Abstract
MicroRNA 143/145 (miR143/145) is restricted to adult smooth muscle cell (SMC) lineages and mediates, in part, the expression of several SMC contractile genes. Although the function of miR143/145 has begun to be elucidated, its transcriptional regulation in response to various signaling inputs is poorly understood. In an effort to define a miR signature for SMC differentiation, we screened human coronary artery SMCs for miRs modulated by TGF-β1, a known stimulus for SMC differentiation. Array analysis revealed a number of TGF-β1-induced miRs, including miR143/145. Validation studies showed that TGF-β1 stimulated miR143/145 expression in a dose- and time-dependent manner. We utilized several chemical inhibitors and found that SB203580, a specific inhibitor of p38MAPK, significantly decreased TGF-β1-induced miR143/145 expression. siRNA studies demonstrated that the effect of TGF-β1 on miR143/145 was dependent upon the myocardin and serum response factor transcriptional switch as well as SMAD4. TGF-β1 stimulated a 580-bp human miR143/145 enhancer, and mutagenesis studies revealed a critical role for both a known CArG box and an adjacent SMAD-binding element for full TGF-β1-dependent activation of the enhancer. Chromatin immunoprecipitation assays documented TGF-β1-mediated enrichment of SMAD3 and SMAD4 binding over the enhancer region containing the SMAD-binding element. Pre-miR145 strongly promoted SMC differentiation, whereas an anti-miR145 partially blocked TGF-β1-induced SMC differentiation. These results demonstrate a dual pathway for TGF-β1-induced transcription of miR143/145, thus revealing a novel mechanism underlying TGF-β1-induced human vascular SMC differentiation.
Keywords: MicroRNA, SMAD Transcription Factor, Smooth Muscle, Transcription Enhancers, Transcription Regulation, Myocardin, Serum Response Factor
Introduction
SMCs3 display remarkable phenotypic adaptation in response to physical, chemical, and biological perturbations. Such altered SMC phenotypes play a major role in the pathogenesis of many human diseases, including asthma, atherosclerosis, restenosis, hypertension, transplant arteriopathy, and Alzheimer angiopathy (1–3). Accumulating evidence has shown SMC differentiation to be tightly regulated by an interacting network of environmental stimuli, signaling pathways, and various transcription factors, most notably SRF and MYOCD (2, 4–8). TGF-β1 is among the most potent soluble growth factors that activate SMC contractile gene expression in both specified SMC and non-SMC types (9–15). Members of the TGF-β1 superfamily transmit signals through both SMAD-dependent and SMAD-independent pathways (16). The classic pathway is through transmembrane serine-threonine kinase receptors, which mediate the phosphorylation of receptor-specific SMAD2 and SMAD3. The phosphorylated SMAD2-SMAD3 complex then interacts with the common SMAD4 to form a heteromeric complex, which translocates to the nucleus and binds to Smad-binding elements (SBE) located in the regulatory region of a number of target genes (16). SMAD-independent pathways, such as MAPK and PI3K, can also be triggered by TGF-β to initiate signal transduction and gene regulation (17, 18). Both SMAD-dependent and SMAD-independent pathways have been demonstrated to contribute to SMC differentiation in a context-dependent manner (11, 14).
MicroRNAs (miRs) represent a class of conserved, small non-coding RNAs that bind to and target mRNA for degradation or a block in translation (19, 20). Aberrant expression of miRs frequently leads to abnormalities in development and the pathogenesis of disease (21–25), and a growing number of miRs have been discovered as powerful modulators in cardiovascular biology. For example, miR21 has been linked to the SMC proliferative phenotype in a balloon-injured rat carotid artery model (26). The miR221/222 gene is up-regulated after vascular injury and appears to mediate the SMC proliferative phenotype as well as intimal hyperplasia (27, 28). miR26a was shown to be elevated with SMC differentiation, but it appears to negatively regulate expression of SMC contractile markers (29). Conversely, miR1 and miR10a seem to promote the SMC differentiated phenotype in embryonic stem cells treated with retinoic acid (30, 31). Interestingly, a growing number of miRs are direct targets of SRF/MYOCD (32, 33), suggesting an important interplay between this pivotal transcriptional switch and miR-mediated events within SMC lineages.
The miR143/145 bicistronic gene was recently demonstrated to be specific for adult SMC lineages and functions to promote the SMC contractile phenotype (34–38). As with a number of other SMC-specific genes (8), miR143/145 is a direct target of SRF and is reduced in the vessel wall after injury or during atherosclerotic lesion development (34–38). Thus, miR143/145 is part of the molecular signature of SMCs and contributes to the acquisition of an SMC differentiation state. Although the expression and function of many SMC miRs have begun to be elucidated, the upstream signaling pathways that converge upon SMC miR promoter/enhancer elements are less understood.
Emerging evidence has demonstrated that TGF-β1 modulates several miRs, which in turn control expression of protein-coding genes associated with such cellular programs as epithelial-mesenchymal transition, skeletal muscle cell differentiation, and cell proliferation (39–41). Because TGF-β1 is such a strong activator of SMC differentiation, we hypothesized that a TGF-β1-induced miR program exists to help coordinate the SMC differentiated phenotype. Here, we report initial results of a TGF-β1-responsive miR signature in human coronary artery smooth muscle cells (HCASM) and show that the miR143/145 gene is among the more dramatically induced miRs. We further show through gain- and loss-of-function studies that TGF-β1-induced p38MAPK-SRF-MYOCD and SMAD pathways activate miR143/145 transcription in HCASM. Importantly, we show that these TGF-β1-dependent pathways converge upon an upstream region of the miR143/145 gene harboring a previously defined CArG box and a newly identified SBE. These results establish a novel mechanism for TGF-β1-induced SMC differentiation through the transcriptional activation of miR143/145.
EXPERIMENTAL PROCEDURES
SMC Culture and Treatments
Multiple independent isolates of HCASM were purchased from Invitrogen and maintained in medium 231 with growth supplements as provided by the manufacturer. Rat aortic SMCs (RASM) were purchased from ATCC and cultured in Dulbecco's modified Eagle's medium (high glucose) supplemented with 10% fetal bovine serum without antibiotics or antimycotics. RASM within 7 passages were used throughout many studies. For TGF-β1 treatment, growing cells were seeded in 6-well plates and cultured until 90% confluence. Following serum starvation overnight, cells were treated with TGF-β1 (1 ng/ml for HCASM and 4 ng/ml for RASM) for 24 h. For inhibitor studies, cells were pretreated with the indicated signaling inhibitors 30 min prior to TGF-β1 treatment. SB203580 and SB202190 (specific inhibitors to p38MAPK) and PD98059 (inhibitor to ERK1/2) were from Calbiochem. The concentration of each inhibitor was 10 μm except for the indicated concentrations used in dose-dependent studies.
MicroRNA Screen
Total RNA was harvested from HCASM treated with and without TGF-β1 for 24 h using RNAeasy (Qiagen). After validating the effect of TGF-β1 on the induction of SMC contractile gene expression by quantitative RT-PCR, RNA samples were applied to a microRNA array provided by LC Sciences (Houston, TX) as indicated (42). Briefly, ∼5 μg of total RNA was used for size fractionation. Only the small RNAs (<300 nucleotides) isolated were 3′-extended with a poly(A) tail using poly(A) polymerase. Two different tags were randomly used for either control or TGF-β1-treated RNA samples. Hybridization was performed overnight on a Paraflo microfluidic human microRNA chip (MRA-1001 LC Sciences). After RNA hybridization, tag-conjugating Cy3 and Cy5 dyes were circulated through the microfluidic chip for dye staining. Fluorescence images were collected using a laser scanner (GenePix 4000B, Molecular Devices) and digitized using Array-Pro image analysis software (Media Cybernetics). The array contained probes only to mature miRs. Data were analyzed by first subtracting the background and then normalizing the signals using a LOWESS filter. The ratio of the two sets of detected signals (log2 transformed, balanced) and p values of the t test were calculated.
RNA Extraction and RT-PCR
Total RNA was extracted from cultured SMCs by miRNeasy as above, and cDNA synthesis was carried out with a first strand cDNA synthesis kit (GE Healthcare). Semiquantitative PCR, SYBR Green-based real-time PCR (MyIQ, Bio-Rad), or Taqman assays (Applied Biosystems) were used to measure mRNA or miR levels. Control housekeeping genes were Gapd (for mRNA) and SnoRNA48 or U6 (for miR). We used the 2−ΔΔCt method for normalization of raw data as described (43). The primers used for different target mRNA and miRs are listed in supplemental Table 1.
Western Blotting
Cells were rinsed with phosphate-buffered saline (PBS) twice, and protein was extracted in cold lysis buffer containing 1% protease inhibitor mixture (Sigma) as described (43). Protein concentration was determined by a detergent-compatible protein assay (Bio-Rad). Equal amounts of protein were resolved by SDS-PAGE, transferred onto nitrocellulose membranes, blocked with 5% nonfat milk for 1 h, and then incubated with the indicated primary antibody overnight at 4 °C. After a 1-h incubation with the appropriate secondary antibody, specific signals were revealed by enhanced chemiluminescence reagent (Pierce). The primary antibodies used were as follows: ACTA2 (Sigma, A2547); CNN1 (DAKO, M3556); CCND1 (BD Pharmingen, 556470); SRF (Santa Cruz Biotechnology, sc-335); p38MAPK (Santa Cruz Biotechnology, sc-535); SMAD3 (Santa Cruz Biotechnology, sc-101154); TUBA (Sigma, T-5168); and SMAD2 (3103), phospho-SMAD2 (3101S), phospho-p38MAPK (9211S), SMAD4 (9515), ERK1/2 (9122), and phospho-ERK1/2 (4376) all from Cell Signaling Technology. The HSP27 and phospho-HSP27 antibodies were a kind gift from Dr. Jun-ichi Abe (University of Rochester).
Immunofluorescence Microscopy
HCASM were dispersed under sterile glass coverslips and grown until 90% confluence. After 24 h of serum starvation, cells were stimulated with TGF-β1 (1.0 ng/ml) for 24 h. Immunofluorescence was conducted as described (44). Briefly, cells were washed twice with PBS (pH at 7.4) and then fixed in freshly prepared 4% paraformaldehyde for 10 min. After being rinsed three times with PBS-Tween 20, cells were permeabilized with 0.1% Triton X-100 for 5 min. A 1:200 dilution of mouse anti-human CNN1 or a 1:300 dilution of anti-ACTA2 and 1:200 diluted goat anti-mouse IgG Texas Red conjugate (Abcam) were used to detect CNN1 or ACTA2 protein expression. Nuclear profiles were revealed with a brief incubation in DAPI (Molecular Probes) prior to microscopic observation. Fluorescence was visualized with an inverted Olympus IX70 fluorescence microscope and photographed for direct importation into Adobe Photoshop. All images were processed in an equivalent manner to faithfully capture the real time images of each sample.
Small Interfering RNA Transfections
siRNA to SRF was purchased from Ambion (4392420). The ON-TARGET plus SMART pool siRNA to human SMAD4 (L-003902–00) and MYOCD (NM_153604) were from Dharmacon siRNA Technologies. A scrambled siRNA duplex was used for negative control. Lipofectamine 2000 (Invitrogen) was used to deliver siRNA according to the manufacturer's instructions. Following overnight siRNA transfection, cells were refed with fresh growth medium for 24 h before treatment as indicated. RNA or protein was extracted 48–72 h after transfection, and qPCR or Western blotting was used to determine knockdown efficiency.
Cloning and Mutagenesis of miR143/145 Enhancer
The human miR143/145 enhancer encompassing a conserved CArG box and a putative SBE were PCR-amplified from genomic DNA derived from HCASM using high fidelity polymerase (Roche Applied Science). Primers are listed in supplemental Table 1. The enhancer was then cloned into a BglII site of the pGL3 luciferase reporter containing a minimal thymidine kinase promoter (Promega). Point mutations of the CArG and SBE sites were made using the QuikChange mutagenesis kit (Stratagene). All cloned promoter constructs were submitted to Cornell University Life Sciences Core Laboratories Center to validate nucleotide sequence fidelity.
Transfection and Luciferase Assays
Due to poor efficiency of gene delivery to HCASM, we used RASM and 10T1/2 cells to perform luciferase assays. Gene pulser electroporation (Bio-Rad) was used to deliver miR143/145 reporter and other indicated plasmids. Briefly, growing RASM were trypsinized from culture plates, rinsed twice with PBS, and mixed with transfection buffer before electroporation. The conditions for electroporation were as follows: voltage at 300 V, capacitance at 500 ohms in a 4-mm cuvette. After electroporation, cells were dispersed in 24-well plates and allowed to adhere overnight before refeeding with fresh growth medium. Lipofectamine 2000 was used to transfect reporters in 10T1/2 cells. Cells were seeded in 24-well plates and grown until 90% confluence. Transfections were done according to the manufacturer's instructions. 6 h after transfection, cells were refed with 10% FBS for 24 h. After serum starvation, cells were treated with TGF-β1 (4 ng/ml) for 24 h. Serum starvation lasted 24 h for 10T1/2 cells and 3 h for RASM. To correct for varying transfection efficiency and the side effect of TGF-β1 on cell proliferation, a Renilla reporter gene (Promega) was included as an internal control. Cell lysates were prepared for the luciferase assay as described by the manufacturer (Promega). All transfections were performed in quadruplicate and repeated in at least three independent experiments. Data were analyzed with GraphPad Prism Software (version 4.0, GraphPad Software Inc.) and expressed as the normalized -fold increase over controls ± S.D.
MicroRNA Overexpression and Knockdown Studies
Growing HCASM were transfected with 30 nm precursor miR mimics or antisense miR inhibitors (also known as anti-miRs) using siPORT NeoFX (Applied Biosystems), following the manufacturer's instructions. RNA or protein was harvested 72 h after transfection, and miR expression level was quantified with TaqMan miR assays (Applied Biosystems). The probes used are listed in supplemental Table 1.
Chromatin Immunoprecipitation (ChIP) Assays
Conventional ChIP assays were carried out with EZ-ChIP (Millipore) as described previously (44). HCASM treated with TGF-β1 for 24 h were used. Chromatin complexes were immunoprecipitated with SMAD4 or SMAD3 antibody or a rabbit IgG control. Primers for amplifying fragments of the miR143/145 locus containing SBE or a remote downstream region (within primary miR143/145) are included in supplemental Table 1. For quantitative ChIP, we subjected equivalent amounts of total DNA from input and precipitated IgG and SMAD4 samples to qPCR. Raw data for the anti-SMAD4 sample from control and TGF-β1-treated HCASM were normalized to its IgG control, and the normalized control sample was set to 1.
RESULTS
TGF-β1 Induces SMC Differentiation Marker Expression in HCASM
TGF-β1 induces SMC differentiation in a variety of cell culture model systems (9–15). However, there are surprisingly no studies that have examined SMC differentiation genes in TGF-β1-treated HCASM, and nothing is known about the miR profile of these cells. We thus sought to initially characterize the phenotypic response of HCASM to TGF-β1 stimulation. Using quantitative PCR, we found that 24-h TGF-β1 treatment evoked increases in SMC contractile genes, including CNN1, TAGLN, and ACTA2 (Fig. 1A). We also confirmed elevations in the expression of SMC contractile proteins by Western blotting (Fig. 1B) and immunofluorescence microscopy (Fig. 1C). Similar results were obtained in independent isolates of HCASM treated with TGF-β1, and the prodifferentiation effects were comparable with those seen with the addition of differentiation medium (data not shown). These results establish HCASM as a reliable in vitro model system for TGF-β1-induced SMC differentiation and the profiling of differentiation-associated miRs.
FIGURE 1.
TGF-β1-induced SMC differentiation in HCASM. HCASM were treated with TGF-β1 (1 ng/ml) or vehicle for 24 h, and the indicated SMC markers were analyzed by qPCR (A), Western blotting (B), and immunofluorescence microscopy (C). Relative mRNA levels in A reflect -fold changes relative to the vehicle control, arbitrarily set to 1. Data are representative of four independent experiments. Error bars, S.D.
TGF-β1 Induces miR143/145 Expression in HCASM
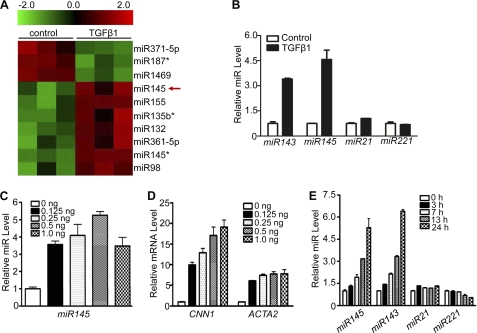
Based on the preceding results, we stimulated HCASM for 24 h with TGF-β1 or vehicle and performed an miR screen. Three independent experiments revealed a consistent array of differentially expressed miRs between vehicle and TGF-β1-treated HCASM, 10 of which showed statistically significant changes (Fig. 2A). Several miRs were shown to be reduced with TGF-β1 and are the subject of ongoing investigation (Fig. 2A). Consistent with a previous report (39), we found TGF-β1-induced expression of miR155 in HCASM (Fig. 2A). Interestingly, miR145, a highly specific miR for SMC lineages (34–38), was found to be among those miRs induced with TGF-β1 (Fig. 2A). Similar array findings were observed for miR143 (data not shown), which is co-expressed with miR145 as a bicistronic miR gene (35, 37). We validated up-regulation of miR143/145 following TGF-β1 treatment by qPCR (Fig. 2B) and Northern blotting (supplemental Fig. 1A). To rule out the possibility that induction of miR143/145 was unique to the sample of HCASM analyzed, three independent HCASM isolates were similarly treated with TGF-β1 and tested for miR143/145 expression by qPCR. Results from all isolates showed strong induction of miR143/145 by TGF-β1 (data not shown). Moreover, RASM treated with TGF-β1 exhibited similar induction of miR143/145 (data not shown). Interestingly, although miR21 was previously shown to be induced by TGF-β1 in human SMC (45), this miR was not among those found to be induced by TGF-β1 in our array. Moreover, qPCR validation studies showed only a mild increase in miR21 following TGF-β1 treatment (Fig. 2B). As a further control, we found that TGF-β1 elicited little effect on expression of miR221 (Fig. 2B). Dose-dependent studies revealed parallel increases in miR145 and the SMC contractile genes CNN1 and ACTA2 with concentrations of TGF-β1 as low as 0.125 ng/ml (Fig. 2, C and D). A similar dose-dependent effect was seen with miR143 (supplemental Fig. 1B). Finally, TGF-β1-induced miR143/145 was time-dependent; elevated expression first emerged at 7 h and increased to the highest level 24 h following TGF-β1 treatment. In contrast, little to no induction of miR21 or miR221 was observed over the time course of TGF-β1 stimulation (Fig. 2E). These data validate our array findings and firmly establish the specific induction of miR143/145 in HCASM treated with TGF-β1.
FIGURE 2.
TGF-β1-induced miR143/145 expression in HCASM. A, heat map illustrating relative expression of a sample of miRs in triplicate control or TGF-β1 (1 ng/ml)-treated HCASM. B, qPCR validation of indicated miRs in HCASM treated with 1 ng/ml TGF-β1 for 24 h. The expression level of each miR reflects the -fold change relative to vehicle control (set to 1). C and D, qPCR analysis of miR145 (C) and two SMC contractile genes (D) in HCASM following 24-h treatment with varying concentrations of TGF-β1. E, HCASM were treated with TGF-β1 (1 ng/ml) for the indicated times, and qPCR was performed to measure normalized expression of miR143/145, miR21, and miR221. Relative miR expression is the -fold change relative to control (time 0) set to 1. All data shown are representative of at least three independent experiments. Error bars, S.D.
TGF-β1-induced SRF and MYOCD Reinforce miR143/145 Expression
Having demonstrated TGF-β1-mediated increases in miR143/145 expression, we next sought to begin elucidating potential mechanisms for such induction. The miR143/145 promoter region contains a conserved CArG box that is responsive to both SRF and MYOCD (35, 37). We therefore asked if TGF-β1 could stimulate SRF and MYOCD expression. Due to the lack of a sensitive MYOCD antibody for detecting low level endogenous MYOCD protein, we were limited to examining MYOCD mRNA. Both semiquantitative and quantitative PCR showed a reproducible and comparable time course of MYOCD mRNA expression following TGF-β1 treatment. The induction was seen as early as 3 h, peaked at 7 h, and declined 13 h after TGF-β1 stimulation (Fig. 3A). In contrast to peak induction of miR143/145 at 24 h (Fig. 2E), TGF-β1-induced MYOCD expression peaked earlier, suggesting that MYOCD may contribute to the later induction of miR143/145. To test this idea, we applied siRNA to MYOCD. As seen in Fig. 3B, a smart pool siRNA specific to human MYOCD effectively knocked down the endogenous MYOCD mRNA in both vehicle and TGF-β1-treated HCASM. Importantly, this MYOCD knockdown abrogated TGF-β1-induced miR145 expression (Fig. 3B). Consistent with a previous report (46), we found that the 67-kDa SRF protein increased upon TGF-β1 treatment (Fig. 3C). To test the importance of SRF in TGF-β1-induced miR143/145 expression, we applied siRNA to SRF. Quantitative PCR showed efficient siRNA knockdown of SRF and a striking decrease in TGF-β1-induced miR143/145 (Fig. 3D). A comparable reduction in CNN1 mRNA expression was also observed (Fig. 3D). Similar results were seen when utilizing a short hairpin RNA (47) targeting a different region of SRF (data not shown). Collectively, these data provide strong evidence supporting a role for MYOCD and SRF in the TGF-β1-mediated increase in miR143/145 expression.
FIGURE 3.
TGF-β1-induced MYOCD and SRF contribute to miR143/145 expression in HCASM. A, qPCR of MYOCD mRNA after the indicated times of TGF-β1 treatment. Inset, a similar study assayed by semiquantitative RT-PCR. B, qPCR of MYOCD and miR145 in the absence or presence of TGF-β with or without siRNA to MYOCD. Please note that the apparent decrease in MYOCD with TGF-β1 alone versus control was a consistent finding, but the time course of stimulation with TGF-β1 in this experiment was 24 h (compare with A). C, Western blot of SRF in HCASM treated with TGF-β1 (1 ng/ml) for the indicated times. D, qPCR of the indicated genes in the absence or presence of TGF-β1 with or without siRNA to SRF. All data are representative of at least three independent experiments. Error bars, S.D.
p38MAPK Is an Important Pathway for TGF-β1-induced miR143/145 Expression
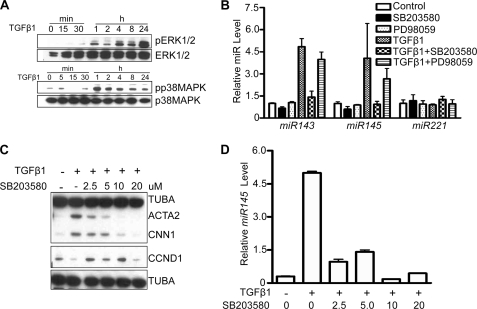
It is well known that SMAD-independent pathways, including ERK1/2 and p38MAPK, transduce TGF-β1 signals (16–18). In HCASM, we found that both ERK1/2 and p38MAPK were activated 1 h and persisted up to 24 h following TGF-β1 treatment (Fig. 4A). We also found that ERK5 and PI3K were activated by TGF-β1 stimulation of HCASM (data not shown). In order to define the pathway(s) mediating TGF-β1-induced miR143/145 expression, we evaluated several inhibitors specific to each activated signal transducer. Among the tested inhibitors, only SB203580, a specific inhibitor for p38MAPK, completely blocked TGF-β1-induced miR143/145 expression (Fig. 4B). In contrast to SB203580, PD98059, a specific ERK1/2 inhibitor, exerted little to no effect on TGF-β1-induced miR143/145 expression (Fig. 4B). A dose-dependent suppressive effect of SB203580 was observed in TGF-β1-induced SMC contractile genes with nearly complete inhibition at a dose of 10 μm (Fig. 4C). This concentration of SB203580 completely blocked the phosphorylation of HSP27, a well known substrate for p38MAPK (48) (supplemental Fig. 2A). A similar dose-dependent effect of SB203580 was noted with respect to TGF-β1-mediated increases in miR143 (supplemental Fig. 2B) and miR145 (Fig. 4D). These effects were specific because the same concentration of SB203580 derepressed the effects of TGF-β1 on cyclin D1 (CCND1) expression (Fig. 4C). To determine whether the inhibitory action of SB203580 on TGF-β1-induced miR143/145 and SMC contractile gene expression was a consequence of some off-target effect, we tested SB202190, another specific inhibitor to p38MAPK, and obtained similar results (data not shown). These findings suggest that p38MAPK is an important pathway for TGF-β1-mediated miR143/145 induction in HCASM.
FIGURE 4.
p38MAPK is critical for TGF-β1-induced miR143/145 expression in HCASM. A, Western blotting of signaling proteins in HCASM stimulated with TGF-β1 (1 ng/ml) for the indicated times. B, qPCR of indicated miRs in HCASM pretreated for 30 min with inhibitor followed by 24-h stimulation with 1 ng/ml TGF-β1 (n = 3/condition ± S.D. (error bars)). Control samples were static cultures without any treatment. C and D, HCASM were pretreated with the indicated dose of SB203580 for 30 min followed by TGF-β1 treatment for 24 h. Samples were then processed for Western blotting (C) or qPCR (D). Results were reproduced in three independent experiments.
p38MAPK Controls MYOCD/SRF Expression
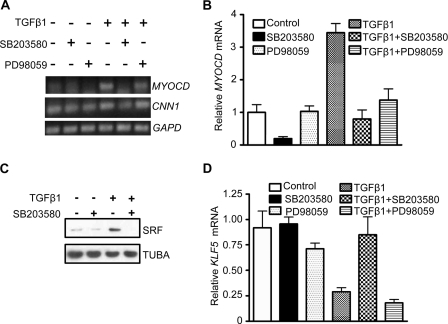
Consistent with the data above (Fig. 3A), TGF-β1 treatment for 7 h resulted in clear increases in MYOCD mRNA expression (Fig. 5A). This induction was completely abolished upon treatment with SB203580 as shown by semiquantitative PCR (Fig. 5A) and quantitative PCR (Fig. 5B). SB203580 also potently reduced TGF-β1-induced SRF expression (Fig. 5C). TGF-β1 repressed the expression of KLF5, a known inhibitor of SMC differentiation (49), but pretreatment with SB203580 antagonized this inhibitory effect (Fig. 5D), suggesting that the suppressive effect of SB203580 on SRF and MYOCD is not a consequence of general repression. There was a milder inhibitory effect of TGF-β1 on KLF4 expression (supplemental Fig. 3). In general, the PD98059 compound had less of an effect on TGF-β1-mediated changes in gene expression (Fig. 5). Taken together, these results suggest that p38MAPK plays an important role in transducing TGF-β1 signaling to the induction of MYOCD/SRF.
FIGURE 5.
p38MAPK is critical for MYOCD and SRF expression in HCASM. A, semiquantitative RT-PCR of the indicated genes under basal (−) or TGF-β1 (+) stimulation for 7 h in the absence or presence of a 30-min pretreatment with each signaling inhibitor. B, qPCR of MYOCD mRNA in HCASM treated as in A (n = 3/condition, ± S.D. (error bars)). C, Western blotting of 67-kDa SRF in HCASM treated with vehicle (water) or TGF-β1 for 24 h in the absence or presence of a 30-min pretreatment with SB203580 compound. D, qPCR analysis of KLF5 mRNA in HCASM treated identically as in B. All data were reproducible in at least two independent experiments.
SMAD4-dependent Regulation of miR143/145 and MYOCD
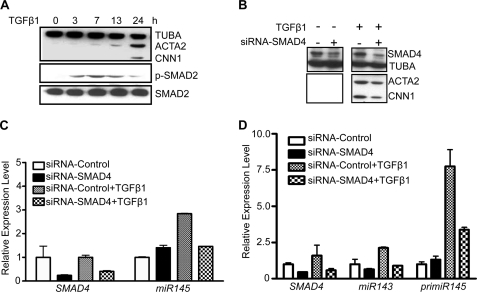
TGF-β1 signals through type I and type II receptors, and the signals are transmitted to the nucleus largely through SMAD proteins (16–18). To determine whether SMAD signaling is involved in TGF-β1-induced miR143/145 and MYOCD expression, we first measured SMAD2 phosphorylation. Phosphorylated SMAD2 appeared 3 h and persisted up to 24 h following TGF-β1 treatment, a time course that paralleled induction of ACTA2 and CNN1 (Fig. 6A) as well as miR143/145 expression (Fig. 2E). To directly assess the role of SMAD signaling in SMC differentiation marker expression, we used siRNA to reduce endogenous SMAD4 and then treated HCASM with TGF-β1. SMAD4 was effectively knocked down by more than 50% in both vehicle and TGF-β1-treated HCASM (Fig. 6B). Lowering steady-state levels of SMAD4 resulted in a partial decrease in TGF-β1-mediated MYOCD mRNA expression (supplemental Fig. 4). Further, SMAD4 knockdown caused a decrease in TGF-β1-induced ACTA2 and CNN1 protein expression (Fig. 6B). Similar knockdown studies resulted in a reduction of TGF-β1-induced miR145 expression (Fig. 6C). Finally, SMAD4 knockdown attenuated TGF-β1-induced miR143 and pri-miR145 expression (Fig. 6D). Collectively, these results support a TGF-β1-directed SMAD pathway in the early stimulation of MYOCD and subsequent miR143/145 and SMC contractile protein expression.
FIGURE 6.
TGF-β1-induced miR143/145 and SMC markers require SMAD4 in HCASM. A, Western blotting of the indicated proteins in HCASM treated with 1 ng/ml TGF-β1 for the indicated times. B, HCASM pretreated with siRNA (30 nm) to SMAD4 for 24 h were stimulated with or without 1 ng/ml TGF-β1 for an additional 24 h, followed by Western blotting of SMAD4, ACTA2, and CNN1. Note that we were unable to detect basal ACTA2 and CNN1 expression by Western blot in growing HCASM due to low level expression under these experimental conditions (compare with Fig. 1B). C, same experiment as in B except that miR145 levels were assessed by qPCR. D, similar experiment as in C except that miR143 and primary miR145 were evaluated by qPCR. Data shown are representative of two independent experiments. Error bars, S.D.
TGF-β1 Signaling Targets an Upstream miR143/145 Enhancer
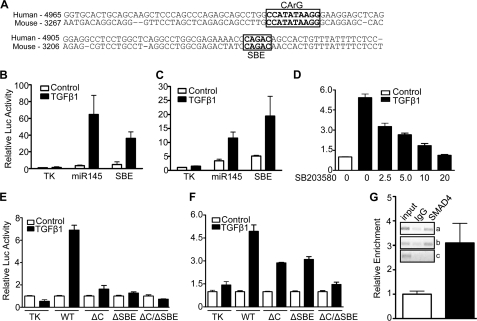
The up-regulation of miR143/145 by TGF-β1 and impaired induction upon knockdown of SRF/MYOCD or SMAD4 suggested that TGF-β1 signaling may converge upon miR143/145 regulatory sequences controlling transcription of this miR gene. Previous work on the mouse miR143/145 promoter showed a functional upstream CArG box that binds SRF and is necessary for activity in transgenic mice (35, 37). A similar CArG box is located ∼5 kb upstream of the human miR143/145 gene (Fig. 7A). Comparative genomics revealed a conserved SBE adjacent to the CArG box (Fig. 7A). To begin to test if these conserved elements are responsive to TGF-β1 stimulation, we cloned a 580-bp region encompassing both the CArG box and SBE into a luciferase reporter and assessed activity following TGF-β1 stimulation of 10T1/2 and RASM cells. In both cell types, TGF-β1 was found to activate the miR143/145 580-bp enhancer (Fig. 7, B and C). Consistent with the expression results above (Fig. 4, B and D, and supplemental Fig. 2B), SB203580 inhibited TGF-β1-induced miR143/145 enhancer activity in a dose-dependent manner (Fig. 7D). To determine the necessity of the CArG box and SBE in TGF-β1 activation of the miR143/145 enhancer, we mutated each site in isolation or together. Each mutant in isolation exhibited an attenuated response to TGF-β1; the CArG/SBE double mutant completely lost the response to TGF-β1 stimulation in 10T1/2 cells and showed less response compared with single site mutants in RASM (Fig. 7, E and F). To determine whether SMAD4 binds the putative SBE, we performed ChIP assays with HCASM treated with TGF-β1 for 24 h. Two separate primer pairs showed an enrichment of DNA containing the SBE following immunoprecipitation with SMAD4 antibody; no such enrichment was seen when primers flanking a region of the pri-miR143/145 transcript were used (Fig. 7G, inset). Additional quantitative ChIP studies showed ∼3-fold enrichment of SMAD4 with TGF-β1 stimulation (Fig. 7G). SMAD3 was also enriched over the SBE following TGF-β1 treatment (supplemental Fig. 5). Taken together, these results suggest that TGF-β1 directly activates the miR143/145 upstream enhancer region through parallel pathways that converge at a CArG box and SBE.
FIGURE 7.
TGF-β1 signaling converges upon a conserved CArG box and SBE in the upstream miR143/145 regulatory region. A, alignment of upstream enhancer region of human and mouse miR143/145 gene with conserved CArG and SBE sites boxed. The numbers to the left indicate the distance of each sequence from the approximate transcription start site in each indicated species. B–D, luciferase activity of the wild type human miR143/145 enhancer (labeled miR145 for simplicity) in 10T1/2 cells (B) or RASM (C) in the absence (Control) or presence of TGF-β1 or in 10T1/2 cells following TGF-β1 stimulation in the absence or presence of a 30-min pretreatment with increasing amounts (μm) of SB compound (D). A synthetic multimerized SBE reporter (SBE in B and C) was included as a control for TGF-β1 responsiveness. E and F, wild type and mutant miR143/145 enhancer activity in 10T1/2 cells (E) or RASM (F) treated in the absence (Control) or presence of TGF-β1 for 24 h. ΔC, point mutant CArG box; ΔSBE, point mutant SBE (see supplemental Table 1 for mutations). Luciferase activity for each construct (B–F) was normalized to the respective control set to 1. Each transfection was assayed in quadruplicate and repeated in multiple independent experiments with results very similar to those shown here. G, quantitative ChIP assay in HCASM treated for 24 h without (open bar) or with (filled bar) TGF-β1. Equal quantities of cross-linked DNA were immunoprecipitated with similar concentrations of a control rabbit IgG or a rabbit anti-SMAD4 antibody and processed for qPCR as described under “Experimental Procedures.” The inset shows gels from similarly processed cultures of HCASM. Note that the images shown represent inversions of the original gels to better illustrate the bands. Primers amplifying a long region (a) or a shorter region (b) encompassing the SBE were used to PCR-amplify the reverse cross-linked and purified DNA. As a negative control, primers flanking a region ∼3 kb downstream from the SBE site (within the primiR143/145 gene) (c) were used for PCR amplification. The ChIP data shown were repeated in two independent experiments. Error bars, S.D.
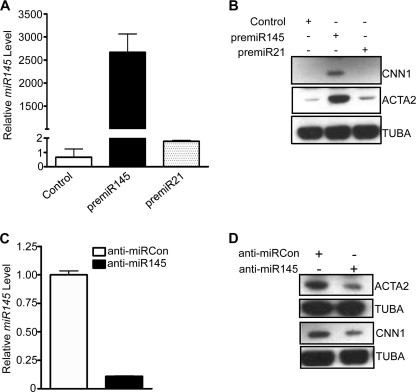
miR145 Contributes to TGF-β1-induced SMC Differentiation
Recently, studies from several independent groups demonstrated that miR143/145 expression is limited to adult SMC lineage and mediates, in part, the ability to direct adult SMC contractile gene expression (34–38). However, it is not known whether induced miR145 is one of the mechanisms involved for TGF-β1-induced SMC differentiation. To address this possibility, we first delivered precursor miR145 mimic to HCASM, and 3 days after delivery, we assessed SMC contractile gene expression. Ectopic pre-miR145 led to an increase in CNN1 and ACTA2 expression as revealed by Western blotting (Fig. 8B). Importantly, anti-miR145 (Fig. 8C) reduced TGF-β1-induced CNN1 and ACTA2 protein (Fig. 8D). Attenuated expression of ACTA2 was further confirmed by immunofluorescence staining (data not shown). These results demonstrate that miR145 contributes, at least partially, to TGF-β1-induced HCASM differentiation.
FIGURE 8.
miR145 contributes to TGF-β1-induced HCASM differentiation. A, growing HCASM were transfected with the indicated pre-miRs or a control miR mimic (30 nm each) for 72 h, and then qPCR was done for miR145 expression. B, Western blotting of the indicated SMC marker proteins in growing HCASM transfected as in A. Note the near absence of SMC markers in the control and miR21-transfected cells. C and D, TGF-β1 (1 ng/ml)-stimulated HCASM following transfection with 30 nm anti-miR to either control (open bar) or miR145 (filled bar) were processed for qPCR of miR145 (C) or Western blotting of the indicated SMC differentiation proteins (D). Note that serum starvation does not induce expression of SMC contractile proteins or miR143/145 (data not shown). Each experiment was replicated in at least two independent studies. Error bars, S.D.
DISCUSSION
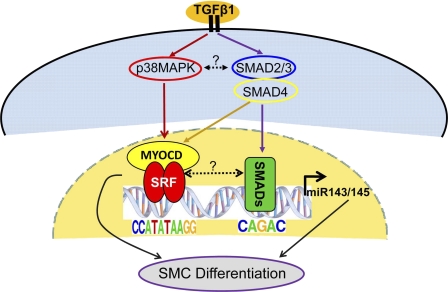
miR145 is a tumor suppressor gene and a critical regulator of human stem cell growth and differentiation (50–52). Recently, several independent groups reported roles for miR145 in the control of SMC phenotypes (34–38). Given the strong functionalities of miR145 as a regulator of cell fate and growth, it is of critical importance to elucidate its molecular regulation. In the present study, we show that TGF-β1 induces miR143/145 expression in HCASM. We further demonstrate that two TGF-β1 signaling pathways (p38MAPK and SMAD) converge upon an upstream miR143/145 enhancer to directly mediate transcription of this bicistronic miR gene (Fig. 9).
FIGURE 9.
Model for TGF-β1-mediated signaling to the miR143/145 regulatory region. A classical (SMAD4) and non-classical (p38MAPK) pathway of TGF-β1 signaling converge at a novel SBE and CArG box, respectively, to induce transcription of miR143/145, which subsequently leads to SMC differentiation marker expression in coordination with SRF/MYOCD. Experimental evidence reported here is depicted with solid lines, whereas hypothetical cross-talk between upstream signaling pathways or transcriptional complexes is indicated with dotted lines.
Emerging evidence has demonstrated that TGF-β1 signaling facilitates its regulatory functions, in part, through direct targeting of miR regulatory sequences. For example, in murine mammary gland epithelial cells, TGF-β1 was found to induce miR155 expression through a functional SBE within its promoter, and induced miR155 was important in TGF-β-induced epithelial-mesenchymal transition, cell migration, and invasion (39). The intronic miR216a and miR217 genes have also been shown to be regulated by TGF-β1 via an E-box located within the host gene's promoter region. Up-regulated miR216a and miR217 directly target PTEN, a repressor of Akt kinase activity, leading to elevated Akt kinase in kidney disorders and other metabolic diseases (40). Here, we provide strong evidence for TGF-β1-mediated transcriptional activation of the miR143/145 gene. We also observed several other miRs to be induced or repressed with TGF-β1 stimulation. Thus, an important future goal will be to functionally characterize the TGF-β1 microRNAome in vascular SMC.
TGF-β/BMP proteins are recognized as powerful stimuli for SMC differentiation (9–15). Thus far, there has been limited information regarding the role of TGF-β/BMP regulated miRs in SMC differentiation. The only report on this subject is one showing TGF-β1-induced miR21 expression through a post-transcriptional mechanism, leading to human pulmonary artery SMC differentiation (45). The latter finding showed peak induction of miR21 following only 2 h of TGF-β1 treatment with levels gradually declining at 20 h (45). In our unbiased array of TGF-β1-treated HCASM, miR21 was absent from the list of significantly induced miRs, possibly because of the different time sampled (24 h). Consistent with this idea, we recently found that TGF-β1-treated pulmonary artery SMCs exhibit only weak induction of miR21 following 24-h stimulation (data not shown). Our qPCR validation study demonstrated a slight increase in miR21 3 h post-TGF-β1 stimulation. It is possible that the apparent weaker induction of miR21 seen in HCASM at this time versus previous data in pulmonary artery SMCs (45) stems from the distinct embryological origins of these two SMC lineages (53). Alternatively, variations in induction of gene expression probably differ between individual cell isolates, reflecting the inherent genetic diversity of humans.
MYOCD is a molecular trigger switch for the vascular SMC differentiation program (4–7). We found a reproducible transient increase in MYOCD mRNA following TGF-β1 treatment. This temporal pattern of MYOCD expression is consistent with a previous report in transformed human cells (54). Because peak TGF-β1-induced MYOCD expression preceded that of miR143/145, we considered the possibility that MYOCD might contribute to the delay in peak miR143/145. Indeed, TGF-β1-induced miR145 was impaired when endogenous MYOCD was knocked down. Interestingly, recent studies have postulated that miR145 may augment MYOCD mRNA to establish a feed-forward mechanism for SMC differentiation (34, 35). We attempted to examine if such a mechanism was operative in HCASM but were unable to observe consistent activation of MYOCD by miR145. These results are consistent with the sustained expression of miR145 following TGF-β1 treatment versus only transient MYOCD mRNA induction. Clearly, the transcriptional and post-transcriptional regulation of MYOCD mRNA warrants deep analysis to uncover its positive regulation by TGF-β1 as well as its well known reduction in expression under conditions of SMC phenotypic adaptation (4, 55, 56).
Several lines of evidence provided here support an essential role for a p38MAPK-SRF-MYOCD axis in regulating TGF-β1-induced miR143/145 expression. First, TGF-β1 induces SRF and MYOCD in HCASM, and this induction is blunted by pretreatment with SB203580. Second, knockdown of SRF or MYOCD decreases TGF-β1-induced miR143/145 expression. Finally, luciferase assays reveal that SB203580 suppresses TGF-β1-induced miR143/145 promoter activity, and mutation of a previously defined conserved CArG box (35, 37) causes a loss in response to TGF-β1. Our results are in agreement with previous studies showing an involvement of p38MAPK in regulating SMC differentiation (14, 57). Importantly, we offer evidence that p38MAPK directly regulates the expression of SRF and MYOCD, thus providing new insight into the mechanism of TGF-β1-p38MAPK in modulating SMC differentiation. Future studies should identify downstream substrates of p38MAPK responsible for SMC differentiation.
The classical pathway for TGF-β1 signaling is through a family of SMAD proteins (16). SMAD-dependent signaling has been implicated in the transcription of smooth muscle contractile genes (11, 58). Here, we provide evidence for the involvement of SMAD signaling in TGF-β1-induced miR143/145 transcription. Mutation of a conserved SBE severely diminishes the response of the miR143/145 upstream enhancer to TGF-β1. Interestingly, this SBE site is 46 bp downstream of the CArG box. Gel shift assays using a probe flanking both elements failed to reveal a higher order complex of SRF-SMAD4 binding (data not shown). However, we cannot rule out an indirect interaction between these factors in the complex milieu of remodeled chromatin. These findings, coupled with the CArG box mutant data showing a similar loss in TGF-β1 response, suggest a dual pathway of miR143/145 transcriptional activation involving p38MAPK-SRF-MYOCD and a SMAD complex converging on their respective cis elements in the upstream miR143/145 regulatory region (Fig. 9). A previous study showed that MYOCD potentiates SMAD3-mediated transcription of the SM22 promoter in a CArG box-independent fashion (58). We did not observe an effect of MYOCD on the miR143/145 enhancer carrying a CArG box mutation, suggesting that, in this context, MYOCD does not transactivate the SBE (data not shown). Finally, p38MAPK regulates the SMAD pathway via controlling the stability of SMAD3 (59). In this context, we noted a mild inhibition of phosphorylated SMAD2 in TGF-β1-stimulated SMC pretreated with the SB203580 compound (data not shown). Further studies should examine the extent to which there is reciprocal cross-talk between the p38MAPK and SMAD pathways in the control of miR143/145 expression.
miR143/145 is a bicistronic locus under control of a common promoter (35, 37). Recently, however, there have been some curious findings that challenge this paradigm of regulation. For example, in a human stem cell model, miR145 was shown to be repressed by OCT4 via a putative OCT4 element located 1.5 kb upstream of pre-miR145 (52). The 1.5 kb promoter used in that study contains the pre-miR143 sequence, which raises the question as to whether a cryptic internal promoter exists driving only miR145 expression. Further, a conserved sequence 3′ to pre-miR145 was demonstrated to be crucial for miR145 expression in vitro (60). Because the 5′ pre-miR143 was not included, the role of this sequence in pre-miR143 expression is unclear. What is clear is that a conserved CArG box upstream of miR143/145 is necessary for specifying SMC-restricted expression of miR143/145 in mice (35, 37). The work presented here extends these findings by demonstrating a dual pathway for the transcriptional activation of miR143/145 by TGF-β1 in human SMC (Fig. 9).
In summary, we have shown that TGF- β1 induces the transcription of miR143/145 in human coronary artery SMC. Evidence supports the involvement of parallel pathways of p38MAPK and SMAD signaling converging upon two adjacent cis elements that bind SRF/MYOCD and SMAD3/4. These studies reveal a novel mechanism underlying TGF-β1-induced SMC differentiation.
Supplementary Material
Acknowledgments
We thank Sarah L. Cowan for expert technical support and the two anonymous reviewers for excellent suggestions.
Addendum
While this paper was under revision, we became aware of a related paper showing similar induction of the miR143/145 gene by TGF-β1 (61).
This work was supported, in whole or in part, by National Institutes of Health Grants HL62572 and HL091168 (to J. M. M.). This work was also supported by American Heart Association Scientist Development Grant 10SDG3670036 (to X. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–5.
- SMC
- smooth muscle cell
- SRF
- serum response factor
- MYOCD
- myocardin
- SBE
- Smad-binding element(s)
- miR
- microRNA
- HCASM
- human coronary artery smooth muscle cell(s)
- RASM
- rat aortic smooth muscle cell(s)
- qPCR
- quantitative PCR.
REFERENCES
- 1. Hirota J. A., Nguyen T. T., Schaafsma D., Sharma P., Tran T. (2009) Pulm. Pharmacol. Ther. 22, 370–378 [DOI] [PubMed] [Google Scholar]
- 2. Owens G. K., Kumar M. S., Wamhoff B. R. (2004) Physiol. Rev. 84, 767–801 [DOI] [PubMed] [Google Scholar]
- 3. Bell R. D., Deane R., Chow N., Long X., Sagare A., Singh I., Streb J. W., Guo H., Rubio A., Van Nostrand W., Miano J. M., Zlokovic B. V. (2009) Nat. Cell Biol. 11, 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen J., Kitchen C. M., Streb J. W., Miano J. M. (2002) J. Mol. Cell. Cardiol. 34, 1345–1356 [DOI] [PubMed] [Google Scholar]
- 5. Du K. L., Ip H. S., Li J., Chen M., Dandre F., Yu W., Lu M. M., Owens G. K., Parmacek M. S. (2003) Mol. Cell. Biol. 23, 2425–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yoshida T., Sinha S., Dandré F., Wamhoff B. R., Hoofnagle M. H., Kremer B. E., Wang D. Z., Olson E. N., Owens G. K. (2003) Circ. Res. 92, 856–864 [DOI] [PubMed] [Google Scholar]
- 7. Wang Z., Wang D. Z., Pipes G. C., Olson E. N. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7129–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miano J. M. (2003) J. Mol. Cell. Cardiol. 35, 577–593 [DOI] [PubMed] [Google Scholar]
- 9. Hirschi K. K., Rohovsky S. A., D'Amore P. A. (1998) J. Cell Biol. 141, 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Masszi A., Di Ciano C., Sirokmány G., Arthur W. T., Rotstein O. D., Wang J., McCulloch C. A., Rosivall L., Mucsi I., Kapus A. (2003) Am. J. Physiol. Renal Physiol. 284, F911–F924 [DOI] [PubMed] [Google Scholar]
- 11. Hu B., Wu Z., Phan S. H. (2003) Am. J. Respir. Cell Mol. Biol. 29, 397–404 [DOI] [PubMed] [Google Scholar]
- 12. Chen S., Lechleider R. J. (2004) Circ. Res. 94, 1195–1202 [DOI] [PubMed] [Google Scholar]
- 13. Sinha S., Hoofnagle M. H., Kingston P. A., McCanna M. E., Owens G. K. (2004) Am. J. Physiol. Cell Physiol. 287, C1560–C1568 [DOI] [PubMed] [Google Scholar]
- 14. Deaton R. A., Su C., Valencia T. G., Grant S. R. (2005) J. Biol. Chem. 280, 31172–31181 [DOI] [PubMed] [Google Scholar]
- 15. Tang Y., Urs S., Boucher J., Bernaiche T., Venkatesh D., Spicer D. B., Vary C. P., Liaw L. (2010) J. Biol. Chem. 285, 17556–17563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Derynck R., Zhang Y. E. (2003) Nature 425, 577–584 [DOI] [PubMed] [Google Scholar]
- 17. Hanafusa H., Ninomiya-Tsuji J., Masuyama N., Nishita M., Fujisawa J., Shibuya H., Matsumoto K., Nishida E. (1999) J. Biol. Chem. 274, 27161–27167 [DOI] [PubMed] [Google Scholar]
- 18. Bakin A. V., Tomlinson A. K., Bhowmick N. A., Moses H. L., Arteaga C. L. (2000) J. Biol. Chem. 275, 36803–36810 [DOI] [PubMed] [Google Scholar]
- 19. Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. (2010) Nature 466, 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hendrickson D. G., Hogan D. J., McCullough H. L., Myers J. W., Herschlag D., Ferrell J. E., Brown P. O. (2009) PLoS Biol. 7, e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iorio M. V., Croce C. M. (2009) J. Clin. Oncol. 27, 5848–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee Y., Samaco R. C., Gatchel J. R., Thaller C., Orr H. T., Zoghbi H. Y. (2008) Nat. Neurosci. 11, 1137–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Small E. M., Olson E. N. (2011) Nature 469, 336–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Couzin J. (2008) Science 319, 1782–1784 [DOI] [PubMed] [Google Scholar]
- 25. Small E. M., Frost R. J., Olson E. N. (2010) Circulation 121, 1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ji R., Cheng Y., Yue J., Yang J., Liu X., Chen H., Dean D. B., Zhang C. (2007) Circ. Res. 100, 1579–1588 [DOI] [PubMed] [Google Scholar]
- 27. Liu X., Cheng Y., Zhang S., Lin Y., Yang J., Zhang C. (2009) Circ. Res. 104, 476–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Davis B. N., Hilyard A. C., Nguyen P. H., Lagna G., Hata A. (2009) J. Biol. Chem. 284, 3728–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leeper N. J., Raiesdana A., Kojima Y., Chun H. J., Azuma J., Maegdefessel L., Kundu R. K., Quertermous T., Tsao P. S., Spin J. M. (2011) J. Cell. Physiol. 226, 1035–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xie C., Huang H., Sun X., Guo Y., Hamblin M., Ritchie R. P., Garcia-Barrio M. T., Zhang J., Chen Y. E. (2011) Stem Cells Dev. 20, 205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang H., Xie C., Sun X., Ritchie R. P., Zhang J., Chen Y. E. (2010) J. Biol. Chem. 285, 9383–9389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao Y., Samal E., Srivastava D. (2005) Nature 436, 214–220 [DOI] [PubMed] [Google Scholar]
- 33. Park C., Hennig G. W., Sanders K. M., Cho J. H., Hatton W. J., Redelman D., Park J. K., Ward S. M., Miano J. M., Yan W., Ro S. (2011) Gastroenterology 141, 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng Y., Liu X., Yang J., Lin Y., Xu D. Z., Lu Q., Deitch E. A., Huo Y., Delphin E. S., Zhang C. (2009) Circ. Res. 105, 158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cordes K. R., Sheehy N. T., White M. P., Berry E. C., Morton S. U., Muth A. N., Lee T. H., Miano J. M., Ivey K. N., Srivastava D. (2009) Nature 460, 705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boettger T., Beetz N., Kostin S., Schneider J., Krüger M., Hein L., Braun T. (2009) J. Clin. Invest. 119, 2634–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xin M., Small E. M., Sutherland L. B., Qi X., McAnally J., Plato C. F., Richardson J. A., Bassel-Duby R., Olson E. N. (2009) Genes Dev. 23, 2166–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Elia L., Quintavalle M., Zhang J., Contu R., Cossu L., Latronico M. V., Peterson K. L., Indolfi C., Catalucci D., Chen J., Courtneidge S. A., Condorelli G. (2009) Cell Death Differ. 16, 1590–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kong W., Yang H., He L., Zhao J. J., Coppola D., Dalton W. S., Cheng J. Q. (2008) Mol. Cell Biol. 28, 6773–6784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kato M., Putta S., Wang M., Yuan H., Lanting L., Nair I., Gunn A., Nakagawa Y., Shimano H., Todorov I., Rossi J. J., Natarajan R. (2009) Nat. Cell Biol. 11, 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun Q., Zhang Y., Yang G., Chen X., Zhang Y., Cao G., Wang J., Sun Y., Zhang P., Fan M., Shao N., Yang X. (2008) Nucleic Acids Res. 36, 2690–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wickramasinghe N. S., Manavalan T. T., Dougherty S. M., Riggs K. A., Li Y., Klinge C. M. (2009) Nucleic Acids Res. 37, 2584–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Long X., Tharp D. L., Georger M. A., Slivano O. J., Lee M. Y., Wamhoff B. R., Bowles D. K., Miano J. M. (2009) J. Biol. Chem. 284, 33671–33682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Long X., Bell R. D., Gerthoffer W. T., Zlokovic B. V., Miano J. M. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 1505–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davis B. N., Hilyard A. C., Lagna G., Hata A. (2008) Nature 454, 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hirschi K. K., Lai L., Belaguli N. S., Dean D. A., Schwartz R. J., Zimmer W. E. (2002) J. Biol. Chem. 277, 6287–6295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Streb J. W., Miano J. M. (2005) J. Biol. Chem. 280, 4125–4134 [DOI] [PubMed] [Google Scholar]
- 48. Landry J., Huot J. (1995) Biochem. Cell Biol. 73, 703–707 [DOI] [PubMed] [Google Scholar]
- 49. Fujiu K., Manabe I., Ishihara A., Oishi Y., Iwata H., Nishimura G., Shindo T., Maemura K., Kagechika H., Shudo K., Nagai R. (2005) Circ. Res. 97, 1132–1141 [DOI] [PubMed] [Google Scholar]
- 50. Shi B., Sepp-Lorenzino L., Prisco M., Linsley P., deAngelis T., Baserga R. (2007) J. Biol. Chem. 282, 32582–32590 [DOI] [PubMed] [Google Scholar]
- 51. Wang S., Bian C., Yang Z., Bo Y., Li J., Zeng L., Zhou H., Zhao R. C. (2009) Int. J. Oncol. 34, 1461–1466 [PubMed] [Google Scholar]
- 52. Xu N., Papagiannakopoulos T., Pan G., Thomson J. A., Kosik K. S. (2009) Cell 137, 647–658 [DOI] [PubMed] [Google Scholar]
- 53. Majesky M. W. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 1248–1258 [DOI] [PubMed] [Google Scholar]
- 54. Milyavsky M., Shats I., Cholostoy A., Brosh R., Buganim Y., Weisz L., Kogan I., Cohen M., Shatz M., Madar S., Kalo E., Goldfinger N., Yuan J., Ron S., MacKenzie K., Eden A., Rotter V. (2007) Cancer Cell 11, 133–146 [DOI] [PubMed] [Google Scholar]
- 55. Hendrix J. A., Wamhoff B. R., McDonald O. G., Sinha S., Yoshida T., Owens G. K. (2005) J. Clin. Invest. 115, 418–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tharp D. L., Wamhoff B. R., Turk J. R., Bowles D. K. (2006) Am. J. Physiol. Heart Circ. Physiol. 291, H2493–H2503 [DOI] [PubMed] [Google Scholar]
- 57. Martin-Garrido A., Brown D. I., Lyle A. N., Dikalova A., Seidel-Rogol B., Lassègue B., San Martín A., Griendling K. K. (2011) Free Radic. Biol. Med. 50, 354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Qiu P., Ritchie R. P., Fu Z., Cao D., Cumming J., Miano J. M., Wang D. Z., Li H. J., Li L. (2005) Circ. Res. 97, 983–991 [DOI] [PubMed] [Google Scholar]
- 59. Hayes S. A., Huang X., Kambhampati S., Platanias L. C., Bergan R. C. (2003) Oncogene 22, 4841–4850 [DOI] [PubMed] [Google Scholar]
- 60. La Rocca G., Shi B., Sepp-Lorenzino L., Baserga R. (2011) J. Cell. Physiol. 226, 602–607 [DOI] [PubMed] [Google Scholar]
- 61. Davis-Dusenbery B. N., Chan M. C., Reno K. E., Weisman A. S., Layne M. D., Lagna G., Hata A. (2011) J. Biol. Chem. 286, 28097–28110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.