Y., Noji H., Bald D., Yasuda R., Kinosita K., Jr., Yoshida M. (2000) J. Biol. Chem. 275, 14260–14263 [DOI] [PubMed] [Google Scholar]
- 15. Gao Y. Q., Yang W., Karplus M. (2005) Cell 123, 195–205 [DOI] [PubMed] [Google Scholar]
- 16. Mao H. Z., Weber J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18478–18483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yasuda R., Noji H., Yoshida M., Kinosita K., Jr., Itoh H. (2001) Nature 410, 898–904 [DOI] [PubMed] [Google Scholar]
- 18. Gao Y. Q., Yang W., Marcus R. A., Karplus M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11339–11344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nishizaka T., Oiwa K., Noji H., Kimura S., Muneyuki E., Yoshida M., Kinosita K., Jr. (2004) Nat. Struct. Mol. Biol. 11, 142–148 [DOI] [PubMed] [Google Scholar]
- 20. Pu J., Karplus M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1192–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Suzuki T., Ueno H., Mitome N., Suzuki J., Yoshida M. (2002) J. Biol. Chem. 277, 13281–13285 [DOI] [PubMed] [Google Scholar]
- 22. Klionsky D. J., Brusilow W. S., Simoni R. D. (1984) J. Bacteriol. 160, 1055–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matsui T., Yoshida M. (1995) Biochim. Biophys. Acta 1231, 139–146 [DOI] [PubMed] [Google Scholar]
- 24. Senior A. E., Latchney L. R., Ferguson A. M., Wise J. G. (1984) Arch. Biochem. Biophys. 228, 49–53 [DOI] [PubMed] [Google Scholar]
- 25. Adachi K., Noji H., Kinosita K., Jr. (2003) Methods Enzymol. 361, 211–227 [DOI] [PubMed] [Google Scholar]
- 26. Mnatsakanyan N., Hook J. A., Quisenberry L., Weber J. (2009) J. Biol. Chem. 284, 26519–26525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Szabo A. G., Rayner D. M. (1980) J. Amer. Chem. Soc. 102, 554–563 [Google Scholar]
- 28. Matsumoto S., Hammes G. G. (1975) Biochemistry 14, 214–224 [DOI] [PubMed] [Google Scholar]
- 29. Lakowicz J. R. (2006) Principles of Fluorescence Spectroscopy, 3rd Edition, Springer, New York [Google Scholar]
- 30. Hudson E. N., Weber G. (1973) Biochemistry 12, 4154–4161 [DOI] [PubMed] [Google Scholar]
- 31. Watanabe T., Inesi G. (1982) Biochemistry 21, 3245–3259 [DOI] [PubMed] [Google Scholar]
- 32. Feniouk B. A., Suzuki T., Yoshida M. (2007) J. Biol. Chem. 282, 764–772 [DOI] [PubMed] [Google Scholar]
- 33. Yagi H., Tsujimoto T., Yamazaki T., Yoshida M., Akutsu H. (2004) J. Am. Chem. Soc. 126, 16632–16638 [DOI] [PubMed] [Google Scholar]
- 34. Yagi H., Kajiwara N., Iwabuchi T., Izumi K., Yoshida M., Akutsu H. (2009) J. Biol. Chem. 284, 2374–2382 [DOI] [PubMed] [Google Scholar]
- 35. Kobayashi M., Akutsu H., Suzuki T., Yoshida M., Yagi H. (2010) J. Mol. Biol. 398, 189–199 [DOI] [PubMed] [Google Scholar]
- 36. Shimabukuro K., Yasuda R., Muneyuki E., Hara K. Y., Kinosita K., Jr., Yoshida M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14731–14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vinogradov A. D. (2000) J. Exp. Biol. 203, 41–49 [DOI] [PubMed] [Google Scholar]
- 38. Feniouk B. A., Suzuki T., Yoshida M. (2006) Biochim. Biophys. Acta 1757, 326–338 [DOI] [PubMed] [Google Scholar]
- 39. Baylis Scanlon J. A., Al-Shawi M. K., Nakamoto R. K. (2008) J. Biol. Chem. 283, 26228–26240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rodgers A. J., Wilce M. C. (2000) Nat. Struct. Biol. 7, 1051–1054 [DOI] [PubMed] [Google Scholar]
- 41. Cingolani G., Duncan T. M. (2011) Nat. Struct. Mol. Biol. 18, 701–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yagi H., Kajiwara N., Tanaka H., Tsukihara T., Kato-Yamada Y., Yoshida M., Akutsu H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11233–11238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gibbons C., Montgomery M. G., Leslie A. G., Walker J. E. (2000) Nat. Struct. Biol. 7, 1055–1061 [DOI] [PubMed] [Google Scholar]
- 44. Iino R., Hasegawa R., Tabata K. V., Noji H. (2009) J. Biol. Chem. 284, 17457–17464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hara K. Y., Kato-Yamada Y., Kikuchi Y., Hisabori T., Yoshida M. (2001) J. Biol. Chem. 276, 23969–23973 [DOI] [PubMed] [Google Scholar]
- 46. Suzuki T., Murakami T., Iino R., Suzuki J., Ono S., Shirakihara Y., Yoshida M. (2003) J. Biol. Chem. 278, 46840–46846 [DOI] [PubMed] [Google Scholar]
- 47. Aggeler R., Capaldi R. A. (1996) J. Biol. Chem. 271, 13888–13891 [DOI] [PubMed] [Google Scholar]
- 48. Bulygin V. V., Duncan T. M., Cross R. L. (1998) J. Biol. Chem. 273, 31765–31769 [DOI] [PubMed] [Google Scholar]
- 49. Kato-Yamada Y., Bald D., Koike M., Motohashi K., Hisabori T., Yoshida M. (1999) J. Biol. Chem. 274, 33991–33994 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.