Abstract
Heat shock protein 70 (Hsp70) is well documented to possess general cytoprotective properties in protecting the cell against stressful and noxious stimuli. We have recently shown that expression of the stress-inducible Hsp70.3 gene in the myocardium in response to ischemic preconditioning is NF-κB-dependent and necessary for the resulting late phase cardioprotection against a subsequent ischemia/reperfusion injury. Here we show that the Hsp70.3 gene product is subject to post-transcriptional regulation through parallel regulatory processes involving microRNAs and alternative polyadenylation of the mRNA transcript. First, we show that cardiac ischemic preconditioning of the in vivo mouse heart results in decreased levels of two Hsp70.3-targeting microRNAs: miR-378* and miR-711. Furthermore, an ischemic or heat shock stimulus induces alternative polyadenylation of the expressed Hsp70.3 transcript that results in the accumulation of transcripts with a shortened 3′-UTR. This shortening of the 3′-UTR results in the loss of the binding site for the suppressive miR-378* and thus renders the alternatively polyadenylated transcript insusceptible to miR-378*-mediated suppression. Results also suggest that the alternative polyadenylation-mediated shortening of the Hsp70.3 3′-UTR relieves translational suppression observed in the long 3′-UTR variant, allowing for a more robust increase in protein expression. These results demonstrate alternative polyadenylation of Hsp70.3 in parallel with ischemic or heat shock-induced up-regulation of mRNA levels and implicate the importance of this process in post-transcriptional control of Hsp70.3 expression.
Keywords: Gene Regulation, Heat Shock Protein, MicroRNA, mRNA, NF-kappa B (NF-KB), Alternative Polyadenylation, Post-transcriptional Regulation
Introduction
Heat shock (HS)3 and the subsequent expression of heat shock proteins have long been known to possess cytoprotective properties and provide cardioprotection against ischemia/reperfusion (I/R) injury (1–3). The Hsp70 family is among the best studied of the heat shock proteins, has been shown to be induced by many cardiac preconditioning stimuli, and plays a necessary role in the second window of protection (3–6). Hsp70.1 and Hsp70.3 are two nearly identical stress-inducible Hsp70 genes present in the murine heart. The two protein products differ by only a single amino acid, but curiously, the sequence of the two genes is divergent in the regulatory regions of the gene promoter (beyond the first 270 amino acids proximal to the transcriptional start site) and within the 3′-untranslated region (3′-UTR) of the mRNA transcript. Historically, these two genes are considered to be functionally redundant.
We have recently shown that NF-κB-dependent expression of heat shock protein 70.3 (Hsp70.3), but not the closely related Hsp70.1, is necessary for the late phase or second window of ischemic preconditioning (IPC) cardioprotection against acute I/R in the heart (6). In fact, Hsp70.3 and Hsp70.1 appear to contribute differing functions to cell survival in the myocardium (6, 7). Thus, it is important to understand the regulation of these two genes independently. We previously reported that NF-κB inhibition suppresses, but does not completely prevent, the IPC-induced increase of Hsp70.3 mRNA levels in the myocardium (6). Here we show that the late IPC-induced increase of Hsp70.3 protein expression is subject to post-transcriptional regulation after late IPC. Given the known importance of the 3′-UTR in post-transcriptional regulatory processes, this region of the Hsp70.3 gene was examined with regard to potential post-transcriptional regulatory elements. In addition to the predicted binding sites of 12 different miRNAs, the 3′-UTR of the Hsp70.3 mRNA sequence was predicted to harbor four distinct strong polyadenylation signals. Although generally attributed as an independent phenomenon, a prominent role for both miRNAs and alternative polyadenylation (APA) in post-transcriptional regulation of gene expression is now widely recognized (8, 9). Thus, we hypothesized that coordinated actions by miRNAs and APA exert post-transcriptional regulation on Hsp70.3 expression in the myocardium.
The results reported here show that IPC induces a decrease in the levels of the Hsp70.3-targeting miRNAs miR-378* and miR-711, both of which were found to exert post-transcriptional suppression on Hsp70.3 in vitro via the 3′-UTR. In addition, the results demonstrate the alternative polyadenylation of Hsp70.3 transcripts in the myocardium resulting in mRNA transcripts with varying 3′-UTR lengths. There is a significant and preferential increase in the levels of Hsp70.3 mRNA transcripts with shortened 3′-UTRs following Hsp70.3-inducing stimuli such as HS, IPC, or I/R. This APA-mediated shortening of the Hsp70.3 3′-UTR contributes to increased protein expression through the removal of a regulatory miRNA binding site and enhanced translational efficiency.
EXPERIMENTAL PROCEDURES
Animals and Surgical Models
All mice were maintained in accordance with institutional guidelines, the Guide for the Care and Use of Laboratory Animals (National Institutes of Health, revised 1996), and the University of Cincinnati Institutional Animal Care and Use Committee. Hsp70.1 knock-out and IκBα dominant-negative mice (2M) have been previously characterized (10–12). All studies were done were using strain- and age-matched mice; no gender differences were observed.
The in vivo I/R surgical model was performed via reversible occlusion of the left anterior descending coronary artery as described previously (6, 13). Late ischemic preconditioning was achieved with six cycles of 4 min of ischemia followed by 4 min of reperfusion. All sham surgeries were performed following a similar protocol but without tightening of the suture of the left anterior descending coronary artery. At the appropriate tissue collection time points, hearts were removed, and left ventricular tissue was dissected away, rinsed in sterile PBS, and flash-frozen in liquid nitrogen.
Protein Isolation and Western Blotting
Isolation of nuclear and cytoplasmic proteins from in vivo left ventricular tissue was performed as described by Brown et al. (10). Total protein was isolated from in vitro cell cultures using radioimmunoprecipitation lysis buffer (Santa Cruz Biotechnology) supplemented with a protease inhibitor mixture tablet (Complete mini; Roche Applied Science). Western blotting was done using 10 μg of protein extract per lane. Blocking was performed for 1 h at room temperature using 5% dry milk in 0.1% Tween 20 TBS (T-TBS). Primary antibodies for Hsp70 (Santa Cruz Biotechnology) or pan-actin (Sigma) were added overnight at 4 °C, and secondary antibodies (anti-mouse, GE Healthcare; anti-rabbit, Santa Cruz Biotechnology) were added for 1–2 h at room temperature in T-TBS. Quantitative assessment was done with the National Institutes of Health ImageJ software using pan-actin as a loading control.
Hsp70.3 Luciferase Reporters
Hsp70.3-luciferase reporter constructs were made using either a CMV promoter or the endogenous Hsp70.3 promoter (3974 bp upstream from the protein coding sequence including the 5′-UTR). The Hsp70.3 promoter was PCR-purified from genomic DNA of C57/Bl6 mice and cloned into the Promega pGL4.10 luciferase reporter vector. The CMV promoter used was isolated from the Promega pGL4.75 vector. The Hsp70.3 3′-UTR sequence (1253 bp from the end of the protein coding sequence) was also PCR-purified from genomic DNA of C57/Bl6 mice. All constructs were amplified in Escherichia coli under the selection of 100 μg/ml ampicillin and sequence-verified prior to use. For luciferase assays, cells were rinsed with sterile PBS, 40 μl of cell culture lysis buffer (Promega) was added per well, and plates were incubated at room temperature for 5 min. 100 μl of luciferase assay reagent (Promega) was then added per well, and luminescence was read immediately. The data (luciferase output) for each 3′-UTR construct was normalized to the same promoter construct lacking the 3′-UTR.
RNA Isolation and qRT-PCR
Left ventricular tissue was collected 3 h following an IPC stimulus as described above. For measurements of miRNA, total RNA was isolated using an acid phenol:chloroform technique and enriched for the small RNA fraction per the manufacturer's instructions (mirVana miRNA isolation kit; Ambion). cDNA was synthesized from the small RNA-enriched fraction using an RT2 first strand cDNA synthesis kit (SABiosciences). Specific miRNA levels were assessed through quantitative real-time-PCR using miRNA-specific primers (SABiosciences). For assessment of Hsp70.3 transcript levels, total RNA was isolated (RNeasy kit; Qiagen), and cDNA was synthesized (high capacity RNA-to-cDNA kit; Applied Biosystems) per the manufacturer's instructions. All samples were performed in triplicate with a minimum of three independent experimental replicates with expression differences calculated using the ΔΔ Ct approximation method using 18 S mRNA as a loading control (14). Corrections for primer efficiency were made where appropriate using the Pfaffl method (15).
3′-RACE and Quantitative Assessment of APA
Qualitative 3′-RACE assessment of the Hsp70.3 3′-UTR population was performed via a PCR reaction with an Hsp70.3-specific primer and a modified oligo(dT)18 primer 5′-NT18-3′, with “N” representing A, G, or C, to allow for more specific binding at the beginning of the poly(A) tail and accurate size assessment of the 3′-UTR sequence. Quantitative measurements of the ratios of 3′-UTR transcripts were done via qRT-PCR using primers specific to sequence regions just proximal to the poly(A) site product being measured. To take into account that these primers would recognize polyadenylation products from any distal poly(A) site (i.e. primers recognizing sequence just proximal to poly(A) site 2 would detect product resulting from polyadenylation at site 2, 3, or 4), the quantitative result from each primer set is expressed as a normalized ratio to the total transcript as measured by primers just distal to the end of the protein coding sequence (proximal to all potential poly(A) sites). The resulting PCR products from both RACE and qRT-PCR were sequence-validated to be the proper Hsp70.3 3′-UTR products.
Cell Culture and Transfections
H9c2 cells were obtained from ATCC and grown under conditions of 5% CO2, 95% air at 37 °C in DMEM supplemented with 10% FBS. Wild-type and Hsp70.1 murine embryonic fibroblasts (MEFs) were isolated from 18-day-old embryos as described previously (16). HSF-1−/− and p65−/− MEFs were kind gifts from Drs. Hector Wong (Cincinnati Children's Hospital) and Denis Guttridge (Ohio State University), respectively. Experiments were performed in either 96-well (reporter assays) or 6-well (Western blots) plates with an initial seed density of 3 × 103 and 1 × 105 cells/well, respectively; cells were transfected 24 h after initial seeding. For reporter assays, 30 ng of plasmid DNA/well was transfected using Lipofectamine 2000 (Invitrogen) at the recommended ratio of 2.5 μl/1.0 μg of plasmid DNA. RNAi (miRNA, siRNA, or negative control RNAi; Qiagen) was transfected at the indicated molar doses using Lipofectamine 2000 at the recommended ratio of 1.0 μl/20 pmol of RNAi. All transfections were done in reduced serum Opti-MEM, an equal volume of DMEM supplemented with 20% FBS was added 4 h following transfection, and cells were then cultured overnight prior to experimental treatment. Heat shock was accomplished by placing cells at 42 °C for 1 h (with all other conditions held constant), and cells were allowed to recover at 37 °C for the indicated times.
Statistical Analysis
All results are reported as means ± S.E. Unpaired Student's t tests were performed to assess differences between groups, and statistical significance was considered at p ≤ 0.05.
RESULTS
Hsp70.3 Expression Is Subject to Post-transcriptional Regulation in the Myocardium
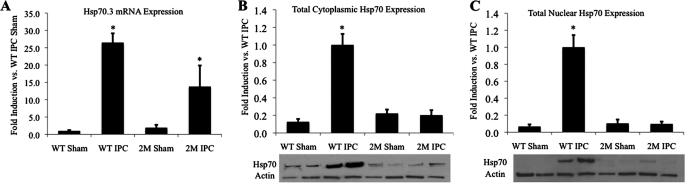
We previously showed that Hsp70.3 mRNA expression was induced 26.5-fold in the myocardium after late IPC in wild-type mice and 13.8-fold in 2M NF-κB dominant-negative mice (6). Assessment of total Hsp70 protein levels after late IPC showed a robust 7.7- and 14.3-fold increase in cytoplasmic and nuclear fractions, respectively, of Hsp70 protein expression in wild-type mice (Fig. 1). However, despite only a partial reduction in Hsp70.3 mRNA levels (26.5–13.8-fold) in 2M IPC relative WT IPC, no significant increase in total cytoplasmic or nuclear Hsp70 protein expression was observed in 2M NF-κB dominant-negative mice, relative to sham controls following IPC (Fig. 1). This result indicates that Hsp70 protein expression is subject to post-transcriptional suppression in 2M mice (with dominant-negative suppression of NF-κB activation) after late IPC in the heart.
FIGURE 1.
Hsp70.3 expression is post-transcriptionally regulated in late IPC. A, IPC strongly induces Hsp70.3 mRNA expression that is partially, but not fully, NF-κB-dependent. B and C, Western blots show that total myocardial Hsp70 protein is increased in both the cytoplasmic and the nuclear fractions 24 h after IPC in WT mice, but not in 2M NF-κB dominant-negative mice, despite a >10-fold mRNA induction. *, p ≤ 0.05 versus WT sham.
Post-transcriptional Regulation of Hsp70.3 Is Mediated by the 3′-UTR
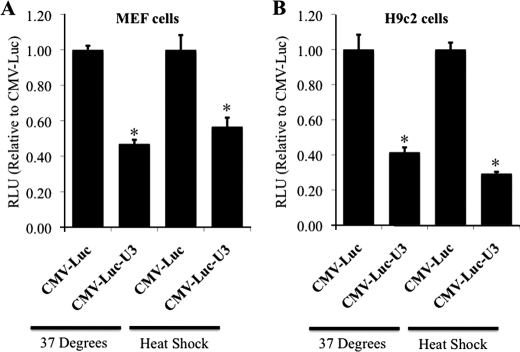
Post-transcriptional regulation of gene expression has most commonly been observed to be mediated through the 3′-UTR of the mRNA transcript (8, 9). Thus, luciferase reporter plasmids containing the 3′-UTR (∼1.25 kb from the end of the protein coding sequence) of the Hsp70.3 gene were used to examine the regulatory role of the Hsp70.3 3′-UTR on expression. Inclusion of the Hsp70.3 3′-UTR reduced reporter expression by ∼50% at basal levels as well as after HS in both MEF and H9c2 cells (Fig. 2). The suppressive effect of the Hsp70.3 3′-UTR sequence was observed with both a CMV promoter (Fig. 2) and the Hsp70.3 promoter, indicating that this suppressive effect is independent of promoter regulation and specific to the 3′-UTR sequence.
FIGURE 2.
The Hsp70.3 3′-UTR contains suppressive elements. The addition of the Hsp70.3 3′-UTR sequence leads to a post-transcriptional suppression of reporter expression (luciferase activity expressed as signal intensity, normalized to the same construct without the 3′-UTR) under both basal and stimulated (HS) conditions in both MEF cells (A) and H9C2 cells (B). *, p ≤ 0.05 versus CMV-Luc control. RLU, relative light units.
Late IPC Reduces the Expression of the Hsp70.3-targeting miRNAs
Analysis of the Hsp70.3 3′-UTR sequence with regard to miRNA binding sites revealed that 12 miRNAs are predicted to bind within the 3′-UTR (as determined via the Sanger miRBase target database) (Table 1) (17). Quantitative RT-PCR assessment of the myocardial expression levels of the 12 Hsp70.3-targeting miRNAs found that IPC induced a suppression of two Hsp70.3-targeting miRNAs: miR-378* and miR-711. Post-IPC, levels of miR-711 were found to be reduced in WT mice, but not 2M mice, indicating that post-IPC suppression of miR-711 is dependent on NF-κB activation in the cardiac myocyte (Table 1). IPC-induced suppression of miR-378* levels was independent of NF-κB (Table 1).
TABLE 1.
IPC induced changes in Hsp70.3 targeting miRNA
qRT-PCR was used to assess IPC-induced changes in expression levels of 12 predicted Hsp70.3 targeting miRNAs (as predicted by the Sanger miRBase target database). miR-378* and miR-711 (bold text) were the only two predicted Hsp70.3 targeting miRNAs whose levels were significantly (p ≤ 0.05) decreased in late IPC. The IPC-induced decrease in miR-711 expression, but not miR-378*, appears to be NF-κB-dependent. DN, dominant-negative.
| Number | miRNA ID | WT IPC vs. WT sham | NF-κB DN vs. WT IPC | NF-κB vs. WT sham |
|---|---|---|---|---|
| 1 | mmu-miR-485* | 1.95 | 1.29 | 2.51 |
| 2 | mmu-miR-224 | −1.03 | −1.45 | −1.49 |
| 3 | mmu-miR-711 | −2.63 | 3.76 | 1.43 |
| 4 | mmu-miR-7a* | 1.13 | 1.25 | 1.41 |
| 5 | mmu-miR-139–5p | −1.16 | −1.63 | −1.89 |
| 6 | mmu-miR-378* | −1.75 | −1.06 | −1.85 |
| 7 | mmu-miR-342–3p | −1.2 | −1.36 | −1.62 |
| 8 | mmu-miR-28 | −1.09 | −1.07 | −1.16 |
| 9 | mmu-miR-301b | 1.04 | 1.06 | 1.1 |
| 10 | mmu-miR-212 | −1.25 | −1.24 | −1.55 |
| 11 | mmu-miR-490 | −1.11 | −1.44 | −1.6 |
| 12 | mmu-miR-301a | 1.01 | −1.16 | −1.16 |
miR-378* and miR-711 Mediate Post-transcriptional Suppression of Hsp70.3 Expression via the 3′-UTR
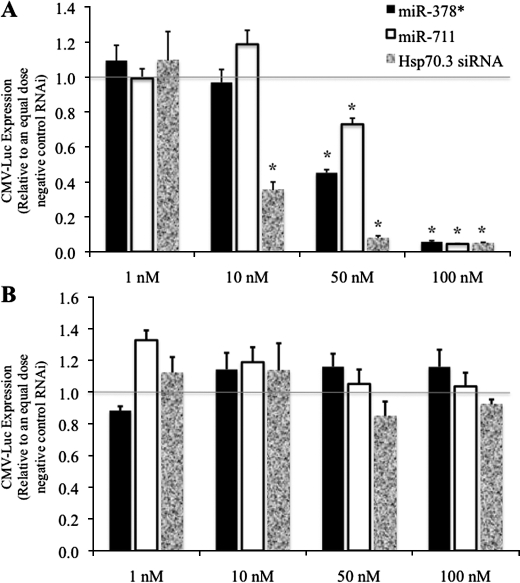
Next, the ability of exogenous miR-378* or miR-711 to inhibit expression of the Hsp70.3 3′-UTR luciferase reporters was examined in H9c2 rat cardiac myoblast cells. To test miR-378*- and miR-711-mediated regulation of the Hsp70.3 3′-UTR, H9c2 cells were transfected with CMV-luciferase-70.3 3′-UTR reporter (CMV-Luc-U3) and increasing doses of miR-378*, miR-711, a non-targeting negative control RNAi, or an Hsp70.3-targeting siRNA (Fig. 3 and supplemental Fig. S1). Results show that treatment with miR-378*, miR-711, or an Hsp70.3-targeting siRNA led to a dose-dependent reduction in CMV-Luc-U3 expression when compared with treatment with an equal dose of non-targeting negative control siRNA (Fig. 3A). The miR-378*- and miR-711-mediated suppression of the CMV-Luc-U3 reporter is sequence-specific to the Hsp70.3 3′-UTR as evidenced by the inability of the miRNAs to reduce expression of a similar reporter lacking the Hsp70.3 3′-UTR sequence (Fig. 3B).
FIGURE 3.
miR-378* and miR-711 suppress Hsp70.3 reporter expression via interaction with the 3′-UTR. H9c2 cardiac myoblast cells were treated with CMV-luciferase reporters with (A) or without (B) the Hsp70.3 3′-UTR sequence and increasing doses of miR-378*, miR-711, or an Hsp70.3 3′-UTR-specific siRNA. Results show a 3′-UTR-specific dose-dependent decrease in expression following treatment with miR-378*, miR-711, or an Hsp70.3 siRNA. *, p ≤ 0.05 versus equal dose negative control RNAi.
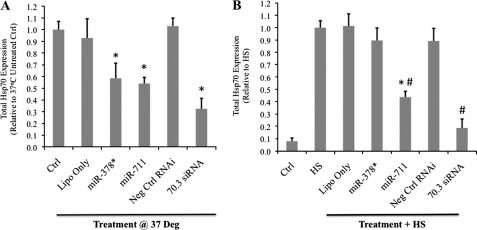
The effects of miR-378* and miR-711 on endogenous protein levels were also examined using Hsp70.1 knock-out MEFs to allow for easy measurement of Hsp70.3 protein levels without interference from Hsp70.1; an antibody able to distinguish between the inducible Hsp70 genes is not available. Results show that treatment with either 100 nm miR-711 or 100 nm miR-378* is able to reduce basal expression of Hsp70.3 protein, as measured by Western blotting (Fig. 4A). However, treatment with miR-711, but not miR-378*, is able to reduce Hsp70.3 protein induction following a 1-h heat shock treatment (Fig. 4B).
FIGURE 4.
miRNA suppression of endogenous Hsp70.3 protein expression. Hsp70.1−/− MEFs were treated with 100 nm doses of miR-378*, miR-711, a non-targeting negative control RNAi (Neg Ctrl RNAi), or an Hsp70.3-targeting siRNA. A, at basal conditions, treatment with either miR-378* or miR-711 resulted in a significant reduction in Hsp70.3 protein expression as measured by Western blotting. Ctrl, control; Lipo only, Lipofectamine 2000 only. B, at heat shock, treatment with miR-711, but not miR-378*, suppressed Hsp70.3 protein induction. *, p ≤ 0.05 versus untreated controls. #, p ≤ 0.05 versus HS cells.
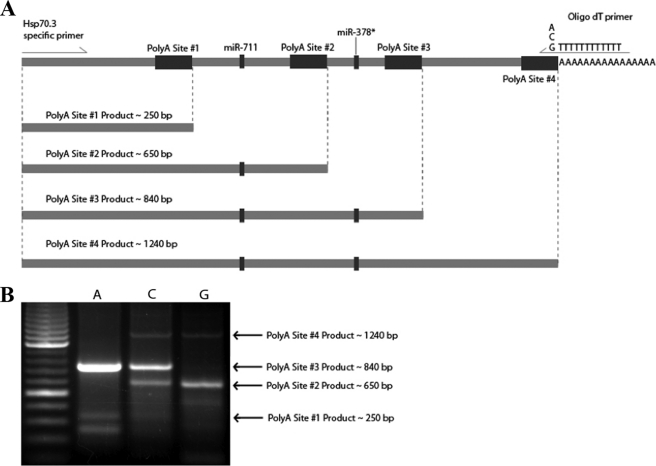
The Hsp70.3 mRNA Transcript Is Expressed with Multiple Distinct 3′-UTR Lengths as a Result of Alternatively Polyadenylated Transcripts
In addition to miRNA binding sites, the Hsp70.3 3′-UTR sequence is also predicted to contain four distinct polyadenylation recognition sites (Fig. 5). 3′-RACE was used to survey for the expression of Hsp70.3 transcripts harboring 3′-UTRs of different sizes as a result of APA. Results using either primary MEF cells or in vivo myocardium show the presence of multiple distinct size populations of Hsp70.3 3′-UTR lengths (Fig. 6). The arrows in the figure depict predicted PCR fragment sizes correlating with APA at each of the four predicted polyadenylation sites. These qualitative RACE results suggest that Hsp70.3 polyadenylation occurs primarily at APA sites 2 and 3. Interestingly, the annotated GenBankTM sequence for Hsp70.3 does not indicate expression of the 3′-UTR past APA site 2. However, a more detailed examination of expressed sequence tags located at least one expressed sequence tag (CO041885.1) that does represent a long variant of the Hsp70.3 3′-UTR supporting the use of APA site 3.
FIGURE 5.
The Hsp70.3 3′-UTR contains multiple polyadenylation sites. Sequence analysis of the 3′-UTR of Hsp70.3 shows that at least four distinct polyadenylation sites are predicted within the first 1250 bases from the end of the protein coding sequence. The predicted binding site for miR-711 is located between the first and second polyadenylation signals, whereas the predicted miR-378* site is located between the third and fourth polyadenylation signals.
FIGURE 6.
The Hsp70.3 mRNA transcript is expressed with multiple 3′-UTR lengths. A, four different size populations of Hsp70.3 3′-UTR (∼250, 650, 840, and 1240 bps) are predicted based on the four distinct polyadenylation (PolyA) sequence recognition sites found within the 3′-UTR. B, 3′-RACE results using primary MEF cells show the endogenous expression of at least four distinct Hsp70.3 3′-UTR size populations corresponding to the predicted alternative polyadenylated 3′-UTRs. The A, C, and G lanes of the gel represent the nucleotide base used at the proximal end of the oligo(dT) primer to increase PCR priming efficiency and specificity at the proximal end of the poly(A) tail.
Induction of Hsp70.3 mRNA Expression Results in Shortening of the Hsp70.3 3′-UTR
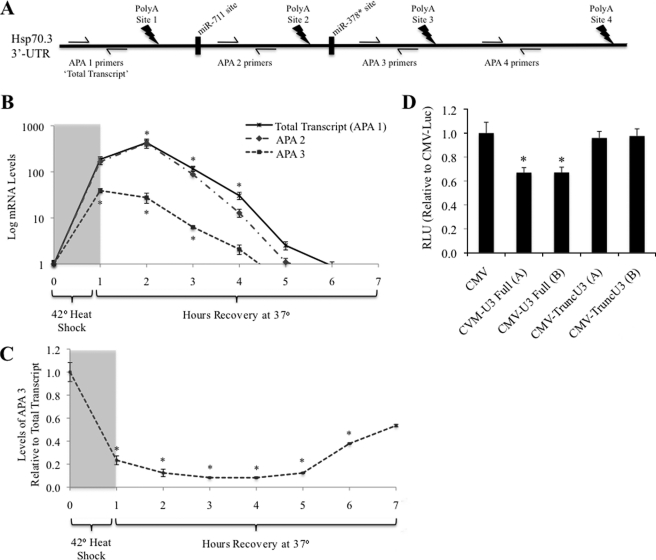
Hsp70.3 mRNA levels were assessed in WT MEF cells following a 1-h HS using qRT-PCR designed to assess the expression level of each Hsp70.3 APA product through amplification of specific regions of the Hsp70.3 transcript relative to the four polyadenylation sites (APA 1–4) (Fig. 7A). Results show that both total Hsp70.3 mRNA transcript and total APA 2 mRNA transcript increase to the same levels and that both peak at 2 h following the onset of HS, indicating that there is no significant truncation or polyadenylation of the mRNA transcript at APA site 1 (Fig. 7B). APA 3 transcript, however, peaks at 1 h following HS, is not up-regulated to the same degree, and is reduced back to baseline more quickly than total or APA 2 transcript (Fig. 7B). In addition, as total post-HS expression of total Hsp70.3 mRNA increases, the ratio of APA 3:total transcript decreases, indicating that the 3′-UTR of the transcript is shortened via APA at site 2 rather than at site 3 (Fig. 7C).
FIGURE 7.
Assessment of the relative transcript levels of the four different Hsp70.3 polyadenylation products. A, expression and alternative polyadenylation (PolyA) of Hsp70.3 mRNA was done via qRT-PCR amplification of specific regions of the Hsp70.3 transcript relative to the four polyadenylation sites (APA 1–4). B and C, as total post-HS expression of Hsp70.3 in MEFs increases (B), the ratio of APA 3:total transcript decreases, indicating that the 3′-UTR of the transcript is shortened via APA at site 2 rather than at site 3 (C). *, p ≤ 0.05 versus baseline control. D, CMV-luciferase reporters harboring a truncated version of the Hsp70.3 3′-UTR at APA site 2 (to mimic the APA-induced shortening of the 3′-UTR) show that post-transcriptional suppression is lost upon truncation of the 3′-UTR. *, p ≤ 0.05 versus CMV-Luc expression. RLU, relative light units.
Negligible expression of the Hsp70.3 mRNA region distal to poly(A) site 3 (APA 4 transcript) was detected, indicating that the vast majority of Hsp70.3 transcripts are polyadenylated at either APA site 2 or APA site 3. This lack of a detected signal from polyadenylation at APA site 4 is supported by predictions showing APA site 4 to be the weakest of the four predicted APA sites (DNA Functional Site Miner).
To confirm a physiological role of APA on expression of Hsp70.3, we compared expression from luciferase reporters with either the full-length Hsp70.3 3′-UTR or a version truncated just past APA site 2 to mimic the APA-induced shortening observed in the endogenous gene. As previously observed in Fig. 2, inclusion of the full-length 3′-UTR results in a significant suppression of reporter expression, but when the shortened 3′-UTR sequence truncated just past the APA 2 was included, this suppressive effect was no longer observed (Fig. 7D). This result supports the hypothesis that APA-mediated truncation of this sequence from the 3′-UTR acts to enhance expression of the gene.
APA-mediated Truncation of the Hsp70.3 3′-UTR Relieves Suppression through Both Enhanced mRNA Stability and Translational Efficiency
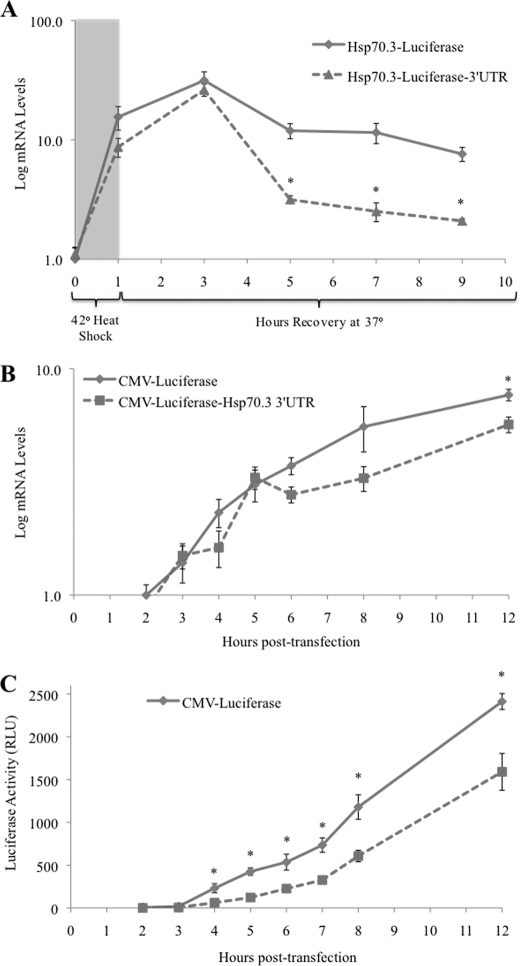
To determine whether the suppressive effect of the 3′-UTR is a post-transcriptional effect exerted directly on the mRNA transcript or a silencing of translation, the levels of expressed mRNA from reporter genes with and without the Hsp70.3 3′-UTR were assessed. First, reporters driven by the endogenous Hsp70.3 promoter were used such that transcriptional induction could be triggered by an HS stimulus. Results show that a 1-h HS triggers an immediate and robust increase in reporter transcripts regardless of the presence of the Hsp70.3 3′-UTR sequence. However, the reporter including the Hsp70.3 3′-UTR shows a much quicker decline from peak levels when compared with a similar reporter lacking the Hsp70.3 3′-UTR (Fig. 8A). This result is in agreement with results from Fig. 7 showing that levels of the endogenous transcript containing the full-length 3′-UTR return to baseline before levels of the APA-shortened transcript and suggests that shortening of the Hsp70.3 3′-UTR may enhance the stability of the mRNA transcript.
FIGURE 8.
Correlation of mRNA and protein production from Hsp70.3 3′-UTR luciferase reporters. A, using qRT-PCR with primers specific to an internal region of luciferase, mRNA levels of a luciferase reporter controlled by the Hsp70.3 promoter with and without the full-length Hsp70.3 3′-UTR were tracked in H9c2 cells following a 1-h HS. B, mRNA levels of CMV-luciferase reporters with and without the full-length Hsp70.3 3′-UTR were assessed in H9c2 cells beginning at 2 h after transfection. C, assessment of protein levels (as measured by luciferase activity) of CMV-luciferase reporters with and without the full-length Hsp70.3 3′-UTR were assessed in H9c2 cells beginning at 2 h after transfection. *, p ≤ 0.05 between reporter plasmids with and without the Hsp70.3 3′-UTR. RLU, relative light units.
Next, the mRNA and protein levels were simultaneously measured from a CMV-luciferase reporter with and without the Hsp70.3 3′-UTR. Unlike the Hsp70.3 promoter-driven reporter, the transcriptional expression of which is acutely stimulated and reduced after a 1-h HS, the CMV promoter-controlled reporters drive a continuously high level of transcriptional expression. We could not detect significant 3′-UTR-dependent differences in mRNA levels from CMV promoter-controlled reporters for the first 8 h following transfection (Fig. 8B). Although a significant difference in mRNA levels is apparent at 12 h after treatment, a significant suppression of protein levels from the reporter with the Hsp70.3 3′-UTR is observed as early as 4 h after treatment and persists throughout the assessed time points (Fig. 8, B and C). The difficulty in detecting differences in mRNA levels related to the presence of the 3′-UTR likely reflects the fact that the CMV promoter drives strong constitutive expression of mRNA. We conclude that, in addition to altering the stability of the mRNA, it appears that the removal of the Hsp70.3 3′-UTR sequence increases protein levels.
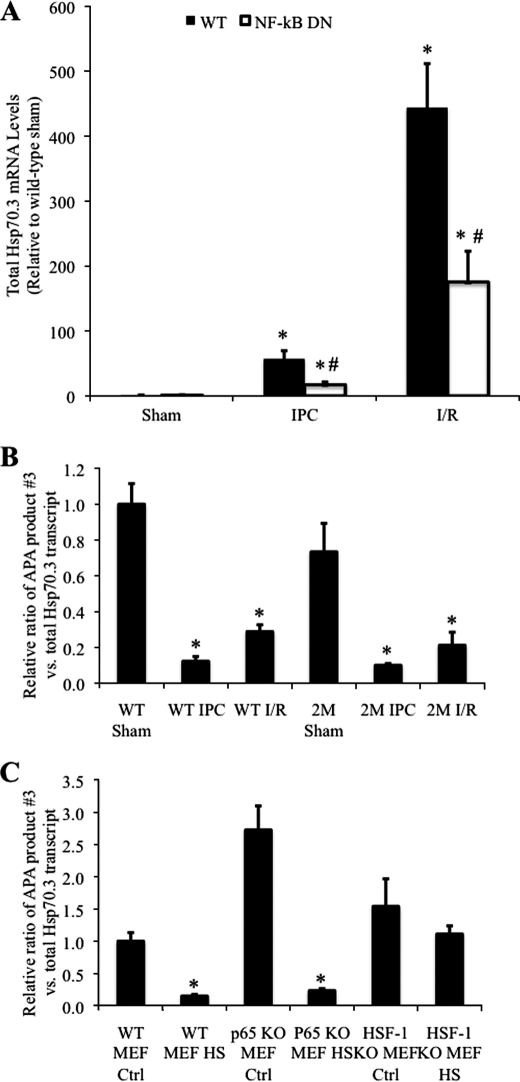
Hsp70.3 APA-mediated 3′-UTR Shortening Is Observed in the Heart in Vivo following IPC or I/R
Total Hsp70.3 transcript levels in the in vivo myocardium are increased in a partially NF-κB-dependent manner following IPC or I/R (Figs. 1 and 9A) (6). Similar to what was observed following HS-induced Hsp70.3 mRNA expression, in both IPC and I/R, the increase in total Hsp70 mRNA is accompanied by an APA-mediated shortening of the Hsp70.3 3′-UTR (Fig. 9B). To investigate the mechanism of Hsp70.3 APA, the role of NF-κB in this process was investigated using 2M IκBα dominant-negative mice. This mouse model of transgenic IκBα dominant-negative inhibition of NF-κB was used because it was in these mice following IPC that the post-transcriptional regulation of Hsp70.3 was first observed (Fig. 1). However, similar to what was observed in the wild-type mice, results show a significant reduction in the ratio of APA product 3 in the 2M mice following both IPC and I/R (Fig. 9B). Thus, upon a stimulus that induces Hsp70.3 mRNA expression, the Hsp70.3 3′-UTR is expressed as a shortened form due to increased APA at polyadenylation site 2. The APA effect was also observed to be unaltered in MEF cells deficient for the p65 (RelA) subunit of NF-κB (Fig. 9C). Taken together, these results indicate that NF-κB activity does not play a regulatory role in alternative polyadenylation of the Hsp70.3 3′-UTR.
FIGURE 9.
APA-mediated shortening of the Hsp70.3 3′-UTR is dependent on HSF-1, but not NF-κB, activity. A, qRT-PCR was used to measure the increase in total Hsp70.3 mRNA following IPC and I/R in wild-type and NF-κB-dominant-negative (NF-κB-DN) mice. B, IPC- and I/R-induced shortening of the Hsp70.3 3′-UTR (APA 3:total transcript ratio) is maintained in NF-κB dominant-negative mice, indicating that this effect is independent of NF-κB activation in the heart. C, similar to WT MEFs, p65−/− MEFs display an HS-dependent shortening of the 3′-UTR. APA of the Hsp70.3 3′-UTR is abrogated in HSF-1−/− MEFs. In B and C, ratios of APA 3:total transcript are expressed relative to WT sham (in vivo controls) or WT MEF 37 °C control (WT MEF Ctrl) (in vitro controls). *, p ≤ 0.05 versus corresponding control group. #, p ≤ 0.05 versus wild-type.
The Transcription Factor HSF-1 Is Needed for Alternative Polyadenylation of the Hsp70.3 3′-UTR
Because the transcriptional regulator of heat shock, HSF-1, has been previously suggested to play a role in mRNA polyadenylation (18), we also examined the role of HSF-1 in HS-mediated APA of the Hsp70.3 transcript. Results using HSF-1−/− MEFs (the homozygous mice are not viable) demonstrate that the Hsp70.3 3′-UTR does not undergo an APA-mediated shortening following HS in MEF cells lacking HSF-1 (Fig. 9C). These results implicate an obligatory role for the transcription factor HSF-1 in APA-mediated shortening of the Hsp70.3 3′-UTR.
DISCUSSION
The results presented here demonstrate that Hsp70 protein expression in the myocardium is subject to post-transcriptional regulation after late IPC. We show that in addition to NF-κB-dependent transcriptional up-regulation of the gene (6), late IPC functions to remove microRNA inhibition via (i) NF-κB-dependent reduction of miR-711 (Hsp70.3-suppressive) levels, (ii) NF-κB-independent reduction of miR-378* (Hsp70.3-suppressive), and (iii) HSF-1-dependent APA, which acts to remove the suppressive miR-378* binding site from the Hsp70.3 3′-UTR. We further provide evidence that the suppressive effects of the miRNAs result in reduction in Hsp70.3 mRNA levels and a decrease in Hsp70.3 protein. Specifically, it appears that the shortening of the 3′-UTR by APA enhances the amount of protein produced, perhaps by increased translation efficiency.
The role of miRNAs in modulating gene expression in response to ischemic stimuli in the heart is coming to light as more and more gene products are being functionally identified as targets of miRNA regulation (19). A direct regulation of Hsp70 expression by miRNAs in the heart has not yet been demonstrated. However, in a recent study by Yin et al. (20), it was shown that IPC increases the expression of the miRNAs miR-23b, miR-483, and miR-1 in the heart and that administration of these miRNAs was sufficient to protect the myocardium from subsequent I/R injury. Although this effect was suggested to be due to an miRNA-mediated increase in endothelial NOS (eNOS), HSF-1, and Hsp70 expression, this is likely to be an indirect effect on Hsp70 expression due to the fact that there are no predicted target sites for these miRNAs in the Hsp70.3 gene sequence (20).
Our previous work showed that NF-κB activation in cardiac myocytes is necessary for Hsp70.3 expression in the heart following IPC and I/R (6). However, NF-κB does not appear to play a role in the alternative polyadenylation of the Hsp70.3 mRNA transcript. The lack of increased Hsp70.3 protein expression following IPC in 2M mice despite only a partial decrease in mRNA levels (Fig. 1) and an NF-κB-independent APA-induced shortening of the 3′-UTR (Fig. 9, B and C) suggests that other processes in addition to APA shortening of the 3′-UTR are involved in post-transcriptional regulation of Hsp70.3 expression. NF-κB-dependent down-regulation of miR-711 levels, as observed in IPC (Table 1), likely represents one of these additional processes. The results presented here are novel in that they demonstrate a stimulus-specific regulation of alternative polyadenylation that has direct consequences on the miRNA-mediated post-transcriptional regulation. Specifically, the removal of the miR-378* binding site in the truncated Hsp70.3 APA product removes the action of miR-378*, rendering regulation of the shorter transcripts relatively more dependent upon NF-κB-dependent changes in miR-711.
Alternative polyadenylation has been previously shown to be a potentially critical post-transcriptional regulator of gene expression (9, 21). The primary means by which this occurs is presumed to be through the removal of regulatory sequence elements within the 3′-UTR of the mRNA transcript, thus affecting miRNA binding, mRNA stability, or binding of other proteins or regulatory factors, including those involved in translation. The loss of regulatory elements in the 3′-UTR due to APA-mediated shortening of the transcript has recently been recognized as means by which oncogenes may escape regulation in cancer cells (21). Similarly, our results show that increased expression of Hsp70.3 mRNA in the myocardium is correlated with a shortening of the 3′-UTR that results in the loss of a predicted binding site for a suppressive miRNA. Experiments with acute 1-h heat shock induction (Fig. 8A) demonstrate that although the induction kinetics and peak mRNA levels are similar, the decrease in mRNA levels after 3 h is faster for the luciferase reporter mRNA that contains the Hsp70.3 3′-UTR. Taken together with data that the Hsp70.3 mRNA with the APA 2 terminus decreases in level more quickly after heat shock than the APA 3 transcript, this supports a reduced stability of the mRNA with the APA 2 termination. In addition, our results suggest that the APA-shortened version of the Hsp70.3 mRNA results in more efficient protein translation than its full-length counterpart (Fig. 8). Note that because these studies were performed with the luciferase reporter, differences in Hsp70.3 protein stability would not play a role in this effect. Other studies have also associated alternative polyadenylation with direct effects on translation efficiency and protein expression. Brain-derived neurotrophic factor (BDNF), chloramphenicol acetyltransferase (CAT), and the apoptosis gene hap have been shown to be expressed as multiple alternative polyadenylation products, with one product displaying a much higher efficiency of protein expression (22–24).
Although as many as 50% of expressed genes have been identified to contain alternative polyadenylation sites (25), the role of alternative polyadenylation in the regulation of gene expression remains an underexplored field. In addition to Hsp70.3, COX-2 is a well known NF-κB-dependent cardioprotective gene product that has been shown to be subject to alternative polyadenylation (26–29). In the case of COX-2 alternative polyadenylation, it has been shown that this process is dependent upon the interaction of a discreet set of RNA-binding proteins (polypyrimidine tract binding protein (PTB), PTB associated splicing factor (PSF), p54 nono-related binding protein (p54nrb), and U1A) with upstream sequence elements in the 3′-UTR that control the efficiency of polyadenylation (29). In addition to the core polyadenylation factors such as cleavage/polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), cleavage factor I (CFI), and cleavage factor II (CFII), additional factors including transcriptional and splicing factors whose specific role in polyadenylation is not yet completely understood have also been identified as mediators of the polyadenylation process (9). Whether or not these or similar RNA-binding proteins play a role in the regulation of Hsp70.3 APA remains to be seen. It is still not entirely clear how the activity of many of these proteins or their modulation of polyadenylation specificity or efficiency is controlled within the cell.
Our results suggest that in the absence of HSF-1, APA of the Hsp70.3 gene is abrogated (Fig. 9C). However, it is unclear from these data whether this is a direct or an indirect role for HSF-1 in APA of Hsp70.3. The notion that a transcription factor may play a regulatory role in polyadenylation is not a new one as it is has been previously shown that mRNA processing is linked to transcription and that polyadenylation factors can directly interact with transcription factors (30, 31). Previous work suggests that HSF-1 can physically interact with the polyadenylation factors symplekin and CstF64, potentially playing a role in polyadenylation of the Hsp70 genes (18). However, at this point, the identity of the regulatory factors controlling polyadenylation site selection within the Hsp70.3 mRNA remains unknown and is the subject of ongoing/future work.
In addition to the identity of the RNA-binding proteins responsible for alternative polyadenylation of the Hsp70.3 transcript, there are other unanswered questions. First, is APA-mediated shortening of the Hsp70.3 3′-UTR directly linked to the same stimuli that lead to a transcriptional induction of the Hsp70.3 gene, or is there differential APA regulation? Second, do miRNAs or other small non-coding RNAs play direct roles in the regulation of alternative polyadenylation? Finally, although we show here how APA plays a direct role in regulating the binding of miR-378*, by removing its binding site, the question of whether miRs or other non-coding RNAs play critical roles in regulating APA remains unanswered. It is possible that co-evolution of the miRNA and APA regulatory processes has resulted in some interesting regulatory interactions in this regard.
In conclusion, our results indicate that alternative polyadenylation plays a critical role as one of multiple integrated regulatory steps controlling Hsp70.3 gene product expression. At basal levels, constitutive expression levels of the mRNA and protein are both readily detectable. Upon an Hsp70.3-inducing stimulus, the protein up-regulation is achieved through the coordinated actions of: 1) transcriptional induction of the gene (partially dependent on NF-κB); 2) decreased expression levels of Hsp70.3-targeting miRNAs (as measured in the in vivo myocardium after late IPC; miR-711 was decreased in an NF-κB-dependent manner, and miR-378* was NF-κB-independent); and 3) alternative polyadenylation-mediated shortening of the 3′-UTR resulting in loss of a suppressive miRNA binding site, mRNA stabilization, and increased protein production. It is interesting that in addition to their roles in transcriptional modulation, NF-κB and HSF-1 both play coordinated roles in post-transcriptional regulation of Hsp70.3: NF-κB through regulation of the Hsp70.3-targeting miR-711 and HSF-1 through regulation of APA. These results enhance the understanding of how miRNAs and alternative polyadenylation contribute to the post-transcriptional regulation of Hsp70.3 and may be extended to additional genes whose expression is subject to similar means of post-transcriptional control through microRNAs and alternative polyadenylation.
Given the emerging role for APA as an important post-transcriptional regulator of gene expression, the significance of these results has implications that stretch far beyond just the understanding of Hsp70.3 gene expression in myocardium. APA represents an underexplored, yet critical, regulatory step in the control of gene expression. The increased mechanistic understanding of APA and its integration with miRNA regulatory networks will greatly increase our knowledge of post-transcriptional gene regulation and may lead to the identification of novel therapeutic targets for the treatment of diverse pathologies such as cancer, diabetes, neurodegenerative diseases, and aging in addition to post-myocardial infarction pathophysiology and cardiomyopathy in the cardiovascular system.
Supplementary Material
Acknowledgments
We thank Jackie Belew for support in maintaining mouse breeding colonies and organizing procedures and for research assistance and Hector Wong (Cincinnati Children's Hospital) and Denis Guttridge (Ohio State University) for providing us with HSF-1−/− and p65−/− MEF cells, respectively.
This work was supported, in whole or in part, by National Institutes of Health Grants HL63034 and HL091478 (to W. K. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- HS
- heat shock
- Hsp70
- heat shock protein 70
- I/R
- ischemia/reperfusion
- IPC
- ischemic preconditioning
- miRNA
- microRNA
- miR
- microRNA
- APA
- alternative polyadenylation
- qRT-PCR
- quantitative RT-PCR
- RACE
- rapid amplification of cDNA ends
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1. Currie R. W., Karmazyn M., Kloc M., Mailer K. (1988) Circ. Res. 63, 543–549 [DOI] [PubMed] [Google Scholar]
- 2. Mestril R., Dillmann W. H. (1995) J. Mol. Cell. Cardiol. 27, 45–52 [DOI] [PubMed] [Google Scholar]
- 3. Donnelly T. J., Sievers R. E., Vissern F. L., Welch W. J., Wolfe C. L. (1992) Circulation 85, 769–778 [DOI] [PubMed] [Google Scholar]
- 4. Marber M. S., Latchman D. S., Walker J. M., Yellon D. M. (1993) Circulation 88, 1264–1272 [DOI] [PubMed] [Google Scholar]
- 5. Marber M. S., Mestril R., Chi S. H., Sayen M. R., Yellon D. M., Dillmann W. H. (1995) J. Clin. Invest. 95, 1446–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tranter M., Ren X., Forde T., Wilhide M. E., Chen J., Sartor M. A., Medvedovic M., Jones W. K. (2010) J. Mol. Cell. Cardiol. 49, 664–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilhide M. E., Tranter M., Ren X., Chen J., Sartor M. A., Medvedovic M., Jones W. K. (2011) J. Mol. Cell. Cardiol. 51, 82–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bartel D. P. (2009) Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lutz C. S. (2008) ACS Chem. Biol. 3, 609–617 [DOI] [PubMed] [Google Scholar]
- 10. Brown M., McGuinness M., Wright T., Ren X., Wang Y., Boivin G. P., Hahn H., Feldman A. M., Jones W. K. (2005) Am. J. Physiol. Heart Circ. Physiol. 289, H466–H476 [DOI] [PubMed] [Google Scholar]
- 11. Dawn B., Xuan Y. T., Marian M., Flaherty M. P., Murphree S. S., Smith T. L., Bolli R., Jones W. K. (2001) J. Mol. Cell. Cardiol. 33, 161–173 [DOI] [PubMed] [Google Scholar]
- 12. Shim E. H., Kim J. I., Bang E. S., Heo J. S., Lee J. S., Kim E. Y., Lee J. E., Park W. Y., Kim S. H., Kim H. S., Smithies O., Jang J. J., Jin D. I., Seo J. S. (2002) EMBO Rep 3, 857–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ren X., Wang Y., Jones W. K. (2004) J. Surg. Res. 121, 120–129 [DOI] [PubMed] [Google Scholar]
- 14. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 15. Pfaffl M. W. (2001) Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wurst W., Joyner A. L. (1993) Gene Targeting: A Practical Approach (Joyner A. L. ed) pp. 36–41, Oxford University Press, New York [Google Scholar]
- 17. Griffiths-Jones S., Saini H. K., van Dongen S., Enright A. J. (2008) Nucleic Acids Res. 36, D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xing H., Mayhew C. N., Cullen K. E., Park-Sarge O. K., Sarge K. D. (2004) J. Biol. Chem. 279, 10551–10555 [DOI] [PubMed] [Google Scholar]
- 19. Ye Y., Perez-Polo J. R., Qian J., Birnbaum Y. (2011) Physiol. Genomics 43, 534–542 [DOI] [PubMed] [Google Scholar]
- 20. Yin C., Salloum F. N., Kukreja R. C. (2009) Circ. Res. 104, 572–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mayr C., Bartel D. P. (2009) Cell 138, 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lau A. G., Irier H. A., Gu J., Tian D., Ku L., Liu G., Xia M., Fritsch B., Zheng J. Q., Dingledine R., Xu B., Lu B., Feng Y. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 15945–15950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qu X., Qi Y., Qi B. (2002) Arch. Biochem. Biophys. 400, 233–244 [DOI] [PubMed] [Google Scholar]
- 24. Yu M., Sha H., Gao Y., Zeng H., Zhu M., Gao X. (2006) Biochem. Biophys. Res. Commun. 345, 479–485 [DOI] [PubMed] [Google Scholar]
- 25. Tian B., Hu J., Zhang H., Lutz C. S. (2005) Nucleic Acids Res. 33, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bolli R., Shinmura K., Tang X. L., Kodani E., Xuan Y. T., Guo Y., Dawn B. (2002) Cardiovasc Res. 55, 506–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hall-Pogar T., Zhang H., Tian B., Lutz C. S. (2005) Nucleic Acids Res. 33, 2565–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harper K. A., Tyson-Capper A. J. (2008) Biochem. Soc, Trans 36, 543–545 [DOI] [PubMed] [Google Scholar]
- 29. Hall-Pogar T., Liang S., Hague L. K., Lutz C. S. (2007) RNA 13, 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McCracken S., Fong N., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S. D., Wickens M., Bentley D. L. (1997) Nature 385, 357–361 [DOI] [PubMed] [Google Scholar]
- 31. Dantonel J. C., Murthy K. G., Manley J. L., Tora L. (1997) Nature 389, 399–402 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.