Abstract
Male sex development and growth occur in response to high affinity androgen binding to the androgen receptor (AR). In contrast to complete amino acid sequence conservation in the AR DNA and ligand binding domains among mammals, a primate-specific difference in the AR NH2-terminal region that regulates the NH2- and carboxyl-terminal (N/C) interaction enables direct binding to melanoma antigen-A11 (MAGE-11), an AR coregulator that is also primate-specific. Human, mouse, and rat AR share the same NH2-terminal 23FQNLF27 sequence that mediates the androgen-dependent N/C interaction. However, the mouse and rat AR FXXLF motif is flanked by Ala33 that evolved to Val33 in primates. Human AR Val33 was required to interact directly with MAGE-11 and for the inhibitory effect of the AR N/C interaction on activation function 2 that was relieved by MAGE-11. The functional importance of MAGE-11 was indicated by decreased human AR regulation of an androgen-dependent endogenous gene using lentivirus short hairpin RNAs and by the greater transcriptional strength of human compared with mouse AR. MAGE-11 increased progesterone and glucocorticoid receptor activity independently of binding an FXXLF motif by interacting with p300 and p160 coactivators. We conclude that the coevolution of the AR NH2-terminal sequence and MAGE-11 expression among primates provides increased regulatory control over activation domain dominance. Primate-specific expression of MAGE-11 results in greater steroid receptor transcriptional activity through direct interactions with the human AR FXXLF motif region and indirectly through steroid receptor-associated p300 and p160 coactivators.
Keywords: Nuclear Receptors, Receptor Structure-Function, Steroid Hormone Receptor, Transcription Coactivators, Transcription Regulation, FXXLF Motif, MAGE-A11, N/C Interaction, Androgen Receptor, Primate-specific
Introduction
Androgen receptor (AR)2 regulation of gene transcription is required for male reproductive development and function. AR is a ligand-dependent transcription factor activated by binding testosterone, a major circulating male steroid hormone, or by dihydrotestosterone (DHT), the more potent 5α-reduced metabolite of testosterone. Evolution of AR among mammals is characterized by complete amino acid sequence conservation in the central DNA binding domain that interacts with androgen response element DNA and in the carboxyl-terminal ligand binding domain that binds androgens with high affinity and specificity (1). Strict sequence conservation in these regions reflects the rigid structural requirements for DNA and hormone binding. In contrast, the AR NH2-terminal region, although also required for AR transcriptional activity, is largely unstructured and less well conserved (2). The human AR NH2-terminal region contains activation function 1 (AF1) between amino acid residues 142–337 (3) preceded by a polymorphic CAG-encoded glutamine repeat that expanded during primate evolution (1). Mechanisms involved in AF1 and glutamine repeat function remain to be established. Expansion of the glutamine repeat length to more than 39 residues results in the adult onset muscle wasting disease known as spinal bulbar muscular atrophy (4). A similar glutamine repeat in rat and mouse AR is shifted in position in the NH2-terminal region relative to human AR (5).
The mammalian AR NH2-terminal region also contains a conserved 23FQNLF27 sequence that binds activation function 2 (AF2) in the AR ligand binding domain to mediate the androgen-dependent AR NH2- and carboxyl-terminal (N/C) interaction (6, 7). An NH2-terminal 433WHTLF437 WXXLF motif contributes to the human AR N/C interaction and transcriptional activity (6–8). The functional importance of the intermolecular AR N/C interaction is indicated by a dependence on high affinity androgen binding, its requirement for optimal gene transcription, and inhibition of the N/C interaction by classical AR antagonists (7, 9–12). There are also naturally occurring human AR single amino acid mutations that cause partial androgen insensitivity and disrupt the AR N/C interaction even though high affinity androgen binding is maintained (13–20). The androgen-dependent human AR N/C interaction reduces p160 coactivator binding to AF2 in the ligand binding domain (21), which suggests regulation of activation domain dominance. p160 coactivator binding to AF2 is increased by melanoma antigen-A11 (MAGE-11), an AR coregulator that binds the human AR FXXLF motif region in a manner competitive with the N/C interaction to expose AF2 for increased p160 coactivator recruitment (22). MAGE-11 also interacts with p300 and p160 coactivators (23, 24).
Studies in this report address the functional consequences of amino acid sequence differences in the AR NH2-terminal region among primates and lower mammals that parallel the evolution of MAGE-11 expression. Our findings suggest that the primate-specific expression of MAGE-11 increases AR transcriptional strength through direct binding to the human AR FXXLF motif region. We provide evidence that MAGE-11 also functions as a more general transcriptional coregulator through interactions with steroid receptor-associated p300 and p160 coactivators.
EXPERIMENTAL PROCEDURES
Expression Vectors
Previously described plasmids include pCMV-hAR that codes for full-length human AR (hAR) (25) and L26A,F27A (6, 23); Δ120–472, 1–660 (3), 1–503 (10), 507–919 (3), and K720A mutants (14); GAL-hAR-(658–919) (26) and -(4–52); pSG5-MAGE-(1–429) that codes for full-length human MAGE-11; GAL-MAGE-(2–429) (GAL-MAGE) and -(112–429); pCMV-FLAG-MAGE-(2–429) (FLAG-MAGE) (27); pVP16-CT-MAGE-(2–429) (VP-MAGE) and VP-MAGE-(112–429) with VP16 activation domain residues 446–490 (Clontech) (22); pNLVP16-hAR-(1–660) (13) and L26A,F27A mutant (6) with VP16 activation domain residues 411–456; pCMV-human glucocorticoid receptor (hGR) (21); pCMV-human progesterone receptor (hPR)-B-(1–688) and pCMV-hGR-(1–550) NH2-terminal and DNA binding domains (22); pSG5-HA-p300 (24); and pSG5-transcriptional intermediary factor 2 (TIF2) (14, 28). Reporter vectors used include human prostate-specific antigen-enhancer-luciferase (PSA-Enh-Luc) (24, 29), 5XGAL4Luc3 (24), rat probasin BH500-Luc (30, 31), and mouse mammary tumor virus (MMTV) luciferase.
pCMV-hARΔ120–472-V33A, -S94A, and pNLVP-hAR-(1–660)-V33A were created by QuikChange site-directed mutagenesis (Stratagene). pCMV-hAR-V33A was created by double PCR mutagenesis. pCMV-GR-(399–795) coding for the rat glucocorticoid receptor (GR) DNA and ligand binding domains was created by PCR amplification and subcloning into EcoRI and BamHI sites of pCMV5. Human progesterone receptor (PR) p5M-hPR-B contains an EcoRI fragment of pSG5-hPR-B (provided by Pierre Chambon) cloned into the EcoRI site of pCMV5 with a HindIII and BamHI deletion in the polylinker region. Human PR-B DNA and ligand binding domains were expressed from pcDNA-myc-PR-B-(486–933) that contains a PCR-amplified EcoRI and internal HindIII fragment of PR-B cloned in pcDNA3-myc-PR-B-(550–933) digested with the same enzymes.
Full-length mouse AR (mAR) pCMV-mAR was created by subcloning full-length C57BL/6 mouse AR sequence from pCMV-HA-mAR (provided by Norman M. Greenberg) into the EcoRI and BamHI sites of pCMV5. pCMV-mAR-L26A,F27A was a triple ligation of pCMV5 EcoRI and BamHI, pCMV-mAR-(1–285)-L26A,F27A EcoRI and MluI, and pCMV5-mAR MluI and BamHI fragments. pCMV-mAR-(1–640) contains a PCR-amplified EcoRI and BamHI fragment of pCMV-mAR cloned in the same sites of pCMV5. pCMV-mAR-(487–899), which corresponds to hAR-(507–919), and pCMV-mAR-(487–899)-K700A contain EcoRI and BamHI fragments from PCR-amplified pCMV-mAR with an added ATG initiating methionine sequence inserted in the same sites of pCMV5. pCMV-mAR-A33V was a triple ligation of pCMV5 EcoRI and BamHI, pCMV-mAR-(1–640)-A33V EcoRI and HindIII, and pCMV5-mAR HindIII and BamHI fragments.
pCMV-mARΔ101–452 corresponds to hARΔ120–472 and was a triple ligation of pCMV-mAR-(1–640)Δ101–452 EcoRI and HindIII, pCMV-mAR HindIII and BamHI, and pCMV5 EcoRI and BamHI fragments. pCMV-mARΔ101–452-L26A,F27A was a triple ligation of PCR-amplified mAR-(1–640)-Δ101–452,L26A,F27A EcoRI and HindIII, pCMV-mAR BamHI and HindIII, and pCMV5 EcoRI and BamHI fragments. pCMV-mARΔ101–452-A33V was a triple ligation of pCMV-mAR-(1–640)Δ101–452-A33V EcoRI and HindIII, pCMV-mAR BamHI and HindIII, and pCMV5 EcoRI and BamHI fragments. pCMV-mARΔ101–452-K700A was a triple ligation of EcoRI- and BamHI-digested pCMV5, an EcoRI and HindIII fragment of pCMV-mAR-(1–640)Δ101–452, and a HindIII and BamHI fragment of pCMV-mAR-(487- 899)-K700A. pCMV-mARΔ101–452-A33V,K700A was the same except with an EcoRI and HindIII fragment of pCMV-mAR-(1–640)Δ101–452-A33V.
GAL-mAR-(4–52) contains an EcoRI and NdeI PCR-amplified fragment from pCMV-mAR cloned in the same sites of GALO. pNLVP16-mAR-(2–640) and rat AR (rAR) pNLVP16-rAR-(2–643) were created by subcloning corresponding PCR-amplified regions of pCMV-mAR and pCMV-rAR (32) into SalI and XbaI sites of pNLVP16. All PCR-amplified regions were verified by DNA sequencing.
DNA Transfection and Gene Expression Studies
Mammalian two-hybrid assays were performed in HeLa cells (5 × 104/well) (26) in 12-well plates transfected using FuGENE 6 (Roche Applied Science), expression vector DNA, and 0.1 μg/well 5XGAL4Luc reporter gene. The day after transfection, cells were transferred to serum-free medium and incubated for 24 h in the absence and presence of DHT. AR transcription assays in HeLa cells utilized wild-type and mutant pCMV-hAR and pCMV-mAR and 0.1 or 0.25 μg/well PSA-Enh-Luc, BH500-Luc that contains the −426 to +28 rat probasin promoter provided by Robert J. Matusik (30, 31), or MMTV-Luc. Cells were harvested in 0.25 ml of lysis buffer containing 1% Triton X-100, 2 mm EDTA, and 25 mm Tris phosphate, pH 7.8. Transcription assays in CV1 cells (4 × 105/6-cm dish) were performed using calcium phosphate precipitation (21). Immediately after transfection and 24 h later, cells were incubated for 24 h in serum-free phenol red-free medium in the absence and presence of DHT and harvested in 0.5 ml of lysis buffer. Luciferase activity was measured using an automated Lumistar Galaxy luminometer (BMG Labtech), and values (mean ± S.E.) are representative of at least three independent experiments. The siRNA oligonucleotide experiments were performed in HeLa cells (2 × 105/well) in 6-well plates and in COS cells (2 × 106 cells/10-cm dish) using p300 siRNAs, nonspecific siRNA-3 (Dharmacon RNA Technologies), and Lipofectamine 2000 (Invitrogen) in the absence of antibiotics (24).
Immunoblot Analysis
Immunoblots were performed by transfecting monkey kidney COS cells (2 × 106/10-cm dish) using DEAE-dextran (26, 33). After 24 h, cells were placed in serum-free, phenol red-free medium with and without DHT. The next day cells were harvested in immunoblot lysis buffer containing 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 0.15 m NaCl, 2 mm EDTA, 0.05 mm NaF, 2 mm sodium vanadate, and 50 mm Tris-HCl, pH 7.5 with 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, and Complete protease inhibitor mixture (Roche Applied Science). In some experiments, cells were incubated for 24 h with 1 μm MG132 proteosome inhibitor (Sigma). Cell extracts were analyzed in polyacrylamide gels containing SDS and probed using the following antibodies: rabbit anti-VP16 activation domain (Abcam ab4809; 1:1000 dilution), rabbit anti-GAL4 DNA binding domain (Santa Cruz Biotechnology sc-577; 1:500 dilution), rabbit anti-human AR32 (0.4 μg/ml) and AR52 (10 μg/ml) anti-peptide immunoglobulin G (25), mouse anti-β-actin (Abcam; 1:5000 dilution); affinity-purified rabbit polyclonal human p300 (C-20) antibody (Santa Cruz Biotechnology sc-585; 1:75–200 dilution), mouse anti-TIF2 (BD Transduction Laboratories; 1:100–250 dilution), rabbit polyclonal GR antibody (Santa Cruz Biotechnology sc-1003; 1:200), and rabbit polyclonal PR antibody (Santa Cruz Biotechnology sc-7208; 1:200). Endogenous MAGE-11 was detected in transblots using 10 μg of rabbit polyclonal antibody-1 prepared against baculovirus-expressed and purified FLAG-tagged human MAGE-11 and 10 μg of MAGE-Ab-94–108 anti-peptide immunoglobulin G (34). Gels were calibrated using EZ-Run prestained Rec protein ladder (Fisher Bioreagents) and analyzed for chemiluminescence (SuperSignal West Dura Extended Duration Substrate, Pierce).
Immunoprecipitation was performed by expressing FLAG empty vector or FLAG-MAGE with pCMV-hAR, pCMV-mAR, p5M-hPR-B, or pCMV-hGR in COS cells (2 × 106/10-cm dish) using DEAE-dextran transfection (26, 33). Cell lysates from two 10-dishes were extracted in 0.25 ml of lysis buffer containing 1% Triton X-100, 0.5% deoxycholate, 0.15 m NaCl, 0.05 m NaF, 2 mm EDTA, 2 mm sodium vanadate, and 50 mm Tris, pH 7.6 with 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, and Complete protease inhibitor mixture (Roche Applied Science); diluted 4-fold with the same buffer without deoxycholate; and precleared for 15 min at 4 °C with 0.1 ml of Sepharose CL-4B (Sigma). Cell extracts were transferred to 15 μl of anti-FLAG M2-agarose (Sigma) and incubated for 2 h at 4 °C. Samples were pelleted; washed with lysis buffer without deoxycholate; resuspended in 0.05 ml of 2× SDS buffer containing 3.3% SDS, 10% 2-mercaptoethanol, 10% glycerol, and 0.12 m Tris-HCl, pH 6.8; incubated for 5 min at 90 °C; and analyzed on immunoblots as above.
Genome Analysis of MAGE Families and Two-hybrid Screens
The human MAGE-11 amino acid sequence was compared with the NCBI BLAST Assembled RefSeq Database for human, Rhesus macaque, mouse and rat genomes. We obtained no experimental evidence for human MAGE-11 isoform B indicated by the human genome database.
The Clontech Mate and Plate yeast two-hybrid screen was performed by preparing mouse testis complementary DNA (cDNA) pool using SMART cDNA synthesis technology, recombination with Matchmaker prey vector pGADT7-Rec, and transformation in yeast strain Y187. Two independent screens of 2 × 108 colonies of a mouse testis library were performed using GAL-mAR-(4–52) or GAL-mAR-(2–285) as bait in yeast strain Y2HGold. Selection parameters included resistance to aureobasidin A, X-α-Gal blue colony formation, and growth in the absence of histidine or adenine. Screening using GAL-mAR-(4–52) as bait yielded 11 positive independent clones compared with 17 positive independent clones using GAL-mAR-(2–285) as bait. None of the identified protein interactions with mouse AR were FXXLF motif-dependent.
Lentivirus Transduction and Real Time RT-PCR
LAPC-4 human prostate cancer cells were maintained in RPMI 1640 medium containing 10% fetal calf serum (Cellgro), 1 nm R1881 (PerkinElmer Life Sciences), 2 mm l-glutamine, penicillin, and streptomycin (Invitrogen). HeLa-AR1C-PSA-Luc-A6 cells stably express human AR and the PSA-Luc reporter gene and were maintained in minimum Essential medium (Invitrogen) containing 10% fetal calf serum, 2 mm l-glutamine, penicillin, streptomycin, 500 μg/ml Geneticin (G418, Invitrogen), and 100 μg/ml hygromycin B (Roche Applied Science). MAGE-11 lentivirus short hairpin RNA (shRNA)-169, -827, -947, and -964; human AR shRNA-5; the empty vector shRNA nonspecific siRNA-1; and 18-bp-spacer nonspecific siRNA-2 were created using the Open Biosystems TRC1 shRNA libraries following standard protocols. LAPC-4 (3 × 106/well) or HeLa-AR-PSA cells (2 × 105/well) in 6-well plates were used without virus or transduced with 0.15 ml (LAPC-4 cells) or 0.025 ml (HeLa-AR cells) of HEK-293 cell supernatant containing ∼106 lentivirus virus particles/ml. After 48 h, cells were passaged into selection medium containing 3 μg/ml puromycin dihydrochloride (Cellgro) in 10-cm dishes for immunoblot analysis and 6-cm dishes for RT-PCR analysis. After 9 days in selection medium, cells for RT-PCR analysis were incubated for 24 h in medium containing 10% charcoal-stripped fetal calf serum with and without DHT. Cells for immunoblots were incubated with and without 10 nm DHT, extracted in lysis buffer, and analyzed.
Total RNA was extracted using 1 ml of TRIzol reagent (Invitrogen)/6-cm dish followed by chloroform/isopropanol extraction. First strand cDNA was prepared using SuperScript II reverse transcriptase (Invitrogen). Real time PCR was performed using an Eppendorf Realplex4 Mastercycler and QuantiTect SYBR Green PCR kit (Qiagen) in 20-μl reactions. Peptidylprolyl isomerase A served as a housekeeping control gene. Peptidylprolyl isomerase A forward primer 5′-ATCTTGTCCATGGCAAATGC-3′ and reverse primer 5′-GCCTCCACAATATTCATGCC-3′ amplify a 134-bp 371–505-nucleotide fragment overlapping the exon 4 and 5 junction coding for amino acid residues 123–169 (GenBankTM accession number NM_021130). PSA forward primer 5′-CTCATCCTGTCTCGGATTGT-3′ and reverse primer 5′-ATGAAACAGGCTGTGCCGAC-3′ amplify a 189-bp 99–287-nucleotide fragment overlapping the exon 2 and 3 junction that codes for amino acid residues 20–82 (GenBank accession number NM_001648). MAGE-11 forward primer 5′-GGAGACTCAGTTCCGCAGAG-3′ and reverse primer 5′-TGGGACCACTGTAGTTGTGG-3′ amplify a 63-bp 123–185-nucleotide fragment coding for amino acid residues 24–43 with the probe centered at nucleotide 154 (GenBank accession number AY747607.1) overlapping the exon 2 and 3 junction (22, 34). PCRs (20 μl) contained 4 μl of cDNA from 0.1 μg of total RNA, 10 μl of SYBR Green Master Mix, 2 μl of 2 μm forward and reverse primers, and 2 μl of RNase-free water. PCR conditions were one cycle at 94 °C for 20 min and 55 cycles at 94 °C for 40 s, 57 °C for 40 s, and 72 °C for 40 s. 10-fold serial dilutions of cDNA were amplified in triplicate to generate standard curves. Unknown mRNA levels were extrapolated based on standard curves and Ct values normalized to peptidylprolyl isomerase A.
HeLa-AR-PSA-Luc-A6 cell growth assays were performed by plating 5000 cells/well of 24-well plates in 0.5 ml of medium containing 10% fetal bovine serum. Cells were incubated for increasing times and assayed using the cell counting kit (Dojindo Laboratories). WST-8 reagent (0.02 ml) was added to 0.2 ml of serum-free medium and incubated for 2.5 h at 37 °C. Optical density measurements were determined at 485 nm.
RESULTS
Species-specific AR FXXLF Motif Interactions
The AR NH2-terminal FXXLF motif sequence 23FQNLF27 is conserved among mammals with the hydrophobic character maintained throughout lower vertebrates including fish (Fig. 1A) (7, 35). However, the sequence flanking the AR FXXLF motif differs with Ala33 in mouse, rat, and other lower vertebrates evolved to Val33 in human, chimpanzee, Rhesus macaque, lemur, and other primates (Fig. 1, A and B). Human AR Val33 is in a predicted α-helical region that extends from the FXXLF motif that interacts with AF2 in the ligand binding domain bound to a high affinity androgen. AR FXXLF motif binding to AF2 competes with AR FXXLF motif binding to MAGE-11, a human AR coregulator (22, 36).
FIGURE 1.
Vertebrate evolution of AR FXXLF motif region and model of AR, MAGE-11, TIF2, and p300 interactions. A, human AR amino acid residues 16–34 (Homo sapiens; GenBank accession no. P10275) are compared with corresponding AR regions from chimpanzee (Pan troglodytes; GenBank accession no. NP001009012), crab-eating macaque (Macaca fascicularis; GenBank accession no. AAC73050), collared brown lemur (Eulemur fulvus collaris; GenBank accession no. U94178), house mouse (Mus musculus; GenBank accession no. NP038504.1), Norway rat (Rattus norvegicus; GenBank accession no. NP036634), African clawed frog (Xenopus laevis; GenBank accession no. NP001084353), goldfish (Carassius auratus; GenBank accession no. AAM09278), Japanese eel (Anguilla japonica; GenBank accession no. BAA75464; AR-α), and rainbow trout (Oncorhynchus mykiss; GenBank accession no.NP001117656; AR-α). Highlighted in red are FXXLF motif residues required for the AR N/C interaction or interaction with MAGE-11. B, human AR NH2-terminal FXXLF motif residues 23–33 (hydrophobic residues underlined) precede AF1. High affinity androgen binding in the human AR ligand binding domain (LBD) initiates FXXLF motif binding to AF2 in the N/C interaction and restricts p160 coactivator LXXLL motif binding to AF2. Val33 flanking the AR FXXLF motif in humans and other primates was required for the N/C interaction inhibition of AF2 and a direct interaction with MAGE-11. AR in lower mammals such as rat and mouse has Ala33 instead of Val33. Ala33 was not compatible with a direct interaction with MAGE-11. In agreement with MAGE family genome sequence analysis that MAGE-11 is primate-specific, MAGE-11 or another FXXLF motif-interacting protein was not identified in a mouse testis expression library using the mouse AR FXXLF motif region as bait. A direct interaction between human AR and MAGE-11 provides a primate-specific gain in function. MAGE-11 increases activity of other steroid hormone receptors in an FXXLF motif-independent manner through interactions with receptor-associated p300 and p160 coactivators. DBD, DNA binding domain.
Species-specific differences in the AR N/C interaction and AR FXXLF motif binding to MAGE-11 were investigated for human, rat, and mouse AR in mammalian two-hybrid assays. The androgen-dependent N/C interaction between the AR FXXLF motif and AF2 in the ligand binding domain (6) was assayed in the absence and presence of DHT using GAL-hAR-(658–919) in which the GAL4 DNA binding domain was expressed as a fusion protein with the AR ligand binding domain conserved in human, mouse, and rat (5).
In the presence of 10 nm DHT, GAL-hAR-(658–919) interacted with human VP-hAR-(1–660), mouse VP-mAR-(2–640), and rat VP-rAR-(2–643), which are corresponding regions of the AR NH2-terminal and DNA binding domains fused to the VP16 activation domain (Fig. 2A). VP-hAR-(1–660) also interacted in two-hybrid assays with full-length GAL-MAGE and the GAL-MAGE-(112–429) carboxyl-terminal fragment that contains the F-box-like binding region for the human AR FXXLF motif (22, 23). However, there was essentially no interaction of GAL-MAGE or GAL-MAGE-(112–429) with mouse VP-mAR-(2–640) or rat VP-rAR-(2–643) (Fig. 2B).
FIGURE 2.
Human, rat, and mouse AR FXXLF motif interaction with AR ligand binding domain and MAGE-11. A, GAL-hAR-(658–919) (0.1 μg) was expressed in HeLa cells with 0.1 μg of 5XGAL4Luc3 and 50 ng of VP16 empty vector (−), human VP-hAR-(1–660), mouse VP-mAR-(2–640), or rat VP-rAR-(2–643) with and without 10 nm DHT. GAL-hAR-(658–919) contains the human AR ligand binding domain sequence conserved in mouse and rat, except hAR Leu659 outside the hormone binding region is methionine in mouse and rat AR. B, GAL-MAGE and GAL-MAGE-(112–429) (50 ng) were expressed in HeLa cells with 0.1 μg of 5XGAL4Luc3 and 50 ng of VP16 empty vector (−), VP-hAR-(1–660), VP-mAR-(2–640), or VP-rAR-(2–643). Inset, VP16 empty vector (−), VP-hAR-(1–660), VP-mAR-(2–640), and VP-rAR-(2–643) (5 μg) were expressed in COS cells incubated with 1 μm MG132 for 24 h prior to harvest. The transblot of cell extracts (80 μg of protein/lane) was probed using VP16 antibody. C, GAL-hAR-(4–52) and GAL-mAR-(4–52) (50 ng) were expressed in HeLa cells with 0.1 μg of 5XGAL4Luc3 and 50 ng of VP16 empty vector (−), VP-MAGE, or VP-MAGE-(112–429). Inset, GALO empty vector (−), GAL-hAR-(4–52) (H), or GAL-mAR-(4–52) (M) (5 μg) were expressed in COS cells incubated with 1 μm MG132 for 24 h prior to harvest. The blot was probed using GAL4 antibody. D, GAL-hAR-(658–919) (0.1 μg), GAL-MAGE (50 ng), or GAL-MAGE-(112–429) (50 ng) was expressed in HeLa cells with 0.1 μg of 5XGAL4Luc3 and 50 ng of VP16 empty vector (−), VP-hAR-(1–660) wild type (WT) or L26A,F27A (LFAA) or V33A (VA33) mutant, or VP-mAR-(2–640) WT or A33V (AV33) or L26A,F27A mutant. Inset, VP16 empty vector (−), VP-hAR-(1–660) WT and V33A and L26A,F27A (LFAA) mutants, and VP-mAR-(2–640) WT and A33V and L26A,F27A mutants (10 μg) were expressed in COS cells and incubated for 24 h with 1 μm MG132 prior to harvest. The transblot of cell extracts (60 μg of protein/lane) was probed using VP16 antibody. E, pCMV-hAR-(507–919) (50 ng) and PSA-Enh-Luc (0.1 μg) were transfected in HeLa cells with 50 ng of pCMV5 empty vector (−), pCMV-hAR-(1–503) WT or V33A or L26A,F27A (LFAA) mutant, or pCMV-mAR-(1–483) WT or A33V or L26A,F27A mutant. Cells were incubated with and without 10 nm DHT. Inset, pCMV5 empty vector (−), pCMV-hAR-(1–503) WT and V33A and L26A,F27A (LFAA) mutants, and pCMV-mAR-(1–483) WT and A33V and L26A,F27A mutants (5 μg) were expressed in COS cells. Cell extracts (20 μg of protein/lane) were analyzed on the transblot using AR32 antibody.
The inability of the mouse and rat AR FXXLF motif region to bind MAGE-11 was also evident using the mouse GAL-mAR-(4–52) FXXLF motif peptide that differs from human GAL-hAR-(4–52) by Ala33 instead of Val33. VP-MAGE and VP-MAGE-(112–429) interacted with GAL-hAR-(4–52), but there was no interaction with GAL-mAR-(4–52) (Fig. 2C). Direct in vitro binding between GST-hAR-(4–52) and MAGE-11 was demonstrated previously (22).
When assayed using VP-hAR-(1–660) that contains human AR NH2-terminal and DNA binding domains, the V33A mutation decreased the human AR N/C interaction and eliminated the interaction with MAGE-11 (Fig. 2D). Conversely, VP-mAR-(2–640)-A33V with a substitution mutation to mimic human AR interacted with GAL-MAGE and GAL-MAGE-(112–919) and to a greater extent with GAL-hAR-(658–919). Dependence on the AR FXXLF motif was demonstrated by the inability of VP-hAR-(1–660)-L26A,F27A to interact with GAL-AR-(658–919), GAL-MAGE, or GAL-MAGE-(112–429).
Dependence of the AR N/C interaction on Val33 was investigated further by expressing corresponding hAR-(1–503) and mAR-(1–483) NH2-terminal fragments that lack the DNA and ligand binding domains with the hAR-(507–919) DNA and ligand binding domain fragment. Greatest androgen-dependent and N/C interaction-dependent intermolecular activation of the PSA enhancer resulted from hAR-(507–919) coexpressed with hAR-(1–503) or mAR-(1–483)-A33V; activation was lower with hAR-(1–503)-V33A or mAR-(1–483) (Fig. 2E). The androgen-dependent intermolecular interaction activation between these fragments was eliminated by L26A,F27A mutations in the human and mouse AR FXXLF motif.
The results suggest that the evolutionary change from alanine to valine at AR residue 33 in the primate lineage strengthens the androgen-dependent AR N/C interaction in primates. The same transition mutation to Val33 also enables human AR to interact directly with MAGE-11.
Greater Transcriptional Activity of Human AR
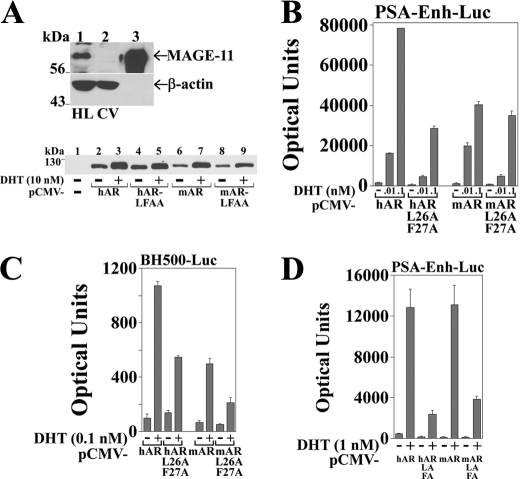
The ability of MAGE-11 to increase human AR transcriptional activity (22–24, 27) together with the inability of mouse AR to interact with MAGE-11 (Fig. 2) and the apparent absence of MAGE-11 in mouse (see Fig. 6) suggested that MAGE-11 may contribute to species-specific differences in AR transcriptional strength. Transcriptional activities of human and mouse AR were compared in HeLa cells that express endogenous MAGE-11 (Fig. 3A, top panel, lane 1) and in CV1 cells with low levels of MAGE-11 mRNA (37) and no detectable MAGE-11 protein (Fig. 3A, top panel, lane 2). Transcriptional activity was determined using the human PSA and rat probasin enhancer-luciferase reporter genes that depend on the AR N/C interaction for maximal activation by human AR (7). In agreement with an androgen-dependent N/C interaction for human and mouse AR (Fig. 2), both receptors were stabilized in the presence of 10 nm DHT (Fig. 3A, lower panel) (21, 38).
FIGURE 6.
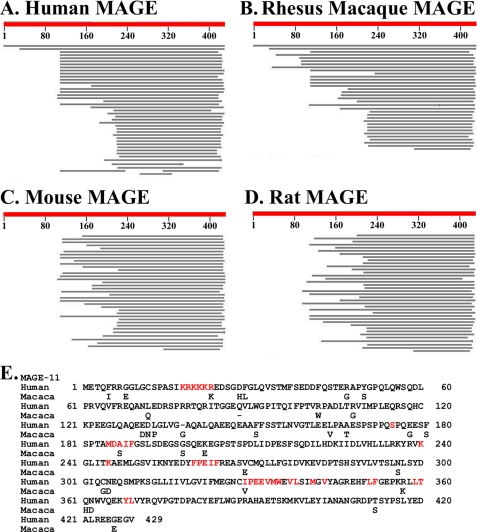
Human, Rhesus macaque, mouse, and rat MAGE gene families and human and macaque MAGE-11 sequence alignment. Shown are schematic diagrams of NCBI BLAST Assembled RefSeq results comparing the human MAGE-11 amino acid sequence with the MAGE gene family of human (A), Rhesus macaque (B), mouse (C), and rat (D) where the top black bar represents MAGE-11 in human and macaque absent in mouse and rat. E, amino acid sequence alignment of human (GenBank accession no. NP-005357) and M. mulatta Rhesus monkey MAGE-11 (GenBank accession no. XM-001090027) indicates 93% homology. Highlighted in red are conserved regions reported for nuclear targeting residues 18–23, mitogen-activated protein kinase phosphorylation site Ser174, 185MXXIF189 interaction site for p300, Lys240 and Lys245 monoubiquitinylation sites, 260FPEIF264 interaction site for TIF2, F-box-like residues 329–369 that bind the human AR FXXLF motif, and cell cycle checkpoint kinase Chk1 phosphorylation site Thr360 in the F-box (22–24, 27).
FIGURE 3.
Comparison of human and mouse AR transcriptional activity. A, top panel, immunoblot of endogenous MAGE-11 in HeLa (HL; lane 1) and CV1 (CV; lane 2) cell extracts (120 μg of protein/lane) was probed using MAGE-11 and β-actin antibodies. A cell extract (0.2 μg of protein) of COS cells expressing pSG5-MAGE served as control (lane 3). Bottom panel, immunoblot of extracts (40 μg of protein/lane) from COS cells transfected with 5 μg of pCMV5 empty vector (−), pCMV-hAR, pCMV-mAR, and L26A,F27A mutants (LFAA). Cells were incubated for 24 h with and without 10 nm DHT. The transblot was probed using AR32 antibody that recognizes the same NH2-terminal epitope in human and mouse AR. B, pCMV-hAR, pCMV-mAR, and L26A,F27A mutants (10 ng) were transfected with 0.25 μg of PSA-Enh-Luc in HeLa cells incubated with and without 0.01 or 0.1 nm DHT. C, pCMV-hAR, pCMV-mAR, and L26A,F27A mutants were transfected with 0.25 μg of BH500-Luc rat probasin reporter in HeLa cells incubated for 24 h with and without 0.1 nm DHT. D, pCMV-hAR, pCMV-mAR, and L26A,F27A mutants (0.1 μg) were transfected with 5 μg of PSA-Enh-Luc in CV1 cells with and without 1 nm DHT.
Androgen-dependent human AR transactivation of PSA-Enh-Luc (Fig. 3B) and probasin BH500-Luc (Fig. 3C) exceeded that of mouse AR in HeLa cells in the presence of 0.1 nm DHT. The activity of human and mouse AR F26A,F27A FXXLF motif mutants decreased to the greatest extent at the lower concentrations of DHT. It was noteworthy that the transcriptional strength of hAR-L26A,F27A in which the FXXLF motif binding site for MAGE-11 was disrupted was similar to that of wild-type mouse AR (Fig. 3, B and C). In CV1 cells that essentially lack MAGE-11 (Fig. 3A), human and mouse AR transactivation of PSA was similar and FXXLF motif-dependent (Fig. 3D).
The results suggest that maximal human and mouse AR transactivation requires the AR N/C interaction. The greater transcriptional strength of human AR relative to mouse in cells expressing MAGE-11 was consistent with the coactivator effects of MAGE-11 interaction with the human AR FXXLF motif region.
Requirement for MAGE-11 in Human AR Transcriptional Activity
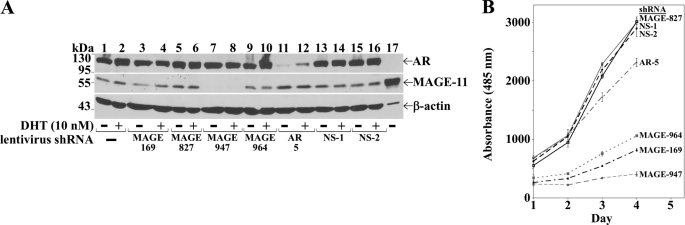
Lentiviruses expressing MAGE-11 shRNAs were tested for their ability to silence MAGE-11 expression. Maximal inhibition of MAGE-11 protein expression was seen with MAGE-11 shRNA-947 in HeLa cells that stably express AR (Fig. 4A, lanes 7 and 8). MAGE-11 shRNA-947 was also more effective in inhibiting HeLa-AR cell growth (Fig. 4B). AR protein expression was reduced using lentivirus AR shRNA-5 (Fig. 4A, lanes 11 and 12).
FIGURE 4.
Silencing MAGE-11 slows growth of HeLa cells stably expressing AR. A, HeLa-AR1C-PSA-Luc-6A cells stably expressing AR were not treated with lentivirus (lane 1 and 2) or transduced with lentivirus MAGE-11 shRNA-169, -827, -947, and -964 (lanes 3–10); human AR shRNA-5 (lanes 11–12); empty vector (NS-1; lanes 13 and 14); or 18-bp-spacer nonspecific shRNA (NS-2; lanes 15 and 16). After puromycin selection, cells were incubated for 24 h with and without 10 nm DHT. Cell extracts (65 μg of protein/lane) were separated on SDS gels. Transblots were probed using AR32 and AR52 antibodies combined; MAGE-Ab-59–79, MAGE-Ab-94–108, and FLAG-MAGE-11 antibodies combined; and β-actin antibody. Extracts from COS cells expressing pSG5-MAGE (5 μg of protein; lane 17) served as control. B, HeLa-AR1C-PSA-Luc-A6 cells were incubated with lentivirus as described in A. After puromycin selection, cell growth assays were performed over several days as described under “Experimental Procedures.” The data are the mean of triplicate measurements.
The requirement for MAGE-11 in androgen-dependent human AR transactivation was investigated by quantitative RT-PCR of endogenous PSA in LAPC-4 prostate cancer cells that have MAGE-11 mRNA levels ∼10-fold greater than HeLa cells, ∼100-fold greater than Ishikawa human endometrial cancer cells, and ∼1000-fold greater than normal human prostate cells (37). As seen with the HeLa-AR cells (Fig. 4), lentivirus AR shRNA-5 and MAGE-11 shRNA-947 were most effective in silencing AR and MAGE-11 expression in LAPC-4 cells, respectively, based on immunoblot (Fig. 5A, lanes 4 and 6) and RT-PCR analyses of MAGE-11 mRNA (Fig. 5B). Human AR shRNA-5 and MAGE-11 shRNA-947 decreased androgen-dependent human AR transactivation of PSA to an extent not seen with the partial silencing of MAGE-11 (Fig. 5C). Specificity of inhibition was suggested by androgen-dependent up-regulation of PSA without lentivirus or with control lentivirus that did not alter AR or MAGE-11 levels (Fig. 5, A–C). MAGE-11 shRNA-947 did not alter AR protein levels (Fig. 5A, lane 4), but knockdown of AR decreased MAGE-11 mRNA (Fig. 5B). In agreement with the low levels of MAGE-11 in most cells and the requirement for maximal silencing of MAGE-11 to inhibit AR activity, the results suggest that MAGE-11 is a low abundance but essential human AR coregulator.
FIGURE 5.
Requirement for MAGE-11 in human AR transactivation. A, LAPC-4 cells were untreated (lane 1) or transduced with lentivirus MAGE-11 shRNA-169, -827, -947, and -964 (lanes 2–5); human AR shRNA-5 (lane 6); empty vector (NS1; lane 7); or 18-bp-spacer nonspecific shRNA (NS2; lane 8). Cell extracts (100 μg of protein/lane) separated on transblots were probed using AR32 and AR52 antibodies combined, MAGE-Ab-94–108 and FLAG-MAGE-11 antibodies combined, and β-actin antibody. B, quantitative real time RT-PCR analysis of MAGE-11 mRNA from LAPC-4 cells was performed as described under “Experimental Procedures” using cells not treated with lentivirus (−) or transduced with lentivirus MAGE-11 shRNA-169, -827, -947, and -964 (M); human AR shRNA-5 (AR); and 18-bp nonspecific shRNA-2 (NS). Cells were incubated for 24 h with 10 nm DHT. MAGE-11 mRNA levels were normalized to peptidylprolyl isomerase A (PPIA) mRNA. C, quantitative real time RT-PCR analysis of PSA mRNA was performed in LAPC-4 cells as in B except that cells were incubated for 24 h with and without 10 nm DHT. Results indicate PSA mRNA -fold induction normalized to peptidylprolyl isomerase A assayed in triplicate ±S.E. relative to activity without DHT.
Absence of MAGE-11 Homologue in Mice
To determine whether less evolved mammals express MAGE-11 or a related protein that interacts with the AR FXXLF motif, genome-wide sequence comparisons (Fig. 6) and two-hybrid screens were performed. Comparison of human, Rhesus macaque (Macaca mulatta), mouse, and rat MAGE gene families suggests divergent evolution between the lower mammals and primates (Fig. 6, A–D). The 429-amino acid human and Rhesus macaque MAGE-11 proteins represented by the top black bar (Fig. 6, A and B) were absent in the mouse and rat genomes (Fig. 6, C and D). Human and Rhesus macaque MAGE-11 have 93% amino acid sequence homology with shared regions important for coregulator function. Conserved functional domains include nuclear localization residues 18–23, Ser174 mitogen-activated protein kinase phosphorylation site, 185MXXIF189 interaction motif for p300, Lys240 and Lys245 monoubiquitinylation sites required to bind the human AR FXXLF motif, 260FPEIF264 FXXIF interaction motif for TIF2, and Thr360 Chk1 cell cycle checkpoint kinase phosphorylation site in the F-box-like 329–369 residues that bind the human AR FXXLF motif (23, 24, 27) (Fig. 6E, highlighted in red). The 7% sequence divergence between human and Rhesus macaque MAGE-11 outside these functional domains is consistent with continuing evolution of the MAGE gene family in primates.
With MAGE-11 absent in the mouse and rat genomes, there remained the possibility of a related AR FXXLF motif-interacting protein in mouse. This idea gained some support from the extensive sequence conservation in the MAGE homology domain region that interacts with the human AR FXXLF motif. Yeast two-hybrid screens were performed using a mouse testis library. Mouse GAL-mAR-(4–52) bait vector contained Ala33 instead of Val33 in GAL-hAR-(4–52) used to identify MAGE-11 in a human testis library (22). A second mouse testis library screen was performed using GAL-mAR-(2–285) that contained additional mouse AR NH2-terminal sequence.
None of the 28 positive mouse AR-interacting proteins were FXXLF motif-dependent or recapitulated the transcription enhancing effects of MAGE-11 seen with human AR. Results from MAGE gene family comparisons and two-hybrid screens provide evidence that the coregulator function of MAGE-11 is primate-specific.
Transcriptional Synergy between MAGE-11 and Receptor-associated p300 and p160 Coactivators
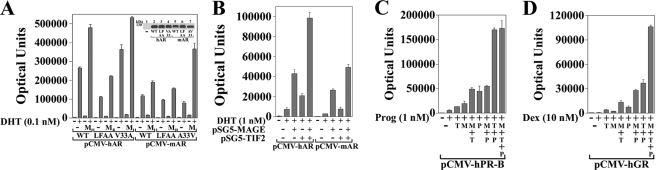
MAGE-11 binds TIF2, a coactivator that interacts with AF2 in the AR ligand binding domain, and p300 (23, 24), an acetyltransferase that interacts with the NH2-terminal region of AR and other steroid receptors (39) (Fig. 1B). This raised the possibility that MAGE-11 has transcriptional effects independent of binding an FXXLF motif by interacting with receptor-associated coactivators. Initial support for this came from the coimmunoprecipitation of FLAG-MAGE in a complex with human and mouse AR (Fig. 7A), human GR (Fig. 7B), and human PR-B (Fig. 7C) expressed in cells treated with receptor agonists and EGF. The experimental conditions were similar to the optimal interaction conditions for nuclear and cytoplasmic MAGE-11 interaction with liganded and unliganded human AR, respectively (22, 27). MAGE-11 also increased the transcriptional activity of mouse AR and the L26A,F27A FXXLF motif mutant and was most effective with mAR-A33V (Fig. 8A). The hAR-L26A,F27A FXXLF motif mutation that disrupts the N/C interaction decreased activity, whereas hAR-V33A with an N/C interaction retained activity that increased with MAGE-11. MAGE-11 functioned synergistically with TIF2 and mouse AR (Fig. 8B) and increased human PR-B and GR activity with TIF2 and p300 (Fig. 8, C and D).
FIGURE 7.
Coimmunoprecipitation of FLAG-MAGE-11 with human AR, mouse AR, human PR-B, and human GR. COS cells were transfected with 3 μg of FLAG empty vector or FLAG-MAGE and 1 μg of pCMV-hAR or 2 μg of pCMV-mAR in cells incubated with and without 10 nm DHT (A), 2 μg of pCMV-hGR in cells incubated with and without 10 nm dexamethasone (DEX; B), and 2 μg of p5M-hPR-B in cells incubated with and without 10 nm progesterone (PROG; C), all in the presence of 0.1 μm EGF. Cell extracts (50 μg of protein/lane) and immunoprecipitated proteins (IP) were analyzed by immunoblot using AR, GR, and PR antibodies.
FIGURE 8.
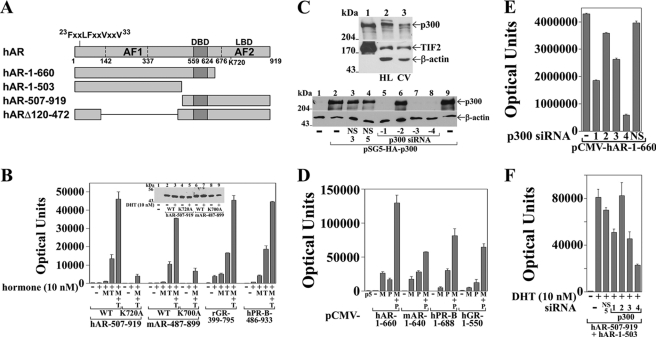
MAGE-11 increases steroid receptor transcriptional activity. A, pCMV-hAR WT and L26A,F27A (LFAA) and V33A (VA33) mutants and pCMV-mAR WT and L26A,F27A and A33V (AV33) mutants (10 ng) were transfected with 25 ng of pSG5 empty vector (−) or 25 ng of pSG5-MAGE (M) and 0.25 μg of PSA-Enh-Luc in HeLa cells. Cells were incubated in the absence and presence of 0.1 nm DHT. (Inset) pCMV5 empty vector (−), wild-type pCMV-hAR (WT hAR), L26A,F27A (LFAA), and V33A mutants, and pCMV-mAR (WT mAR), L26A,F27A, and A33V mutants (5 μg) were expressed in COS cells. The transblot of cell extracts (40 μg protein/lane) was probed using AR32 antibody. B, pCMV-hAR and pCMV-mAR (0.1 μg) were transfected in CV1 cells with 1.5 μg of pSG5 empty vector (−), 0.5 μg of pSG5-MAGE, and/or 2 μg of pSG5-TIF2 and 3 μg of PSA-Enh-Luc. Cells were incubated in the absence and presence of 1 nm DHT. C, p5M-hPR-B (0.1 μg) and PSA-Enh-Luc (2.5 μg) were transfected in CV1 cells with and without 1.5 μg of pSG5 (−), 1.5 μg of pSG5-TIF2 (T), 0.5 μg of pSG5-MAGE (M), and 1.5 μg of pSG5-HA-p300 (P). Cells were incubated in the absence and presence of 1 nm progesterone (Prog). D, pCMV-hGR (0.1 μg) and PSA-Enh-Luc (2.5 μg) were transfected as in C with cells incubated in the absence and presence of 10 nm dexamethasone (Dex).
Because the transcriptional effects of TIF2 are mediated predominantly by LXXLL motif binding to AF2 in the ligand binding domain (21, 36), the effects of MAGE-11 and TIF2 were evaluated further using corresponding DNA and ligand binding domain regions of mouse and human AR, human PR, and rat GR. Deletion of the NH2-terminal region in hAR-(507–919) (Fig. 9A) removed the inhibitory effect of the AR N/C interaction on AF2 activation by TIF2 (21). MAGE-11 itself had little effect on the transcriptional activity of hAR-(507–919), mAR-(487–899), rGR-(399–795), or hPR-B-(486–933) (Fig. 9B). In contrast, TIF2 increased the activity of each of these fragments and functioned synergistically with MAGE-11. The decrease in activity of hAR-(507–919)-K720A and the corresponding mAR-(487–899)-K700A charge clamp mutant demonstrated a dependence on TIF2 binding to AF2 (14, 40).
FIGURE 9.
Steroid receptor synergism with MAGE-11, TIF2, and p300. A, full-length human AR amino acid residues 1–919 and hAR-(1–660), -(1–503), -(507–919), and Δ120–472 deletion mutants contain the NH2-terminal 23FXXLFXXVXXV33 motif, AF1, DNA binding domain (DBD), and AF2 in the ligand binding domain (LBD) as indicated. Human AR-(1–660) corresponds to mAR-(1–640), hPR-B-(1–688), and hGR-(1–550). Human AR-(507–919) corresponds to mAR-(487–899), rGR-(399–795) and hPR-B-(486–933). Human ARΔ120–472 corresponds to mARΔ101–452. B, pCMV-hAR-(507–919) WT or K720A mutant, corresponding pCMV-mAR-(487–899) and K700A mutant, pCMV-rGR-(399–795), and pcDNA-myc-PR-B-(486–933) (0.1 μg) were transfected with 3 μg of MMTV-Luc and 0.5 μg of pSG5 empty vector (−), pSG5-MAGE (M), and/or pSG5-TIF2 (T) in CV1 cells. Cells were incubated with and without 10 nm DHT for AR, 10 nm dexamethasone for GR, and 10 nm progesterone for PR. Inset, pCMV5 empty vector (−), pCMV-hAR-(507–919) and K720A mutant, and corresponding pCMV-mAR-(487–899) and K700A mutant (5 μg) were expressed in COS cells. The transblot of cell extracts (50 μg of protein/lane) was probed using AR52 antibody. C, top panel, endogenous protein extracts (100 μg/lane) from HeLa (HL; lane 2) and CV1 (CV; lane 3) cells were compared with extracts of COS cells expressing pSG5-HA-p300 and pSG5-TIF2 as control (10 μg of protein; lane 1). The blot was probed using p300, TIF2, and β-actin antibodies. Bottom panel, pSG5 empty vector (lane 1) and pSG5-HA-p300 (lanes 2–9) (2 μg) were expressed in COS cells with and without 10 nm nonspecific siRNA-3 (NS3), -5 (NS5), or p300-siRNA-1–4 using Lipofectamine 2000. Cell extracts (15 μg of protein/lane) on the transblot were probed overnight at 4 °C using p300 and β-actin antibodies. D, CV1 cells were transfected with 3 μg of PSA-Enh-Luc and 10 ng of human pCMV-hAR-(1–660), pCMV-mAR-(1–640), p5M-hPR-B-(1–688), or pCMV-hGR-(1–550) with and without 1.5 μg of pSG5 (p5) empty vector (−), 0.5 μg of pSG5-MAGE (M), and 1 μg of pSG5-HA-p300 (P). E, pCMV-hAR-(1–660) (25 ng) was transfected into HeLa cells using Lipofectamine 2000 without siRNA (−) or with 10 nm p300 siRNA-1–4, nonspecific siRNA-3 (NS), and 0.1 μg of PSA-Enh-Luc. F, pCMV-hAR-(507–919) (50 ng) and pCMV-hAR-(1–503) (25 ng) were transfected in HeLa cells with 0.1 μg of PSA-Enh-Luc as above except with 5 nm p300 siRNAs and nonspecific siRNA-5 (NS5). Cells were incubated with and without 10 nm DHT.
Synergistic effects of MAGE-11 and p300 in activating the PSA-enhancer-luciferase reporter gene were analyzed using human AR-(1–660) (Fig. 9A) and corresponding NH2-terminal and DNA binding regions of mouse AR and human PR-B and GR that lack the ligand binding domain. Studies were performed in CV1 cells that have 2–3-fold less endogenous p300 than HeLa cells (Fig. 9C, top panel) and in HeLa cells to assess the effects of silencing p300 (Fig. 9, E and F). MAGE-11 and p300 synergistically increased the activity of hAR-(1–660), mAR-(1–640), hPR-B-(1–688), and hGR-(1–550) (Fig. 9D). A requirement for endogenous p300 was demonstrated with hAR-(1–660) expressed in HeLa cells and p300 siRNA-1, -3, and -4 (Fig. 9E) that reduced p300 levels (Fig. 9C, lower panel). p300 siRNAs also inhibited transcriptional activity of the androgen-dependent intermolecular AR N/C interaction between hAR-(507–919) and hAR-(1–503) (Fig. 9, A and F) (24). Specificity of inhibition was suggested by persistent activation in the presence of nonspecific siRNA or p300 siRNA-2 that did not silence p300 (Fig. 9, E and F). The results suggest that MAGE-11 can increase steroid receptor transcriptional activity also by an FXXLF motif-independent mechanism through its interaction with receptor-associated TIF2 and p300.
Human and Mouse AR AF2 Activity
The inhibitory effect of the human AR N/C interaction on AF2 activity can be relieved either by mutating the AR FXXLF motif to FXXAA or by MAGE-11 binding to the human AR FXXLF motif (21, 23). The inability of mouse AR to interact directly with MAGE-11 raised the possibility that mouse AR AF2 activity may be inhibited to a greater extent than human AR by the N/C interaction. On the other hand, a weaker mouse AR N/C interaction (Figs. 2E and 8A) may not inhibit AF2 activity. To test this further, the effect of the AR N/C interaction on human and mouse AR AF2 activation by TIF2 was investigated using hARΔ120–472, an AF1 deletion mutant (Fig. 9A) that depends on AF2 for transcriptional activity (23), and the corresponding mARΔ101–452 mouse mutant.
In agreement with previous studies (23), transactivation of MMTV-Luc by hARΔ120–472 required the expression of both TIF2 and MAGE-11 (Fig. 10A). The inhibitory effect of the human AR N/C interaction on AF2 activity that was overcome by MAGE-11 was also indicated by TIF2 activation of the hARΔ120–472-L26A,F27A FXXLF motif mutant in the absence of MAGE-11. Dependence on TIF2 LXXLL motif binding to AF2 was shown by loss of transcriptional activity by the K720A charge clamp mutant. However, hARΔ120–472-V33A in which Val33 was changed to Ala33 to mimic mouse AR was inactive with and without TIF2 or MAGE-11. This suggests that the human AR N/C interaction inhibition of AF2 activity in hARΔ120–472-V33A could not be rescued by MAGE-11 consistent with the requirement for AR Val33 to interact directly with MAGE-11.
FIGURE 10.
Species differences in AR N/C interaction inhibition of AF2. A, CV1 cells were transfected with 3 μg of MMTV-Luc and 0.1 μg of human pCMV-hARΔ120–472 WT or L26A,F27A (LFAA), S94A (SA94), V33A (VA33), or K720A (KA720) mutant with 1.5 μg of pSG5 empty vector (−) and 2 μg of pSG5-TIF2 (T) with and without 1 μg of pSG5-MAGE (M). Cells were incubated in the absence and presence of 1 nm DHT. Inset, upper panel, pCMV5 empty vector (p5) (−) and pCMV-hARΔ120–472 WT and L26A,F27A, V33A, K720A, and S94A mutants (5 μg) were expressed in COS cells in the presence of 10 nm DHT. Lower panel, pCMV5 empty vector (−); pCMV-hARΔ120–472 WT, Q5, and S94A; and pCMV-mARΔ101–452 (5 μg) were expressed in COS cells. Cell extracts (60 μg of protein/lane) were analyzed by immunoblot using AR32 antibody. B, CV1 cells were transfected with 3 μg of MMTV-Luc and 0.1 μg of corresponding WT mouse pCMV-mARΔ101–452 or L26A,F27A, A33V, K700A, or A33V,K700A mutants with 1.5 μg of pSG5 empty vector (−) and 2 μg of pSG5-TIF2 (T) with or without 1 μg of pSG5-MAGE (M). Cells were incubated in the absence and presence of 1 nm DHT. Inset, pCMV5 empty vector (p5) (−) or pCMV-mARΔ101–452 WT, L26A,F27A, A33V (AV33), K700A (KA700), and A33V,K700A mutants (5 μg) were expressed in COS cells incubated with 10 nm DHT. Cell extracts (50 μg of protein/lane) were analyzed by immunoblot using AR32 antibody.
In contrast, the corresponding mouse mARΔ101–452 and L26A,F27A FXXLF motif mutants were strongly activated by TIF2 with or without the expression of MAGE-11 (Fig. 10B). When Ala33 was changed to Val33 in mARΔ101–452-A33V to mimic human AR, transcriptional activity required TIF2 and MAGE-11. Partial retention of activity by the mARΔ101–452-K700A charge clamp mutant suggested that mouse AR may be activated to some extent by TIF2 outside the mouse AR AF2 site. However, similar to human AR, transcriptional activity was lost by combining mutations in mARΔ101–452-A33V,K700A.
We noted that hARΔ120–472 migrated as a double band on immunoblots, whereas mARΔ101–452 was a single band (Fig. 10, A and B, top insets). In agreement with previous studies showing that human AR is phosphorylated on Ser94 (41, 42), the hARΔ120–472-S94A mutant migrated as a single band similar to wild-type mouse AR. However, the inhibitory effect of the human AR N/C interaction on AF2 activity not seen with mouse AR could not be attributed to differences in Ser94 phosphorylation. Human AR and mouse AR share the same NH2-terminal EDGSPQAH sequence that contains human AR Ser94,Pro95 (underlined), which is Ser74,Pro75 in mouse AR. Human AR Ser94 is shifted in position relative to mouse AR by glutamine repeat residues 58–78 (5). In addition, hARΔ120–472-S94A required MAGE-11 for AF2 activation by TIF2 similar to hARΔ120–472 (Fig. 10A). When the glutamine repeat length of hARΔ120–472 was reduced from 24 to 5 residues to mimic mouse AR, the double band migration was lost (Fig. 10A, bottom inset), but TIF2 and MAGE-11 were required for transactivation (data not shown).
The results suggest fundamental differences in the regulation of human and mouse AR transcriptional activity. Val33 in human AR resulted in a stronger N/C interaction and inhibition of AF2 that was released by the competitive binding of MAGE-11. Ala33 in mouse AR caused a weaker AR N/C interaction that did not require MAGE-11 for AF2 activity in an animal model that does not express MAGE-11.
DISUCSSION
Convergent Evolution of AR NH2-terminal Sequence and MAGE-11
Our findings suggest species-specific differences in AR transactivation mediated by the effects of the AR N/C interaction and MAGE-11. The androgen-dependent AR N/C interaction between the NH2-terminal FXXLF motif and AF2 hydrophobic binding surface of the ligand binding domain appears to occur in all mammals although to a greater extent in primates due to the evolutionary transition mutation from AR Ala33 to Val33. Evidence for the androgen-dependent AR N/C interaction among all mammals was supported by androgen-dependent stabilization of human and mouse AR, an interaction that slows the dissociation rate of bound androgen from human AR (6, 10). Previous studies confirmed a rat AR N/C interaction that increased activity in association with transcriptional effects of p160 coactivators (43). However, the evolutionary change from alanine to valine at residue 33 flanking the 23FQNLF27 motif in a predicted extended α-helical region not only strengthens the N/C interaction in primates but enables human AR to interact directly with MAGE-11, a coregulator that coevolved in the primate lineage. Studies using AR AF1 deletion mutants that depend on AF2 for activity showed that human AR Val33 contributes to the inhibitory effect of the human AR N/C interaction on AF2 activation by TIF2 that is relieved by MAGE-11.
Convergent evolution of the AR NH2-terminal FXXLF motif flanking sequence with the expression of MAGE-11 in primates together with the ability of MAGE-11 to increase human AR transcriptional activity and the dual functions of the human AR FXXLF motif compared with mouse AR suggests that human and nonhuman primates have acquired increased regulatory control over AR activation domains in association with a primate-specific gain in function (Fig. 11). Our previously proposed model for an evolutionary transition from AF2 to AF1 among different steroid hormone receptors (36) appears to apply also to the evolution of mammalian AR where primate AR acquired the ability to regulate AF2 activity. The functional importance of MAGE-11 in human AR transactivation of androgen-responsive genes was supported by the effects of lentivirus shRNA silencing MAGE-11. The results suggest that primates evolved increased regulatory control over AR activation domain usage to increase transcriptional strength.
FIGURE 11.
Model of MAGE-11 interaction with steroid receptors. MAGE-11 F-box residues interact with the extended human AR FXXLFXXVXXV motif sequence 23FQNLFQSVREV33 that ends with Val33. Both hAR and mAR undergo the androgen-dependent N/C interaction. However, human AR Val33 that is Ala33 in mouse AR is required for a direct interaction with MAGE-11 and for the AR N/C interaction inhibition of AF2 activity that is relieved by MAGE-11. MAGE-11 MXXIF motif interaction with p300 (24) enhances transcriptional activity from the NH2-teminal regions of human AR, PR-B, and GR. MAGE-11 FXXIF motif interaction with p160 coactivators (23) enhances AF2 activity from the human AR, PR-B, and GR carboxyl-terminal ligand binding domains (LBD). Although mouse AR is activated by p300 and p160 coactivators, MAGE-11 is not present in mouse, and the mouse AR NH2-terminal FXXLFXXVXXA motif sequence would not interact directly with MAGE-11. Human and nonhuman primate AR transcriptional activity is regulated by the primate-specific coevolution of MAGE-11 and the AR NH2-terminal FXXLFXXVXXV interaction site for MAGE-11. DBD, DNA binding domain.
Species-specific AR Function
Complete amino acid sequence conservation in the AR DNA and ligand binding domains throughout mammalian evolution preserves the structural constraints necessary for high affinity androgen and DNA binding. In contrast is the unstructured AR NH2-terminal region, which is also important for AR function but less well conserved. Human and mouse AR NH2-terminal regions share ∼84% amino acid sequence homology with the most highly conserved regions at human AR amino acid residues 1–53, 234–247, and 360–429 (1, 35). Human AR residues 1–53 contain the FXXLF motif that mediates the N/C interaction (6, 10) and serves as the binding site for MAGE-11 (22). This region precedes the polymorphic glutamine repeat that expanded during primate evolution to an average length of 20–22 residues (1). Human AR glutamine repeat residues 58–78 are shifted in mouse AR to residues 174–193 (1, 5, 25). Expansion to more than 39-glutamine repeat length in human AR causes spinal bulbar muscular atrophy or Kennedy disease, an adult onset muscle wasting disease associated with an AR gain in function that requires the N/C interaction (4, 44). Human AR amino acid residues 234–247 are within the binding region for CHIP (carboxyl terminus of the Hsp70-interacting protein), an E3 ubiquitin ligase that targets AR for degradation (35). Human AR residues 360–429 were implicated in transactivation (45).
Amino acid sequence conservation in the AR DNA binding domain throughout evolution suggests that interaction sites in chromatin may be shared across mammalian species. However, recent studies have shown surprisingly low sequence conservation for the majority of mammalian AR DNA binding sites associated with androgen-regulated genes (46). This suggests that species-specific differences in AR gene regulation may be attributed to differences in coregulatory proteins such as MAGE-11.
Genome-wide sequence analysis and two-hybrid screens showed that the AR coregulator function of MAGE-11 is primate-specific. This is consistent with differences in the rat and mouse AR FXXLF motif flanking sequence where Ala33 in mouse AR is not compatible with direct binding to MAGE-11. However, absence of an FXXLF motif that binds MAGE-11 in mouse AR or other steroid receptors did not rule out transcriptional effects. MAGE-11 coexpressed with mouse AR or human PR-B or GR was present in a complex presumably through its interaction with p300 and p160 coactivators. In agreement with this, MAGE-11 increased steroid receptor transcriptional activity also by an FXXLF motif-independent mechanism. Evidence that MAGE-11 can increase steroid receptor activity through an association with p300 and p160 coactivators broadens its potential impact in human reproductive physiology. MAGE-11 interaction with receptor-associated p300 and p160 coactivators may have been the evolutionary progenitor for the primate-specific direct interaction with human AR (Fig. 11).
Species-specific differences in AR activity have been reported previously. Rat AR activation of MMTV-chloramphenicol acetyltransferase reporter gene in CV1 cells was less than that of human AR with differences attributed to the AR NH2-terminal region (47). However, we did not detect MAGE-11 protein in CV1 cells, and mRNA levels were low (37). This suggests that additional mechanisms involving the NH2-terminal region increase human AR transcriptional strength. Attempts to mimic human AR in a humanized mouse model made use of a human AR NH2-terminal region 5′ SmaI site fragment that begins at human AR amino acid residue 37 so that the chimera retained mouse Ala33 (48). Even with the absence of MAGE-11 in mouse, the unregulated p160 coactivator access to AF2 that characterizes mouse AR would be expected to be retained in the presence of AR Ala33.
CpG dinucleotides are hot spots for mutation that account for approximately one-third of disease-causing germ line mutations (49, 50) and a significant proportion of human AR mutations that cause the androgen insensitivity syndrome (51). CpG dinucleotides can become methylated, and 5-methylcytosine can undergo spontaneous deamination that results in a C→T transition mutation in genomic DNA. Our findings suggest that mutations at CpG dinucleotides also contribute to the evolution of new gene sequences. Ala33 in mouse and rat AR is coded by GCG, which in primates changed to GTG coding for Val33. Rat and mouse AR GCG codon 33 is followed by A in the sequence GCGA, and CGA was preferentially associated with mutations in the retinoblastoma gene (50). Methylation and deamination of CpG in AR codon 33 in the less evolved mammals may have caused the evolutionary transition from Ala33 to Val33 needed for the AR N/C interaction inhibition of AF2 activity and the binding to MAGE-11 that releases that inhibition in primates. MAGE-11 may have evolved to regulate AR transcriptional activity in part by relieving the inhibitory effects of the AR N/C interaction associated with the AR Ala33 to Val33 mutation.
Species-specific MAGE Gene Families
The rapid evolution of the MAGE gene family among mammals results from gene duplication and retrotransposition (52). The MAGE-11 gene is part of a MAGE-A subfamily expressed at Xq28 on the human X chromosome and contains three short unique 5′ coding exons that precede a major, more highly conserved 3′ coding exon (22, 53). A two-hybrid interaction screen of a human testis library using the human AR FXXLF motif region as bait identified MAGE-11 as a human AR coregulator (22). A similar screen of a mouse testis library using the corresponding mouse AR FXXLF motif region as bait suggested the absence of a related AR FXXLF motif-interacting protein in mouse. This finding was consistent with genome-wide sequence analysis of MAGE gene families that no other mouse MAGE family protein corresponds to human MAGE-11. It was also consistent with the absence of an inhibitory effect of the mouse AR N/C interaction on AF2 activity. The results support the absence of MAGE-11 in mammals outside the primate lineage and a primate-specific AR gain in function that increases transcriptional strength of AR and other steroid hormone receptors. The absence of MAGE-11 in lower mammals suggests its coevolution with the AR FXXLF motif flanking sequence and increased regulatory control of AR activation domain dominance through evolution.
The ability of MAGE-11 to increase human AR transcriptional activity places it among a growing list of AR coactivators (54). However, the precise mechanisms that underlie its coregulator function remain to be established. MAGE-11 is a multifunctional protein that interacts with p300 and p160 coactivators (23, 24) and undergoes post-translational modification that includes phosphorylation by mitogen-activated protein kinase and cell cycle checkpoint kinase Chk1 and monoubiquitinylation at lysine residues required to interact with the human AR FXXLF motif (23, 27). A role for MAGE-11 in androgen-dependent cell cycle regulation is suggested by a number of MAGE family proteins that influence cell cycle progression and apoptosis in humans (55). A MAGE-A subfamily binding partner is a RING E3 ubiquitin ligase whose activity increases in association with MAGE proteins (56). Some MAGE-A proteins inhibit p53 tumor suppressor activity during tumor development (57). MAGE-11 was shown to be an interaction partner and inhibitor of the major hypoxia-inducible factor HIF-1α-hydroxylating enzyme prolyl hydroxylase 2 and stabilizes HIF-1α in a potential tumor-associated regulatory mechanism (58). It remains to be established whether these functions are important for MAGE-11 activity as a steroid receptor coregulator.
The ability of MAGE-11 to increase transcriptional strength of human AR through a direct interaction with the AR FXXLF motif has important implications in normal human reproductive physiology and cancer. MAGE-11 mRNA levels increase more than 50-fold in the mid-secretory glandular epithelium of human endometrium during the window of receptivity for implantation, suggesting a role in human embryo implantation or survival (34). MAGE-11 mRNA levels are acutely up-regulated by cyclic AMP, a second messenger signaling response to the preovulatory luteinizing hormone surge. However, the physiological effects of AR and MAGE-11 in human endometrium during the window of implantation remain to be established. MAGE-11 is coexpressed with AR in human ovarian granulosa cells3 where it may function in oocyte survival (34). MAGE-11 mRNA levels increase more than 100-fold in a subset of patients with castration-recurrent prostate cancer undergoing androgen deprivation therapy. The increase in MAGE-11 during prostate cancer growth results from progressive hypomethylation of a CpG island at the transcription start site (37). Increased levels of MAGE-11 that increase human AR signaling during prostate cancer progression may promote prostate cancer cell survival (59). In normal men, increased AR signaling by MAGE-11 may improve physical strength and survival, and in women, it may be important for reproductive success.
Acknowledgments
We are grateful to Andrew T. Hnat and K. Michelle Cobb for technical assistance and Frank S. French for reviewing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant HD16910, United States Public Health Service grant from NICHD; Grant P01-CA77739 from the NCI; Grant 5D43-TW000627-15, a Fogarty International Center Training and Research in Population and Health grant; and Cooperative Agreement U54-HD35041 from the Eunice Kennedy Shriver NICHD as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research.
E. M. Wilson, unpublished results.
- AR
- androgen receptor
- MAGE-11
- melanoma antigen-A11
- DHT
- dihydrotestosterone
- AF
- activation function
- N/C
- NH2- and carboxyl-terminal
- hAR
- human AR
- TIF2
- transcriptional intermediary factor 2
- PSA
- prostate-specific antigen
- Enh
- enhancer
- Luc
- luciferase
- GR
- glucocorticoid receptor
- PR
- progesterone receptor
- mAR
- mouse AR
- rAR
- rat AR
- cDNA
- complementary DNA
- hGR
- human glucocorticoid receptor
- hPR
- human progesterone receptor
- MMTV
- mouse mammary tumor virus
- Ab
- antibody
- rGR
- rat glucocorticoid receptor.
REFERENCES
- 1. Choong C. S., Kemppainen J. A., Wilson E. M. (1998) J. Mol. Evol. 47, 334–342 [DOI] [PubMed] [Google Scholar]
- 2. Lavery D. N., McEwan I. J. (2008) Biochemistry 47, 3360–3369 [DOI] [PubMed] [Google Scholar]
- 3. Simental J. A., Sar M., Lane M. V., French F. S., Wilson E. M. (1991) J. Biol. Chem. 266, 510–518 [PubMed] [Google Scholar]
- 4. La Spada A. R., Wilson E. M., Lubahn D. B., Harding A. E., Fischbeck K. H. (1991) Nature 352, 77–79 [DOI] [PubMed] [Google Scholar]
- 5. Charest N. J., Zhou Z. X., Lubahn D. B., Olsen K. L., Wilson E. M., French F. S. (1991) Mol. Endocrinol. 5, 573–581 [DOI] [PubMed] [Google Scholar]
- 6. He B., Kemppainen J. A., Wilson E. M. (2000) J. Biol. Chem. 275, 22986–22994 [DOI] [PubMed] [Google Scholar]
- 7. He B., Lee L. W., Minges J. T., Wilson E. M. (2002) J. Biol. Chem. 277, 25631–25639 [DOI] [PubMed] [Google Scholar]
- 8. Dehm S. M., Regan K. M., Schmidt L. J., Tindall D. J. (2007) Cancer Res. 67, 10067–10077 [DOI] [PubMed] [Google Scholar]
- 9. Callewaert L., Verrijdt G., Christiaens V., Haelens A., Claessens F. (2003) J. Biol. Chem. 278, 8212–8218 [DOI] [PubMed] [Google Scholar]
- 10. Langley E., Zhou Z. X., Wilson E. M. (1995) J. Biol. Chem. 270, 29983–29990 [DOI] [PubMed] [Google Scholar]
- 11. Klokk T. I., Kurys P., Elbi C., Nagaich A. K., Hendarwanto A., Slagsvold T., Chang C. Y., Hager G. L., Saatcioglu F. (2007) Mol. Cell. Biol. 27, 1823–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schaufele F., Carbonell X., Guerbadot M., Borngraeber S., Chapman M. S., Ma A. A., Miner J. N., Diamond M. I. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9802–9807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Langley E., Kemppainen J. A., Wilson E. M. (1998) J. Biol. Chem. 273, 92–101 [DOI] [PubMed] [Google Scholar]
- 14. He B., Kemppainen J. A., Voegel J. J., Gronemeyer H., Wilson E. M. (1999) J. Biol. Chem. 274, 37219–37225 [DOI] [PubMed] [Google Scholar]
- 15. Ghadessy F. J., Lim J., Abdullah A. A., Panet-Raymond V., Choo C. K., Lumbroso R., Tut T. G., Gottlieb B., Pinsky L., Trifiro M. A., Yong E. L. (1999) J. Clin. Investig. 103, 1517–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thompson J., Saatcioglu F., Jänne O. A., Palvimo J. J. (2001) Mol. Endocrinol. 15, 923–935 [DOI] [PubMed] [Google Scholar]
- 17. Ghali S. A., Gottlieb B., Lumbroso R., Beitel L. K., Elhaji Y., Wu J., Pinsky L., Trifiro M. A. (2003) J. Clin. Endocrinol. Metab. 88, 2185–2193 [DOI] [PubMed] [Google Scholar]
- 18. Quigley C. A., Tan J. A., He B., Zhou Z. X., Mebarki F., Morel Y., Forest M. G., Chatelain P., Ritzén E. M., French F. S., Wilson E. M. (2004) Mech. Ageing Dev. 125, 683–695 [DOI] [PubMed] [Google Scholar]
- 19. He B., Gampe R. T., Jr., Hnat A. T., Faggart J. L., Minges J. T., French F. S., Wilson E. M. (2006) J. Biol. Chem. 281, 6648–6663 [DOI] [PubMed] [Google Scholar]
- 20. Jääskeläinen J., Deeb A., Schwabe J. W., Mongan N. P., Martin H., Hughes I. A. (2006) J. Mol. Endocrinol. 36, 361–368 [DOI] [PubMed] [Google Scholar]
- 21. He B., Bowen N. T., Minges J. T., Wilson E. M. (2001) J. Biol. Chem. 276, 42293–42301 [DOI] [PubMed] [Google Scholar]
- 22. Bai S., He B., Wilson E. M. (2005) Mol. Cell. Biol. 25, 1238–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Askew E. B., Bai S., Hnat A. T., Minges J. T., Wilson E. M. (2009) J. Biol. Chem. 284, 34793–34808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Askew E. B., Bai S., Blackwelder A. J., Wilson E. M. (2010) J. Biol. Chem. 285, 21824–21836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lubahn D. B., Joseph D. R., Sar M., Tan J., Higgs H. N., Larson R. E., French F. S., Wilson E. M. (1988) Mol. Endocrinol. 2, 1265–1275 [DOI] [PubMed] [Google Scholar]
- 26. Askew E. B., Gampe R. T., Jr., Stanley T. B., Faggart J. L., Wilson E. M. (2007) J. Biol. Chem. 282, 25801–25816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bai S., Wilson E. M. (2008) Mol. Cell. Biol. 28, 1947–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Voegel J. J., Heine M. J., Tini M., Vivat V., Chambon P., Gronemeyer H. (1998) EMBO J. 17, 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang W., Shostak Y., Tarr P., Sawyers C., Carey M. (1999) J. Biol. Chem. 274, 25756–25768 [DOI] [PubMed] [Google Scholar]
- 30. Rennie P. S., Bruchovsky N., Leco K. J., Sheppard P. C., McQueen S. A., Cheng H., Snoek R., Hamel A., Bock M. E., MacDonald B. S., Nickel B. E., Chang C., Liao S., Cattini P. A., Matusik R. J. (1993) Mol. Endocrinol. 7, 23–36 [DOI] [PubMed] [Google Scholar]
- 31. Kasper S., Rennie P. S., Bruchovsky N., Sheppard P. C., Cheng H., Lin L., Shiu R. P., Snoek R., Matusik R. J. (1994) J. Biol. Chem. 269, 31763–31769 [PubMed] [Google Scholar]
- 32. Tan J. A., Joseph D. R., Quarmby V. E., Lubahn D. B., Sar M., French F. S., Wilson E. M. (1988) Mol. Endocrinol. 2, 1276–1285 [DOI] [PubMed] [Google Scholar]
- 33. He B., Minges J. T., Lee L. W., Wilson E. M. (2002) J. Biol. Chem. 277, 10226–10235 [DOI] [PubMed] [Google Scholar]
- 34. Bai S., Grossman G., Yuan L., Lessey B. A., French F. S., Young S. L., Wilson E. M. (2008) Mol. Hum. Reprod. 14, 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He B., Bai S., Hnat A. T., Kalman R. I., Minges J. T., Patterson C., Wilson E. M. (2004) J. Biol. Chem. 279, 30643–30653 [DOI] [PubMed] [Google Scholar]
- 36. He B., Gampe R. T., Jr., Kole A. J., Hnat A. T., Stanley T. B., An G., Stewart E. L., Kalman R. I., Minges J. T., Wilson E. M. (2004) Mol. Cell 16, 425–438 [DOI] [PubMed] [Google Scholar]
- 37. Karpf A. R., Bai S., James S. R., Mohler J. L., Wilson E. M. (2009) Mol. Cancer Res. 7, 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kemppainen J. A., Lane M. V., Sar M., Wilson E. M. (1992) J. Biol. Chem. 267, 968–974 [PubMed] [Google Scholar]
- 39. Frønsdal K., Engedal N., Slagsvold T., Saatcioglu F. (1998) J. Biol. Chem. 273, 31853–31859 [DOI] [PubMed] [Google Scholar]
- 40. He B., Wilson E. M. (2003) Mol. Cell. Biol. 23, 2135–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou Z. X., Kemppainen J. A., Wilson E. M. (1995) Mol. Endocrinol. 9, 605–615 [DOI] [PubMed] [Google Scholar]
- 42. Gioeli D., Ficarro S. B., Kwiek J. J., Aaronson D., Hancock M., Catling A. D., White F. M., Christian R. E., Settlage R. E., Shabanowitz J., Hunt D. F., Weber M. J. (2002) J. Biol. Chem. 277, 29304–29314 [DOI] [PubMed] [Google Scholar]
- 43. Ikonen T., Palvimo J. J., Jänne O. A. (1997) J. Biol. Chem. 272, 29821–29828 [DOI] [PubMed] [Google Scholar]
- 44. Orr C. R., Montie H. L., Liu Y., Bolzoni E., Jenkins S. C., Wilson E. M., Joseph J. D., McDonnell D. P., Merry D. E. (2010) J. Biol. Chem. 285, 35567–35577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jenster G., van der Korput H. A., Trapman J., Brinkmann A. O. (1995) J. Biol. Chem. 270, 7341–7346 [DOI] [PubMed] [Google Scholar]
- 46. Hu S., Yao G., Guan X., Ni Z., Ma W., Wilson E. M., French F. S., Liu Q., Zhang Y. (2010) Mol. Endocrinol. 24, 2392–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chamberlain N. L., Driver E. D., Miesfeld R. L. (1994) Nucleic Acids Res. 22, 3181–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Albertelli M. A., Scheller A., Brogley M., Robins D. M. (2006) Mol. Endocrinol. 20, 1248–1260 [DOI] [PubMed] [Google Scholar]
- 49. Cooper D. N., Youssoufian H. (1988) Hum. Genet. 78, 151–155 [DOI] [PubMed] [Google Scholar]
- 50. Mancini D., Singh S., Ainsworth P., Rodenhiser D. (1997) Am. J. Hum. Genet. 61, 80–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Quigley C. A., De Bellis A., Marschke K. B., el-Awady M. K., Wilson E. M., French F. S. (1995) Endocr. Rev. 16, 271–321 [DOI] [PubMed] [Google Scholar]
- 52. Delbridge M. L., Graves J. A. (2007) Soc. Reprod. Fertil. Suppl. 65, 1–17 [PubMed] [Google Scholar]
- 53. Irvine R. A., Coetzee G. A. (1999) Immunogenetics 49, 585. [DOI] [PubMed] [Google Scholar]
- 54. Heemers H. V., Tindall D. J. (2007) Endocr. Rev. 28, 778–808 [DOI] [PubMed] [Google Scholar]
- 55. Matsumoto K., Taniura H., Uetsuki T., Yoshikawa K. (2001) Gene 272, 173–179 [DOI] [PubMed] [Google Scholar]
- 56. Doyle J. M., Gao J., Wang J., Yang M., Potts P. R. (2010) Mol. Cell 39, 963–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Marcar L., Maclaine N. J., Hupp T. R., Meek D. W. (2010) Cancer Res. 70, 10362–10370 [DOI] [PubMed] [Google Scholar]
- 58. Aprelikova O., Pandolfi S., Tackett S., Ferreira M., Salnikow K., Ward Y., Risinger J. I., Barrett J. C., Niederhuber J. (2009) Cancer Res. 69, 616–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wilson E. M. (2010) Ther. Adv. Urol. 2, 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]