Abstract
Heterotrimeric G protein complexes are conserved from plants to mammals, but the complexity of each system varies. Arabidopsis thaliana contains one Gα, one Gβ (AGB1), and at least three Gγ subunits, allowing it to form three versions of the heterotrimer. This plant model is ideal for genetic studies because mammalian systems contain hundreds of unique heterotrimers. The activation of these complexes promotes interactions between both the Gα subunit and the Gβγ dimer with enzymes and scaffolds to propagate signaling to the cytoplasm. However, although effectors of Gα and Gβ are known in mammals, no Gβ effectors were previously known in plants. Toward identifying AGB1 effectors, we genetically screened for dominant mutations that suppress Gβ-null mutant (agb1-2) phenotypes. We found that overexpression of acireductone dioxygenase 1 (ARD1) suppresses the 2-day-old etiolated phenotype of agb1-2. ARD1 is homologous to prokaryotic and eukaryotic ARD proteins; one function of ARDs is to operate in the methionine salvage pathway. We show here that ARD1 is an active metalloenzyme, and AGB1 and ARD1 both control embryonic hypocotyl length by modulating cell division; they also may contribute to the production of ethylene, a product of the methionine salvage pathway. ARD1 physically interacts with AGB1, and ARD enzymatic activity is stimulated by AGB1 in vitro. The binding interface on AGB1 was deduced using a comparative evolutionary approach and tested using recombinant AGB1 mutants. A possible mechanism for AGB1 activation of ARD1 activity was tested using directed mutations in a loop near the substrate-binding site.
Keywords: Arabidopsis, Enzyme Mechanisms, Heterotrimeric G Proteins, Iron, Plant, Signal Transduction, Acireductone Dioxygenase, Effector, Ethylene
Introduction
One way that cells communicate with one another and perceive and respond to both intercellular and extracellular signals is through heterotrimeric guanine nucleotide-binding protein (G protein) signaling. As known for animal cells, G protein signaling begins when an extracellular ligand binds to a seven-transmembrane receptor that is physically coupled to an inactive heterotrimeric complex consisting of Gα-GDP, Gβ, and Gγ subunits located on the cytoplasmic side of the cell membrane. Upon activation by the receptor, the Gα exchanges GDP for GTP and dissociates from the receptor and the obligate Gβγ dimer, allowing free Gα and free Gβγ to participate in downstream signaling processes through concomitant interactions with proteins called effectors. This signaling ceases when Gα hydrolyzes GTP to GDP and the heterotrimer reforms (1, 2). Mammalian species possess a complex array of possible G protein heterotrimer combinations as there are 16 Gα, 5 Gβ, and 12 Gγ genes, making genetic studies difficult. However, the Arabidopsis thaliana genome encodes one Gα (GPA1), one Gβ (AGB1), and three Gγ (AGG1, AGG2, and AGG3) genes (3) and thus three heterotrimeric combinations (2). As a result, all G protein signaling in Arabidopsis can be abolished by mutating only a few genes, making this an ideal genetic model for G protein signaling in a multicellular context. Additionally, because there is high similarity between the G protein components of plants and metazoans, findings from Arabidopsis are applicable to signaling in humans.
Arabidopsis plants lacking AGB1 mRNA transcript (agb1-2) have a wide array of developmental phenotypes. Mature agb1-2 plants display aberrant leaf shape, silique morphology, and increased root mass (4) and are hypersensitive to infection by the necrotrophic pathogens Plectosphaerella cucumerina and Fusarium oxysporum (5, 6). Two-day-old etiolated agb1-2 seedlings have a shorter, thicker hypocotyl and a more open apical hook than wild type seedlings (4, 7). The shortened hypocotyl in agb1-2 seedlings is due to fewer cells (4, 7) and therefore a decrease in cell division. The apical hook morphology is controlled by a number of factors, including the hormone ethylene. Increased ethylene production is one factor that promotes the closure of the apical hook (8).
Although we previously identified proteins that interact genetically (SGB1 (9)) and physically (NDL1 (10)) with AGB1, to date no AGB1 effectors (i.e. proteins whose activities are regulated by AGB1) have been identified in plants. This is in stark contrast to the wide array of studies regarding mammalian Gβ signaling; in mammals, many Gβ effectors have been identified, including adenylyl cyclase 2 (AC2), phospholipase C β2 (PLC-β2), and cation channels (11). However, no protein that interacts with AGB1 in plants has been shown to play a direct role in the modulation of cell division (12). To identify potential effectors in the AGB1 signaling pathway, we utilized an activation tagging approach to randomly create dominant suppressors of agb1-2 (13). Specifically, we screened genes whose increased expression would restore the cell division phenotype displayed in the 2-day-old etiolated agb1-2 hypocotyl (9).
EXPERIMENAL PROCEDURES
Screen for Suppressors of agb1-2 and Plasmid Rescue
sgb3-1D was identified in the activation tagging suppressor screen that was described previously by Wang et al. (9).
Plant Materials
All Arabidopsis seeds were in the Columbia-0 ecotype. The following tDNA insertion mutant alleles were used: ard1-1 (SALK_119327), ard1-2 (GABI_595C04) (14), and ard1-3 (SALK_034308). tDNA insertions were confirmed by full-length genotyping PCR using primers listed in supplemental Table S1. mRNA transcript levels were identified by RT-PCR. Total RNA was isolated from rosette leaves using the RNeasyTM plant mini kit (Qiagen, Valencia, CA), and cDNA synthesis was performed using a poly(dT) primer. RT-PCR was performed with the SuperScript III RT-PCRTM kit (Invitrogen). PCR was performed for 25 cycles (94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min) using the primers listed in supplemental Table S1. agb1-2 has been described previously (4).Gene accession numbers are as follows: AGB1, At4g34460; ARD1, At4g14716; ARD2, At4g14710; ARD3, At2g24600; ARD4, At5g43850.
Hypocotyl and Hook Assays and Epidermal Cell Counting
Seeds were sterilized in 70% ethanol for 1 min and 30% bleach plus 0.01% Tween 20 for 20 min and rinsed 3–5 times with sterile distilled water. Seeds were sown on square Petri plates containing ½× Murashige and Skoog (MS) salts, 1% sucrose, and 0.6% phytagel. The plates were incubated at 4 °C in the dark for 2–4 days, exposed to light for 2 h, and grown vertically in the dark for 2 days (52 h) or 4 days at 22 °C. Upon opening, the plates were immediately scanned and imaged. Apical hook angles and hypocotyl lengths were quantified using ImageJ software. For epidermal cell counting, 52 h seedlings were incubated overnight in a solution of 8:2:1 chloral hydrate (1 mg/ml), glycerol, and water. Epidermal cells were imaged using Nomarski optics as described previously (7).
Stomatal Index
Seven-day-old seedlings were stained with 1 mg/ml propidium iodide for 15 min and rinsed briefly with distilled water before visualization under a laser confocal scanning microscope (Zeiss 710). Stomatal index (SI)2 was calculated using the following equation: SI = number of stomata/(number of stomata + number of pavement cells). SI was calculated for each cotyledon individually, and the means ± S.D. were calculated from five cotyledons from five seedlings.
Ethylene Biosynthesis
Seeds were sterilized as reported above and sown in a 0.06% agarose suspension in 22-ml gas chromatography vials containing 3 ml of 1× MS salts, 1% sucrose, and 0.6% agar. 100–200 seeds were sown for ethylene measurements at 2 days. The vials were placed at 4 °C in the dark for 2–4 days for seed stratification, exposed to light for 2 h at 22 °C, and then capped and grown in the dark at 22 °C for 2 days. Ethylene was measured by gas chromatography as described previously (15). At least three vials were measured for each genotype. The experiment was repeated once.
Statistics
Statistical analyses for the plant growth measurements were performed using a type three two-tailed Student's t test.
Phylogenies and Bioinformatics
Representative ARD sequences were collected via a BLAST analysis across the species indicated using ARD1 as a query. The alignment was generated using ClustalX (16). MrBayes (17) was run using a fixed equalin model, using the inverse γ rate, and sampling 1,000,000 generations at a frequency of 100 for three independent runs with a burn in of 250,000 generations to generate a consensus phylogenetic tree.
Modeling
A homology model of ARD1 was created using MmADI1 (PDB code 1VR3); this structure was identified by a BLAST search and chosen for its high sequence similarity. The model was created using the Insight II software program from Accelrys, Inc. (San Diego). The homology model was evaluated with the Verify three-dimensional function of Insight II, and the resulting normalized score was 0.74, indicating a viable structure.
ARD1-GFP Transient Expression in Arabidopsis Protoplasts
The coding region of ARD1 was cloned into the YFP expression vector pDH51. Arabidopsis mesophyll protoplasts were generated from wild type and agb1-2 plants as described previously (18). Cells were imaged using a Zeiss LSM 710 confocal laser scanning microscope equipped with a C-Apochromat X40 (NA 1.2) water immersion objective and standard Zeiss software (ZEN).
Yeast Three-hybrid (Y3H) Protein Interaction
AGB1 and AGG1 were cloned into the pBridgeTM vector (Clontech). ARD1 was cloned into the p-ENTR/D-TOPOTM vector (Invitrogen) and then recombined into the pACTGW-attR Gateway vector that contains an activation domain and is compatible with the pBridgeTM vector. The bait (AGB1-AGG1) and prey (ARD1) were transformed separately or together into yeast strain AH109. All three strains (bait alone, prey alone, or bait and prey) were grown on nutritionally selective media. The presence of the bait and prey was confirmed by the expression of nutritional markers (positive growth on media lacking tryptophan and leucine, respectively). Interaction was confirmed by the expression of an additional nutritional marker (positive growth on media lacking histidine). For Y3H experiments involving AGB1 mutants, the coding sequence of wild type AGB1 and of each mutant was cloned into the pAS vector, which is compatible with the ARD1-pACTGW-attr vector.
Protein Expression (ARDs and AGB1 Used for Pulldowns and Enzymatic Assays)
The coding regions of ARD1–4 were cloned into pDEST17TM and pDEST15TM (Invitrogen) containing an N-terminal poly-His tag and an N-terminal GST tag, respectively, and were expressed in BL21-RosettaTM Escherichia coli cells. Protein expression was induced at OD = 0.6 for 2 h at 37 °C. ARD-His was purified on a Talon column (Clontech). ARD-GST was purified on an immobilized glutathione column (Pierce). Protein concentrations were determined using the Coomassie PlusTM reagent (Pierce). To purify Gβγ protein from insect cells, AGB1 was cloned into pDEST8 (untagged vector), and AGG1 containing a C-terminal mutation of the CAAX (where A is any aliphatic amino acid and X is any amino acid) box prenylation site (C95*) was cloned into pDEST10TM (N-terminal His6 tag) (Invitrogen). Baculoviruses against each protein were generated and propagated using the Bac-to-BacTM baculovirus expression system (Invitrogen). Viruses were propagated separately in Sf9 cells, co-expressed in Hi-5 cells, and purified on a TalonTM column (Clontech), and concentrations were determined as described above.
AGB1 Polyclonal Antibody
A peptide consisting of 18 N-terminal residues (Thr-14 to Leu-31) of AGB1 was synthesized and conjugated to the keyhole limpet hemocyanin. This peptide carrier was used to raise an antibody in rabbits (Open Biosystems, Huntsville, AL). In total, five immunizations were administered. The terminal serum was incubated at a dilution of 1:20,000 in phosphate-buffered saline containing 5% powdered milk and 0.01% Tween 20 for 2 h to detect purified proteins. The antiserum lot used here was designated AGB1–1607-Ext.
Co-purification via Affinity Pulldown
Bacterial cells expressing a GST-tagged ARD protein (ARD1–4) were lysed (sonicated in 25 mm NaP, 300 mm NaCl, 0.2 mm PMSF, and 1× protease inhibitor mixture (Sigma)) and then incubated with glutathione beads. After washing, purified AGB1-AGG1 protein was added to the ARD bead mixture. After incubation, the slurry was washed several times, and the protein complexes were eluted from the beads by boiling in SDS loading buffer. A subset of the elution product was run on an SDS-polyacrylamide gel. The proteins were transferred to a nitrocellulose membrane and probed with the polyclonal AGB1 antibody described above.
Bimolecular Fluorescence Complementation (BiFC)
BiFC was performed according to Grigston et al. (19) with several modifications. The coding sequences of ARD1, ARD2, ARD3, ARD4, and AHP2 were cloned into the BiFC vectors pCL112_JO (YFP-n) and pCL113_JO (YFP-c). The coding sequence of AGB1 was cloned into pBatL-sYFP-C (YFP-c). Constructs were co-infiltrated with mitochondrial red fluorescent protein (mt-Rk obtained from the Arabidopsis Biological Resource Center, CD3-991) as a positive transformation control. Samples were imaged using a Zeiss LSM 710 confocal laser scanning microscope equipped with a C-Apochromat X40 (NA 1.2) water immersion objective and standard Zeiss software (ZEN). Visualization of YFP was achieved using a 514-nm argon laser line for excitation, and the photomultiplier detector was set to collect emission bandwidth at 526–569 nm. Visualization of red fluorescent protein was achieved using a 560-nm diode laser for excitation, and the photomultiplier detector was set to collect emission bandwidth at 565–621 nm.
AGB1 and ARD1 Mutagenesis
The coding sequences of AGB1 and ARD1 were mutagenized using the QuikChange Multi and QuikChange Lightning Site-directed MutagenesisTM kits, respectively (Agilent Technologies, Santa Clara, CA). Mutagenesis primers were designed using the QuikChange primer design program. Mutations were confirmed by sequencing and were expressed and purified as described above.
Inductively Coupled Plasma-Mass Spectroscopy
Measurements were performed in the Mass Spectrometry Facility of the Department of Chemistry, University of North Carolina, Chapel Hill, using a Varian 820 inductively coupled plasma-MS (Palo Alto, CA), now part of Bruker Daltonics (Billerica, MA). Protein samples were prepared by adding 1 ml of 70% HNO3 to 40 μg of protein (sample 1) and 24 μg of protein (sample 2) for ∼10 h. Each sample was then diluted with 2% HNO3 solution to a final volume of 10 ml. All solutions were made with 18 megohms of deionized water. A 5 ppb indium solution (in 2% HNO3) was used as the instrument internal standard during the runs. Isotopes 24Mg, 55Mn, 57Fe, 59Co, and 60Ni were quantitatively monitored in a peak hopping mode with a total of 100 scans per point per isotope (50,000 μs of dwell time and five replicates of 20 scans).
Carbon Monoxide Assay
The carbon monoxide (CO) assay was performed according to Sundin and Larsson (20). The formation of CO was monitored and quantified using a Hewlett-Packard 6890 gas chromatograph equipped with an Agilent G2747A nickel catalyst system with a flame ionization detector. The injector temperature was set at 60 °C, the oven temperature at 35 °C, the nickel catalyst temperature at 375 °C, and the detector temperature at 250 °C. The enzyme-catalyzed solution was placed in a gas tight sealed vial with 1 ml of CO liberating solution (7.5 g of saponin in 1 m sulfuric acid). This vial was then vortexed for 1 min and shaken for 40 min at 250 rpm at 37 °C. An aliquot of 100 μl from the headspace gas phase was injected with a gas-tight syringe into the inlet of the GC column.
Enzymatic Assay
The enzymatic assays to probe for activity of ARD1 and mutant proteins were performed according to Zhang et al. (21) with some modifications. All experiments were performed in three consecutive steps in an anaerobic cuvette. First, the substrate acireductone was built to the final concentration of about 125 μm by 75 nm E1 enzyme and in the presence of 200 μg/ml catalase. The buffer saturated with molecular oxygen (280 mm) was then added, and the rate of acireductone decay was measured at 308 nm. The average oxygen-induced decay rate was 8.5 × 10−11 ± 1.5 × 10−11 mol of substrate/s. This rate was accounted for in the calculations of the initial rates of the enzymes. Finally, a controlled amount of ARD1 variant was added, and the depletion of acireductone was monitored at 308 nm for at least 300 s. The initial rates were calculated by selecting the linear portion of the graph and calculating the linear fit in this region.
RESULTS
Overexpression of the Coding Region of At4g14716 (Formally SGB3) Suppresses the agb1-2 Etiolated Phenotype
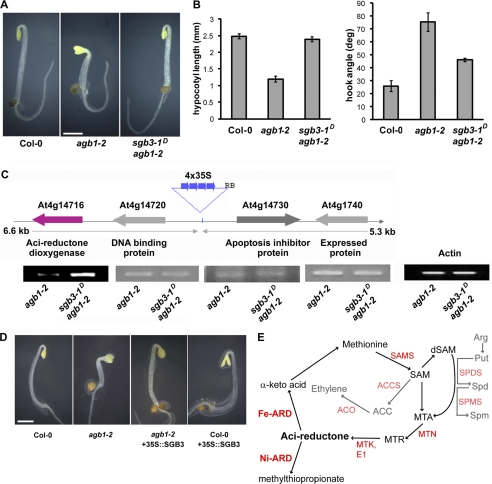
To identify dominant mutations that suppress a null (agb1-2) phenotype, agb1-2 seeds were transformed with a transcriptional enhancer element from the cauliflower mosaic virus 35S promoter that was randomly inserted into the genome. The resulting transgenic population was screened for genes whose overexpression suppressed the shortened hypocotyls and open apical hook present in 2-day-old, etiolated agb1-2 seedlings (Fig. 1, A and B) (4, 13). Eight genetic loci suppressed this phenotype to varying degrees, and these loci were named Suppressor of G Beta 1–8. The suppressor screen and one of the resulting genetic suppressors, SGB1, was described previously (9). This study focuses on SGB3, which fully rescued the hypocotyl length phenotype and partially rescued the apical hook opening phenotype. The genomic fragment containing the enhancer was isolated by plasmid rescue, and the genetic position of the enhancer was determined by sequencing. Because the 35S enhancer can stimulate expression of genes within a 10-kb range (5 kb in each direction of the enhancer insertion position) (13), the transcript levels of the four adjacent genes (within the 10-kb region surrounding the enhancer) were tested to determine which genes had higher expression levels due to the enhancer (Fig. 1C). The only gene displaying an elevated mRNA transcript was At4g14716 (Arabidopsis Genome Initiative accession). To show that increased expression of At4g14716 was responsible for the rescued agb1-2 phenotype that was observed in the enhancer population, we ectopically expressed the coding region of At4g14716 under the control of the strong 35S promoter into both agb1-2 and wild type backgrounds. As shown in Fig. 1D, overexpression and ectopic expression of the At4g14716 coding region rescued the etiolated hook and hypocotyl phenotypes observed in agb1-2 seedlings. To determine whether any other phenotypes were rescued by the ectopic overexpression of At4g14716 in agb1-2 plants, SI was measured in 7-day-old seedlings. Although the SI in agb1-2 is significantly higher than that of Col-0 plants (22), overexpression of At4g14716 in agb1-2 rescued the wild type phenotype (supplemental Fig. S1A). The overexpression of the At4g14716 coding region in the presence of AGB1 (wild type plants) did not affect the 2-day-old etiolated apical hook opening or hypocotyl length (quantified in supplemental Fig. S1B), indicating that the At4g14716 overexpression phenotype requires the loss of AGB1. These data show that AGB1 and At4g14716 interact genetically to control hypocotyl length and apical hook opening.
FIGURE 1.
sgb3-1D is a genetic suppressor of agb1-2. A, 2-day-old etiolated phenotype of wild type, agb1-2, and sgb3-1D/agb1-2 seedlings. Bar, 1 mm. B, quantification of the hypocotyl lengths and apical hook angles observed in A. Error bars, standard error. C, enhancer locus identification and confirmation of enhanced SGB3 transcript levels by RT-PCR. PCR products were run on two separate gels in nonadjacent lanes. Predicted functions of the proteins encoded by each gene are indicated below the gene name. D, recapitulation of the sgb3-1D/agb1-2 phenotype by overexpressing SGB3 in the presence and absence of AGB1. E, methionine salvage pathway as defined by experiments in bacterial and plant systems (28, 41). AdoMet is recycled into methionine via the intermediates MTA, 5-methylthioribose (MTR), and acireductone. Ni-ARD catalyzes an off-pathway reaction, although Fe-ARD catalyzes an on-pathway reaction and promotes the recycling of methionine. Characterized enzymes are indicated in red (SAMS, AdoMet synthase; MTN, MTA nucleosidase; MTK, MTA kinase; E1, E1 enolase/phosphatase). Ethylene is produced from AdoMet via 1-aminocyclopropane-1-carboxylate (ACC) by 1-aminocyclopropane-1-carboxylate synthase (ACS), and 1-aminocyclopropane-1-carboxylate oxidase (ACO). AdoMet is also decarboxylated (dSAM) and facilitates the synthesis of the polyamines spermidine (Spd, via Spd synthases, SPDS) and spermine (Spm, via Spm synthases, SPMS) from putrescin (Put).
At4g14716 Encodes ARD1, an Acireductone Dioxygenase
A BLAST analysis revealed that SGB3 encodes a gene previously annotated as ARD1, an acireductone dioxygenase-like protein. ARD1 has high similarity to previously identified eukaryotic acireductone dioxygenase proteins (supplemental Fig. S2A). ARD1 is 79% identical and 92% similar to Oryza sativa (rice) ARD1 (OsARD1, e value = 4.0 × 10−91) and 63% identical and 77% similar to Mus musculus (mouse) ARD (MmADI1, e value = 5.0 × 10−65). ARD1 is 28% identical and 49% similar to a prokaryotic ARD from the bacterium Klebsiella oxytosa (KoARD, e value = 1.0 × 10−7). There are two available ARD atomic structures, one of KoARD and one of MmADI1. Based on the higher sequence similarity between ARD1 and MmADI1, we utilized the MmADI1 crystal structure (PDB code 1VR3) (23) as a template and threaded ARD1 onto it to generate a homology model (supplemental Fig. S2B). The structure was robust (homology score = 0.74), and as shown in supplemental Fig. S2B, most of the surface and active site residues were conserved between plants and mammals (pink residues). Additionally, the three histidine residues and one glutamic acid required for metal binding in the ARD1 active site (24) were fully conserved (supplemental Fig. S2B, dark blue residues).
The enzymatic activity of ARD proteins was previously characterized in bacteria, plants, and mammals. ARD catalyzes a committed step in the methionine salvage pathway, which recycles methionine from S-adenosylmethionine (AdoMet) (Fig. 1E) (25). This pathway is important in that it provides the organism with a source of methionine under limiting conditions, regulates the production of polyamines, and in plants allows for the production of ethylene (26). KoARD proteins catalyze two distinct reactions dependent upon which divalent metal ion is bound in the active site (27, 28). Fe-bound ARD catalyzes the on-pathway reaction that converts acireductone to the keto acid α-keto-γ-methylthiobutyrate (the methionine precursor). Ni-ARD catalyzes an off-pathway reaction whose products are γ-(methylthio)propionate, carbon monoxide, and formate (29). The different ARD activities are measured in vitro by monitoring the accumulation of CO (off-pathway) or keto acid (on-pathway) (30–32).
ARD1 Function in Vivo
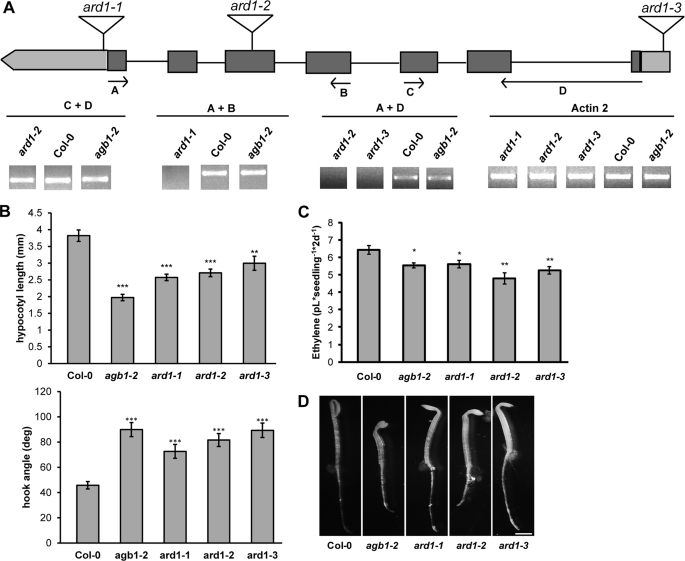
ARD1 tDNA insertion mutants were obtained to determine the loss-of-function, hypocotyl phenotype. The mRNA transcript levels were determined by RT-PCR (Fig. 2A). Two of the three ARD1 alleles, ard1-1 and ard1-3, were transcript-null. ard1-2 likely created a truncated transcript because the RT-PCR results detected the presence of a fragment upstream of the tDNA insertion site, but no full-length product was present (Fig. 2A, reactions C + D and A + D, respectively). Two-day-old etiolated seedlings lacking a functional ARD1 transcript mimicked the agb1-2 phenotype in that they had short hypocotyls, and they showed an apical hook opening that was intermediate to agb1-2 and wild type plants (Fig. 2B). We compared the estimated number of epidermal cells in the 2-day-old etiolated seedlings and found that ard1-1, like agb1-2, contained fewer cells (∼10 and ∼9 cells, respectively) than wild type plants (∼20 cells). agb1-2 plants overexpressing ARD1 (Fig. 1D, agb1-2 + 35S::SGB3) contained the wild type number of cells (data not shown); this indicates that ARD1 overexpression rescues the agb1-2 hypocotyl length phenotype by restoring cell division.
FIGURE 2.
ARD1 and AGB1 share etiolated hypocotyl phenotypes. A, location of three tDNA insertions in ARD1. Arrows indicate genotyping primers used to amplify the mRNA transcript. RT-PCR was performed using the primers indicated. Actin2 was used as a reference transcript. PCR products were run on two separate gels in nonadjacent lanes. B, hypocotyl lengths and apical hook angles of 2-day-old etiolated seedlings. *, p < 0.05; **, p < 0.005; ***, p < 0.0005. Results were an average of two independent experiments and are representative of three additional experiments performed. Error bars, means ± S.E. C, ethylene biosynthesis of wild type and ard1 or agb1 mutant plants grown in the dark for 2 days. Each measurement is an average of at least three vials, and the experiments were repeated with similar results. *, p < 0.05; **, p < 0.005. Error bars, means ± S.E. D, etiolated phenotype of 2-day-old seedlings. Bar, 1 mm.
These results prompted the hypothesis that AGB1 regulates ARD1 activity to control cell division. Regulation could occur at different levels, for example, by control of ARD1 subcellular location or by direct control of ARD1 catalytic properties. To test the former, the localization of ARD1-GFP was determined in both the wild type and agb1-2 backgrounds, and no major difference in patterns was detected using transient co-expression in protoplasts (supplemental Fig. S3A).
One of the products of the methionine salvage pathway in plants is the gaseous hormone ethylene (Fig. 1E). When etiolated seedlings are grown in the presence of ethylene, they display a set of phenotypes called the triple response, shorter hypocotyls, exaggerated closure of the apical hook, and radially expanded hypocotyls (8). The steady-state level of ethylene in agb1 and ard1 mutant seedlings was determined because of the following: 1) ARD1 may operate in the methionine salvage pathway based on its homology with known ARD proteins (supplemental Fig. S2); 2) AGB1 positively regulates ARD1 genetically, and 3) 2-day-old etiolated agb1-2 and ard1 mutants display some aspects of the triple response. ard1 null mutants display one triple response trait (short hypocotyls), leading to the prediction that ethylene levels in ard1 mutants may be elevated. Conversely, another ard1 triple response trait (open hooks) leading to the prediction that ethylene levels are lower than wild type seedlings. The steady-state level of ethylene in 2-day-old etiolated seedlings was slightly reduced in agb1-2, ard1-1, ard1-2, and ard1-3 compared with wild type (Fig. 2C). These data correlate with the 2-day-old apical hook phenotypes of these plants (Fig. 2D).
ARD1 Physically Interacts with AGB1
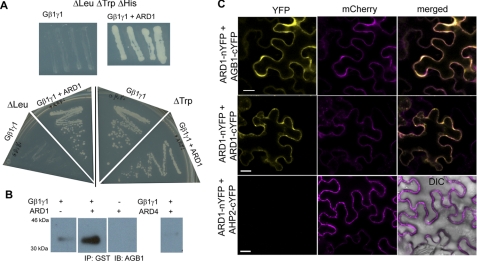
We showed that ARD1 genetically interacts with AGB1 because ARD1 overexpression rescues the agb1-2 etiolation phenotype (Fig. 1). To test for physical interaction, Y3H genetic complementation was performed. To confirm that each construct could be expressed in yeast cells, single transformants were grown on an appropriate nutritional dropout medium (Δleucine for ARD1 and Δtryptophan for AGB1) (Fig. 3A). ARD1 was expressed in AH109 yeast cells in the presence or absence of the AGB1-AGG1 heterodimer. When grown on nutritional media selecting for a positive interaction between the three proteins (Δhistidine), only yeast cells containing both AGB1-AGG1 and ARD1 were able to grow (Fig. 3A). Yeast containing AGB1-AGG1 alone or ARD1 alone failed to grow, indicating that neither construct alone was responsible for the growth observed on Δhistidine media. These data suggest that ARD1 physically interacts with the AGB1-AGG1 heterodimer.
FIGURE 3.
AGB1 and ARD1 interact physically. A, growth of yeast strain AH109 containing the proteins indicated (AGB1-AGG1 (AtGβγ) alone or AtGβγ + ARD1) on yeast dropout media missing leucine, tryptophan, and histidine. This selects for a positive interaction between each of the two genes, resulting in no growth for the strain containing AtGβγ alone and positive growth for the strains containing both AtGβγ and ARD1. Four independent colonies were cultured on the triple dropout media to confirm the interaction. Positive growth on media lacking leucine or tryptophan confirms the presence of ARD1 or AtGβγ, respectively. B, His6-tagged Gβγ was pulled down with ARD1-GST on a glutathione resin and detected by immunoblotting (IB) with anti-AGB1. Gβγ was not pulled down with ARD4-GST. Proteins were separated on one gel in nonadjacent lanes. IP, immunoprecipitation. C, bimolecular fluorescence complementation in N. benthamiana leaf epidermal cells (error bar, 20 μm). ARD1 interacts with AGB1 and homodimerizes. ARD1 does not interact with cytosolic protein AHP2 (negative control, merged panel includes differential interference contrast image to show the cell borders). Mitochondrial red fluorescent protein expression (mCherry) confirms positive transformation of all cells.
To test the validation of the Y3H results, we determined if recombinant ARD1 protein interacted with the AGB1-AGG1 heterodimer using co-purification via an affinity pulldown assay. Lysed bacterial cells expressing a GST-tagged ARD1 protein were incubated with glutathione beads, and after washing the ARD1-bead complexes, purified AGB1-AGG1 protein was added. The protein complexes were eluted from the beads and were subjected to SDS-PAGE. A polyclonal anti-AGB1 raised against an AGB1 peptide from the N-terminal helix of AGB1 was used (see “Experimental Procedures” and supplemental Fig. S3B) to detect the proteins by immunoblot analysis. As seen in Fig. 3B, AGB1 was pulled down with ARD1, corroborating the Y3H data and showing that these proteins interact physically in vitro. However, a separate GST fusion protein (ARD4-GST) did not pull down AGB1, indicating that ARD1, and not GST, was responsible for this positive interaction.
To determine whether these proteins interact physically in vivo, we utilized BiFC. Each bait and prey combination (ARD1 versus AGB1, ARD1, or AHP2) was cloned into vectors containing either the C- or N-terminal half of YFP, and all combinations were expressed in Nicotiana benthamiana leaf epidermal cells. A positive interaction was scored by the reconstitution of YFP fluorescence as visualized under scanning confocal microscopy. ARD1 was able to reconstitute the fluorescent signal with AGB1, confirming the positive interaction between the two proteins (Fig. 3C, top panel). Additionally, ARD1 was confirmed to form a homodimer or oligomer, which was shown for the rice protein OsARD1 (31). As a negative control, we showed that ARD1 does not interact with a cytokinin-pathway cytosolic protein AHP2 (Fig. 3C, bottom panel).
ARD1 Has Acireductone Dioxygenase Enzymatic Activity That Is Stimulated by AGB1
Recombinant His6-tagged ARD1 was purified to perform enzymatic assays. To determine which metal or metals were bound in the active site, ARD1 protein was analyzed using inductively coupled plasma mass spectrometry (Table 1). ARD in the on-pathway binds Fe2+ or Mg2+, whereas ARD in the off-pathway binds Ni2+, Co2+, or Mn2+ (29). Therefore, we assayed for the presence of each of the five divalent metal ions. Fe(II) constituted 80% of the metal ions in the protein sample. The remaining ions in the sample were Mg2+ (13%), Co2+ (5%), and Ni2+ (2%). Together, these data suggest that ARD1 primarily performs the on-pathway reaction and that the purified ARD1 displayed 100% metal occupancy. The equal molar concentration of Fe2+ and ARD1 indicate this metal is iron.
TABLE 1.
Quantification of metals bound to ARD1
An inductively coupled plasma-mass spectrometry analysis was utilized to detect the presence of 24Mg, 55Mn, 57Fe, 59Co, and 60Ni. Elemental concentration is expressed as parts/billion (ppb) and mole/mol.
| Sample | Elemental concentration | Metal/protein |
|---|---|---|
| ppb | mol/mol | |
| EF-01 | ||
| 24Mg | 1.56 | 0.37 |
| 55Mn | 0.00 | 0.00 |
| 57Fe | 9.34 | 0.96 |
| 59Co | 0.63 | 0.06 |
| 60Ni | 0.21 | 0.02 |
| EF-02 | ||
| 24Mg | 0.62 | 0.24 |
| 55Mn | 0.00 | 0.00 |
| 57Fe | 7.26 | 1.25 |
| 59Co | 0.44 | 0.07 |
| 60Ni | 0.08 | 0.01 |
A polyhistidine-tagged ARD1 protein was used to measure the enzymatic activity with acireductone. Because acireductone is short lived and reactive with atmospheric oxygen, the reaction was performed anaerobically, and acireductone was generated in situ immediately prior to the addition of ARD1 using a previously published protocol (21). ARD1 depleted the acireductone substrate, but no CO production was observed as detected by gas chromatography, suggesting that ARD1 does not significantly catalyze the off-pathway reaction. Although significant enzymatic activity was observed with a freshly prepared sample of ARD1 (Fig. 4, A, white squares, and B), its activity decreased over time, making it difficult to kinetically characterize the enzyme. After 7 days at 4 °C, all activity was lost; if the enzyme was stored at −20 °C, the observed rates were 50% or less than the originally recorded numbers. Because of these factors, all rate comparison assays were performed on the same day and with the same batch of ARD1 that was used as a reference.
FIGURE 4.
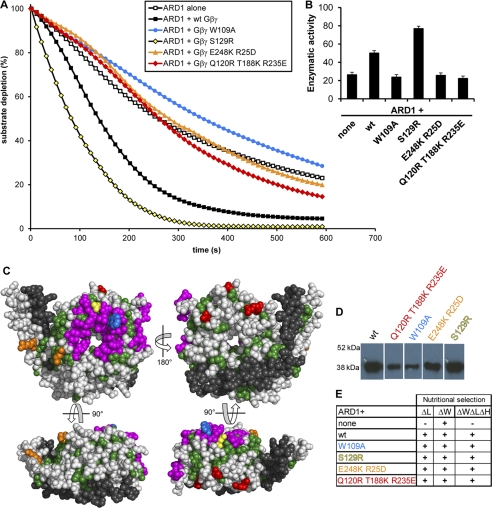
AGB1-AGG1 stimulates ARD1, and this stimulation is reduced in several AGB1 mutants. A, ARD1 enzymatic activity in the presence and absence of wild type AGB1-AGG1 (Gβγ) and various AGB1 mutants in a 1:1 molar ratio. B, enzymatic rates of ARD1 activity in the presence of each AGB1-AGG1 wild type or mutant construct indicated (none indicates ARD1 alone). The rates are expressed in moles of substrate/mol of enzyme/s ± S.E. and were recorded as initial rates. These rates account for the average oxygen-induced decay rate (base line, see “Experimental Procedures”). C, four views of the AGB1 protein surface. All colored residues compose a region strictly conserved between plant and mammalian species. Green residues are conserved, but they have no previously known function and may be required for structural integrity. Magenta residues form the Gα binding interface. Colored residues indicate point mutations created (W109A, blue; S129R, yellow; E248K/R25D, orange; Q120R, T188K, R235E, red; colors are consistent with those in A, D, and E). Gray residues are not conserved between plants and mammals. Black residues are the Gγ protein. D, AGB1 mutant proteins were co-expressed in E. coli with AGG1 and purified via the His tag on AGG1 by affinity column chromatography (immunoblot, anti-AGB1). Proteins were separated on one gel in nonadjacent lanes. E, Y3H growth of strains containing ARD1 in the presence or absence of wild type and mutant AGB1 proteins. Cells expressing the genes encoding mutant AGB1 proteins were able to grow on media lacking histidine, indicating that the mutations do not disrupt the physical interaction between AGB1 and ARD1.
Because ARD1 and AGB1 interact physically (Fig. 3), we tested whether AGB1 directly affected ARD1 enzymatic activity. We performed the same assay described above in the presence or absence of purified AGB1-AGG1 protein. In multiple experiments and independently of the ARD1 batch or the day on which the experiment was performed after its purification, AGB1-AGG1 consistently stimulated ARD1 activity about 2-fold when mixed in a 1:1 ARD1/AGB1-AGG1 molar ratio (Fig. 4, A, black squares versus open squares, and B). The error was estimated based on the maximum error calculated in similar experiments. Although these experiments were repeated several times, the instability of the enzyme and therefore the difference in initial rates from experiments performed on different days or with different preparations of enzymes made it impossible to directly compare rates from different experiments.
To determine the extent to which AGB1-AGG1 stimulates ARD1 enzymatic activity, we measured ARD1 activity in the absence or presence of increasing concentrations of AGB1-AGG1. The activity of ARD1 increased with an increasing molar ratio of AGB1-AGG1/ARD1 (supplemental Fig. S4A) and was so rapid that an initial rate could not be calculated once the molar ratio exceeded 2:1 (supplemental Fig. S4B). This level of stimulation is in the same range (3–6-fold) as the stimulation of the effector PLC-β2 that was previously reported (33).
Identification of a Potential AGB1-ARD1 Interface
To further characterize the interaction between AGB1 and ARD1, we sought to identify key residues that could form critical contacts at an interface between the two proteins. Because neither AGB1 nor ARD1 was crystallized alone or in complex, an evolutionary approach to predict binding regions between the two was necessary. For this analysis, our previously published prediction of novel Gβ interfaces based on the evolutionary history of Gβ proteins was used (33). We identified surface regions of the AGB1 structure that are invariant between plants and mammals. This region was reduced by culling residues that formed a hydrogen bond with either the polypeptide backbone or with Gγ (which is required to stabilize Gβ) because some conserved surface residues are critical for structural maintenance, although others are required for protein-protein interactions that first evolved in plants and are maintained throughout eukaryotic evolution (see “plant ancestor” of Fig. 4A of Ref. 33). Rather, residues with solvent-exposed functionality were assumed to be available for protein-protein interactions. Based on these criteria, a limited number of residues were chosen, and four sets of corresponding mutant AGB1 proteins were created as follows: the single mutations W109A and S129R, the double mutant E248K/R25D, and the triple mutant Q120R/T188K/R235E (Fig. 4C). Trp-109 and Ser-129 are both located in the conserved surface that is known from mammalian studies to bind Gα (33, 34). Additionally, Trp-109 is an interaction “hot spot” in mammalian proteins and is critical for interaction of Gβ with not only Gα but PLC-β2, AC2, and cation channels (34). The remaining two regions had no previously identified functions in mammals and thus represented key contacts of potential interaction interfaces with ARD1 (an interaction identified in plants but not yet identified in mammals). Mutant proteins were co-expressed with AGG1 (Gγ) containing a His tag for affinity purification and a mutated CAAX box for improved solubility. The AGB1-AGG1 heterodimer was purified from Hi5 insect cells by affinity column chromatography. The His-tagged AGG1 selected AGB1-AGG1 dimers (Fig. 4D) ensuring that the mutant AGB1 proteins were properly folded, as their interaction with AGG1 was necessary for purification. In addition, the mutant proteins physically interacted with ARD1 in a Y3H assay (Fig. 4E) indicating that the mutant AGB1 proteins were still able to at least weakly bind to ARD1.
The mutated AGB1-AGG1 dimers were tested for their ability to activate ARD1 in vitro. The mutant S129R fully activated ARD1 to the same or a greater level than wild type AGB1-AGG1, suggesting that Ser-129 is not critical for stimulation or that the S129R mutation confers a slight enhancement of AGB1-AGG1 stimulation of ARD1. The mutants W109A, E248K/R25D, and Q120R/T188K/R235E abolished ARD1 stimulation, suggesting that these residues form critical contacts for ARD1 stimulation (Fig. 4, A and B). Together, these data suggest that these three groups of residues are required for AGB1-AGG1 to stimulate ARD1 enzymatic activity. These results suggest that AGB1-AGG1, and not an Sf9 Gβγ contaminant that was not detected by anti-AGB1 (see supplemental Fig. S3B for lack of reactivity between uninfected Sf9 cells and anti-AGB1), was responsible for this stimulation; because wild type and mutant AGB1 constructs were purified together, the lack of stimulation in some AGB1 mutants confirms that the stimulation by the remaining constructs was not due to contamination.
ARD1 Activation Mechanism
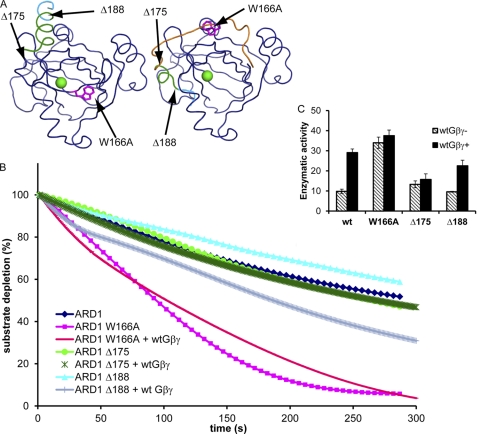
From the modeled ARD1 structure (supplemental Fig. S2), we predicted that a region of ARD1 would interact with Gβγ and move in response to its binding, changing the accessibility of the ARD1 active site (Fig. 5A). The C-terminal α-helix (α-5) and preceding loop of ARD1 partially block the opening of the active site occupied by the metal ion. This obstruction likely lowers basal ARD1 activity, and by displacement of this helix and/or loop, Gβγ may stimulate ARD1 activity. In addition, a conserved tryptophan residue exists in the middle of the loop, and it is likely involved in the activation mechanism. To test the proposed mechanism of activation, we created and purified three versions of mutant ARD1 protein. One protein contained an alanine instead of the conserved tryptophan (W166A). The remaining proteins contained either a deletion of the C-terminal portion of α-5 (Δ188) or a deletion of all of α-5 and a portion of the preceding loop (Δ175). In the absence of AGB1-AGG1 protein, only the W166A mutant had higher enzymatic activity than wild type ARD1 (Fig. 5, B and C). However, when AGB1-AGG1 was added to the reaction, neither W166A nor Δ175 was stimulated by AGB1-AGG1 as seen in wild type and in Δ188. These data suggest that Trp-166 represses the enzymatic activity of ARD1 and that Trp-166 and the region between residues 175 and 188 are required for stimulation by AGB1-AGG1. The C-terminal portion of α5 was not required for stimulation by AGB1-AGG1 and did not negatively regulate ARD1 enzymatic activity.
FIGURE 5.
Mechanism of ARD1 stimulation by AGB1. A, structural model of ARD1 derived from two crystal structures (MmADI1, PDB code 1VR3; KoARD, PDB code 1ZRR) depicting the proposed mechanism of stimulation by AGB1-AGG1. Green sphere represents the bound metal ion in the active site. Left structure is 1VR3, with truncated and mutated regions indicated in magenta, green, and cyan. Right structure is a chimera of 1VR3 and 1ZRR; navy residues were derived from 1VR3, and orange loop and green/cyan helix were derived from 1ZRR. The location of the C-terminal loop and helix indicates the inactive and active conformations of the protein. The three mutations made on ARD1 are indicated in the structure. B, enzyme activity of wild type versus mutant ARD1 in the presence or absence of wild type AGB1-AGG1 (Gβγ). The final enzyme concentration was adjusted to 24 nm, and the substrate was built up to about 125 μm in all cases. The molar ratio of ARD1/Gβγ was 0.65:1. C, enzymatic rates of ARD1 wild type and mutants in the presence or absence of wild type Gβγ as indicated in B. The rates are expressed in moles of substrate/mol of enzyme/s ± S.D. and were recorded as initial rates on the 3rd day after purification. wtGβγ− indicates the rate for the enzyme alone, and wtGβγ+ indicates the rate of the enzyme with the addition of wild type Gβγ. These rates account for the average oxygen-induced decay rate (base line, see “Experimental Procedures”).
DISCUSSION
AGB1 physically interacts with ARD1 to stimulate catalytic activity. Several lines of evidence support direct interaction and regulation, including site-directed mutagenesis to map roughly the protein-protein interface. The region of Gβγ that stimulates ARD1 activity is conserved between plants and mammals, raising the possibility that the activation mechanism is ancient.
ARD proteins are present in bacterial species although heterotrimeric G protein components are not. How is it possible that Gβ proteins stimulate ARD enzymatic activity in eukaryotes but that this stimulation is not required for prokaryotic activity? To answer this question, we created a model predicting the molecular mechanism of activation of ARD1 by AGB1-AGG1 (Fig. 5). The three ARD1 binding regions on AGB1 are spread throughout one surface as shown in the top left panel of Fig. 4C, and the surface area is large enough that binding to the whole region would require a conformational change of ARD1. Upon examination of the modeled structure of ARD1 (supplemental Fig. S2) and of its parent structure (mADI1, PDB code 1VR3 (23)), we noticed several features that would allow such an interaction to occur. First, the C-terminal α-5 helix of ARD1 and the loop directly preceding it pack tightly against the opening to the pocket that the metal ion and substrate occupy, thus obscuring the opening. We propose that this packing creates a low basal activity of ARD1. Binding of AGB1-AGG1 would displace the helix and loop, opening the ARD1 pocket to promote faster substrate/product movement into and out of the active site. Furthermore, we propose that the large hydrophobic tryptophan residue in the middle of the loop regulates this movement.
We tested these predictions by creating two truncation mutations (Δ175 and Δ188) and one substitution mutation (W166A). The higher basal activity of W166A indicates that Trp-166 restricts the basal enzymatic activity of ARD1. However, it is also a critical point of regulation by AGB1-AGG1, as AGB1-AGG1 was unable to further stimulate this mutated protein. The portion of the loop and α-5 helix between residues 175 and 188 did not restrict basal ARD1 activity but was critical for stimulation by AGB1-AGG1 (the C terminus of α-5 was not involved in this process). Therefore, we suggest that Trp-166 serves as a latch that maintains ARD1 in a state of low basal enzymatic activity (designated inactive state). Upon binding by AGB1-AGG1, this latch is unlocked, and the enzyme activity is stimulated. AGB1-AGG1 utilizes both Trp-166 and at least a subset of the residues 175–188 to achieve this stimulation.
These predictions are supported by several structural observations. First, although the crystallized mammalian structure was in the inactive conformation, with the loop and C-terminal helix obscuring the pocket, the Ni-ARD bacterial structure (PDB code 1ZRR (35)) showed a displaced loop and helix and a much more accessible pocket, and the Fe-ARD bacterial structure (PDB code 2HJI (28)) showed a disordered C terminus that indicates mobility of the C-terminal loop and helix. Thus, the structural differences observed agree with our proposed method of stimulation; because the bacterial structures exist in a more active state, their only regulation may be from the divalent metal-bound active site. However, the inactive state of the eukaryotic protein suggests the need for regulation in this case by Gβγ.
The best understood function of ARD proteins is to operate in the methionine salvage pathway, utilizing one of two classes of divalent metal ions to affect the structure and thus to convert acireductone into either a methionine precursor or an off-pathway product. The high sequence similarity between ARD1 and the previously characterized KoARD and MmADI1 within the regions required for enzymatic activity (supplemental Fig. S5A) suggested that ARD1 might function in the methionine salvage pathway, and our results confirmed this prediction. However, it is also known that mammalian ARD enzymes are moonlighting proteins (36), performing diverse functions in addition to (and independently of) the enzymatic activity. For example, human ARD (hADI1) serves as a cancer suppressor in prostate cells. When its levels were increased in tumor cells, hADI1 was found to induce programmed cell death independent of its enzymatic function (37). hADI1 has also been shown to move between the cytoplasm and nucleus via an undefined nuclear localization signal and a noncanonical nuclear export signal. One of its nuclear functions is to facilitate mRNA splicing; this function is also independent of methionine salvage enzymatic activity (38).
ard1 null mutants have shorter hypocotyls than wild type due to fewer cells but the reason is unresolved. This may involve intermediates of the methionine pathway or some other function of ARD1 (discussed below). The methionine salvage pathway recovers the thiomethyl group of methylthioadenosine (MTA, Fig. 1E), a by-product of polyamine biosynthesis, and it has been shown that polyamines are critical for cell division and cell cycle maintenance (39). In addition, MTA, a by-product of the methionine salvage cycle, inhibits polyamine synthesis via spermine and spermidine synthases (40). Previous studies showed that mutants of several methionine salvage pathway components displayed altered polyamine levels (41, 42). Therefore, it is possible that a decrease in the polyamine pool contributes to the reduced cell division observed in ard1 and agb1-2 hypocotyls.
Another hallmark of 2-day-old etiolated seedlings is a closed apical hook; this phenotype protects the apical meristem from damage as the seedling grows through the soil, and ethylene production contributes to the maintenance of the hook (8). Because 2-day-old etiolated ard1 and agb1-2 seedlings display an opened apical hook, we speculate that the decrease in ethylene production in these mutants may contribute to this tissue-specific phenotype. It is important to note that the decrease in ethylene production measured at this developmental stage is, although statistically significant, quite small (15–25%), although we are aware that such a small change in ethylene has been correlated to a significant alteration in hypocotyl length (43).
Arabidopsis and other plants (but no nonplant organisms) contain multiple ARD proteins; the Arabidopsis genome encodes AtARD1–4. AtARD1 is 96% identical to AtARD2 and 80% identical to AtARD3 but only 67% identical to AtARD4. These Arabidopsis ARD orthologs are more similar to ARD proteins from rice than to ARDs from nonplant species, but the ARD1, ARD2, and ARD3 of different plants are more closely related to each other (between species) than to the ARD4 of their own species (supplemental Fig. S5B). Each of the four Arabidopsis ARD proteins can interact with Gβγ in vitro and in vivo (supplemental Fig. S6); G protein signaling may also utilize these ARD paralogs.
Although there is a high degree of similarity between ARD1, -2, and -3, ARD4 is divergent, and we speculate it could possess a different subset of functions than the other three. The duplication of ARD proteins in plants is of particular interest in that it occurred after the divergence of plants from the ancestral lineage but did not occur in any other family. Additionally, ARD1 likely diverged from ARD4 before the divergence of rice and Arabidopsis and then duplicated twice to yield ARD1, ARD2, and ARD3 after the split (supplemental Fig. S5B). This pattern and lack of subsequent divergence indicates a functional necessity for multiple ARD family members. Although plant ARD proteins display conserved metal-binding residues, suggesting enzymatic activity, Arabidopsis ARD proteins also possess the conserved noncanonical nuclear export signal identified in the mammalian ARD proteins (supplemental Fig. S2B, underlined region), suggesting a nuclear role as well.
It is still not yet clear what other processes ARD proteins might regulate in plants and which (if any) of these additional functions are also regulated by G protein signaling. We identified a role of ARD1 in cell division and that role depends on the ability of AGB1 to stimulate ARD1 enzymatic activity. Here, one clear picture of regulation of ARD1 activity by AGB1 emerges, but other functions and mechanisms may follow.
Supplementary Material
Acknowledgments
We thank Arwen Frick-Cheng for technical assistance in creating the baculoviruses used to generate the various forms of AGB1 protein; Tony Perdue for microscopic sample preparation and imaging expertise; and Gyeong Mee Yoon and Joe Kieber for the use of their gas chromatograph and advice in measuring ethylene production.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM065989 from NIGMS (to the A.M.J. laboratory) and Grant R01-GM44191 (to the T. C. P. laboratory). This work was also supported by Department of Energy Grant DE-FG02-05er15671 and National Science Foundation Grants MCB-0723515 and MCB -0718202.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Table S1.
- SI
- stomatal index
- ARD
- acireductone dioxygenase
- AdoMet
- S-adenosylmethionine
- MTA
- S-methylthioadenosine
- BLAST
- basic local alignment search tool
- Y3H
- yeast-three hybrid
- BiFC
- bimolecular fluorescence complementation
- PDB
- Protein Data Bank.
REFERENCES
- 1. McCudden C. R., Hains M. D., Kimple R. J., Siderovski D. P., Willard F. S. (2005) Cell. Mol. Life Sci. 62, 551–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Temple B. R., Jones A. M. (2007) Annu. Rev. Plant Biol. 58, 249–266 [DOI] [PubMed] [Google Scholar]
- 3. Chakravorty D., Trusov Y., Zhang W., Acharya B. R., Sheahan M. B., McCurdy D. W., Assmann S. M., Botella J. R. (2011) Plant J., in press [DOI] [PubMed] [Google Scholar]
- 4. Ullah H., Chen J. G., Temple B., Boyes D. C., Alonso J. M., Davis K. R., Ecker J. R., Jones A. M. (2003) Plant Cell 15, 393–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Llorente F., Alonso-Blanco C., Sánchez-Rodriguez C., Jorda L., Molina A. (2005) Plant J. 43, 165–180 [DOI] [PubMed] [Google Scholar]
- 6. Trusov Y., Sewelam N., Rookes J. E., Kunkel M., Nowak E., Schenk P. M., Botella J. R. (2009) Plant J. 58, 69–81 [DOI] [PubMed] [Google Scholar]
- 7. Ullah H., Chen J. G., Young J. C., Im K. H., Sussman M. R., Jones A. M. (2001) Science 292, 2066–2069 [DOI] [PubMed] [Google Scholar]
- 8. Woeste K., Kieber J. J. (1998) Philos. Trans. R Soc. Lond B. Biol. Sci. 353, 1431–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang H. X., Weerasinghe R. R., Perdue T. D., Cakmakci N. G., Taylor J. P., Marzluff W. F., Jones A. M. (2006) Mol. Biol. Cell 17, 4257–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mudgil Y., Uhrig J. F., Zhou J., Temple B., Jiang K., Jones A. M. (2009) Plant Cell 21, 3591–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smrcka A. V. (2008) Cell. Mol. Life Sci. 65, 2191–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen J. G., Gao Y., Jones A. M. (2006) Plant Physiol. 141, 887–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weigel D., Ahn J. H., Blázquez M. A., Borevitz J. O., Christensen S. K., Fankhauser C., Ferrándiz C., Kardailsky I., Malancharuvil E. J., Neff M. M., Nguyen J. T., Sato S., Wang Z. Y., Xia Y., Dixon R. A., Harrison M. J., Lamb C. J., Yanofsky M. F., Chory J. (2000) Plant Physiol. 122, 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rosso M. G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003) Plant Mol. Biol. 53, 247–259 [DOI] [PubMed] [Google Scholar]
- 15. Vogel J. P., Woeste K. E., Theologis A., Kieber J. J. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4766–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huelsenbeck J. P., Ronquist F., Nielsen R., Bollback J. P. (2001) Science 294, 2310–2314 [DOI] [PubMed] [Google Scholar]
- 18. Yoo S. D., Cho Y. H., Sheen J. (2007) Nat. Protoc. 2, 1565–1572 [DOI] [PubMed] [Google Scholar]
- 19. Grigston J. C., Osuna D., Scheible W. R., Liu C., Stitt M., Jones A. M. (2008) FEBS Lett. 582, 3577–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sundin A. M., Larsson J. E. (2002) J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 766, 115–121 [DOI] [PubMed] [Google Scholar]
- 21. Zhang Y., Heinsen M. H., Kostic M., Pagani G. M., Riera T. V., Perovic I., Hedstrom L., Snider B. B., Pochapsky T. C. (2004) Bioorg. Med. Chem. 12, 3847–3855 [DOI] [PubMed] [Google Scholar]
- 22. Zhang L., Hu G., Cheng Y., Huang J. (2008) Dev. Biol. 324, 68–75 [DOI] [PubMed] [Google Scholar]
- 23. Xu Q., Schwarzenbacher R., Krishna S. S., McMullan D., Agarwalla S., Quijano K., Abdubek P., Ambing E., Axelrod H., Biorac T., Canaves J. M., Chiu H. J., Elsliger M. A., Grittini C., Grzechnik S. K., DiDonato M., Hale J., Hampton E., Han G. W., Haugen J., Hornsby M., Jaroszewski L., Klock H. E., Knuth M. W., Koesema E., Kreusch A., Kuhn P., Miller M. D., Moy K., Nigoghossian E., Paulsen J., Reyes R., Rife C., Spraggon G., Stevens R. C., van den Bedem H., Velasquez J., White A., Wolf G., Hodgson K. O., Wooley J., Deacon A. M., Godzik A., Lesley S. A., Wilson I. A. (2006) Proteins 64, 808–813 [DOI] [PubMed] [Google Scholar]
- 24. Chai S. C., Ju T., Dang M., Goldsmith R. B., Maroney M. J., Pochapsky T. C. (2008) Biochemistry 47, 2428–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pirkov I., Norbeck J., Gustafsson L., Albers E. (2008) FEBS J. 275, 4111–4120 [DOI] [PubMed] [Google Scholar]
- 26. Lin T., He X., Yang L., Shou H., Wu P. (2005) Gene 360, 27–34 [DOI] [PubMed] [Google Scholar]
- 27. Dai Y., Wensink P. C., Abeles R. H. (1999) J. Biol. Chem. 274, 1193–1195 [DOI] [PubMed] [Google Scholar]
- 28. Ju T., Goldsmith R. B., Chai S. C., Maroney M. J., Pochapsky S. S., Pochapsky T. C. (2006) J. Mol. Biol. 363, 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pochapsky T. C., Pochapsky S. S., Ju T., Mo H., Al-Mjeni F., Maroney M. J. (2002) Nat. Struct. Biol. 9, 966–972 [DOI] [PubMed] [Google Scholar]
- 30. Dai Y., Pochapsky T. C., Abeles R. H. (2001) Biochemistry 40, 6379–6387 [DOI] [PubMed] [Google Scholar]
- 31. Sauter M., Lorbiecke R., Ouyang B., Pochapsky T. C., Rzewuski G. (2005) Plant J. 44, 718–729 [DOI] [PubMed] [Google Scholar]
- 32. Wray J. W., Abeles R. H. (1995) J. Biol. Chem. 270, 3147–3153 [DOI] [PubMed] [Google Scholar]
- 33. Friedman E. J., Temple B. R., Hicks S. N., Sondek J., Jones C. D., Jones A. M. (2009) J. Mol. Biol. 392, 1044–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ford C. E., Skiba N. P., Bae H., Daaka Y., Reuveny E., Shekter L. R., Rosal R., Weng G., Yang C. S., Iyengar R., Miller R. J., Jan L. Y., Lefkowitz R. J., Hamm H. E. (1998) Science 280, 1271–1274 [DOI] [PubMed] [Google Scholar]
- 35. Pochapsky T. C., Pochapsky S. S., Ju T., Hoefler C., Liang J. (2006) J. Biomol. NMR 34, 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jeffery C. J. (2003) Trends Genet. 19, 415–417 [DOI] [PubMed] [Google Scholar]
- 37. Oram S. W., Ai J., Pagani G. M., Hitchens M. R., Stern J. A., Eggener S., Pins M., Xiao W., Cai X., Haleem R., Jiang F., Pochapsky T. C., Hedstrom L., Wang Z. (2007) Neoplasia 9, 643–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gotoh I., Uekita T., Seiki M. (2007) Genes Cells 12, 105–117 [DOI] [PubMed] [Google Scholar]
- 39. Oredsson S. M. (2003) Biochem. Soc. Trans. 31, 366–370 [DOI] [PubMed] [Google Scholar]
- 40. Avila M. A., García-Trevijano E. R., Lu S. C., Corrales F. J., Mato J. M. (2004) Int. J. Biochem. Cell Biol. 36, 2125–2130 [DOI] [PubMed] [Google Scholar]
- 41. Bürstenbinder K., Waduwara I., Schoor S., Moffatt B. A., Wirtz M., Minocha S. C., Oppermann Y., Bouchereau A., Hell R., Sauter M. (2010) Plant J. 62, 977–988 [DOI] [PubMed] [Google Scholar]
- 42. Kim J. H., Kim H. S., Lee Y. H., Kim Y. S., Oh H. W., Joung H., Chae S. K., Suh K. H., Jeon J. H. (2008) Plant Cell Physiol. 49, 1627–1632 [DOI] [PubMed] [Google Scholar]
- 43. Tsuchisaka A., Yu G., Jin H., Alonso J. M., Ecker J. R., Zhang X., Gao S., Theologis A. (2009) Genetics 183, 979–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.