Abstract
NAD kinase catalyzes the phosphorylation of NAD+ to synthesize NADP+, whereas NADH kinase catalyzes conversion of NADH to NADPH. The mitochondrial protein Pos5 of Saccharomyces cerevisiae shows much higher NADH kinase than NAD kinase activity and is therefore referred to as NADH kinase. To clarify the structural determinant underlying the high NADH kinase activity of Pos5 and its selectivity for NADH over NAD+, we determined the tertiary structure of Pos5 complexed with NADH at a resolution of 2.0 Å. Detailed analysis, including a comparison of the tertiary structure of Pos5 with the structures of human and bacterial NAD kinases, revealed that Arg-293 of Pos5, corresponding to His-351 of human NAD kinase, confers a positive charge on the surface of NADH-binding site, whereas the corresponding His residue does not. Accordingly, conversion of the Arg-293 into a His residue reduced the ratio of NADH kinase activity to NAD kinase activity from 8.6 to 2.1. Conversely, simultaneous changes of Ala-330 and His-351 of human NAD kinase into Ser and Arg residues significantly increased the ratio of NADH kinase activity to NAD kinase activity from 0.043 to 1.39; human Ala-330 corresponds to Pos5 Ser-272, which interacts with the side chain of Arg-293. Arg-293 and Ser-272 were highly conserved in Pos5 homologs (putative NADH kinases), but not in putative NAD kinases. Thus, Arg-293 of Pos5 is a major determinant of NADH selectivity. Moreover, Ser-272 appears to assist Arg-293 in achieving the appropriate conformation.
Keywords: Crystal Structure, Mitochondria, NAD, NADH, Yeast, Mycobacterium tuberculosis, NAD Kinase, NADH Kinase, Human
Introduction
NAD kinase (NADK; EC 2.7.1.23)3 catalyzes the phosphorylation of NAD+, yielding NADP+; NADK is a vital enzyme required for the regulation of cellular concentrations of NAD+ and NADP+ (1, 2). All organisms contain at least one NADK ortholog gene in their genome, whereas fungi and plants generally have several (1). In the genome of fungus Saccharomyces cerevisiae, three NADK ortholog genes (UTR1, YEF1, and POS5) are found. Yef1 and cytosolic Utr1 are NADKs with high NADK activity, but low NADH kinase (NADHK, i.e. NADH phosphorylating) activity; however, the mitochondrial enzyme Pos5 exhibits a higher NADHK activity (3–6). In the genome of the plant Arabidopsis thaliana, three NADK ortholog genes (NADK1, NADK2, and NADK3) exist (7, 8); NADK3 also exhibits higher NADHK activity than NADK activity (8). Thus, Pos5 and NADK3 have been referred to as NADHKs (6, 8). In contrast to these eukaryotic NADHKs, NADKs from Gram-negative bacteria Escherichia coli, Sphingomonas sp. A1, and Salmonella enterica exhibit no NADHK activity (3, 6, 9, 10).
Phylogenetic tree analysis indicates that fungal Pos5 homologs are distinguishable from other NADKs, including Utr1, Yef1, and human NADK, and also from NADK3 (supplemental Fig. S1) (6). Almost all of fungal Pos5 homologs have a mitochondria-targeting sequence, implying that fungal Pos5 homologs encode mitochondrial NADHKs (6). Among the fungal Pos5 homologs, only the NADHK activity of Pos5 has been experimentally demonstrated (6, 11).
The ability of an enzyme to express NADK or NADHK activity has a significant impact on the intracellular balance of NAD(H) and NADP(H). Previously, we compared the primary structures of NADKs with low NADHK activity, such as Mycobacterium tuberculosis NADK (Ppnk), with those of NADKs that have no NADHK activity, such as E. coli NADK (YfjB) (3). On the basis of the tertiary structure of Ppnk complexed with NAD+ (Ppnk-NAD+), we have demonstrated that Arg-170 of YfjB is one structural determinant that confers a strict specificity for NAD+ alone. We succeeded in relaxing this specificity by changing the Arg-170 into Gly, Thr, Gln, or His (3). However, the mechanism by which an NADHK such as Pos5 exhibits higher NADHK activity than NADK activity and the structural basis for discrimination of NADH from NAD+ remain to be elucidated.
To address these issues, we have determined the tertiary structure of Pos5 complexed with NADH (Pos5-NADH). Comparison of the structure of Pos5-NADH with that of Ppnk-NAD+ or the recently released apo-form of human NADK (Protein Data Bank code 3pfn) provided a clue regarding a major structural determinant responsible for discrimination of NAD+ from NADH.
EXPERIMENTAL PROCEDURES
Plasmids
The plasmid pMK2159 carries POS5ΔMTS, encoding a Pos5 in which the N-terminal 16 residues (residues 2–17) have been replaced by an N-terminal sequence, 1M18STLDSHS24. The gene is cloned into the NdeI/BamHI sites of pET-28b (Novagen, San Diego, CA) (6). The plasmid pMK2784 carries cDNA encoding human NADK lacking its N-terminal 87 residues, cloned into the BamHI site of pQE-30 (Qiagen) (12). To alter the selected amino acid residues, site-directed mutagenesis was performed by PCR using the pMK2159 or pMK2784 as a template. The mutated plasmids were confirmed by DNA sequencing.
Expression and Purification
Pos5 was expressed in MK2162 cells (E. coli RosettaBlue(DE3) (Novagen) carrying pMK2159) as described previously (6). Briefly, MK2162 cells were grown at 37 °C in 4 liters of LB medium (1 liter/2-liter flask) containing 25 μg/ml kanamycin and 34 μg/ml chloramphenicol to an A600 of 0.6–0.7. Then isopropyl thio-β-galactoside was added to a final concentration of 0.2 mm, and the cultivation was continued at 16 °C for 72 h. The cells were collected, suspended in lysis buffer (11), and disrupted by sonication. After centrifugation, the clear supernatant was used as a cell extract, from which Pos5 was purified by using nickel-nitrilotriacetic acid-agarose (1.8 × 9 cm; Qiagen) as described previously (6). The purified Pos5 was concentrated by ultrafiltration using an Amicon model 8200 (Amicon, Lexington, MA) equipped with an Ultrafiltration Membrane (nominal molecular weight limit: 10,000) (Millipore, Danvers, MA). The concentrated solution was applied to a HiLoadTM16/60 SuperdexTM 200-pg column (1.6 × 60 cm; GE Healthcare) equilibrated with 50 mm Tris-HCl, pH 7.5, containing 200 mm NaCl, 2.0 mm 2-mercaptoethanol, and 250 mm imidazole. Pos5 was eluted with the same buffer. Fractions containing Pos5 were combined to a total volume of 12 ml and dialyzed overnight against 50 mm Tris-HCl, pH 7.5. After dialysis, Pos5 was concentrated to 11.5 mg/ml by ultrafiltration in a Centriprep tube (molecular weight cut-off, 10,000; Millipore). The concentration of Pos5 was determined using A280. A solution containing purified Pos5 with A280 of 1.0 was calculated to contain 1.7 mg/ml from the molar extinction coefficient (27,180) and the calculated molecular mass (46,284.6 Da) of Pos5 using ExPASy. The final yield of Pos5 was 26 mg. Mutated Pos5 was expressed in E. coli RosettaBlue(DE3) and purified as described above. Human NADK and mutated human NADK were expressed in E. coli Rosetta-gami(DE3)pLysS (Novagen) and purified as described (12).
NADK activity was assayed in a 1.0-ml reaction mixture containing 2.0 mm NAD+, 10 mm ATP, 10 mm MgCl2, 5.0 mm glucose 6-phosphate, 0.5 unit glucose 6-phosphate dehydrogenase, and 200 mm Tris-HCl, pH 8.0 (6). NADHK activity was assayed as described (6) in a 1.0-ml reaction mixture containing 2.0 mm NADH, 5.0 mm ATP, 5.0 mm MgCl2, and 100 mm Tris-HCl, pH 8.0.
Crystallization
Crystals of Pos5-NADH were prepared using the sitting drop vapor diffusion method in a 24-well VDX plate (Hampton Research, Aliso Viejo, CA) at 20 °C. 10 μl of protein solution (10.4 mg/ml Pos5 and 5.0 mm NADH in 50 mm Tris-HCl, pH 7.5) was mixed with 10 μl of the reservoir solution (15% (v/v) 2-methyl 2,4-pentanediol, 5% (w/v) polyethylene glycol 4000 and 100 mm imidazole-HCl, pH 8.0) to form the drop. In 2 weeks, the crystals grew to a size larger than 0.4 mm.
Data Collection
Crystals of Pos5-NADH on a nylon loop (Hampton Research) were placed directly in a cold nitrogen-gas stream at −173 °C, and x-ray diffraction images of the crystals were collected at −173 °C under the nitrogen-gas stream using a Jupiter 210 charge-coupled device detector and synchrotron radiation of wavelength 1.0 Å at the BL-38B1 station of SPring-8 (Hyogo, Japan). 240 diffraction images, with 1.0° oscillation, were collected as a series of consecutive data sets. Diffraction data were processed using the HKL2000 program package (13).
Structure Determination and Refinement
The crystal structure of Pos5-NADH was solved by molecular replacement using Molrep (14) from the CCP4 package (15); the structures of five NADKs (Protein Data Bank codes 2i1w, 1y3h, 1z0z, 1yt5, and 2an1) were used as reference models. Coot (16) was used for manual modification of the initial model. Initial rigid body refinement, and several rounds of restrained refinement against the data set were performed using Refmac5 (17). Water molecules were incorporated where the difference in density exceeded 3.0 σ above the mean, and the Fo − Fc omit map showed a density of >3.0 σ. An NADH molecule was also incorporated. The final model was refined at 2.00 Å resolution. Refinement was performed using 50.0–2.00 Å resolution data and 2.00–1.93 Å data were truncated to get R-factor below 20. Figures for the protein structure were prepared using PyMOL (18). Electric charge on the molecular surface of Pos5 or NADKs was calculated using Adaptive Poisson-Boltzmann Solver (19).
Sequence and Structure Homology
The structure of Pos5-NADH was compared with other protein structures using DALI (20). The primary structure of Pos5 was compared with those in the database using Blastp (21) or the ClustalW (22). Mitochondrial signal sequence was predicted using TargetP (23).
Phylogenetic tree and multiple alignment were constructed as follows. Among 1,533 proteins detected by Blastp search using the primary structure of Pos5 as a query, NADK3 was detected as the 1,483th most similar protein. Other plant NADK3 homologs showed weaker similarity with Pos5 than NADK3. Using the top 100 proteins homologous with Pos5 and also the NADK3 homologs including NADK3 (8 proteins), we constructed a phylogenetic tree using N-J tree with branched length on the Kyoto Encyclopedia of Genes and Genomes website. We also constructed a multiple alignment of the top 100 proteins using ClustalW (22). Primary structures of NADK3, NADK (MJ0917) from Methanococcus jannaschii, Ppnk, YfjB, and NADK from Sphingomonas sp. A1 were also aligned based on the results of Blastp.
Protein Data Bank Accession Number
Coordinates have been deposited in the Protein Data Bank as accession code 3AFO.
RESULTS AND DISCUSSION
Structure Determination of Pos5-NADH
Pos5 was crystallized in the presence of NADH, and crystals of Pos5-NADH were obtained in 2 weeks at 20 °C. The Pos5-NADH crystal belonged to group P21212. The tertiary structure of Pos5-NADH was determined at 2.00 Å resolution; this was the first structure solution for an NADHK. The data collection statistics are summarized in Table 1. The final R-factor was 19.7%, and Rfree was 24.4%. The refined model of Pos5-NADH consisted of 701 residues, two NADH, and 478 water molecules per two subunits of Pos5-NADH (Pos5-NADH-A/B) in an asymmetric unit. The DNA sequence of the POS5ΔMTS encodes a polypeptide of 398 residues, that lacks the N-terminal 16 residues (residues 2–17) of the wild-type protein; the N terminus of a polypeptide encoded on POS5ΔMTS is 1M18STLDSHS24 (6). The N-terminal sequence of the purified Pos5 was determined to be 27LQSGSK32 by N-terminal amino acid sequence analysis, indicating that the His tag and N-terminal 10 residues (1M18STLDSHSLK26) were absent from purified Pos5. Taken together, electron density was present for all but residues 53–56, 358–375, and 409–414 of Pos5-NADH-A and all but residues 27–33, 315–322, 350–375, and 409–414 of Pos5-NADH-B.
TABLE 1.
Data collection and structure refinement statistics
| Data collection | |
|---|---|
| Wavelength (Å) | 1.000 |
| Resolution range (Å) | 50−1.93(2.00−1.93) |
| Space group | P21212 |
| Cell dimension (Å) | a = 98.7, b = 132.8, c = 59.6 |
| Molecules in asymmetric unit | 2 |
| Unique reflections | 59,896 |
| Completeness (%) | 99.5 (99.0) |
| Rmerge (%) | 6.4 (39.9) |
| I/σ | 16.8 (5.9) |
| Redundancy | 8.0 (7.0) |
| Refinement | |
| Determination method | Molecular replacement |
| Resolution range (Å) | 50.0–2.00 (2.05–2.00) |
| Number of reflections | 50,821 (3,709) |
| R-factor/Rfree (%) | 19.7/24.4 (22.9/30.4) |
| Average B-factor (Å2) | |
| Protein | 30.3 |
| NADH | 24.2 |
| Root mean square deviations | |
| Bond (Å) | 0.009 |
| Angle (°) | 1.187 |
| Ramachandran plot | |
| Most favored regions (%) | 91.5 |
| Additional allowed regions (%) | 8.2 |
| Generously allowed regions (%) | 0.3 |
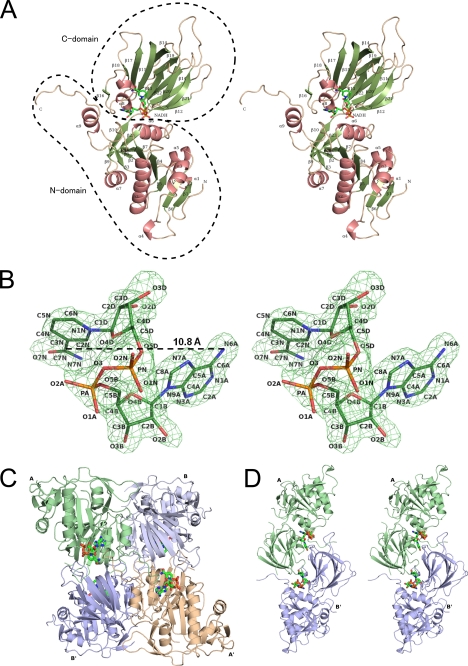
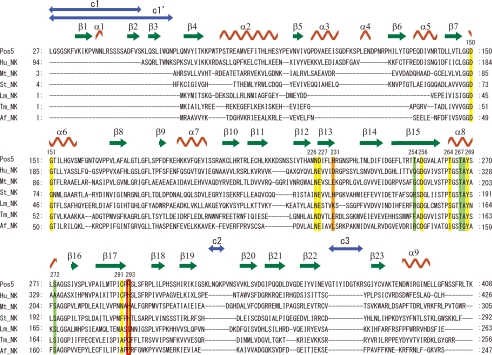
The overall structure of Pos5-NADH consists of an N-domain (residues 1–204 and 376–414) and a C-domain (residues 205–375). The overall structure of Pos5-NADH is similar to that of Ppnk (24) (Fig. 1A). Secondary structural elements of Pos5, corresponding to Fig. 1A, are shown on the primary structure of Pos5 (Fig. 2). Electron density corresponding to NADH was also observed (Fig. 1B). As with NAD+ in other solved NADK structures, NADH in Pos5-NADH displayed a twisted or bent conformation, such that the distance between the adenine six-amino group and the nicotinamide amide carbon is 10.8 Å (Fig. 1B and supplemental Fig. S2A) (24–27).
FIGURE 1.
A, stereo view of a ribbon model of Pos5-NADH. The α-helices and β-strands are colored in salmon pink and green, respectively. These features are numbered from their N termini and are also shown in Fig. 2. NADH is denoted in green (nitrogen, blue; oxygen, red; phosphorus, orange). B, the stereo view of the electron density of NADH bound to Pos5. The electron density is a Fo − Fc omit map contoured at 3 σ. NADH is colored as in A. C, ribbon model of quaternary (tetramer) structure of Pos5-NADH. The asymmetric unit contains two subunits (Pos5-NADH-A/B and Pos5-NADH-A′/B′). NADH is indicated as in A. D, stereo view of the ribbon model of the Pos5-NADH-A/B′ in Fig. 1C.
FIGURE 2.
Multiple alignment of primary structures of NADKs and Pos5 based on their tertiary structures. Alignment was conducted using Dali (20). Hu_NK, Mt_NK, Af_NK, Lm_NK, Tm_NK, and St_NK correspond to NADKs from human (Protein Data Bank code 3pfn, A chain), M. tuberculosis (Protein Data Bank code 1u0r, C chain), A. fulgidus (Protein Data Bank code 1z0s, B chain), L. monocytogenes (Protein Data Bank code 2i2b, A chain), T. maritima (Protein Data Bank code 1yt5, A chain), and S. typhimurim (Protein Data Bank code 2an1, A chain) (24–26, 28). The secondary structural features of Pos5 are labeled as in Fig. 1A. The residues interacting with NADH in Pos5 (Fig. 3A, Table 2) are denoted above the primary structure of Pos5 and shown in yellow in the aligned sequences. The aligned residues corresponding to Arg-293 of Pos5 are also boxed in red. The residues interacting with the Arg-293 of Pos5 (Table 5) are shown in green in the aligned sequences. The aligned residues corresponding to His-231 of Pos5 are indicated in orange. Residues that are not observed in their tertiary structures are not shown in this alignment: these include residues 1–26, 53–56, 358–375, and 409–414 of Pos5; residues 1–4, 41–75, and 307 of Mt_NK; residues 110–114 of Lm_NK; residues 257–258 of Tm_NK; and residues 1–3, 14–20, and 50 of St_NK. The three additional Pos5-specific structures (c1, c2, and c3) comprising residues 27–52, 316–320, and 350–377, and the c1′ structure, comprising residues 27–65 and overlapping with c1, are indicated by blue arrows.
The two subunits of Pos5-NADH (Pos5-NADH-A/B) in the asymmetric unit formed a dimer. The dimer contacts an adjacent dimer (Pos5-NADH-A′/B′), forming a tetramer in the quaternary structure (Fig. 1C). These four monomers create six different contacts, which can be then grouped into three types according to the size of the contact area. The most extensive interaction, as judged by the size of the contact area, is created by pairs A-B or A′-B′ and has an area of ∼2000 Å2 (Fig. 1C and supplemental Fig. S2B). Less extensive contacts are created by pairs A-B′ and B-A′, which create an NADH-binding site as described below and have an area of ∼930 Å2 (Fig. 1, C and D). The A-A′ or B-B′ contact represents the weakest interaction and account for ∼780 Å2 of the surface area (Fig. 1C and supplemental Fig. S2C).
Comparison of the Tertiary Structure of Pos5 with Those of Other NADKs
Homology analysis on Dali (20) showed that Pos5 resembled NADKs from human (Protein Data Bank code 3pfn; Z-score: 34.2, root mean square deviation: 2.4 Å, compared Cαs of 294); bacteria, M. tuberculosis (Protein Data Bank code 1u0t, 27.1, 3.0 Å, Cαs of 265), Listeria monocytogenes (Protein Data Bank code 2q5f, 26.4, 2.1 Å, Cαs of 251), Thermotoga maritima (Protein Data Bank code 1yt5, 26.1, 2.2 Å, Cαs of 253), and Salmonella typhimurim (Protein Data Bank code 2an1, 28.9, 2.6 Å, Cαs of 279), and an archaeon, Archaeoglobus fulgidus (Protein Data Bank code 1suw, 25.8, 2.1 Å, Cαs of 244) (24–26, 28).
Multiple alignment of the primary structures of Pos5 and those six NADKs was performed based on their tertiary structures, using Dali (20) (Fig. 2). The primary structure of Pos5 contains three additional Pos5-specific structures (c1, c2, and c3) comprising residues 27–52, 316–320, and 350–377; these structures are not present in other NADKs (Fig. 2). c1′, which comprises residues 27–65 and overlaps with c1, was also identified as a Pos5-specific structure when compared with bacterial and archaeal NADKs (Fig. 2). These c1′, c2, and c3 structures are respectively located in the N terminus, a loop structure in C-domain, and a linker-loop structure between the N- and C-domains (Fig. 2 and supplemental Fig. S3A). Blastp analysis using the primary structures of c1′, c2, and c3 as query sequences showed that only c1′ was homologous with N-terminal regions of Pos5 homologs from yeasts (Candida glabrata, Lachancea thermotolerans, Kluyveromyces lactis, Zygosaccharomyces rouxii, Vanderwaltozyma polyspora, and Pichia pastoris) belonging to the Saccharomycetes family. Multiple alignment of these homologous N-terminal regions using ClustalW (22) and predictions of mitochondria targeting sequences using TargetP (23) indicated that the predicted N-terminal mitochondria-targeting sequences in these proteins are followed by N-terminal regions that are homologous with c1′ (supplemental Fig. S3B). It is tempting to speculate that c1′ structure functions to guarantee the correct mitochondrial targeting of these Pos5 homologs.
NADH-binding Site in Pos5
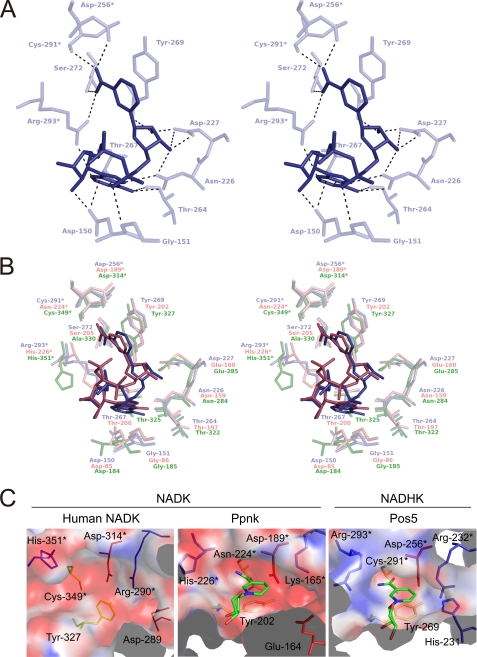
As with the NAD+-binding site in other solved NADK structures (24–27), the NADH-binding site of Pos5 was formed in a deep cleft between the N- and C-domains and in the dimer interface between the two subunits (A-B′ or A′-B) (Fig. 1, A and D). The NADH-binding site of Pos5-NADH-A and the interactions with NADH are shown in Table 2 and Fig. 3A. The Asp-256, Cys-291, and Arg-293 in Pos5 come from another subunit (B′). The amino acid residues forming the NADH-binding site of Pos5 are highly conserved (except for Cys-291 and Arg-293, as discussed below) relative to the sequences of NADKs whose tertiary structures have been solved. These NADKs include Ppnk complexed with NAD+ (Ppnk-NAD+) and human NADK (apo-form); the conservation is evident with respect to both the primary (Fig. 2) and tertiary structures (Fig. 3B). The Cys-291 of Pos5 corresponds to Cys-349 of human NADK in both primary and tertiary structures (Figs. 2 and 3B). The Arg-293 of Pos5 corresponds to His-226 of Ppnk and His-351 of human NADK (Figs. 2 and 3B). Among the NADKs shown in Fig. 2 whose tertiary structures have been solved, it should be noted that catalytic activities have been determined experimentally only for Ppnk, human NADK, and Pos5 (6, 11, 29–31). Thus, in light of the conservation of Cys-291 in Pos5 with Cys-349 in human NADK, we selected Arg-293 of Pos5 as the first candidate for a structural determinant of the high NADH kinase activity of Pos5 and its ability to discriminate NADH from NAD+.
TABLE 2.
Selected contacts between Pos5 and NADH
Selected contacts between NADH and residues of Pos5-NADH-A molecule, except for Arg-293, Asp-256, and Cys-291, are shown.
| NADH atoms | Hydrogen bond (< 3.5 Å) |
||
|---|---|---|---|
| Target atoms | Distance | ||
| Å | |||
| Nicotinamide | |||
| N7N | Ser-272 | OG | 2.55 |
| Arg-293a | NH1 | 3.49 | |
| O7N | Asp-256a | OD1 | 3.12 |
| Cys-291a | O | 3.08 | |
| Ribose (nicotinamide side) | |||
| O2D | Asp-227 | OD2 | 2.45 |
| Tyr-269 | N | 3.08 | |
| O3D | Asn-226 | OD1 | 3.09 |
| Asp-227 | OD1 | 2.71 | |
| Asp-227 | OD2 | 3.35 | |
| Pyrophosphoric acid | |||
| O1N | Gly-151 | N | 3.24 |
| Ribose (adenine side) | |||
| O2B | Asp-150 | OD2 | 2.67 |
| Adenine | |||
| N1A | Thr-267 | OG1 | 2.67 |
| N3A | Arg-293a | NH2 | 3.26 |
| N6A | Asn-226 | ND2 | 3.00 |
| Thr-264 | O | 3.04 | |
| N7A | Asn-226 | OD1 | 2.93 |
| N9A | Asp-150 | OD2 | 3.31 |
a These residues come from another subunit (Pos5-NADH-B′ molecule).
FIGURE 3.
A, stereo diagram of the binding site for NADH in Pos5-NADH-A. NADH in Pos5-NADH-A is indicated in deep blue. Asp-256*, Cys-291*, and Arg-293* come from another subunit (Pos5-NADH-B′). B, stereo diagram of the binding site for NADH in Pos5-NADH-A as in Fig. 3A, the binding site for NAD+ in Ppnk-NAD+ (salmon pink; Protein Data Bank code 1u0r, A chain), and the corresponding region of human NADK (lime green; 3pfn, A chain). The residues denoted by asterisks come from another subunit. C, electric charge on the molecular surfaces of NAD(H)-binding sites of NADKs [human NADK (Protein Data Bank code 3pfn, A chain) and Ppnk-NAD+ (Protein Data Bank code 1y3h, B chain)] and NADHK (Pos5). Positive and negative charges at pH 8.0 are colored in blue and red, respectively. NAD+ and NADH are colored as in Fig. 1A. Residues are colored in blue (His, Lys, and Arg), red (Asp and Glu), and yellow (Cys, Asn, and Tyr). The residues denoted by asterisks come from another subunit in each structure.
Conservation of Arg-293 of Pos5 in the Primary Structures of Other Proteins
To determine whether the Arg-293 of Pos5 is conserved in the primary structures of other Pos5 homologs, NADKs, and plant NADK3, we constructed a phylogenetic tree and a multiple alignment as described under “Experimental Procedures.” The phylogenetic tree indicated that NADK3 homologs are distinguishable from other proteins, which can be further divided into Pos5 homologs (41 proteins) and other eucaryotic NADK homologs including Utr1 and human NADK (supplemental Fig. S1). The NADK3 and Pos5 homologs are putative NADHKs, whereas others are putative NADKs (supplemental Fig. S1). Multiple alignment demonstrated that the Arg-293 of Pos5 is conserved in the primary structures of 39 of 41 Pos5 homologs and also in NADK3. In contrast, in 64 of 64 NADK homologs, Arg-293 of Pos5 corresponds to a His residue (supplemental Fig. S4). The Cys-291 of Pos5 is highly conserved in both Pos5 and NADK homologs (supplemental Fig. S4). Thus, we revealed that the Arg-293 of Pos5 is highly conserved among the primary structures of the Pos5 homologs, where it corresponds to a His residue that is conserved among the NADK homologs.
Arg-293 of Pos5 Is the Determinant for Discrimination of NAD+ and NADH
To develop hypotheses about the role of Arg-293, we compared the electrostatic level on the molecular surface of NADH-binding site of Pos5 (NADHK) with those of NADKs (Ppnk and human NADK). Obviously, the surface of the NADH-binding site of Pos5 (NADHK) was positively charged at pH 8.0 (Fig. 3C) and pH 9.5 (data not shown), whereas those of NADKs were negatively charged at pH 8.0 (Fig. 3C), pH 7.5 (for human NADK; data not shown), and pH 6.5 (for Ppnk; data not shown). The optimum pH for NADHK activity of Pos5 is pH 9.5, whereas the optima for NADK activity of Pos5, human NADK, and Ppnk are pH 8.0, 7.5, and 6.5, respectively (6, 29, 31). The positively charged surface of the NADH-binding site of Pos5 was attributed to Arg-293 (pKa of 12). Accordingly, artificial replacement of the Arg-293 with the His residue (pKa of 6.0) using COOT (16) resulted in a negatively charged surface (data not shown).
To further explore whether the Arg-293 is a structural determinant of Pos5 high NADH kinase activity and the ability to discriminate NADH from NAD+, we purified a Pos5 mutant, R293H, in which Arg-293 was converted to a His residue. We examined the catalytic properties of R293H and compared the mutant with wild-type Pos5. For the wild-type protein, the ratio of NADHK activity to NADK activity was 8.6 (Table 3). As expected, the NADK activity of Pos5 R293H increased by 2.4-fold and NADHK activity deceased by 40%, resulting in the decrease of the ratio of NADHK activity to NADK activity to 2.1 (Table 3). To get more detailed information, we determined kinetic values (Table 4). Catalytic activities of Pos5 were characterized by high NADHK activity, i.e. by high kcat and low Km for NADH, and also by low NADK activity, i.e. low kcat and high Km for NAD+ (Table 4). In the case of Pos5 R293H, kcat for NADH decreased by 52%, and Km for NAD+ was also lowered by 71%, as expected. These kinetic values indicated that the Arg-293 is at least one of the structural determinants of the high NADHK activity of Pos5 and for the discrimination of NADH from NAD+.
TABLE 3.
NADK and NADHK activities of purified enzymes
| NADK |
NADHK |
NADHK/NADK | |||
|---|---|---|---|---|---|
| Specific activity | Fold | Specific activity | Fold | ||
| units/mg | units/mg | ||||
| Pos5 | 0.7 | 1.0 | 6.0 | 1.0 | 8.6 |
| Pos5 R293H | 1.7 | 2.4 | 3.6 | 0.6 | 2.1 |
| Pos5 H231D | NDa | 0.4 | 0.067 | ||
| Pos5 R293H/H231D | NDa | NDa | |||
| Human NADK | 14 | 1.0 | 0.6 | 1.0 | 0.043 |
| Human NADK H351R | NDa | NDa | |||
| Human NADK H351R A330S | 0.00192 | 0.000137 | 0.00266 | 0.00443 | 1.39 |
a ND, not detected. NADK activity was not detected even after a 10-min reaction in the presence of the purified proteins (0.31 mg of Pos5 H231D, 0.42 mg of Pos5 R293H H231D, and 0.35 mg of human NADK H351R). The purified Pos5 (0.31 mg) and human NADK (0.35 mg) theoretically yield ΔA340 values of 13 and 250, respectively, after a 10-min NADK reaction. NADHK activity was not detected even after a 10-min reaction in the presence of the purified proteins (2.4 μg of Pos5 R293H H231D and 0.18 mg of human NADK H351R). Purified Pos5 (2.4 μg) yield ed ΔA340 values of 0.45 and human NADK (0.18 mg) yields theoretically 7.4 after a 10-min NADHK reaction.
TABLE 4.
Kinetic values for NAD+ and NADH of Pos5 and Pos5 R293H
|
kcata |
Km |
kcat/Km |
||||
|---|---|---|---|---|---|---|
| NAD+ | NADH | NAD+ | NADH | NAD+ | NADH | |
| s−1 | mm | |||||
| Pos5 | 7.4 ± 0.9 | 16.1 ± 2.8 | 4.5 ± 0.6 | 0.190 ± 0.04 | 1.6 | 85 |
| Pos5 R293H | 6.2 ± 1.2 | 7.7 ± 1.9 | 1.3 ± 0.3 | 0.032 ± 0.01 | 4.8 | 241 |
a Pos5 was taken as homotetramer. kcat was calculated using 185,138.4 Da as the molecular mass of the homotetrameric Pos5.
From the viewpoint of the electrostatics of the relevant molecular surfaces (Fig. 3C), Pos5 would hinder positively charged NAD+ from coming close to the NADH-binding site, which is also positively charged because of Arg-293. However, this is not the case in NAD+-binding sites of NADKs (Ppnk and human NADK), which are negatively charged. The surfaces of NAD+-binding sites of other NADKs whose tertiary structures have been solved, except for NADK of A. fulgidus, are also negatively charged (supplemental Fig. S5), although the activities of these NADKs have not been reported (25, 26, 28). In the primary structures of these negatively charged NADKs, the residues corresponding to the Arg-293 of Pos5 are His, Ile, or Gln (Fig. 2). Thus, we speculate that these NADKs are not also NADHKs. With regard to the NADK of A. fulgidus, the putative NAD(H)-binding site is positively charged because of Arg-143 and Lys-126, although the residue corresponding to Pos5 Arg-293 in the A. fulgidus protein is Phe-182 (Fig. 2 and supplemental Fig. S5). Thus, the possibility remains that the NADK of A. fulgidus also displays high NADHK activity.
The Km for NADH of Pos5 R293H was decreased by 83% (Table 4) relative to the wild-type protein. Based on the decreased Km, we speculated that Arg-293 decreases the ability of even NADH to approach the NADH-binding site and that Pos5 substituted Arg-293 for His to completely deny access to NAD+ at the sacrifice of optimal affinity for NADH.
Role of His-231 of Pos5
Although we revealed the significance of Arg-293 in Pos5, the mutant R293H still exhibits high NADHK activity and low NADK activity (Table 3), indicating that the other determinants of specificity must exist. Upon further inspection of the NADH-binding site of Pos5, we found that His-231 also contributes to the positive-charged surface of the NADH-binding site (Fig. 3C). In contrast, the corresponding positions of NADKs are acidic residues (Glu-164 of Ppnk and Asp-289 of human NADK; Fig. 3C). Notably, His-231 of Pos5 was completely conserved in all 41 Pos5 homologs, whereas the corresponding residue is Asp in 60 of the 65 NADK homologs (supplemental Fig. S4). Taken together, these observations imply that His-231 is also a structural determinant of specificity. However, both NADK and NADHK activities of the purified Pos5 mutants H231D and Pos5 R293H H231D were significantly diminished or below the detection limit (Table 3).
Increasing the Ratio of NADHK Activity to NADK Activity in Human NADK
Based on our finding that Pos5 Arg-293, which corresponds to the His-351 of human NADK, is one of the determinants that we sought, it was expected that we might confer high NADHK activity on human NADK by converting the His-351 of human NADK into Arg residue. However, purified human NADK H351R lacked both NADK and NADHK activities (Table 3).
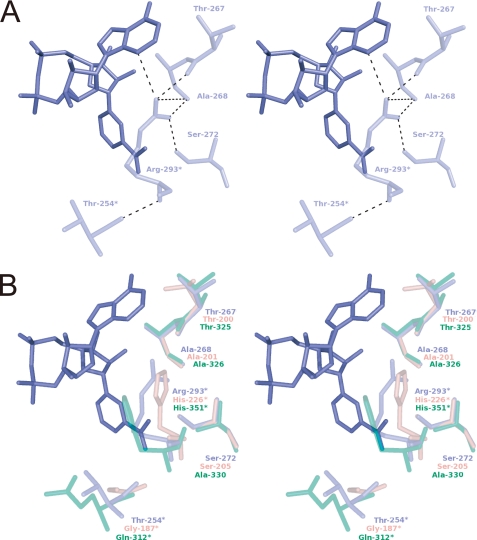
We hypothesized that the side chain of Pos5 Arg-293 requires other specific residues to attain a suitable conformation. There are four residues (Thr-254, Thr-267, Ala-268, and Ser-272) that interact with Arg-293 (Table 5 and Fig. 4A). Among these, Thr-267, Ala-268, and Ser-272 interact with the side chain of Arg-293, whereas Thr-254 contacts the N atom of main chain (Table 5 and Fig. 4A). Thr-267 also interacts with the NADH molecule (Fig. 3, A and B, and Table 2).
TABLE 5.
Selected contacts between Pos5 Arg-293 and Pos5
Selected contacts between Arg-293 of Pos5-NADH-A molecule and residues of Pos5-NADH-B′ molecule, except for Thr-254 of Pos5-NADH-A molecule.
| Hydrogen bond (<3.5 Å) |
|||
|---|---|---|---|
| Target atoms | Distance | ||
| Å | |||
| N | Thr-254 | O | 2.78 |
| NH1 | Thr-267 | O | 3.20 |
| Ala-268 | O | 2.76 | |
| Ser-272 | OG | 2.96 | |
| NH2 | Thr-267 | O | 2.82 |
FIGURE 4.
The residues interacting with Arg-293 of Pos5. A, stereo diagram of the residues that interact with Arg-293 in Pos5-NADH-A. NADH in Pos5-NADH-B′ is shown in blue. The residues in Pos5-NADH-B′ are shown. The residues denoted by asterisks come from subunit A. B, stereo diagram of the residues shown in Fig. 4A and the corresponding superimposed residues of Ppnk-NAD+ (pink, Protein Data Bank code 1u0t, A chain) and human NADK (translucent green, Protein Data Bank code 3pfn, A chain). The residues denoted by asterisks come from another subunit.
Among the residues that interact with the side chain of Arg-293, Thr-267 and Ala-268 are highly conserved in both Pos5 and NADK homologs, including Ppnk and human NADK (Fig. 4B and supplemental Fig. S4). However, Ser-272 of Pos5 is conserved in Pos5 homologs but can be either Ser (e.g. the Ser-205 of Ppnk) or Ala (e.g. the Ala-330 of human NADK) in NADK homologs (Fig. 4B and supplemental Fig. S4). Hence, we attributed the inactivation of human NADK H351R to Ala-330, which does not interact with the converted Arg-351. Accordingly, simultaneous conversions of both His-351 and Ala-330 into Arg and Ser residues resulted in recovery of measurable NADK and NADHK activities. Furthermore, the ratio of NADHK activity to NADK activity was markedly increased from 0.043 to 1.39, although the absolute activity levels were lower than wild type (Table 3). These results indicate that the side chain of the converted Arg-351 of human H351R A330S requires Ser-330 to assist Arg-351 in attaining a suitable side chain conformation. Furthermore, the data imply that in Pos5, Ser-272 has a critical role in assisting Arg-293 in attaining a suitable side chain confirmation. The significance of Ser-272 is in good agreement with the high level of conservation of this Ser residue in Pos5 homologs (supplemental Fig. S4).
Significance of the Intersubunit Contact
In NADKs, the residue that corresponds to Thr-254 of Pos5 is also critical in recognition of substrate (e.g. E. coli NADK YfjB) and conferring strict specificity for NAD+ (3). Because the residue is derived from another subunit, the intersubunit contact is important for substrate recognition in NADKs (3). In this study, we revealed the significance of Arg-293 to the high NADHK activity of Pos5 and its ability to discriminate NADH from NAD+. The Arg-293 also comes from another subunit (Fig. 3A), again demonstrating the significance of the intersubunit contact.
Supplementary Material
This work was supported in part by Grant-in-Aid for Scientific Research 21780069 from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to S. K.) and by the Program for Promotion of Basic Research Activities for Innovative Biosciences.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
The atomic coordinates and structure factors (code 3AFO) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- NADK
- NAD kinase
- NADHK
- NADH kinase
- Pos5-NADH
- Pos5 complexed with NADH
- Ppnk
- NADK of M. tuberculosis
- YfjB
- NADK of E. coli
- Ppnk-NAD+
- Ppnk complexed with NAD+.
REFERENCES
- 1. Kawai S., Murata K. (2008) Biosci. Biotechnol. Biochem. 72, 919–930 [DOI] [PubMed] [Google Scholar]
- 2. Pollak N., Dölle C., Ziegler M. (2007) Biochem. J. 402, 205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mori S., Kawai S., Shi F., Mikami B., Murata K. (2005) J. Biol. Chem. 280, 24104–24112 [DOI] [PubMed] [Google Scholar]
- 4. Shi F., Kawai S., Mori S., Kono E., Murata K. (2005) FEBS J. 272, 3337–3349 [DOI] [PubMed] [Google Scholar]
- 5. Bieganowski P., Seidle H. F., Wojcik M., Brenner C. (2006) J. Biol. Chem. 281, 22439–22445 [DOI] [PubMed] [Google Scholar]
- 6. Miyagi H., Kawai S., Murata K. (2009) J. Biol. Chem. 284, 7553–7560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turner W. L., Waller J. C., Vanderbeld B., Snedden W. A. (2004) Plant Physiol. 135, 1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Turner W. L., Waller J. C., Snedden W. A. (2005) Biochem. J. 385, 217–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grose J. H., Joss L., Velick S. F., Roth J. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7601–7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ochiai A., Mori S., Kawai S., Murata K. (2004) Protein Expr. Purif. 36, 124–130 [DOI] [PubMed] [Google Scholar]
- 11. Strand M. K., Stuart G. R., Longley M. J., Graziewicz M. A., Dominick O. C., Copeland W. C. (2003) Eukaryot. Cell 2, 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohashi K., Kawai S., Koshimizu M., Murata K. (2011) Mol. Cell. Biochem., in press [DOI] [PubMed] [Google Scholar]
- 13. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 14. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 15. Collaborative Computational Project, N. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 16. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 17. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 18. DeLano W. L. (2004) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 19. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Holm L., Rosenström P. (2010) Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000) J. Mol. Biol. 300, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 24. Mori S., Yamasaki M., Maruyama Y., Momma K., Kawai S., Hashimoto W., Mikami B., Murata K. (2005) Biochem. Biophys. Res. Commun. 327, 500–508 [DOI] [PubMed] [Google Scholar]
- 25. Liu J., Lou Y., Yokota H., Adams P. D., Kim R., Kim S. H. (2005) J. Mol. Biol. 354, 289–303 [DOI] [PubMed] [Google Scholar]
- 26. Poncet-Montange G., Assairi L., Arold S., Pochet S., Labesse G. (2007) J. Biol. Chem. 282, 33925–33934 [DOI] [PubMed] [Google Scholar]
- 27. Petrelli R., Sham Y. Y., Chen L., Felczak K., Bennett E., Wilson D., Aldrich C., Yu J. S., Cappellacci L., Franchetti P., Grifantini M., Mazzola F., Di Stefano M., Magni G., Pankiewicz K. W. (2009) Bioorg. Med. Chem. 17, 5656–5664 [DOI] [PubMed] [Google Scholar]
- 28. Oganesyan V., Huang C., Adams P. D., Jancarik J., Yokota H. A., Kim R., Kim S. H. (2005) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 640–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kawai S., Mori S., Mukai T., Suzuki S., Yamada T., Hashimoto W., Murata K. (2000) Biochem. Biophys. Res. Commun. 276, 57–63 [DOI] [PubMed] [Google Scholar]
- 30. Raffaelli N., Finaurini L., Mazzola F., Pucci L., Sorci L., Amici A., Magni G. (2004) Biochemistry 43, 7610–7617 [DOI] [PubMed] [Google Scholar]
- 31. Lerner F., Niere M., Ludwig A., Ziegler M. (2001) Biochem. Biophys. Res. Commun. 288, 69–74 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.