Abstract
Population genetic analyses of bacterial genes whose products interact with host tissues can give new understanding of infection and disease processes. Here we show that strains of the genetically diverse gastric pathogen Helicobacter pylori from Amerindians from the remote Peruvian Amazon contain novel alleles of cagA, a major virulence gene, and reveal distinctive properties of their encoded CagA proteins. CagA is injected into the gastric epithelium where it hijacks pleiotropic signaling pathways, helps Hp exploit its special gastric mucosal niche, and affects the risk that infection will result in overt gastroduodenal diseases including gastric cancer. The Amerindian CagA proteins contain unusual but functional tyrosine phosphorylation motifs and attenuated CRPIA motifs, which affect gastric epithelial proliferation, inflammation, and bacterial pathogenesis. Amerindian CagA proteins induced less production of IL-8 and cancer-associated Mucin 2 than did those of prototype Western or East Asian strains and behaved as dominant negative inhibitors of action of prototype CagA during mixed infection of Mongolian gerbils. We suggest that Amerindian cagA is of relatively low virulence, that this may have been selected in ancestral strains during infection of the people who migrated from Asia into the Americas many thousands of years ago, and that such attenuated CagA proteins could be useful therapeutically.
Keywords: Bacterial Genetics, Cancer Therapy, Cell Differentiation, Inflammation, Signal Transduction, Amerindian, CagA, Helicobacter pylori
Introduction
Population and molecular genetic studies of genes of bacterial pathogens whose products interact with host tissues can give important insights into evolutionary forces, enhance understanding of infection and disease processes, and potentially lead to improvements in human health. Helicobacter pylori (Hp)3 chronically infects the gastric epithelial surface and a narrow band of overlying mucus in billions of people worldwide (1–3). Most Hp infections begin in infancy and can last for decades despite features such as inflammatory responses, mucosal shedding, and peristalsis that make the gastric mucosal niche hostile to nearly all other microbes. Hp constitutes a major risk factor for several gastroduodenal diseases including peptic ulcer and gastric cancer even though most Hp infections are asymptomatic.
DNA sequence analysis of representative housekeeping genes (multilocus sequence typing) has shown that Hp is extremely diverse genetically and that different genotypes predominate in different geographic regions or human populations (4–7). Hp prevalence and multilocus sequence typing studies have indicated that Hp transmission occurs preferentially within families and local communities, not in sweeping epidemics. Such localized transmission fosters accumulation of genetic diversity by genetic drift and selection for adaptation to local conditions including differences in human physiology. Multilocus sequence typing analyses have indicated that probably Hp has infected humans for eons and was carried by the people who migrated into Eurasia from Africa some 60,000 years ago and by the ancestors of modern Amerindians who migrated from Asia into Beringia and then into the Americas some 20,000 years ago (4–7). Geographic differences among alleles of several genes whose products interact with host tissues (such as cagA, vacA, and babA2) are even greater than those of housekeeping genes (4–6, 8). Much of this extra diversity may stem from differences in selective forces operating in different human populations and geographic regions (9, 10).
Prominent among the proteins that Hp uses to manage its special gastric mucosal niche is CagA, a major virulence factor that is injected via a type IV secretion system into gastric epithelium, where it hijacks multiple signaling pathways such as mitogen-activated protein kinases (MAPKs) and phosphoinositide 3-kinase (PI3K)/Akt/IκB kinase (IKK) pathways and thereby alters the actin cytoskeleton, intercellular junctions, and cell polarity and proliferation, and induces inflammation (11–14). Of particular interest are two distinct classes of motifs used by CagA for interaction with host proteins: EPIYA (Glu-Pro-Ile-Tyr-Ala) and CRPIA (conserved repeats responsible for phosphorylation-independent activity) motifs (15). EPIYA motifs undergo tyrosine phosphorylation and mediate tyrosine phosphorylation-dependent interactions with host molecules such as Shp2, Grb2, Crk, Csk, phospholipase Cγ, and PI3K p85 (15–21), whereas CRPIA mediates phosphorylation-independent interactions with Par1, Met, and Tak1 (15, 22, 23).
Recent epidemiological studies revealed that some Hp strains from Amerindians in the Amazon tend to be of a novel genotype, distinct from those of Western and East Asian strains (9, 24–27). Here we describe novel alleles of the cagA gene in strains from the remote Peruvian Amazon and the distinctive properties of the CagA proteins they encode. These Amerindian strains are unusual in causing only attenuated cell proliferation and inflammation responses, resulting in lower virulence and decreased risk of severe pathologies including gastric cancer both in in vitro and in vivo infection models. Much of this attenuation was traced to the low potency of their CRIPA motifs. Furthermore, the attenuated Amerindian cagA alleles diminished the strongly proinflammatory response provoked by more pathogenic strains during mixed infection of gerbils. We suggest that this CagA attenuation may have been selected during ancestral human migrations and that it might be useful therapeutically.
EXPERIMENTAL PROCEDURES
Clinical Specimens
Hp strains from the Machiguenga-speaking residents of Shimaa village in the remote Peruvian Amazon were cultured under standard microaerobic conditions with informed consent in protocols approved by local and international human studies committees (25). Hp strains from separate Ashaninka-speaking Amazonian villages in the province of Satipo were cultured under the same protocols. Gastric biopsies were fixed in pH 7.2 buffered formalin, paraffin-embedded, sectioned, hematoxylin/eosin-stained, and graded histologically using the Sidney System (25). Strains from Amerindian Lima shantytown residents were cultured similarly. All Peruvian strains used in these analyses were obtained with informed consent. Other strains used here were from the collections of Sasakawa, Berg, Kamiya, and Zou laboratories. The cagA sequences were determined by CagTF and CagTR primer-mediated PCR amplifying the PY region from Hp genomic DNAs (28) and then submitted to the DDBJ/EMBL/GenBankTM databases (accession nos. AB587140-AB587258). Although some Amerindian isolates such as Shi470 have a second apparently defective copy of cagA inserted between cag14 and cag15 in the middle of the cag pathogenicity island (PAI) in addition to cagA in the usual cag PAI right end location (27), our PCR primers were designed only to detect, clone, and analyze the conventionally placed cagA gene.
Sequence and Bioinformatic Analysis
Sequence analysis was performed with programs and data in Blast homology search programs (www.ncbi.nlm.nih.gov) and genome sequence databases. Multiple Sequence Alignments and Unrooted Trees constructed by Neighbor-Joining were performed with programs in ClustalW. Sequence logos analysis was performed with the program in WebLogo 3.
Hp Infection in Cell Culture
Reference Hp strains such as 26695 and ATCC43504 (also often referred to as NCTC11637) and their isogenic ΔcagA and ΔvirD4 mutants were previously described (15, 16, 29). ATCC43504 ΔcagA derivatives containing chromosomal FLAG-tagged cagA (W, W/EA, W/AM-I, or W/AM-II) genes were constructed for this study. Briefly, FLAG-tagged cagA genes were ligated with kanamycin-resistant genes as part of the same transcription unit, and then these fragments were ligated between 1000-bp segments that flank the 5′ and 3′ ends of the ATCC43504 cagA gene. These DNA constructs were then used for transformation of ΔcagA strains with selection for kanamycin resistance and thereby chromosomal placement of our engineered cagA genes. Hp strains were cultured according to standard procedures (15, 16). AGS, HEK293, and 293T cells were maintained in Dulbecco's modified Eagle's medium (Sigma) containing 10% fetal bovine serum. Cultured cells were infected with Hp at a multiplicity of infection of 100.
Hp Infection in Cell Culture and Animals
Gerbil infection experiments were approved by the University of Tokyo ethics committee and carried out as described (15, 29). Briefly, 6-week-old male MGS/Sea Mongolian gerbils (CLEA Japan Inc.) were intragastrically inoculated with 109 colony forming units (CFU) of Hp strain ATCC43504 or its derivatives. After 8 weeks, the stomachs were examined to determine bacterial load and analyzed by immunohistology. A competition assay was performed by co-inoculating wild-type ATCC43504 and derivatives with engineered cagA alleles (109 CFU of each strain). The competitive index was calculated as the ratio of the wild-type strain CFU:mutant strain CFU. The two genotypes were distinguished by colony PCR of PY regions in cagA. If no CFU were recovered from the stomach, a CFU count of one at the lowest dilution was assigned. Data were analyzed statistically using a Mann-Whitney U test for unpaired groups.
Plasmid Construction and Transfection
NF-κB luciferase reporter was from Promega and serum response element (SRE) and p53 luciferase reporters were from Stratagene. Myc-Par1 and Tpr-Met (dominant-active Met) plasmids were kindly provided by Drs. Miki (Osaka University) (30) and Park (McGill University), respectively; the K1110A-Tpr-Met mutant has been described (15). The PY regions of the cagA genes from the W (26695), EA (JpnTK1003), AM-I (Shi257), and AM-II (Shi193) strains were cloned into pGEX-6P-1 (Amersham Biosciences) to perform the glutathione S-transferase (GST) pulldown assays. The 26695 CagA, ΔPY (Δ871–1026), ΔPY2 (Δ871–976), PR (Y899F/Y918F/Y972F), R952A/R986A, and PR-RA (PR-R952A/R986A) plasmids were previously described (15). The cagA genes from the EA (JpnTK1003), AM-I (Shi257), and AM-II (Shi193) strains were cloned into pEGFP-C1 (Clontech), and their mutants and 26695 ΔPY2 CagA (Δ871–976) mutants were generated using the site-directed mutagenesis kit (Stratagene). The human IL-8 (−1498/+44) and MUC2 promoters (−2631/+111) were amplified from genomic DNA of AGS cells and cloned into pGL4.20 (Promega). The 26695 ΔPY cagA and Shi193 cagA genes were Myc-tagged at their 3′ ends and cloned into pcDNA3.1 (Invitrogen), and the 26695 cagA gene was FLAG- or T7-tagged at its 3′ end and cloned into pC4-Fv1E (ARIAD Pharmaceuticals, Inc.) for Fv-mediated homodimerization assays. Culture cells were transiently transfected with appropriate plasmids using FuGENE 6 (Roche Applied Science), Lipofectamine 2000, or Lipofectamine LTX (Invitrogen).
RNA Transfection
To knockdown endogenous host genes, cells were transfected for 48 h with appropriate small interfering RNAs (siRNAs, 100 nm) using Lipofectamine RNAiMAX (Invitrogen). The siRNAs were synthesized, purified, and duplexed by RNAi Co., Ltd. The siRNAs for the luciferase (Luc) control, Par1, Met, and p85 have been described (15).
GST Pulldown Assay
GST pulldown assays were performed as previously described (15, 16) using GST-fused CagA PY region proteins from W (26695), EA (JpnTK1003), AM-I (Shi257), and AM-II (Shi193) strains, which each harbor EPIYA tyrosine phosphorylation motifs. These proteins were purified from Escherichia coli expressing an inducible Elk6-tyrosine kinase domain (strain TKB1; Stratagene) according to the manufacturer's instructions.
Immunoprecipitation, Immunoblotting, and Immunostaining
Immunoprecipitation, immunoblotting, and immunostaining were performed using appropriate antibodies as previously described (15, 16). After immunostaining, specimens were examined using a confocal laser-scanning microscope (Carl Zeiss LSM510), and fluorescent images were analyzed using LSM510 Version 3.2 software (Carl Zeiss). Akt kinase assays using purified GSK3β protein were performed using an Akt kinase assay kit (Cell Signaling Technology) according to the manufacturer's instructions. IKK kinase assays using purified GST-IκB protein were performed as described (31).
Antibodies and Reagents
Rabbit anti-CagA, anti-p-CagA, and anti-UreA polyclonal antibodies (pAbs) were previously described (15, 16). Mouse anti-Crk monoclonal antibody (mAb) was from BD Transduction Laboratories. Mouse anti-phospholipase Cγ and anti-Par1 mAbs were from Upstate Biotechnology. Rabbit anti-p- extracellular signal-regulated kinase 1/2 (Erk1/2; Thr-202/Tyr-204), anti-Erk1/2, anti-p-p38 (Thr-180/Tyr-182), anti-p38, anti-p-Akt (Ser-473), anti-Akt, anti-p-Met (Tyr-1349), anti-p-IκB (Ser-32), and anti-p-Src (Try-416) pAbs and mouse anti-Csk, anti-IKKγ, anti-IKKβ, anti-Tyr(P) (PY20), and anti-Myc mAbs were from Cell Signaling Technology. Rabbit anti-Met, anti-Shp2, anti-Grb2, anti-Stat3, and anti-MUC2 pAbs and mouse anti-p85 mAb were from Santa Cruz Biotechnology. Rabbit anti-p-Tau (Ser-262) pAb and mouse anti-Src and anti-Tau pAbs were from Calbiochem. Rabbit anti-GFP pAb was from Medical and Biological Laboratories. Mouse anti-actin mAb was from Chemicon International. Mouse anti-T7 mAb was from Novagen. Mouse anti-FLAG mAbs and Anguilla anguilla agglutinin (for staining gastric pit cells) were from Sigma. Rhodamine phalloidin, Alexa Fluor 633 phalloidin, and TO-PRO-3 were from Molecular Probes.
Luciferase Assay
Luciferase assays were performed using the Dual-luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. The firefly luciferase levels were measured and normalized to the activity of phRL-TK-derived Renilla luciferase. Data were analyzed statistically using Student's t test for unpaired groups and expressed as the means ± S.E. of triplicate experiments.
Homodimerization Assay
Homodimerization assays were performed using the Regulated Homodimerization kit (ARIAD Pharmaceuticals, Inc.) according to the manufacturer's instructions. Briefly, 293T cells were co-transfected for 24 h with the Fv-W-CagA-FLAG, Fv-W-CagA-T7, and W/AM-II-CagA-Myc plasmids in the presence or absence of AP20187 (100 nm), a small molecule that induces homodimerization of Fv domain-containing fusion proteins.
RESULTS
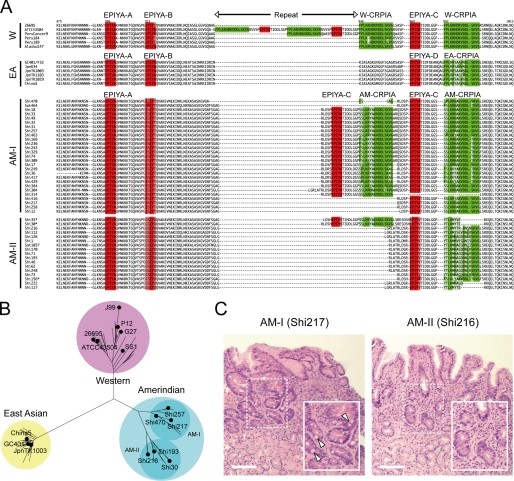
Histopathologic examination of gastric biopsy specimens from 39 Hp-infected Amerindian residents of Shimaa, a small village in the remote Peruvian Amazon (25, 27), indicated that most villagers had active gastritis and mild atrophy, and several had intestinal metaplasia. None had peptic ulcer disease or gastric cancer (Fig. 1C and supplemental Table S1). We next examined the cagA genes and encoded CagA proteins of Hp cultured from these Shimaa villagers. Prior studies had shown that CagA of European and North American (Western (W)) and East Asian (EA) strains differed markedly in their C-terminal region (PY region) and that these differences are functionally important (11–14). PY regions contain several EPIYA tyrosine phosphorylation motifs: EPIYA-A and EPIYA-B motifs characteristic of both W and EA strains and EPIYA-C and EPIYA-D motifs characteristic of W and of EA strains, respectively (Fig. 1A) (11, 14). PY regions also contain CRPIA motifs, which are responsible for separate phosphorylation-independent activities of CagA (Fig. 1A) (15).
FIGURE 1.
Amerindian CagA is distinct from Western and East Asian CagA. A, shown is multiple sequence alignment of the PY regions of CagA from Hp isolates. The EPIYA (red) and CRPIA motifs (green) are highlighted. CagA was categorized Western (W), East Asian (EA), and Amerindian (AM-I and AM-II) based on ClustalW analysis. Amerindian isolates, such as Shi35, Shi30, and Shi156, had marginal CRPIA genotypes between AM-I and AM-II (shown with asterisks). B, shown is a neighbor-joining tree of amino acid sequences of the PY regions. C, shown is hematoxylin/eosin staining of gastric antral biopsies from the indicated Hp-infected individuals. Bar = 100 μm. Arrowheads indicate goblet cells in sites of intestinal metaplasia.
By sequencing, we found that CagA proteins from most strains from urban Peruvians are of the W type (Fig. 1A and supplemental Fig. S1). In contrast, those of Shimaa strains are distinct from both W and EA types and could be placed in two main groups designated AM-I and AM-II (AM, Amerindian) (Fig. 1B). Although the overall sequences of AM and W CagAs are similar, AM CagAs have altered or degenerate “EPIYA-B” motifs: ESIYT and GSIYD in AM-I and AM-II CagAs, respectively (Fig. 1A). In addition, CagA sequences in some nominally AM-II strains such as Shi30, Shi35, and Shi156 have AM-I type CRPIA motifs (Fig. 1A), suggesting that they are actually hybrids and thus might differ functionally from purely AM-I or AM-II type CagA proteins. Most CagA sequences found in strains from the distinct Peruvian Amazonian region of Satipo, which is far from Shimaa and whose residents are of another language group, also fit into this AM-I and AM-II classification, as did CagA of several isolates from Colombian and Venezuelan Amazon (supplemental Fig. S1) (9, 24, 26).
The CagA N terminus, which is far from its PY region, is needed for efficient CagA translocation into target epithelial cells, subsequent plasma membrane anchoring, and modulation of C-terminal region activities (32–34). The N-terminal regions of each of four AM-I CagAs that we characterized were similar to those of prototype CagA, whereas each of three AM-II CagAs lacked two large internal segments totaling 180 amino acids (supplemental Fig. S2) and thus may well differ functionally from other CagA proteins.
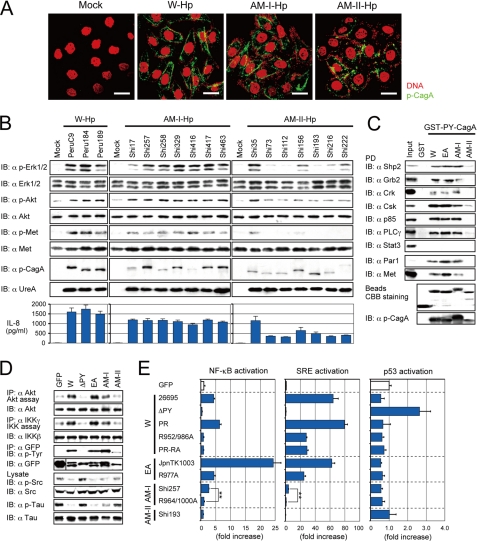
To begin functional testing of AM CagA proteins, we examined CagA tyrosine phosphorylation during Hp infection of AGS cells. CagA from representative W, EA, AM-I, and AM-II strains each underwent such phosphorylation (Fig. 2A and supplemental Fig. S3A). However, AM-II strains were less effective than W and EA strains in inducing subsequent phosphorylation of the host Erk/MAPK, Akt, and Met and production of IL-8 and cell scattering. AM-I strains, although more potent than AM-II strains, were also less active than W and EA strains in these activities (Fig. 2B, supplemental Fig. S3, B and C), each of which is involved in Hp pathophysiology and associated diseases (15).
FIGURE 2.
Amerindian CagA has a reduced ability to induce host responses. A, shown is immunostaining of AGS cells infected for 6 h with the indicated Hp strains (W, 26695; AM-I, Shi257; AM-II, Shi193). Bar = 20 μm. B, immunoblotting (IB) is shown of lysates 4 h post-infection (upper) and ELISAs for IL-8 in the culture supernatants 8 h post-infection (lower). C, shown is immunoblotting of lysates (Input) and pulled-down proteins (PD) by the indicated GST-fused PY regions of CagA (W, 26695; EA, JpnTK1003; AM-I, Shi257; AM-II, Shi193). Coomassie brilliant blue (CBB) stains of the purified GST-proteins (lower) are shown. D, shown are kinase assays and immunoblotting of lysates from 293T cells transfected with the indicated GFP-CagA (W, 26695; ΔPY, 26695 ΔPY; EA, JpnTK1003; AM-I, Shi257; AM-II, Shi193) plasmids and then subjected to immunoprecipitation (IP). E, shown are luciferase assays on lysates from HEK293 cells co-transfected with the indicated reporters and GFP-CagA plasmids (**, p < 0.01). 26695, R952A/R986A, JpnTK1003, R977A, and Shi257 R964A/R1000A are loss-of-function the CagA mutants of CRPIA motifs.
CagA-host cell interactions were further examined in several ways. First, GST pulldown assays indicated that PY regions of W, EA, and AM-I CagAs interacted with known CagA-target proteins more strongly than did AM-II CagA (Fig. 2C). Second, CagA-p85 and CagA-Met interactions are known to mediate phosphorylation of Akt and IKK kinases (15), and CagA-Csk and CagA-Par1 interactions are known to mediate dephosphorylation of Src and Tau kinases, respectively (17, 23). Ectopic GFP-CagA expression in 293T cells showed that AM CagA was less potent than W or EA CagA and that AM-II CagA was less potent than AM-I CagA in Akt and IKK kinase activation and Src and Tau down-regulation (Fig. 2D).
Subcellular localization studies showed that AM-II CagA was distributed diffusely and not strictly localized to the plasma membrane, as were other CagA proteins during Hp infection (Fig. 2A) or plasmid transfection (supplemental Fig. S4A). A W CagA derivative containing internal deletions matching those of AM-II CagA N-terminal regions (called ΔN) was also distributed diffusely (supplemental Fig. S5A). These deletions resulted in attenuation of CagA-dependent NF-κB and SRE activation and cell scattering activities (supplemental Fig. S5B), which reflects the importance of the CagA N terminus for plasma membrane localization, PY region-mediated intracellular target activation, and general modulation of CagA activities (32–34).
Several CagA chimaeras were constructed and studied to further test these ideas. The PY region of W CagA was replaced by that of EA, AM-I, or AM-II CagA (chimaeras called W/EA, W/AM-I, and W/AM-II) (supplemental Fig. S5D). We found that, although these chimeric CagAs localized to plasma membrane microdomains (supplemental Fig. S5C), W/AM-I and W/AM-II CagAs activated NF-κB less effectively than did their W parent or W/EA CagA (supplemental Fig. S5D). The CRPIA domain promotes cell proliferation and inflammation independent of CagA phosphorylation. The CRPIA domain fifth arginine residue is well conserved in W, EA, and AM-I CagA but not AM-II CagA (Fig. 1A and supplemental Fig. S1) (15). We generated W, EA, and AM-I CagAs in which this arginine replaced by alanine (W, R952A/R986A; EA, R977A; AM-I, R964A/R1000A). We found these replacement derivatives to be much less able than the corresponding wild-type CagA proteins to activate NF-κB, SRE, T-cell factor/β-catenin, and activator protein-1 (Fig. 2E and supplemental Fig. S4B). Together these five sets of results indicate that the AM-II CagA protein N-terminal deletion and variant CRPIA sequences each contribute importantly to low potency during Hp infection.
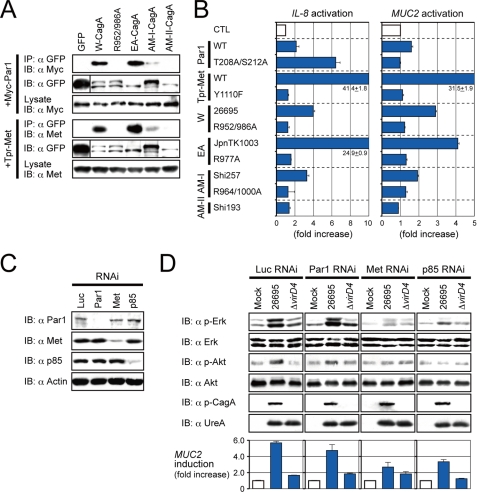
The CRPIA motifs of W and EA CagA are known to interact with Par1 and Met (15, 23). Consistent with the data in Fig. 2C, AM-II CagA interacted less with these proteins (Fig. 3A) and stimulated less Met-dependent expression of IL-8 and MUC2 than did AM-I, W, or EA CagA proteins (Fig. 3B). Mucin 2 (MUC2) is a major marker of gastric intestinal metaplasia that itself is a preneoplastic condition closely linked to chronic Hp infection (35, 36). In support of these results, knockdown of endogenous Met or p85 expression (Fig. 3C) diminished the ability of Hp to activate Erk and Akt and to stimulate MUC2 transcription (Fig. 3D). We thus interpret that AM CRPIA motifs cause decreased CagA-Met interactions and epithelial cell responses to Hp infection.
FIGURE 3.
The CRPIA motif is important for CagA-mediated MUC2 expression. A, immunoblotting (IB) is shown of lysates from 293T cells co-transfected with the indicated GFP-CagA (W, 26695; R952/986A, 26695 R952A/R986A; EA, JpnTK1003; AM-I, Shi257; AM-II, Shi193) and Myc-Par1 or Tpr-Met expression plasmids and then subjected to immunoprecipitation (IP). B, shown are luciferase assays on lysates from AGS cells co-transfected with the indicated reporters (IL-8 or MUC2 promoter) and the indicated GFP-CagA, Myc-Par1 (wild type or kinase-dead T208A/S212A), or Tpr-Met (wild type or kinase-dead Y1110F) plasmids. C and D, AGS cells were transfected for 48 h with the indicated siRNAs (C) and then infected for 4 h with the indicated Hp strain 26695 or its ΔvirD4 derivative (type IV secretion system-defective) (D). Immunoblotting of lysates (C and D, upper) and RT-PCR assays for MUC2 mRNA expression using total RNAs (D, lower) is shown.
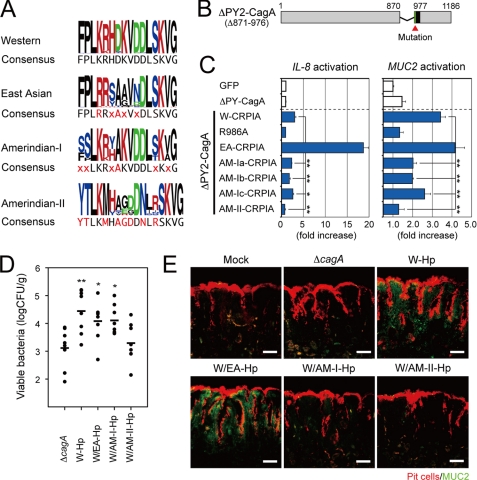
Comparison of several W CagA derivatives showed that the strength of NF-κB and SRE activation varied among CagA proteins and that, although W CagA could contain multiple CRPIA motifs, the strength of activation was not strictly proportional to CRPIA copy number (supplemental Fig. S6, A and B). Given sequence differences among CRPIA motifs of a given type superimposed on consensus motif differences among W, EA, and AM-I and AM-II CagAs (Fig. 4A), we hypothesized that these differences in potency stem from differences in CRPIA sequence per se, overall PY context, or both.
FIGURE 4.
The CRPIA motif in Amerindian CagA is associated with attenuated Hp pathogenesis. A, sequence logos and consensus sequences of the indicated CRPIA motifs of CagA are shown. B, schema of ΔPY2 CagA derivative of Western strain 26695 and sites of point mutation changes are shown. C, luciferase assays of lysates from AGS cells co-transfected with the indicated reporters (IL-8 or MUC2 promoter) and GFP-ΔPY2-CagA plasmids (*, p < 0.05; **, p < 0.01) are shown. Sequences of W, R986A, AM-Ia, AM-Ib, AM-Ic, and AM-II CRPIA constructs are shown in supplemental Figs. S6–S8. D, viable counts of bacteria in the indicated ATCC43504 Hp-infected gerbil stomachs 8 weeks after infection (*, p < 0.05; **, p < 0.01). E, immunostaining of gastric antral sections from infected gerbils. Bar = 50 μm.
To test this we constructed derivatives of ΔPY2 CagA (Δ871–976) that contain just one W CRPIA motif (Fig. 4B and supplemental Fig. S6C) (15) and in which individual residues were changed, often to those of EA CRPIA (supplemental Figs. S6D and S7A). In parallel, we also made EA CagA derivatives in which individual CRPIA residues were replaced by residues that are well conserved in W motifs (Fig. supplemental Fig. S7, B and C). Most single substitutions had only subtle effects on the strength of NF-κB activation (supplemental Fig. S7, A and C). Stronger effects were obtained by multiple replacements that changed a W CRPIA to an EA CRPIA motif and vice versa (supplemental Fig. S7, A and C); an ≤2-fold decrease in NF-κB and SRE activation if individual ΔPY2 CRPIA residues were replaced with the corresponding AM-I residues or replacement of W by AM-I motifs (supplemental Fig. S8A) but an ∼5-fold decrease if the W motif was replaced by an AM-II CRPIA motif (supplemental Fig. S8B). These chimaeras also revealed differences between W, EA, and AM CRPIA motifs in strengths of induction of IL-8 and MUC2 expression, with AM-II being the weakest of all CRPIA motifs tested (Fig. 4C).
Experimental animal infections were used to test if particular CagA types could affect Hp fitness. First, Hp infection studies confirmed that chimeric CagA proteins could be delivered from our engineered Hp strains (containing plasmid-borne TEM-1 β-lactamase tagged, chimeric cagA genes (W, W/EA, W/AM-I, and W/AM-II)) into AGS cells (supplemental Fig. S9). In addition, in gerbil infections, strain ATCC43504 derivatives with W, W/EA, and W/AM-I cagA genes achieved gastric bacterial titers that were similar to one another and, significantly, ∼10-fold higher than those of isogenic strains with ΔcagA and W/AM-II alleles (Fig. 4D). This implies that W, EA, and AM-I alleles contribute to fitness, whereas an AM-II allele may not. In addition, W and W/EA alleles caused strong gastric epithelial induction of MUC2 expression, whereas W/AM-I caused intermediate, and W/AM-II and ΔcagA alleles caused negligible levels of induction (Fig. 4E). Thus these experiments indicate that CRPIA motifs of AM, especially AM-II, strains are relatively impotent and further implicate CRPIA as a key determinant of gastric mucosal changes after Hp infection, which affects Hp fitness and disease risk.
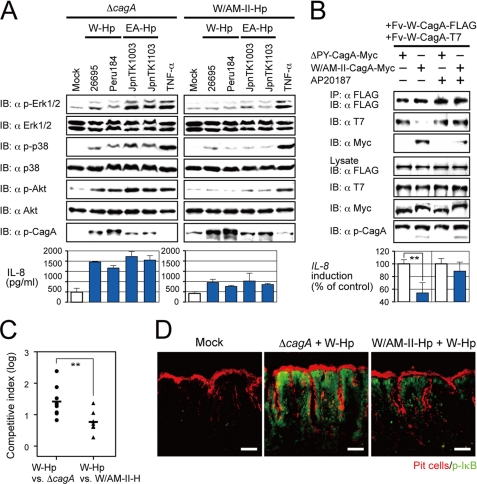
Mixed infection experiments provided important additional insight into these issues. W/AM-II Hp infection of AGS cells strongly suppressed the ability of superinfecting W or EA strains to cause Erk, p38, and Akt phosphorylation or IL-8 production, whereas initial infection with an isogenic ΔcagA strain did not (Fig. 5A). Furthermore, coexpression of W/AM-II and W CagA caused less induction of IL-8 transcription than did ΔPY and W coexpression (Fig. 5B). Because intracellular CagA forms multimers (22, 23), these results suggested interference of W CagA function in mixed complexes that also contained W/AM-II protein.
FIGURE 5.
Amerindian CagA competitively inhibits Western and East Asian CagA activities. A, shown is immunoblotting (IB) of lysates from AGS cells infected for 2 h with ΔcagA or W/AM-II cagA containing derivatives of strain ATCC43504 along with W (26695, Peru184) or EA (JpnTK1003, JpnTK1103) Hp or treated with TNF-α (upper) and ELISAs for IL-8 in the culture supernatants (lower). B, immunostaining (upper) and luciferase assays (lower) are shown on lysates from AGS cells co-transfected with the indicated CagA (ΔPY-CagA-Myc or W/AM-II-CagA-Myc), Fv-W-CagA-FLAG, and Fv-W-CagA-T7 plasmids with or without the dimerizer AP20187 (**, p < 0.01). IP, immunoprecipitate. C, shown is a competitive index of viable counts of bacteria from the stomachs of gerbils co-infected for 8 weeks with the indicated ATCC43504 strains (wild type and ΔcagA or wild type and ΔcagA complemented with W/AM-II cagA) (**, p < 0.01). D, immunostaining for IκB phosphorylation in gastric antral sections from infected gerbils is shown. Bar = 50 μm.
Indeed, cotransfection studies showed that W/AM-II CagA could form heteromultimers with W CagA and inhibit its activity in 293T cells (Fig. 5B). In support, the inhibitory effect of W/AM-II was avoided when W CagA was forcibly premultimerized using AP20187, a small-molecule protein dimerizer (Fig. 5B). In a further illustration of AM-II CagA poisoning in mixed CagA multimers with implications for fitness, we found that W/AM-II CagA-expressing strains interfered with the ability of coinfecting W strains to achieve high titers in gerbil stomachs and to fully induce IKK-mediated IκB phosphorylation (Fig. 5, C and D).
DISCUSSION
Our study of geographically and genetically distinct types of cagA genes established that the CagA protein CRPIA motif is a key determinant of Hp virulence and fitness. We found that AM type CagAs, which are characteristic of Hp strains from Amerindians in the remote Peruvian Amazon village of Shimaa, were significantly attenuated in abilities to stimulate gastric epithelial proliferation and inflammation during infection.
A recent study indicated that such AM strains descend from Asian strains that arrived in the Americas with the ancestral Asian people some 15,000–20,000 years ago and that AM strains are less fit than and substantially displaced by hybrid or W strains in less isolated communities (27). Our in vitro and in vivo experiments indicated AM CagA proteins are less potent and contribute less to virulence and fitness than do W and EA CagA proteins. We, therefore, propose that AM versus W differences in CagA proteins contributed importantly to the apparent displacement of AM by W Hp strains in urban communities.
Intriguingly, residents of Satipo-region villages from elsewhere in the Peruvian Amazon watershed contained both AM and W strains, in contrast to only AM strains in Shimaa residents (Fig. 1A and supplemental Fig. S1). This might reflect more contact with people from outside, e.g. during guerrilla wars of the late 20th century (37, 38). Continued studies of the genetics, virulence, fitness, and associated diseases of Hp from Amazonian and other Latin American populations promises to provide further insights into mechanisms of infection, persistence, and disease, especially in developing country at-risk human populations.
A mutational analysis of CRPIA motifs indicated that they could be key determinants of geographical differences in CagA-mediated host carcinogenic responses (Fig. 4C and supplemental Figs. S6–S8). Indeed, AM strains had less CagA activity than W and EA strains, with AM-II strains having markedly less than AM-I strains. Several nominally AM-II strains (Shi30, Shi35, Shi156) contained AM-I CRPIA motifs (Fig. 1A). These natural hybrids had activities similar to AM-I CagAs and higher than those of AM-II CagAs, as scored by induced Erk, Akt, and Met phosphorylation and IL-8 production (Fig. 2B). These results emphasize the importance of the CagA CRPIA motif in the development of Hp-related gastric illnesses.
Finally, we discovered that AM-II CagA can interfere with other CagA proteins both in vitro and in vivo (Fig. 5), an outcome suggesting new therapeutic approaches. Because current therapies for Hp infection are antibiotic-based and often compromised by antimicrobial resistance and also high rates of reinfection in developing country settings (39), new therapeutic strategies are needed to control Hp infections and diminish risks of associated gastric diseases (40). We propose considering AM-II strains such as Shi193 as novel live vaccines because these strains are highly human-adapted, well attenuated, and yet protective against infection by other more highly pathogenic strains.
Supplementary Material
Acknowledgment
We thank H. Miki for Par1 expression plasmids and all the members of our laboratory for helpful discussions and suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 DK63041, R21 AI078237, R21 AI088337, T53 1007646, and D43TW006581. This work was also supported by grants for Specially Promoted Research (23000012), Scientific Research (S) (20229006), Scientific Research (B) (23390102), and Scientific Research on Priority Areas (18073003) from the Japanese Government (Ministry of Education, Culture, Sports, Science, and Technology).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S9.
- Hp
- H. pylori
- CagA
- cytotoxin-associated gene A
- CRPIA
- conserved repeats responsible for phosphorylation-independent activity
- W
- Western
- EA
- East Asian
- AM
- Amerindian
- IKK
- IκB kinase
- MUC2
- Mucin 2
- NF-κB
- nuclear factor-κB
- SRE
- serum response element
- CFU
- colony forming unit(s)
- mAb and pAb
- monoclonal and polyclonal antibodies, respectively
- PY
- phosphotyrosine.
REFERENCES
- 1. Vogelmann R., Amieva M. R. (2007) Curr. Opin. Microbiol. 10, 76–81 [DOI] [PubMed] [Google Scholar]
- 2. Suerbaum S., Michetti P. (2002) N. Engl. J. Med. 347, 1175–1186 [DOI] [PubMed] [Google Scholar]
- 3. Montecucco C., Rappuoli R. (2001) Nat. Rev. Mol. Cell Biol. 2, 457–466 [DOI] [PubMed] [Google Scholar]
- 4. Atherton J. C., Blaser M. J. (2009) J. Clin. Invest. 119, 2475–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yamaoka Y., Kato M., Asaka M. (2008) Intern. Med. 47, 1077–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suerbaum S., Josenhans C. (2007) Nat. Rev. Microbiol. 5, 441–452 [DOI] [PubMed] [Google Scholar]
- 7. Linz B., Balloux F., Moodley Y., Manica A., Liu H., Roumagnac P., Falush D., Stamer C., Prugnolle F., van der Merwe S. W., Yamaoka Y., Graham D. Y., Perez-Trallero E., Wadstrom T., Suerbaum S., Achtman M. (2007) Nature 445, 915–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Higashi H., Tsutsumi R., Fujita A., Yamazaki S., Asaka M., Azuma T., Hatakeyama M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Olbermann P., Josenhans C., Moodley Y., Uhr M., Stamer C., Vauterin M., Suerbaum S., Achtman M., Linz B. (2010) PLoS Genet. 6, e1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ogura M., Perez J. C., Mittl P. R., Lee H. K., Dailide G., Tan S., Ito Y., Secka O., Dailidiene D., Putty K., Berg D. E., Kalia A. (2007) PLoS Comput. Biol. 3, e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Backert S., Tegtmeyer N., Selbach M. (2010) Helicobacter 15, 163–176 [DOI] [PubMed] [Google Scholar]
- 12. Fischer W., Prassl S., Haas R. (2009) Curr. Top. Microbiol. Immunol. 337, 129–171 [DOI] [PubMed] [Google Scholar]
- 13. Mimuro H., Berg D. E., Sasakawa C. (2008) Bioessays. 30, 515–520 [DOI] [PubMed] [Google Scholar]
- 14. Hatakeyama M. (2008) Curr. Opin. Microbiol. 11, 30–37 [DOI] [PubMed] [Google Scholar]
- 15. Suzuki M., Mimuro H., Kiga K., Fukumatsu M., Ishijima N., Morikawa H., Nagai S., Koyasu S., Gilman R. H., Kersulyte D., Berg D. E., Sasakawa C. (2009) Cell Host Microbe. 5, 23–34 [DOI] [PubMed] [Google Scholar]
- 16. Suzuki M., Mimuro H., Suzuki T., Park M., Yamamoto T., Sasakawa C. (2005) J. Exp. Med. 202, 1235–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsutsumi R., Higashi H., Higuchi M., Okada M., Hatakeyama M. (2003) J. Biol. Chem. 278, 3664–3670 [DOI] [PubMed] [Google Scholar]
- 18. Churin Y., Al-Ghoul L., Kepp O., Meyer T. F., Birchmeier W., Naumann M. (2003) J. Cell Biol. 161, 249–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mimuro H., Suzuki T., Tanaka J., Asahi M., Haas R., Sasakawa C. (2002) Mol. Cell 10, 745–755 [DOI] [PubMed] [Google Scholar]
- 20. Higashi H., Tsutsumi R., Muto S., Sugiyama T., Azuma T., Asaka M., Hatakeyama M. (2002) Science 295, 683–686 [DOI] [PubMed] [Google Scholar]
- 21. Selbach M., Paul F. E., Brandt S., Guye P., Daumke O., Backert S., Dehio C., Mann M. (2009) Cell Host Microbe 5, 397–403 [DOI] [PubMed] [Google Scholar]
- 22. Lamb A., Yang X. D., Tsang Y. H., Li J. D., Higashi H., Hatakeyama M., Peek R. M., Blanke S. R., Chen L. F. (2009) EMBO Rep. 10, 1242–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saadat I., Higashi H., Obuse C., Umeda M., Murata-Kamiya N., Saito Y., Lu H., Ohnishi N., Azuma T., Suzuki A., Ohno S., Hatakeyama M. (2007) Nature 447, 330–333 [DOI] [PubMed] [Google Scholar]
- 24. Mane S. P., Dominguez-Bello M. G., Blaser M. J., Sobral B. W., Hontecillas R., Skoneczka J., Mohapatra S. K., Crasta O. R., Evans C., Modise T., Shallom S., Shukla M., Varon C., Mégraud F., Maldonado-Contreras A. L., Williams K. P., Bassaganya-Riera J. (2010) J. Bacteriol. 192, 3078–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kersulyte D., Lee W., Subramaniam D., Anant S., Herrera P., Cabrera L., Balqui J., Barabas O., Kalia A., Gilman R. H., Berg D. E. (2009) PLoS One 4, e6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yamaoka Y., Orito E., Mizokami M., Gutierrez O., Saitou N., Kodama T., Osato M. S., Kim J. G., Ramirez F. C., Mahachai V., Graham D. Y. (2002) FEBS Lett. 517, 180–184 [DOI] [PubMed] [Google Scholar]
- 27. Kersulyte D., Kalia A., Gilman R. H., Mendez M., Herrera P., Cabrera L., Velapatiño B., Balqui J., Paredes Puente de la Vega F., Rodriguez Ulloa C. A., Cok J., Hooper C. C., Dailide G., Tamma S., Berg D. E. (2010) PLoS One 5, e15076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamaoka Y., Osato M. S., Sepulveda A. R., Gutierrez O., Figura N., Kim J. G., Kodama T., Kashima K., Graham D. Y. (2000) Epidemiol. Infect. 124, 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mimuro H., Suzuki T., Nagai S., Rieder G., Suzuki M., Nagai T., Fujita Y., Nagamatsu K., Ishijima N., Koyasu S., Haas R., Sasakawa C. (2007) Cell Host Microbe 2, 250–263 [DOI] [PubMed] [Google Scholar]
- 30. Terabayashi T., Itoh T. J., Yamaguchi H., Yoshimura Y., Funato Y., Ohno S., Miki H. (2007) J. Neurosci. 27, 13098–13107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koga K., Takaesu G., Yoshida R., Nakaya M., Kobayashi T., Kinjyo I., Yoshimura A. (2009) Immunity 30, 372–383 [DOI] [PubMed] [Google Scholar]
- 32. Murata-Kamiya N., Kikuchi K., Hayashi T., Higashi H., Hatakeyama M. (2010) Cell Host Microbe 7, 399–411 [DOI] [PubMed] [Google Scholar]
- 33. Jiménez-Soto L. F., Kutter S., Sewald X., Ertl C., Weiss E., Kapp U., Rohde M., Pirch T., Jung K., Retta S. F., Terradot L., Fischer W., Haas R. (2009) PLoS Pathog. 5, e1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pelz C., Steininger S., Weiss C., Coscia F., Vogelmann R. (2011) J. Biol. Chem. 286, 8999–9008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yuasa Y. (2003) Nat. Rev. Cancer 3, 592–600 [DOI] [PubMed] [Google Scholar]
- 36. Uemura N., Okamoto S., Yamamoto S., Matsumura N., Yamaguchi S., Yamakido M., Taniyama K., Sasaki N., Schlemper R. J. (2001) N. Engl. J. Med. 345, 784–789 [DOI] [PubMed] [Google Scholar]
- 37. Werner L. (2001) Diary of a Poll Watcher in Peru, Americas (English Ed.), Washington, D. C [Google Scholar]
- 38. Pedro García Hierro S. H., Gray A. (1998) Liberation through Land Rights in the Peruvian Amazon, International Work Group for Indigenous Affairs, Copenhagen, Denmark [Google Scholar]
- 39. Soto G., Bautista C. T., Roth D. E., Gilman R. H., Velapatiño B., Ogura M., Dailide G., Razuri M., Meza R., Katz U., Monath T. P., Berg D. E., Taylor D. N. (2003) J. Infect. Dis. 188, 1263–1275 [DOI] [PubMed] [Google Scholar]
- 40. Gerrits M. M., van Vliet A. H., Kuipers E. J., Kusters J. G. (2006) Lancet Infect Dis. 6, 699–709 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.