Abstract
Specific regulation of target genes by transforming growth factor-β (TGF-β) in a given cellular context is determined in part by transcription factors and cofactors that interact with the Smad complex. In this study, we determined Smad2 and Smad3 (Smad2/3) binding regions in the promoters of known genes in HepG2 hepatoblastoma cells, and we compared them with those in HaCaT epidermal keratinocytes to elucidate the mechanisms of cell type- and context-dependent regulation of transcription induced by TGF-β. Our results show that 81% of the Smad2/3 binding regions in HepG2 cells were not shared with those found in HaCaT cells. Hepatocyte nuclear factor 4α (HNF4α) is expressed in HepG2 cells but not in HaCaT cells, and the HNF4α-binding motif was identified as an enriched motif in the HepG2-specific Smad2/3 binding regions. Chromatin immunoprecipitation sequencing analysis of HNF4α binding regions under TGF-β stimulation revealed that 32.5% of the Smad2/3 binding regions overlapped HNF4α bindings. MIXL1 was identified as a new combinatorial target of HNF4α and Smad2/3, and both the HNF4α protein and its binding motif were required for the induction of MIXL1 by TGF-β in HepG2 cells. These findings generalize the importance of binding of HNF4α on Smad2/3 binding genomic regions for HepG2-specific regulation of transcription by TGF-β and suggest that certain transcription factors expressed in a cell type-specific manner play important roles in the transcription regulated by the TGF-β-Smad signaling pathway.
Keywords: Chromatin Immunoprecipitation (ChiP), Gene Transcription, HNF-4, Smad Transcription Factor, Tissue-specific Transcription Factors
Introduction
Transforming growth factor-β (TGF-β) has multiple roles in growth arrest, apoptosis, differentiation, epithelial-to-mesenchymal transition, and extracellular matrix deposition in various types of cells and is related to embryonic development and various human diseases (1). In cancer cells, TGF-β is known to possess conflicting tumor-suppressive and pro-metastatic functions; TGF-β inhibits cancer progression by cell cycle arrest and apoptosis, although it also helps cancer cells to evade anti-tumor immune response and metastasize to distant organs by epithelial-to-mesenchymal transition. Understanding the precise regulatory mechanisms downstream of this signaling pathway is required for the control of diseases.
Smad family proteins are the principal and specific molecules that transduce signals from the ligand-bound active receptor complexes on the cell surface membrane to the nucleus (2–4). Smad2 and Smad3 form hetero-oligomers with Smad4 after phosphorylation of the C terminus of Smad2 or Smad3 by the receptor complex, and the Smad complex serves as a transcription factor on the genome. The Smad complex was reported to bind to the sequences containing “GTCT” (Smad-binding element) by in vitro screening of the binding sequences and structural analysis of the Smad complex bound to the DNA (5, 6). However, this very simple motif is present everywhere in the genome. It has also been suggested that the binding affinity of the Smad complex to Smad-binding elements is not high. Interaction with other transcription factors and cofactors has been shown to be important to provide functional specificity of TGF-β signaling, and these transcription factors and cofactors facilitate binding of the Smad complex to the favorable positions in the genome. Expressions of these transcription factors and cofactors are often regulated in a cell- or tissue-specific manner, and a subset of these molecules indeed has been shown to be important for the context-dependent Smad binding to the genome and transcriptional regulation of target genes. Target genes of TGF-β that are regulated by the same cofactors are designated as a synexpression group (7), as reported in the regulation of several genes such as CDKN1A/p21 and GADD45A by FOXO family proteins (8).
High throughput analyses of transcription factor binding regions using either an oligonucleotide tiling microarray or massively parallel sequencing are now widely used to understand the roles of transcription factors (9, 10). We have identified Smad2/3 binding regions and Smad4 binding regions using a promoter tiling array in the HaCaT normal human epidermal keratinocyte cell line (11, 12). We found Smad2/3 binding regions at the previously analyzed regions as well as many unrecognized binding regions. Activator protein-1 (AP-1), v-Ets erythroblastosis virus E26 oncogene homolog, and transcription factor AP-2-binding motifs were identified as enriched motifs in the Smad2/3 binding regions in HaCaT cells (11). However, it remains to be determined whether the identified Smad2/3 binding regions are shared with those in other cells and tissues.
Hepatocyte nuclear factor 4α (HNF4α)4 is a member of the hepatocyte nuclear factor family, which includes well conserved nuclear receptors among mammals. HNF4α is expressed in the liver, kidney, small intestine, and pancreas and is essential for the organogenesis of the liver (13, 14). HNF4α is also required for the differentiation of hepatocytes and is engaged in hepatocyte-specific gene regulation related to the synthesis of apolipoproteins, acute phase reactive proteins, and other secreted proteins. HNF4α is located in the nucleus, forms a homodimer, and functions as a transcription factor by binding to DR1 elements in the genome (15).
Several groups have identified a functional relationship between HNF4α and TGF-β signaling. TGF-β down-regulates the expression of variant 1 of HNF4α, one of the transcriptional variants of HNF4α, which has an AF1 transcriptional activation domain in their N terminus (16). On the contrary, the expression of the transcriptional activation domain lacking variant 8 is repressed by TGF-β in normal murine mammary gland (NMuMG) epithelial cells (17). TGF-β has also been reported to regulate the HNF4α expression by proteasome-dependent degradation (16). The effect of HNF4α on TGF-β-induced transcription has also been analyzed for the APOC3 promoter, where HNF4α interacts with Smad3 and Smad4 to induce the APOC3 expression (18, 19). The HNF4α-binding motif in the APOC3 promoter has been shown to be important for TGF-β-induced transcriptional activity, and a mutant of Smad3 that lacks the DNA binding property to Smad-binding elements still interacts with HNF4α to synergistically transactivate the APOC3 promoter (19). Because HNF4α binds to the MH1 DNA binding domain of Smad3 through both its N and C termini (19), Smads may indirectly bind to the APOC3 promoter through HNF4α. However, it is still unclear whether the reported interaction with Smads and mechanisms of transcriptional regulation are generally important for the function of both HNF4α and Smads in hepatocytes.
Here, we identified Smad2/3 binding regions in the HepG2 hepatoblastoma cell line and compared them with the binding regions in HaCaT cells and hepatocellular carcinoma Hep3B cells to elucidate the mechanisms of context-dependent regulation of TGF-β-induced transcription. We found HNF4α as an important factor for HepG2-specific Smad2/3 binding regions and analyzed its regulatory mechanism using a new target gene of HNF4α, MIXL1, under TGF-β stimulation.
EXPERIMENTAL PROCEDURES
Cell Culture
Human hepatoblastoma HepG2 cells and hepatocellular carcinoma Hep3B cells were obtained from the American Type Culture Collection and were cultured in minimum essential medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Thermo Scientific, Rockford, IL), 0.1 mm nonessential amino acids, 1 mm sodium pyruvate, 100 units/ml penicillin G, and 100 μg/ml streptomycin. HaCaT cells were maintained in Dulbecco's modified Eagle's medium (DMEM; catalog no. 11965; Invitrogen) supplemented with 10% FBS, 100 units/ml penicillin G, and 100 μg/ml streptomycin. Cells were grown in a humidified atmosphere with 5% CO2 at 37 °C.
Antibodies and Chemicals
We used the following commercially available antibodies: mouse anti-Smad2/3 (BD Biosciences), anti-α-tubulin (DM1A) (Sigma), rabbit anti-phospho-Smad2 (Cell Signaling Technology, Beverly, MA), anti-phospho-Smad3 (Cell Signaling Technology), and anti-HNF4α (Santa Cruz Biotechnology, Santa Cruz, CA). TGF-β3 was from R & D Systems (Minneapolis, MN).
RNA Interference and Oligonucleotides
Stealth small interfering RNA (siRNA) targeting HNF4α (5′-AAAGCGGCCACGCGAGUCAUACUGG-3′) was purchased from Invitrogen. As a negative control, we used a predesigned siRNA (12935–200, sequence not available). siRNAs were introduced into HepG2 cells using the Lipofectamine RNAi MAX reagent (Invitrogen) according to the manufacturer's instructions (reverse transfection method), using 3 nm siRNA per 1 × 105 cells/ml per well of 12-well plates.
Immunoblotting
SDS-gel electrophoresis and immunoblotting were performed as described, using a LAS-4000 lumino-image analyzer (Fujifilm, Tokyo, Japan) (20). RIPA buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 150 mm NaCl, 50 mm Tris-HCl (pH 8.0), 5 mm EDTA) was used for cell lysis.
Chromatin Immunoprecipitation (ChIP), ChIP-chip, and ChIP Sequencing (Seq)
Sample preparation for ChIP-chip analysis was performed as described previously, using anti-Smad2/3 (BD Biosciences) (11, 21, 22). Briefly, ChIP and control input DNA samples were amplified by two cycles of in vitro transcription and hybridized on separate Affymetrix human promoter 1.0R oligonucleotide tiling arrays (Affymetrix, Santa Clara, CA). Enrichment values (ChIP/control input DNA) were calculated using the MAT algorithm, and Smad2/3 binding regions were determined using detection p values of 10−4 (23). For conventional quantitative anti-Smad2/3 ChIP-quantitative PCR (ChIP-qPCR) analyses, cells were cross-linked with 10 mm dimethyl adipimidate (Thermo Scientific) for 30 min at room temperature before formaldehyde fixation. Bioruptor UCW-301 (output: H, 2 cycles of 30 s of sonication with 30-s intervals; Cosmobio, Tokyo, Japan) was used for sonication of ChIP-qPCR samples. Sample preparation for ChIP-seq was performed according to the manufacturer's instructions (Illumina, San Diego). The obtained read tags were mapped on the NCBI/hg18 human genome assembly using ELAND (Illumina). CisGenome was used for the calculation of significant HNF4α binding regions (one-sample analysis, with default parameters and a false discovery rate (FDR) of less than 0.1%) (24).
Motif Prediction and Mapping
CisGenome was used for both de novo motif prediction and motif mapping of Smad2/3 ChIP-chip and HNF4α ChIP-seq binding regions. Cis-regulatory element annotation system (CEAS) was also used for identification of known motifs in Smad2/3 and HNF4α binding regions (25).
Quantitative PCR Analysis
Quantitative real time PCR analysis was performed as described previously (26). Amplification data were quantified using the standard curve method. Detected signals were confirmed to be specific by a dissociation protocol. All samples were run in duplicate or triplicate, and the results were averaged. Sequences of the primers are available in supplemental Tables S1 and S2.
Reverse Transcription-PCR and Expression Microarray Analysis
Total RNAs were extracted as described previously (26). First strand cDNAs were synthesized using the PrimeScript2 reverse transcriptase (TakaraBio, Shiga, Japan). The experimental procedures for GeneChip (Affymetrix) were performed as described previously (11) using the GeneChip human U133 plus 2.0 oligonucleotide array (Affymetrix). Microarray Suite software 5.0 (Affymetrix) was used with a target intensity of 100. Data from one array were obtained for each sample.
Promoter Reporter Constructs and cDNA Constructs
The human MIXL1 promoter reporter (MIXL1-WT-luc, −583 to −8) and its mutants were constructed by a PCR-based approach and cloned into the pGL4.10 (Promega, Fitchburg, WI) vector with a minimal luciferase promoter sequence (11). Primer sequences used for the construction of MIXL1-mut1-luc were 5′-GCAGGGGTGGTAAATAAATTTAGGGTTATCGGGACAGACGGGAC-3′ and 5′-GTCCCGTCTGTCCGATAACCCTAAATTTATTTACCACCCCTGC-3′. The primer sequences for the construction of MIXL1-mut2-luc were 5′-TCCCCGAGCCCTTAGGGTATTACACCGCCCCGCCTTC-3′ and 5′-GAAGGCGGGGCGGTGTAATACCCTAAGGGCTCGGGGA-3′. MIXL1-luc reporters with mutations in Smad-binding elements were also constructed by PCR (see Fig. 6D for sequences of the mutants). HNF4α and its C115R mutant were constructed by a PCR-based approach using the first strand cDNA of HepG2 cells as a template. The sequences of all cDNAs constructed were verified by sequencing.
FIGURE 6.
Identification of regulatory elements important for TGF-β-induced transactivation of the MIXL1 promoter. A, schematic representation of HNF4α-binding motifs in the Smad2/3 and HNF4α binding region of the MIXL1 promoter. Promoter reporters with mutations in their HNF4α-binding motifs are shown in the lower panel. B, activation of the MIXL1 gene promoter by TGF-β and effects of mutations in putative HNF4α-binding motifs. HepG2 cells were transfected with the MIXL1 promoter and its mutants and treated with TGF-β for 24 h. C, conserved Smad-binding elements (SBEs) of the MIXL1 promoter. Four Smad-binding elements that were conserved between mouse and human (SBE 1–4) are shown with their relative positions from the transcription start site. Nucleotide sequences of Smad-binding elements and their mutations used in D are also shown. WT, wild-type; mut, mutant. D, effect of mutations in Smad-binding elements on TGF-β-induced transcriptional activity of MIXL1 promoter. Cells were treated as in B, and luciferase activities were determined. *, p < 0.05 compared with WT without TGF-β; **, p < 0.05 compared with SBE4 mutant, without TGF-β; n.s., not significant compared with WT and SBE2 mutant, without TGF-β; error bars, S.D.
Luciferase Assay
Cells in 24-well plates were transfected with different combinations of promoter reporter constructs and expression plasmids by using Lipofectamine LTX (Invitrogen). The total amount of transfected DNA was adjusted to the same amount using an empty vector. Twenty four hours later, cells were treated with TGF-β for an additional 24–48 h and lysed. Luciferase activities in the lysates were measured using the Dual-Luciferase® reporter system (Promega) as described previously. For normalization, cells were co-transfected with pGL4.75-SV40-hRluc. Where indicated, siRNAs were transfected 24 h before reporter transfection. All samples were prepared in triplicate, and results were averaged.
Statistical Analysis
The Tukey-Kramer test of R program was used for multiple comparisons of data. The t test was used for two-sample analysis. p values of less than 0.05 were considered to indicate significance for each experiment.
RESULTS
Identification of HNF4α-binding Motif in HepG2-specific Smad2/3 Binding Regions
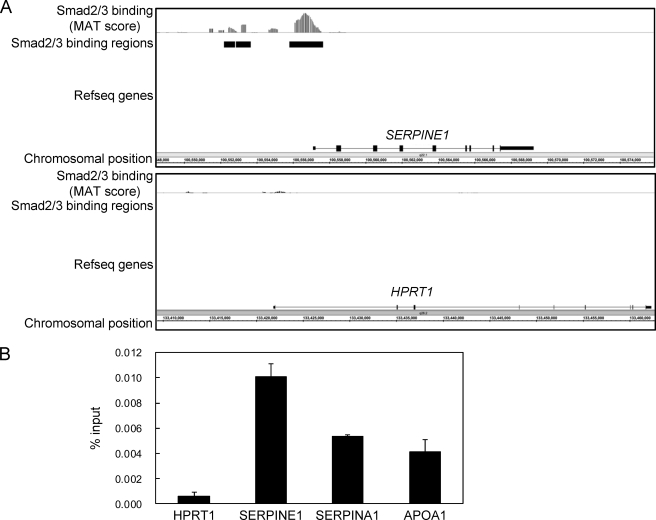
To determine Smad2/3 binding regions in HepG2 hepatoblastoma cells, we stimulated the cells with TGF-β for 1.5 h and fixed them with formaldehyde to cross-link genome-bound molecules to DNA. ChIP on microarray analysis (ChIP-chip) was performed according to the established protocol using an Affymetrix human promoter array (22). Obtained image data were analyzed using the MAT algorithm, which provided enrichment values of the ChIP samples for every promoter region, compared with the input genomic sample. There was significant Smad2/3 binding to the promoter region of the SERPINE1/PAI-1 gene as observed previously in the ChIP-chip analysis of Smad2/3 bindings in HaCaT cells (Fig. 1A, upper panel) (11). In contrast, there was no significant Smad2/3 binding to the HPRT1 locus that served as a negative control region (Fig. 1A, lower panel). We confirmed significant enrichment of Smad2/3 binding to the SERPINE1, SERPINA1/α-antitrypsin, and APOA1 loci using ChIP-qPCR (Fig. 1B). We identified 3,636 significant Smad2/3 binding regions that had detection p values of less than 10−4 within the promoter regions of known genes.
FIGURE 1.
Identification of Smad2/3 binding regions in HepG2 cells. A, Smad2/3 binding to the SERPINE1/PAI-1 locus in HepG2 cells. MAT scores were plotted at the SERPINE1 and HPRT1 loci to obtain a graphical representation of Smad2/3 binding in these regions. Significant Smad2/3 binding regions as determined by detection of p values of 10−4 are shown by black bars. B, percent input values of Smad2/3 binding compared with input genome as determined by ChIP-qPCR. Cells were treated with 120 pm TGF-β for 1.5 h. Cells were cross-linked sequentially with dimethyl adipimidate and formaldehyde. Error bars represent S.D.
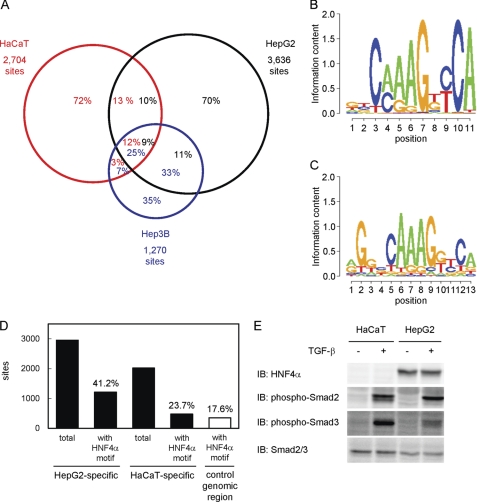
Next, we compared the identified Smad2/3 binding regions in HepG2 cells to those in HaCaT cells (11). We also obtained 1,270 Smad2/3 binding regions in Hep3B hepatocellular carcinoma cells to determine their overlaps (Fig. 2A). We found that only 25.2% of the Smad2/3 binding regions in HaCaT cells (n = 682) were shared with those in HepG2 cells. In contrast, 58.3% of the binding regions in Hep3B cells overlapped with those identified in HepG2 cells, although the number of overlapping binding regions (n = 741) was similar to that between HaCaT and HepG2 cells. Many of the Smad2/3 binding regions were thus unique to each cell type. We determined the candidate target genes of Smad2/3 using the dataset of Smad2/3 binding regions that were either common to HepG2 and HaCaT, HepG2-specific, or HaCaT-specific (supplemental Tables S3-S5), and performed gene ontology analysis of each category by DAVID (27). We did not observe remarkable differences in the top five enriched annotation clusters between the common Smad2/3 binding regions and HepG2-specific Smad2/3 binding region. Conversely, enrichment of cell death and cytoskeleton-related annotations was found in HaCaT-specific binding regions (supplemental Table S6).
FIGURE 2.
Comparison of the Smad2/3 binding regions among different cell lines. A, Venn diagrams showing the overlaps of Smad2/3 binding regions in HaCaT, HepG2, and Hep3B cells. Numbers in the circles indicate percentages of the Smad2/3 binding region of each cell line (red, HaCaT; black, HepG2; blue, Hep3B). B, identification of a motif conserved in HepG2-specific Smad binding regions. Partial genomic sequences within 250 bp from the peak positions of HepG2-specific Smad2/3 binding regions (n = 2,955) were analyzed using the CisGenome Gibbs motif sampler. Default parameters were used for the calculation, except for the numbers of motifs to be identified (n = 10). Matrix datum of the motif calculated by CisGenome was graphically shown using the SegLogo function of the R software. C, HNF4α-binding motif that matched the predicted motif in HepG2-specific Smad2/3 binding regions. The JASPAR CORE data base was used to identify known transcription factor binding motifs similar to the calculated matrix data in B. An HNF4α motif (ID: MA0114.1) was identified as the most similar motif with a comparison score of 21.3, which reached 96.9% of the potential maximal score. D, frequencies of the HNF4α-binding motif in Smad2/3 binding regions. Presence of the HNF4α-binding motif in each Smad2/3 binding region (within 250 bp from the peak signal position) was determined using CisGenome. Frequencies of the motif in either HepG2- or HaCaT-specific Smad2/3 binding regions were then calculated. As a control, matched genomic regions to HaCaT-specific Smad2/3 binding regions were obtained using CisGenome, and the frequency of the HNF4α motif was determined. E, expression of the HNF4α protein and phosphorylation of Smad2/3 in HaCaT and HepG2 cells. Cells were treated with TGF-β for 1.5 h, and the expression of each protein was determined by immunoblotting (IB).
To identify specific motifs in the Smad2/3 binding regions in HepG2 cells, de novo motif prediction was performed using the CisGenome Gibbs motif sampler (supplemental Fig. S1A). We searched for known motifs that had similarity to the calculated motifs using the JASPAR data base (28). As shown in Fig. 2C, we found that one predicted motif (Fig. 2B) was strongly similar to the HNF4α-binding motif (Fig. 2C, 96.9% score). The frequency of the HNF4α motif in HepG2-specific Smad2/3 binding regions was 41.2%, although that in HaCaT-specific Smad2/3 binding regions and its matched random genomic regions was 23.7 and 17.6%, respectively (Fig. 2D). We also analyzed the sequences in Smad2/3 binding regions using the CEAS analysis tool as we did in our previous report (11, 12, 25), and the HNF4α-binding motif was identified as one of the top three enriched motifs in the binding sequences (supplemental Fig. S1B) (25). It should be noted that canonical Smad-binding element (M00974.SMAD, “CAGAC”) was also identified as enriched motif through CEAS analysis and present in 40.6% of the HepG2-specific Smad2/3 binding regions (data not shown). These findings suggested that the HNF4α motif was enriched in HepG2-specific Smad2/3 binding regions and had roles for cell type specificity of Smad2/3 binding and TGF-β-induced transcription in HepG2 cells. HNF4α is one of the “master genes” of hepatocytes and is essential for hepatocyte-specific gene expressions and functions. Because HNF4α was not expressed in HaCaT cells (Fig. 2E), we decided to determine HNF4α binding regions in vivo in the presence of TGF-β.
HNF4α Binding to Its Binding Regions Is Not Extensively Altered by TGF-β
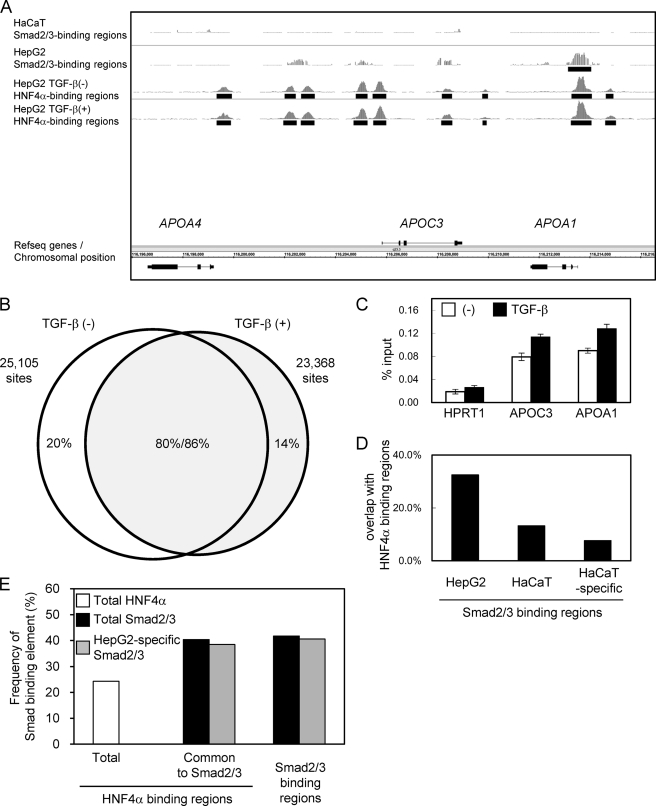
ChIP-chip and ChIP-seq studies of HNF4α binding regions have been reported using HepG2 cells (29–31). We retrieved data (30) from the data base and found that 20.7% of the Smad2/3 binding regions were common to HNF4α binding regions in vivo. However, no reports have yet determined the changes in the binding of HNF4α by extracellular stimulations, including that by TGF-β. We therefore acquired the HNF4α binding data using the newly available ChIP-seq technology to compare the Smad2/3 binding with the HNF4α binding under TGF-β stimulation. We identified 25,105 significant HNF4α binding regions in the absence of TGF-β and 23,368 regions in the presence of TGF-β, at an FDR of less than 0.1%. The APOA4/APOC3/APOA1 gene cluster that is a known target of HNF4α is shown in Fig. 3A. We observed significant HNF4α binding to several of these regions in the absence of TGF-β, and the binding was not extensively changed following stimulation. We also found that there was significant Smad2/3 binding to the APOA1 promoter, which was absent in HaCaT cells. Our data showed that Smad2/3 binding to the APOC3 promoter was not significant.
FIGURE 3.
Identification of HNF4α binding regions in the presence and absence of TGF-β stimulation. A, graphical representation of HNF4α binding to the APOA4/APOC3/APOA1 gene loci. Sequence read numbers of 100-bp sliding window were plotted for HNF4α ChIP-seq samples. Smad2/3 bindings as determined by ChIP-chip analysis were shown in the upper two panels as in Fig. 1A. Black bars represent significant binding regions (FDR, <0.1%). B, Venn diagrams showing overlap between TGF-β-treated and untreated HNF4α binding regions. HNF4α binding regions were determined for each sample (FDR, <0.1%). HNF4α binding regions that have overlapping regions within 500 bp from their positions of maximum read numbers were considered as shared binding regions. C, changes in the HNF4α binding to APOC3 and APOA1 loci were quantitatively determined by ChIP-qPCR analysis. Error bars, S.D. D, frequencies of in vivo HNF4α binding to the Smad2/3 binding regions. Percentages of HNF4α binding within 250 bp from the peak signal position of Smad2/3 binding regions were calculated for the indicated Smad2/3 binding groups. E, frequencies of canonical Smad-binding elements in HNF4α binding regions compared with Smad binding regions in HepG2 cells. A Smad-binding element, M00974.SMAD that was identified as an enriched motif in HepG2-specific Smad2/3 binding regions using CEAS (see text), was selected for calculation. CisGenome was used for mapping of the motif. Presence of the motif for each HNF4α binding region and Smad2/3 binding region was determined using PerlScript.
We then examined the changes in HNF4α binding by TGF-β stimulation. More than 80% of the HNF4α binding regions overlapped between TGF-β-treated and untreated cells. However, there were also specific binding regions for both TGF-β-treated and untreated cells (Fig. 3B). In addition, we calculated the changes in the normalized read numbers within the HNF4α binding regions by TGF-β stimulation and found that some regions indeed had either decreased or increased sequence reads following TGF-β stimulation (data not shown). Percent input values of HNF4α binding to the APOC3 and APOA1 loci were also up-regulated to some extent (Fig. 3C), suggesting that there were some, if limited, roles of TGF-β for HNF4α binding. Using the HNF4α binding data with TGF-β stimulation, we determined the frequency of HNF4α binding in Smad2/3 binding regions in vivo. We found that 32.5% of the Smad2/3 binding regions in HepG2 cells were indeed common to HNF4α binding regions. In contrast, only 13.2% of Smad2/3 binding regions in HaCaT cells were common, and the frequency decreased to 7.7% when HaCaT-specific Smad2/3 binding regions were examined (Fig. 3D). These results suggested that HNF4α and Smad2/3 binding regions are located in close proximity to each other in HepG2 cells, although we could not determine whether HNF4α- and Smad2/3-binding “elements” overlapped within the binding regions because of the limited resolution of ChIP-chip- and ChIP-seq-based assays. We then calculated the frequency of Smad-binding element CAGAC in HNF4α binding regions. 40.4% of the binding regions common to HNF4α and Smad2/3 had Smad-binding elements, compared with 24.3% in the total HNF4α binding regions (Fig. 3E).
Effect of HNF4α on the Expression of Smad2/3 Target Genes
To elucidate the effect of HNF4α on TGF-β-induced transcriptional regulation, we knocked down HNF4α by using siRNA (Fig. 4, A and B). The phosphorylation of Smad2 and Smad3 was not affected by the siRNA under the applied condition (Fig. 4C). We obtained expression microarray data and calculated the changes in the expression of genes with binding regions shared by Smad2/3 and HNF4α in the presence of TGF-β and siRNA. We first analyzed the data of cells transfected with control siRNA. Twenty four hours after TGF-β stimulation, 4.3 and 21.1% of the genes with Smad2/3 binding regions were regulated (either up- or down-regulated) more than 2- and 1.5-fold, respectively (Table 1). We observed that Smad2/3 binding regions were weakly enriched in the genes up-regulated by TGF-β at 1.5 h (supplemental Fig. S2). Many of the genes with Smad2/3 binding regions were not transcriptionally regulated by TGF-β, and these findings were essentially similar to those in our previous analysis in HaCaT cells (11). We then found that HNF4α siRNA inhibited the expression changes of common target genes of HNF4α and Smad2/3 by TGF-β 1.5 h after stimulation (Fig. 4D). This result underscored the general roles of HNF4α in hepatocyte-specific transcriptome regulation by TGF-β. In contrast, the effect of HNF4α silencing was not so obvious in the TGF-β-induced expression changes 24 h after stimulation, compared with the setting after 1.5 h of TGF-β stimulation, although TGF-β-induced expression changes of a subset of genes appeared to be rather enhanced by HNF4α knockdown (Fig. 4E). We focused on the changes at 1.5 h, when we obtained Smad2/3 and HNF4α binding data, and we listed target genes of TGF-β and the effect of HNF4α knockdown (Table 2). We identified MIXL1 as both TGF-β- and HNF4α-regulated gene with no Smad2/3 binding regions in HaCaT cells.
FIGURE 4.
Effect of knockdown of HNF4α on TGF-β-induced gene expression in HepG2 cells. A, confirmation of HNF4α-knocked down samples for microarray analysis. HepG2 cells were transfected with HNF4α siRNA and treated with 120 pm TGF-β for the indicated times and harvested. Expression of HNF4α was determined by RT-qPCR and normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH). siNC, negative control siRNA. B, down-regulation of HNF4α protein expression by siRNA. C, phosphorylation levels of Smad2/3 by using HNF4α siRNA. The top two panels show phosphorylation of Smad2 and Smad3. The 3rd panel indicates the expression of total Smad2/3, and the bottom panel is a loading control. IB, immunoblot. D, heat map of the TGF-β-induced expression of target genes of Smad2/3 and HNF4α and effect of HNF4α siRNA. Target genes that have overlapping binding regions of Smad2/3 and HNF4α were sorted by their induction of probe signal values by TGF-β stimulation for 1.5 h and are represented by color bars in the 1st column, using the TM4 microarray software (60). Relative expression of these genes in HNF4α siRNA samples to the control siRNA is shown in the 2nd column. In addition, a list of genes whose expressions changed more than 1.5-fold is shown in the right panel with their expression changes. E, heat map of target genes of TGF-β with Smad2/3 binding regions common to HNF4α at 24 h after TGF-β stimulation are shown as in D. Genes whose expressions were changed more than 2-fold are shown in the right panel.
TABLE 1.
TGF-β-induced changes in gene expression in relation to Smad2/3 binding
Expression array data transfected with control siRNA and stimulated with TGF-β were compared with Smad2/3 ChIP-chip data. A total of 8,653 genes that had values of more than 100 at least at one time point for one of their probes (n = 13,720) was used for the analysis. Up-regulated or down-regulated genes were determined compared with 0-h values. The positions of peak signals of Smad binding regions (SBRs) relative to the nearby RefSeq genes were first determined, and regions within 5 kb upstream from the transcription start site and the first intron were selected. *a indicates number of genes analyzed by microarray. *b indicates number of genes which have Smad2/3 binding regions.
| All genes |
Genes with SBRs |
*b/*a (%) | ||||
|---|---|---|---|---|---|---|
| *a | % | *b | % | |||
| Total | 8653 | 100.0 | 1941 | 100.0 | 22.4 | |
| Increase | ||||||
| >2-Fold | 1.5 h | 25 | 0.3 | 14 | 0.7 | 56.0 |
| 24 h | 223 | 2.6 | 59 | 3.0 | 26.5 | |
| >1.5-Fold | 1.5 h | 273 | 3.2 | 89 | 4.6 | 32.6 |
| 24 h | 837 | 9.7 | 250 | 12.9 | 29.9 | |
| Decrease | ||||||
| >2-Fold | 1.5 h | 16 | 0.2 | 2 | 0.1 | 12.5 |
| 24 h | 174 | 2.0 | 25 | 1.3 | 14.4 | |
| >1.5-Fold | 1.5 h | 217 | 2.5 | 47 | 2.4 | 21.7 |
| 24 h | 877 | 10.1 | 160 | 8.2 | 18.2 | |
TABLE 2.
TGF-β-induced genes with Smad2/3 and HNF4α binding at 1.5 h
Target genes of TGF-β in HepG2 cells that were induced more than 2-fold at 1.5 h and that have common binding regions for Smad2/3 and HNF4α were sorted by their expression changes in the presence or absence of HNF4α siRNA. Presence of Smad2/3 binding regions in HaCaT cells is also shown in the 2nd column.
| Gene symbol | Smad2/3 binding in HaCaT cells | Relative expression (siNC/siHNF4α) | Induction by TGF-β |
|---|---|---|---|
| -fold | -fold | ||
| CMIP | + | 0.3 | 2.1 |
| REPIN1 | + | 0.3 | 2.2 |
| MIXL1 | − | 0.3 | 2.3 |
| ERRFI1 | + | 0.4 | 2.7 |
| FASTK | + | 0.5 | 2.1 |
| ZNF48 | − | 0.5 | 2.6 |
| CBX4 | − | 0.6 | 2.2 |
| TMEM49 | − | 0.8 | 2.1 |
| ZFP36L2 | − | 0.9 | 2.6 |
| JUNB | + | 1.0 | 9.8 |
| DDIT4 | + | 1.1 | 5.6 |
| GADD45B | + | 1.3 | 4.9 |
| RND1 | − | 1.4 | 5.5 |
HNF4α Provides a New Mechanism of TGF-β-induced MIXL1 Expression
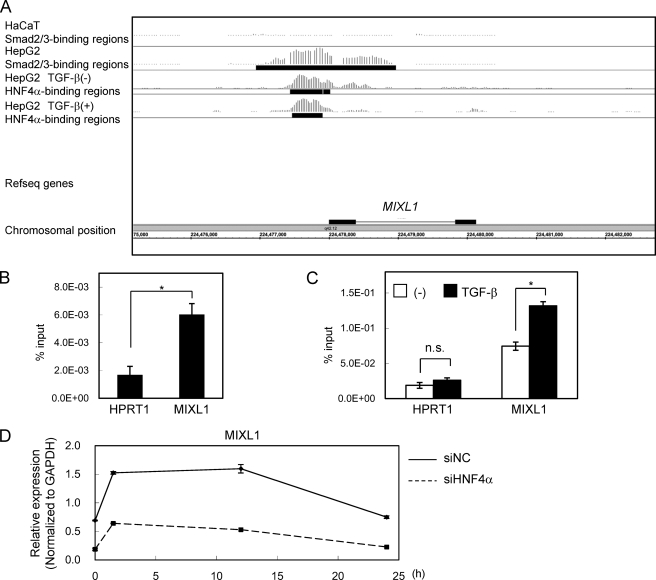
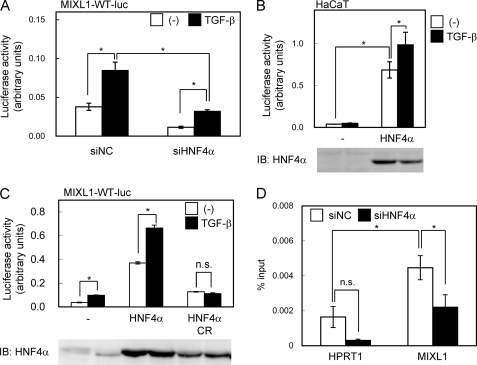
As shown in Fig. 5A, significant binding of Smad2/3 and HNF4α to the MIXL1 promoter was observed (Fig. 5A). We confirmed the binding of these transcription factors by ChIP-qPCR (Fig. 5, B and C) and changes in the expression of MIXL1 by HNF4α siRNA by RT-qPCR (Fig. 5D). We then determined the sequence of the MIXL1 promoter bound by Smad2/3 and HNF4α. We first found two possible HNF4α-binding motifs (Fig. 6A). Using a promoter reporter assay, we found that the transcriptional activity of the reporter containing the Smad2/3-HNF4α binding regions was up-regulated by TGF-β, which was significantly repressed by mutations in the HNF4α-binding sequences (Fig. 6B). We next searched for canonical Smad-binding elements conserved between mouse and human. We identified three Smad-binding elements between the two HNF4α motifs, and one just upstream of the distal HNF4α motif, termed SBE1 to -4 (Fig. 6C). Of them, only a mutation in SBE2 lost TGF-β-induced transcription (Fig. 6D). These results suggested that both HNF4α-binding motifs and SBE2 are required for MIXL1 reporter activity induced by TGF-β.
FIGURE 5.
Smad2/3 and HNF4α bindings in the MIXL1 locus. A, Smad2/3- and HNF4α-enriched regions in the MIXL1 locus are shown as in Fig. 3A. B, HepG2 cells were treated with 120 pm TGF-β for 1.5 h, fixed in formaldehyde, and harvested. Smad2/3 binding to the MIXL1 locus was verified by ChIP-qPCR. HPRT1 served as a negative control. C, HepG2 cells were treated with or without 120 pm TGF-β for 1.5 h, and ChIP-qPCR analysis of the MIXL1 locus using anti-HNF4α was performed as in B. n.s., not significant. D, effects of knockdown of HNF4α on TGF-β-induced expression changes of MIXL1. HepG2 cells were transfected with control siRNA (siNC) or siHNF4α, treated with 3 ng/ml TGF-β for the indicated times, and harvested. HNF4α expression was quantified by RT-qPCR. *, p < 0.05; error bars, S.D.
The transcriptional activity of the reporter was inhibited by HNF4α siRNA, which was observed even without TGF-β, suggesting that preceding binding of HNF4α to its binding motifs as observed in Fig. 5A was important both for the basal and TGF-β-induced transcriptional activation of MIXL1 promoter (Fig. 7A). We also investigated the effect of forced HNF4α expression in HaCaT cells to determine whether HNF4α was able to activate the MIXL1 transcriptional activity in these cells. As shown in Fig. 7B, HNF4α induced the transcriptional activity of the MIXL1 promoter reporter in HaCaT cells. We then examined the effect of a mutant of HNF4α that cannot bind to DNA (HNF4α CR mutant) (32) and found that DNA binding activity was required for its TGF-β-induced transcriptional activation (Fig. 7C). Finally, the effect of HNF4α siRNA on Smad2/3 binding to the MIXL1 promoter was determined. HNF4α siRNA inhibited the TGF-β-induced Smad2/3 binding to the MIXL1 promoter, indicating that the recruitment of Smad2/3 was one of the mechanisms of transcriptional regulation by HNF4α under TGF-β stimulation (Fig. 7D).
FIGURE 7.
Roles of HNF4α on Smad2/3 binding and transcriptional activity of MIXL1 promoter. A, effects of HNF4α knockdown on transactivation of the MIXL1 promoter. HepG2 cells were transfected with siRNAs 1 day before transfection with the reporter constructs. siNC, negative control siRNA. B, effects of exogenous HNF4α on transactivation of the MIXL1 promoter. HNF4α was exogenously expressed in HaCaT cells, and transcriptional activity of MIXL1 reporter was determined. The lower panel shows the protein expression of HNF4α. IB, immunoblot. C, HNF4α (variant 2, RefSeq ID: NM_000457) or its C115R (CR) mutant, which does not bind to DNA, was overexpressed in HepG2 cells. The lower panel shows the protein expression of HNF4α and its mutant. D, effect of HNF4α siRNA on Smad2/3 binding to the MIXL1 locus. HepG2 cells were transfected with siRNAs 24 h before TGF-β stimulation. Cells were fixed 1.5 h after treatment, and ChIP-qPCR was performed as in Fig. 1B. Error bars, S.D.; *, p < 0.05; n.s., not significant.
Taken together, these findings propose that the preceding binding of HNF4α on MIXL1 promoter enables the recruitment of Smad2/3 to this promoter after TGF-β stimulation and confers TGF-β-mediated HepG2-specific MIXL1 induction.
DISCUSSION
Recent technological advances, including ChIP-chip and ChIP-seq, provide a functional platform for comprehensive understanding of transcriptional regulation. This study revealed that Smad2/3 binding regions specifically observed in HepG2 cells were enriched in HNF4α binding regions. HNF4α was also expressed in Hep3B cells, and HNF4α-binding motif was identified in Smad2/3 binding regions in Hep3B cells by CEAS analysis (data not shown), which suggests that the functional relation between Smad2/3 and HNF4α is commonly observed in hepatocyte-derived cells. Based on the findings on the HNF4α-Smad interaction (18), physical interaction between HNF4α and Smads is important, at least in part, for TGF-β-induced Smad2/3 binding and transcriptional activation in HepG2 cells. It is also possible that HNF4α has additional indirect interactive functions for TGF-β signaling. Many regulatory mechanisms control the expression of a proper set of genes in various cells and tissues. At the genome level, CpG methylation plays a central role to avoid unintended expression of genes that are not suitable for the given tissue (33). Modification of the histone tail is also well known to lead to the formation of either euchromatin or heterochromatin. These modifications of the genome or histones allow transcription factors and cofactors to access the cell- and tissue-specific genomic loci to exert their actions. Modifications of the genome and histones are sometimes induced by trans factors during differentiation of the cells and tissues (34, 35). HNF4α physically interacts with the histone acetyltransferase complex and chromatin remodeling complex (29), and it is thus possible that HNF4α induces such epigenomic changes in the liver and indirectly provides Smad2/3 to access to hepatocyte-specific binding regions.
Identification of Smad binding regions downstream of the TGF-β/activin signaling by ChIP-chip analysis has been performed using several cell lines. Recently, Fei et al. (36) reported promoter analysis of Smad2 binding regions in mouse embryonic stem cells by ChIP-chip. We and Qin et al. (12, 37) analyzed Smad4 binding regions under TGF-β stimulation using HaCaT and ovarian surface epithelial cells, respectively. It has been reported that transcription factor binding regions in the same target gene loci differ among the five vertebrate species (38); it is thus difficult to compare the ChIP-chip or ChIP-seq data obtained from mouse and human. Differences in the ChIP efficiencies of the antibodies also make the comparison of the data difficult (12). Importantly, we used the same antibody and sample preparation procedures for HaCaT cells and HepG2 cells. Our present analysis thus revealed for the first time that Smad binding regions greatly differ among cell lines. Analysis of HaCaT-specific trans factors will facilitate our understanding of cell type-specific TGF-β-induced transcription in the future. However, comparison of the number of binding regions in different cell types is still difficult. We found a greater number of Smad2/3 binding regions in HepG2 cells than in HaCaT cells. Because the phosphorylation of Smad3 was weaker and the percent input value of the Smad2/3 ChIP sample was smaller in HepG2 than HaCaT cells, we cannot conclude that HepG2 cells have more Smad2/3 binding regions than HaCaT. It should also be noted that we cannot fully exclude that the antibody recognizes unknown genome-bound molecules in addition to Smad2/3.
Comparison of ChIP-chip and ChIP-seq data of the same transcription factor has been reported (39). In general, ChIP-seq is reported to be more sensitive and specific than ChIP-chip. Oligonucleotide-based array analysis has a potential risk of cross-hybridization and false discovery. Conversely, ChIP-seq also has difficulty in identifying GC-rich sequences (10, 39). We primarily focused on the comparison of our previously reported Smad2/3 binding regions to those of different cell types by the same platform. However, based on the known problems as described above, comparison of the Smad2/3-HNF4α binding regions will be more accurately performed by the ChIP-seq in the future.
Interaction of several transcription factors at the same enhancer positions has been recognized, and the complex is called “enhanceosome.” Structure of such complex and their binding DNA motifs have been analyzed in the interferon-β promoter as reviewed by Panne (40). In enhanceosome, each transcription factor physically interacts with others to provide its adequate surface that can bind to the series of their corresponding DNA motifs. Several reports have identified HNF4α binding regions by ChIP-chip and ChIP-seq analyses (29–31, 38, 41–43). Many transcription factors, e.g. FOXA2, GABP, HNF1α, HNF4γ, HNF6, cohesin, and CDX2, were identified to co-localize with HNF4α through these analyses. Other reports also revealed interaction of FOXO1 or retinoic acid receptor/retinoid X receptor with HNF4α on specific promoters (44, 45). These findings clearly revealed steady-state binding regions of HNF4α on the genome and suggested that transcription factors that co-localize or interact with HNF4α may form enhanceosome with HNF4α. Changes in the HNF4α binding regions were found during differentiation of an intestinal epithelial cell line CaCo2 (43); however, to our knowledge, the effect of single extracellular stimulation on genome-wide HNF4α binding regions has not yet been elucidated. Our present analysis provides the data of HNF4α binding regions following TGF-β stimulation, which were compared with the Smad2/3 binding regions in HaCaT cells that lack the expression of HNF4α. We have found that large proportions of HNF4α binding regions in HepG2 cells were unchanged by TGF-β stimulation. However, some changes in HNF4α binding regions were observed with regard to their positions and their strength, suggesting that TGF-β might regulate a subset of HNF4α binding regions. de Boussac et al. (46) reported that hepatocyte growth factor inhibited HNF4α binding to the ABCC6 promoter, which together suggest the importance of changes in the HNF4α binding positions by external stimuli. We also found that the effect of HNF4α on the TGF-β-induced gene expression after 24 h of TGF-β stimulation was different from that after 1.5 h of TGF-β stimulation. Studies on the changes in the genome-wide HNF4α and Smad2/3 binding after TGF-β stimulation at several time points and ChIP-seq analysis of HNF4α with other interactive factors in relation to their binding DNA sequences will reveal new mechanisms of the regulation of HNF4α-induced transcription in the context of the enhanceosome.
MIXL1 is an ortholog of Xenopus Mix.1, a transcription factor rapidly induced by activin during the early stage of Xenopus development (47). There are six known homologs that have been identified in Xenopus to engage in the formation of mesoderm and endoderm (48, 49). However, only one ortholog of Mix.1 is known in human and mouse (50). MIXL1 is required for the development of the chordamesoderm, heart, and gut in mouse (51). Forced expression of MIXL1 in embryonic stem cells resulted in the differentiation of the cells to endoderm (52). TGF-β is reported to induce Mix.2 promoter activity by formation of a Smad2/Smad4/FAST-1 (FoxH1) complex (53). In mouse, Smads and FAST-1 interact to up-regulate the transcriptional activity of the MIXL1 promoter (54, 55). However, FAST-1 is not expressed in HepG2 cells (56). Our finding of TGF-β-induced MIXL1 expression in HepG2 cells suggests a previously unrecognized regulatory mechanism of its expression by HNF4α in the absence of FAST-1. During development, HNF4α is expressed in the visceral endoderm during the gastrulation stage and plays a role in the differentiation of the embryonic mesoderm (57). MIXL1 is also expressed in the visceral endoderm and induces migration of the embryonic endoderm. HNF4α-null mice embryo showed impaired development of mature visceral endoderm, indicating that HNF4α acts upstream of MIXL1, at least in the visceral endoderm. Notably, both HNF4α and MIXL1 positively regulate the E-cadherin expression (52, 58), and the HNF4α expression was repressed in a model of progression of hepatocellular carcinoma (59). Functional analysis of MIXL1 in liver fibrosis and hepatocellular carcinoma in relation to TGF-β signaling might reveal the roles of MIXL1 in the adult liver in the future.
Supplementary Material
Acknowledgments
We thank Keiko Yuki, Hiroko Meguro, and Kaori Shiina for technical assistance.
This work was supported by KAKENHI Grant-in-Aid for Scientific Research on Innovative Areas, Integrative Research on Cancer Microenvironment Network, 22112002 (to K. M.) Grants-in-Aid for Scientific Research (S) 20221009 (to H. A.) and for Young Scientists (B) 10014456 (to D. K.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, a grant from the Genome Network Project from Ministry of Education, Culture, Sports, Science, and Technology of Japan (to H. A.), and the Global Center of Excellence Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms) from the Japan Society for the Promotion of Science.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1–S6.
- HNF4α
- hepatocyte nuclear factor 4α
- qPCR
- quantitative PCR
- ChIP-seq
- ChIP sequencing
- FDR
- false discovery rate
- CEAS
- cis-regulatory element annotation system.
REFERENCES
- 1. Ikushima H., Miyazono K. (2010) Nat. Rev. Cancer 10, 415–424 [DOI] [PubMed] [Google Scholar]
- 2. Heldin C. H., Miyazono K., ten Dijke P. (1997) Nature 390, 465–471 [DOI] [PubMed] [Google Scholar]
- 3. Feng X. H., Derynck R. (2005) Annu. Rev. Cell Dev. Biol. 21, 659–693 [DOI] [PubMed] [Google Scholar]
- 4. Miyazono K., Kamiya Y., Morikawa M. (2010) J. Biochem. 147, 35–51 [DOI] [PubMed] [Google Scholar]
- 5. Shi Y., Wang Y. F., Jayaraman L., Yang H., Massagué J., Pavletich N. P. (1998) Cell 94, 585–594 [DOI] [PubMed] [Google Scholar]
- 6. Zawel L., Dai J. L., Buckhaults P., Zhou S., Kinzler K. W., Vogelstein B., Kern S. E. (1998) Mol. Cell 1, 611–617 [DOI] [PubMed] [Google Scholar]
- 7. Ikushima H., Miyazono K. (2010) Cancer Sci. 101, 306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gomis R. R., Alarcón C., He W., Wang Q., Seoane J., Lash A., Massagué J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12747–12752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eeckhoute J., Lupien M., Brown M. (2009) Methods Mol. Biol. 556, 155–164 [DOI] [PubMed] [Google Scholar]
- 10. Park P. J. (2009) Nat. Rev. Genet. 10, 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koinuma D., Tsutsumi S., Kamimura N., Taniguchi H., Miyazawa K., Sunamura M., Imamura T., Miyazono K., Aburatani H. (2009) Mol. Cell. Biol. 29, 172–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koinuma D., Tsutsumi S., Kamimura N., Imamura T., Aburatani H., Miyazono K. (2009) Cancer Sci. 100, 2133–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sladek F. M., Zhong W. M., Lai E., Darnell J. E., Jr. (1990) Genes Dev. 4, 2353–2365 [DOI] [PubMed] [Google Scholar]
- 14. Si-Tayeb K., Lemaigre F. P., Duncan S. A. (2010) Dev. Cell 18, 175–189 [DOI] [PubMed] [Google Scholar]
- 15. Jiang G., Nepomuceno L., Hopkins K., Sladek F. M. (1995) Mol. Cell. Biol. 15, 5131–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lucas Sd S., López-Alcorocho J. M., Bartolomé J., Carreño V. (2004) Biochem. Biophys. Res. Commun. 321, 688–694 [DOI] [PubMed] [Google Scholar]
- 17. Ishikawa F., Nose K., Shibanuma M. (2008) Exp. Cell Res. 314, 2131–2140 [DOI] [PubMed] [Google Scholar]
- 18. Kardassis D., Pardali K., Zannis V. I. (2000) J. Biol. Chem. 275, 41405–41414 [DOI] [PubMed] [Google Scholar]
- 19. Chou W. C., Prokova V., Shiraishi K., Valcourt U., Moustakas A., Hadzopoulou-Cladaras M., Zannis V. I., Kardassis D. (2003) Mol. Biol. Cell 14, 1279–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mizutani A., Saitoh M., Imamura T., Miyazawa K., Miyazono K. (2010) J. Biochem. 148, 733–741 [DOI] [PubMed] [Google Scholar]
- 21. Wendt K. S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., Yahata K., Imamoto F., Aburatani H., Nakao M., Imamoto N., Maeshima K., Shirahige K., Peters J. M. (2008) Nature 451, 796–801 [DOI] [PubMed] [Google Scholar]
- 22. Kaneshiro K., Tsutsumi S., Tsuji S., Shirahige K., Aburatani H. (2007) Genomics 89, 178–188 [DOI] [PubMed] [Google Scholar]
- 23. Johnson W. E., Li W., Meyer C. A., Gottardo R., Carroll J. S., Brown M., Liu X. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12457–12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ji H., Jiang H., Ma W., Johnson D. S., Myers R. M., Wong W. H. (2008) Nat. Biotechnol. 26, 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ji X., Li W., Song J., Wei L., Liu X. S. (2006) Nucleic Acids Res. 34, W551–W554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nagano Y., Koinuma D., Miyazawa K., Miyazono K. (2010) J. Biochem. 147, 545–554 [DOI] [PubMed] [Google Scholar]
- 27. Huang da W., Sherman B. T., Lempicki R. A. (2009) Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 28. Portales-Casamar E., Thongjuea S., Kwon A. T., Arenillas D., Zhao X., Valen E., Yusuf D., Lenhard B., Wasserman W. W., Sandelin A. (2010) Nucleic Acids Res. 38, D105–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Daigo K., Kawamura T., Ohta Y., Ohashi R., Katayose S., Tanaka T., Aburatani H., Naito M., Kodama T., Ihara S., Hamakubo T. (2011) J. Biol. Chem. 286, 674–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wallerman O., Motallebipour M., Enroth S., Patra K., Bysani M. S., Komorowski J., Wadelius C. (2009) Nucleic Acids Res. 37, 7498–7508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmidt D., Schwalie P. C., Ross-Innes C. S., Hurtado A., Brown G. D., Carroll J. S., Flicek P., Odom D. T. (2010) Genome Res. 20, 578- 588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taylor D. G., Haubenwallner S., Leff T. (1996) Nucleic Acids Res. 24, 2930–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bernstein B. E., Meissner A., Lander E. S. (2007) Cell 128, 669–681 [DOI] [PubMed] [Google Scholar]
- 34. Kim M. S., Kondo T., Takada I., Youn M. Y., Yamamoto Y., Takahashi S., Matsumoto T., Fujiyama S., Shirode Y., Yamaoka I., Kitagawa H., Takeyama K., Shibuya H., Ohtake F., Kato S. (2009) Nature 461, 1007–1012 [DOI] [PubMed] [Google Scholar]
- 35. Lan F., Nottke A. C., Shi Y. (2008) Curr. Opin. Cell Biol. 20, 316–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fei T., Zhu S., Xia K., Zhang J., Li Z., Han J. D., Chen Y. G. (2010) Cell Res. 20, 1306–1318 [DOI] [PubMed] [Google Scholar]
- 37. Qin H., Chan M. W., Liyanarachchi S., Balch C., Potter D., Souriraj I. J., Cheng A. S., Agosto-Perez F. J., Nikonova E. V., Yan P. S., Lin H. J., Nephew K. P., Saltz J. H., Showe L. C., Huang T. H., Davuluri R. V. (2009) BMC Syst. Biol. 3, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schmidt D., Wilson M. D., Ballester B., Schwalie P. C., Brown G. D., Marshall A., Kutter C., Watt S., Martinez-Jimenez C. P., Mackay S., Talianidis I., Flicek P., Odom D. T. (2010) Science 328, 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ho J. W., Bishop E., Karchenko P. V., Nègre N., White K. P., Park P. J. (2011) BMC Genomics 12, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Panne D. (2008) Curr. Opin. Struct. Biol. 18, 236–242 [DOI] [PubMed] [Google Scholar]
- 41. Boyd M., Bressendorff S., Møller J., Olsen J., Troelsen J. T. (2009) BMC Gastroenterol. 9, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Odom D. T., Zizlsperger N., Gordon D. B., Bell G. W., Rinaldi N. J., Murray H. L., Volkert T. L., Schreiber J., Rolfe P. A., Gifford D. K., Fraenkel E., Bell G. I., Young R. A. (2004) Science 303, 1378–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Verzi M. P., Shin H., He H. H., Sulahian R., Meyer C. A., Montgomery R. K., Fleet J. C., Brown M., Liu X. S., Shivdasani R. A. (2010) Dev. Cell 19, 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ganjam G. K., Dimova E. Y., Unterman T. G., Kietzmann T. (2009) J. Biol. Chem. 284, 30783–30797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mosialou I., Zannis V. I., Kardassis D. (2010) J. Biol. Chem. 285, 30719–30730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. de Boussac H., Ratajewski M., Sachrajda I., Köblös G., Tordai A., Pulaski L., Buday L., Váradi A., Arányi T. (2010) J. Biol. Chem. 285, 22800–22808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rosa F. M. (1989) Cell 57, 965–974 [DOI] [PubMed] [Google Scholar]
- 48. Ecochard V., Cayrol C., Rey S., Foulquier F., Caillol D., Lemaire P., Duprat A. M. (1998) Development 125, 2577–2585 [DOI] [PubMed] [Google Scholar]
- 49. Vize P. D. (1996) Dev. Biol. 177, 226–231 [DOI] [PubMed] [Google Scholar]
- 50. Robb L., Hartley L., Begley C. G., Brodnicki T. C., Copeland N. G., Gilbert D. J., Jenkins N. A., Elefanty A. G. (2000) Dev. Dyn. 219, 497–504 [DOI] [PubMed] [Google Scholar]
- 51. Hart A. H., Hartley L., Sourris K., Stadler E. S., Li R., Stanley E. G., Tam P. P., Elefanty A. G., Robb L. (2002) Development 129, 3597–3608 [DOI] [PubMed] [Google Scholar]
- 52. Lim S. M., Pereira L., Wong M. S., Hirst C. E., Van Vranken B. E., Pick M., Trounson A., Elefanty A. G., Stanley E. G. (2009) Stem Cells 27, 363–374 [DOI] [PubMed] [Google Scholar]
- 53. Watanabe M., Whitman M. (1999) Development 126, 5621–5634 [DOI] [PubMed] [Google Scholar]
- 54. Hart A. H., Willson T. A., Wong M., Parker K., Robb L. (2005) Biochem. Biophys. Res. Commun. 333, 1361–1369 [DOI] [PubMed] [Google Scholar]
- 55. Izzi L., Silvestri C., von Both I., Labbé E., Zakin L., Wrana J. L., Attisano L. (2007) EMBO J. 26, 3132–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hayashi H., Abdollah S., Qiu Y., Cai J., Xu Y. Y., Grinnell B. W., Richardson M. A., Topper J. N., Gimbrone M. A., Jr., Wrana J. L., Falb D. (1997) Cell 89, 1165–1173 [DOI] [PubMed] [Google Scholar]
- 57. Chen W. S., Manova K., Weinstein D. C., Duncan S. A., Plump A. S., Prezioso V. R., Bachvarova R. F., Darnell J. E., Jr. (1994) Genes Dev. 8, 2466–2477 [DOI] [PubMed] [Google Scholar]
- 58. Späth G. F., Weiss M. C. (1998) J. Cell Biol. 140, 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lazarevich N. L., Cheremnova O. A., Varga E. V., Ovchinnikov D. A., Kudrjavtseva E. I., Morozova O. V., Fleishman D. I., Engelhardt N. V., Duncan S. A. (2004) Hepatology 39, 1038–1047 [DOI] [PubMed] [Google Scholar]
- 60. Saeed A. I., Bhagabati N. K., Braisted J. C., Liang W., Sharov V., Howe E. A., Li J., Thiagarajan M., White J. A., Quackenbush J. (2006) Methods Enzymol. 411, 134–193 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.