Abstract
Confocal microscopy images revealed that the tetratricopeptide repeat motif (TPR) domain immunophilin FKBP51 shows colocalization with the specific mitochondrial marker MitoTracker. Signal specificity was tested with different antibodies and by FKBP51 knockdown. This unexpected subcellular localization of FKBP51 was confirmed by colocalization studies with other mitochondrial proteins, biochemical fractionation, and electron microscopy imaging. Interestingly, FKBP51 forms complexes in mitochondria with the glucocorticoid receptor and the Hsp90/Hsp70-based chaperone heterocomplex. Although Hsp90 inhibitors favor FKBP51 translocation from mitochondria to the nucleus in a reversible manner, TPR domain-deficient mutants of FKBP51 are constitutively nuclear and fully excluded from mitochondria, suggesting that a functional TPR domain is required for its mitochondrial localization. FKBP51 overexpression protects cells against oxidative stress, whereas FKBP51 knockdown makes them more sensitive to injury. In summary, this is the first demonstration that FKBP51 is a major mitochondrial factor that undergoes nuclear-mitochondrial shuttling, an observation that may be related to antiapoptotic mechanisms triggered during the stress response.
Keywords: Apoptosis, Heat-shock Protein, Immunophilin, Protein-Protein Interactions, Trafficking, Glucocorticoid Receptor, Hsp70, Hsp90, PPIase, TPR Protein
Introduction
Immunophilins comprise a family of intracellular proteins classified by their ability to bind immunosuppressant drugs. Thus, cyclophilins bind cyclosporine A, whereas FK506-binding proteins (FKBPs)3 bind FK506. The signature domain of the family is the PPIase domain, which shows peptidylprolyl-cis/trans-isomerase enzymatic activity in most of the members of the family and is also the binding site of the drug. The low molecular weight immunophilins FKBP12 and CyPA are related to the immunosuppressive effect when the drug·immunophilin complex inhibits the Ser/Thr-phosphatase activity of PP2B (calcineurin). On the other hand, high molecular weight immunophilins do not play a significant role in immunosuppression. This subfamily shows the FKBP12-like PPIase domains 1 and 2 (also called FK1 and FK2) and the tetratricopeptide repeat motif (TPR) domains (1, 2), which contain degenerative sequences of 34 amino acids repeated in tandem through which they bind to Hsp90. The FK1 domain is responsible for the ability of the proteins to bind FK506 and to confer enzymatic activity (3), and it is also the primary regulatory domain required for steroid hormone receptor regulation (4).
TPR domain proteins exhibit a large degree of sequence diversity, but the structural comparison reveals a highly conserved three-dimensional structure. Individual TPR domains are composed of two antiparallel α-helices separated by a turn. Multiple TPR domains arrange at regular angles and form a right-handed superhelix. This creates a groove with a large amount of surface area available for ligand binding (5, 6). Multiprotein complexes are assembled via this 34-amino acid scaffold motif. In addition to high molecular weight immunophilins, the TPR family includes a large variety of several factors such as the anaphase-promoting complex, the peroxisomal import receptor, NADPH oxidase complexes, the Ser/Thr phosphatase PP5, cell division cycle proteins, and the mitochondrial import receptor complex, translocase of outer mitochondrial membrane (Tom), among many others. In fact, there are more than 20,000 TPR proteins listed in the SMART database.
The biological function of TPR domain immunophilins is not well characterized and still remains poorly understood. Perhaps one of the most relevant interactions is with the TPR acceptor site of Hsp90, whose core is the MEEVD sequence of the chaperone, because via this interaction, immunophilins have been recovered bound to steroid receptor complexes (7). Most of what is known about Hsp90-binding immunophilins is derived from studies performed with steroid receptors. It was first reported that the dynein·dynactin complex binds to FKBP52, favoring the glucocorticoid receptor (GR) retrotransport (8). More recently, this property was also extended for other transcription factors such as the androgen receptor (9), the mineralocorticoid receptor (10), p53 (11), the AIF·Rac3 heterocomplex (12), and adeno-associated virus 2 (13). All these observations suggest that the association of FKBP52 with Hsp90 is relevant for the regulation of the subcellular localization of most soluble Hsp90 client proteins (14). FKBP52 is also associated with other steroid receptors such as progesterone receptor (15) and estrogen receptor (16) and to channels such as the epithelial calcium channel (17) and transient receptor potential channels (18), and it is an important positive regulator of androgen, glucocorticoid, and progesterone receptor function (for a recent review, see Ref. 19). FKBP52 knock-out mice show phenotypes consistent with hormone insensitivity syndromes, in particular androgen insensitivity syndrome (20) and glucocorticoid insensitivity syndrome (21). In prostate cancer cell lines, increased levels of FKBP52 and FKBP51 were also reported, as well as an inhibitory effect of FK506 on androgen-stimulated cell growth (22). Gene knock-out strategies revealed FKBP52, but not FKBP51, as an important facilitator of the physiological androgen receptor activity.
FKBP51 shares 60% identity and 75% similarity with FKBP52. FKBP51 has generally been regarded to be a negative regulator of steroid hormone receptor activity as its overexpression prevents positive receptor regulation by FKBP52 (10, 23, 24). Moreover, FKBP51 does not bind dynein efficiently (25), and its inhibitory actions on steroid receptor function are abrogated by mutations of its TPR domain. Recently, our laboratory has also demonstrated that FKBP52, but not FKBP51, forms complexes with structures of the nuclear pore complex (26), suggesting a role for immunophilins in the passage of client proteins through the pore. In contrast, FKBP51 favors the nuclear exclusion of corticosteroid receptors (27).
Recent studies demonstrated that in both undifferentiated neuroblastoma cells and embryonic hippocampal neurons, the early subcellular relocalization of FKBP51 and FKBP52 relates to neuronal differentiation and neurite outgrowth (28). During these studies on the subcellular redistribution of immunophilins, we noted that FKBP51 shows a peculiar cytoplasmic pattern compatible with mitochondrial localization. In the current study, we demonstrate unequivocally that FKBP51 is primarily mitochondrial in various cell lines and rat organs, that it undergoes nuclear-mitochondrial shuttling, and that it also shows antiapoptotic action when cells are exposed to oxidative stress.
EXPERIMENTAL PROCEDURES
Materials
The AC88 mouse monoclonal IgG against Hsp90 and the N27F3-4 anti-72/73-kDa heat-shock protein monoclonal IgG (anti-Hsp70) were from StressGen (Ann Arbor, MI). Rabbit polyclonal IgG against FKBP51 and the BuGR2 mouse monoclonal IgG against the GR were from Affinity BioReagents (Golden, CO). The MG19 mouse monoclonal IgG against FKBP51 was generated in the laboratory. The UPJ56 rabbit antiserum against FKBP52 was a kind gift from Dr. William Pratt. Mouse monoclonal IgG against cytochrome c (Cyt c), rabbit polyclonal IgG anti-Tom-20, and goat polyclonal IgG anti-lamin B were from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal IgGs against Cox-IV, calnexin A, or actin were from Abcam (Cambridge, UK). Rabbit polyclonal IgG against caspase 3 was from Cell Signaling (Danvers, MA). Mouse monoclonal IgG against the FLAG peptide (M2 clone), proteinase K, dexamethasone, radicicol, and alkaline phosphatase were from Sigma. Secondary antibodies labeled with Alexa Fluor Dyes (488, 546 and 647), and MitoTracker-633 dye were purchased from Molecular Probes (Eugene, OR). HRP-conjugated goat anti-rabbit was from Pierce, and the HRP-conjugated donkey anti-mouse was from Sigma. The siRNAs for FKBP51 and control siRNA were purchased from Thermo Scientific Dharmacon. All cell lines were from ATCC, except the E82.A3 (GR−/−) cell line derived from L929 fibroblasts, which was kindly gifted by Dr. Edwin Sanchez. Plasmids encoding for the PPIase domain of FKBP52 (pSG5PL-FKBP52 (Gly-32–Lys-138) and pCIneo-FLAG-hFKBP12 were generously provided by Dr. Jack M. Renoir. The plasmids pCIneo-FLAG-hFKBP51, pCIneo-FLAG-hFKBP51(K352A), and pCIneo-FLAG-hFKBP51(ΔTPR) were generously gifted by Dr. Theo Rein. The pCMV6 FLAG-TPR was a kind gift from Dr. Michael Chinkers.
Mitochondria Isolation and Fractionation
3T3-L1 fibroblasts were grown in Dulbecco's modified Eagle's medium supplemented with 10% bovine calf serum and antibiotics. When the culture reached 70% of confluence, cells were harvested by trypsinization and washed with saline phosphate buffer. Cell fractionation was achieved by following a standard protocol described in the literature (29), as well as the isolation of whole mitochondria from rat liver (30), mitochondrial fractionation (31), and proteinase K treatment of whole mitochondria (32). Treatments of FKBP51 with alkaline phosphatase followed a previously described procedure (33) in TEG buffer (Tes, pH 7.6, 50 mm NaCl, 4 mm EDTA, 10% (v/v) glycerol) supplemented with one tablet of Complete-Mini protease inhibitor mixture from Roche Diagnostics (Mannheim, Germany) per 2 ml of solution.
Microscopy Studies
Cells were grown on coverslips and processed for indirect immunofluorescence as described in previous studies (34, 35). Briefly, the cells were fixed and permeabilized with methanol at −20 °C for 10 min. They were incubated overnight at 4 °C with 1/100 dilution of primary antibody and for 1 h at room temperature with 1/200 dilution of secondary antibody. The coverslips were mounted in a glycerol-based medium with an antifade solution. Confocal microscopy images were acquired in Pascal-LSM or LSM 510 Meta confocal microscopes (Carl Zeiss, Oberkochen, Germany), using C-Apochromat 40×/1.2 NA and 63×/1.2 NA water-immersion objectives for the LSM and an EC Plan-Neofluar 40×/1.30 oil and Plan-Apochromat 63×/1.4 oil for the LSM510 Meta. Electron microscopy studies were performed in the Department of Electron Microscopy and Applied Biochemistry at the Instituto Nacional de Tecnología Agropecuaria from Castelar, Buenos Aires, using a Jeol 1200XE II microscope (magnification equal to 120,000×). For this purpose, cells were washed with PBS and fixed at room temperature for 1 h with 2% paraformaldehyde, 3% glutaraldehyde in PBS. Cell pellets were dehydrated in ethanol and propylene oxide, embedded in LR White (Polysciences, Inc. Warrington, PA), and polymerized. After thin sectioning (700 Å), samples were collected on carbon-Formvar-coated nickel grids and processed using the immunogold technique. Briefly, grids were sequentially blocked with 0.02 m glycine and 1% bovine serum albumin (BSA) in PBS and labeled with primary antibodies for 45 min at room temperature (rabbit IgG anti-FKBP51 from Affinity BioReagents and BuGR2 mouse IgG anti-GR from StressGen). Grids were washed and incubated with anti-mouse and anti-rabbit IgG conjugated to colloidal gold particles of 15 and 6 nm, respectively (Electron Microscopy Sciences, Hatfield, PA). Control labeling was performed with non-immune IgGs instead of the primary antibodies. Sections were postfixed with 2% paraformaldehyde, 0.5% glutaraldehyde, washed, and stained with 2% uranyl acetate (Sigma).
Coimmunoadsorption of FKBP51 with Mitochondrial GR
Mitochondrial extracts were prepared from rat liver (30). Purified mitochondria (100 μg) were resuspended in BLH buffer (20 mm Hepes, pH 7.9, 1 mm EDTA, 1 mm EGTA, 20 mm Na2MoO4) supplemented with one tablet of Complete-Mini protease inhibitor mix/2 ml of buffer, incubated 30 min on ice, and lysed by six freezing and thawing cycles. After centrifugation (10,000 × g for 30 min), the GR was immunoadsorbed from 250-μl aliquots of supernatant by rotation for 2.5 h at 4 °C with 14 μl of protein A-Sepharose and 3 μl of BuGR2. The immune pellets were washed four times with 1 ml of TEGM buffer (TEG buffer supplemented with 20 mm Na2MoO4). Proteins were resolved in 10% SDS-PAGE, transferred to Immobilon-P membranes, and probed with 0.2 μg/ml BuGR2 for GR, 1 μg/ml AC88 for Hsp90, 0.1% IgG anti-FKBP51, and 0.1% of N27F3-4 anti-72/73-kDa heat-shock protein monoclonal IgG. The immunoblots were then reincubated with the appropriate HRP-conjugated counter antibody, and proteins were visualized by enhanced chemiluminescence.
Cell Death Assays
Cells were transfected with 2 μg of pCIneo-hFKBP51 with TransFast reagent (Promega, Madison, WI). After 24 h, the cells were exposed to H2O2 for 16 h. Viable cells were double-counted by trypan blue exclusion in a Neubauer camera and also quantified by spectrometry at 570 nm after staining with 0.5% crystal violet as described in a previous work (12). The knockdown of FKBP51 was achieved using the commercial kit from Dharmacon following the manufacturer's instructions. Cyt c release to the cytoplasm and cleavage of procaspase 3 were evaluated by Western blotting in cells exposed to H2O2 as described for each figure.
Western Blot Scanning Densitometry
To estimate the relative population of FKBP51 in different cellular fractions or in mitochondria (within the organelle versus associated with the outer membrane), at least four Western blots were scanned and quantified using the Image J program version 1.44 from the National Institutes of Health. Results are expressed as the mean value ± S.D. Where appropriate, FKBP51 bands were normalized against the loading control protein.
RESULTS
FKBP51 Localizes in Mitochondria in Several Cell Types
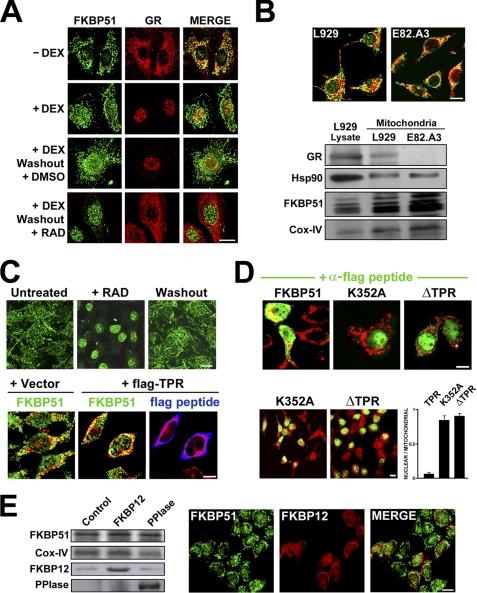
Fig. 1A shows confocal microscopy images of FKBP51 in 3T3-L1 fibroblasts suggesting the mitochondrial localization of this immunophilin. The same cytoplasmic pattern of the FKBP51 signal was obtained with a commercial rabbit IgG and a mouse monoclonal IgG (clone MG19), and the signal was abolished after knocking down FKBP51 with a specific siRNA (Fig. 1A, upper panels). FKBP51 also shows colocalization with the specific mitochondrial marker MitoTracker and the mitochondrial proteins Cyt c and cyclooxygenase IV (Cox-IV) and Tom-20 (Fig. 1A, lower panels).
FIGURE 1.
FKBP51 is localized in mitochondria. A, images by confocal microscopy of FKBP51 (green) in 3T3-L1 fibroblasts whose mitochondria were stained with MitoTracker (MT, red). The immunofluorescence studies were performed with a commercial rabbit IgG (FKBP51) or a mouse IgG clone (MG19). The expression of the immunophilin was silenced with a specific interference RNA (iRNA), and the GR (red) is shown in the same cell. The inset shows a Western blot (upper band, FKBP51; lower band, actin) for control cells (−) and siRNA-treated cells (+). B, various cells types were stained with antibody against FKBP51 (green) and MitoTracker (red). C, confocal microscopy z-scan image series for an indirect immunofluorescence of FKBP51 in NIH-3T3 fibroblasts also treated with MitoTracker. D, purified mitochondria from 3T3-L1 cells were pretreated on ice for 10 min with 1% Triton X-100 followed by an incubation of 15 min with 15 ng/ml proteinase K (+PK) in PBS or with buffer alone (−PK). The Western blot shows the partial resistance of FKBP51 to the digestion unless mitochondria are totally permeabilized. Hsp90 was used as a standard marker of a mitochondrial protein partially associated with the outer membrane, and Cox-IV was used as an inner membrane and matrix mitochondrial marker. Confocal fluorescence images for the outer membrane marker Tom-20, the intermembrane space marker Cyt c, and FKBP51 are shown. The profiles shown below correspond to the intensity of fluorescence of FKBP51 (or the background fluorescence given by a non-immune rabbit IgG). Bar in all panels = 10 μm.
Fig. 1B shows the same localization in other cell types such as L929 mouse fibroblasts, 293T human embryonic kidney cells, Cos-7 green monkey cells, and BHK-1 baby kidney hamster cells, and Fig. 1C shows a z-stack scan for NIH-3T3 cells. Such mitochondrial localization for FKBP51 has not been reported before and is totally unexpected because the analysis of the amino acid sequence of FKBP51 using several web-based algorithms (MitoPROT II, TARGET 1.1, iPSORT, WoLFPSORT, ProteinProwler, pTARGET, CELO, Pence-PA, and LOCkey) does not predict any known sequence compatible with mitochondrial targeting.
To determine whether FKBP51 is within mitochondria rather than associated with the outer membrane, purified mitochondria from 3T3-L1 fibroblasts were pretreated on ice for 10 min with 1% Triton X-100 followed by incubation with proteinase K. Fig. 1D shows that treatment with proteinase K could not abolish the presence of FKBP51 in mitochondria as the protease did for the outer membrane marker Tom-20 (see indirect immunofluorescence for purified mitochondria in the bottom panel). Similarly, the intermembrane space marker Cyt c was not affected. Western blot analysis shows that neither FKBP51 nor Cox-IV was affected unless mitochondria were previously permeabilized with detergent. Western blots indicate that a substantial amount of mitochondrial FKBP51 is localized within the organelle of these cells (64.5 ± 2.5%, a value normalized by Cox-IV signal, n = 4). About one-third of FKBP51 appears to be associated with the outer membrane facing the cytoplasm, as is suggested by the relatively faded signal observed in proteinase K-treated mitochondria. This is also seen by indirect immunofluorescence (bottom panel). In this regard, FKBP51 behaves as has recently been shown for Hsp90 (32), a chaperone that was consequently used as a comparative protein in these Western blots. By using the same methodology, intramitochondrial Hsp90 represents 66.1 ± 3.6% of the organelle pool.
Mitochondrial FKBP51 Forms Complexes with GR
To confirm the mitochondrial localization of FKBP51, a biochemical fractionation of 3T3-L1 cell extracts was accomplished, and 30 μg of proteins of each fraction were resolved by electrophoresis. Western blots of Fig. 2A show that FKBP51 is a ubiquitous protein expressed not only in the soluble fraction (as was expected) but also in the nucleus and mitochondria. Note that both GR and Hsp90, factors with which FKBP51 associates in cytosol, are also expressed in mitochondria. A scanning densitometry of the Western blot permits estimations that in 3T3-L1 cells, FKBP51 is distributed as follows: 32.4 ± 3.7% in the fraction containing microsomes and cytosol, 42.4 ± 5.1% in mitochondria, and 25.2% ± 4.7% in nuclei (100% being the addition of the three individual fractions). In other words, the mitochondrial fraction of FKBP51 is noteworthy. It should be noted that these percentages may vary with cell type and, in the same cell line, according to the conditions in which cells were grown, the number of passages, and certain stimuli (see below).
FIGURE 2.
FKBP51 forms mitochondrial heterocomplexes with the GR. A, subcellular fractionation of 3T3-L1 fibroblasts. This experiment is representative of five equivalent fractionations. μsomes, microsomes; Mito, mitochondria. B, subcellular fractionation of rat liver. C, 20 μg of protein in 100 μl of 3T3-L1 extracts were incubated at room temperature for 15 and 30 min with 1 μl (100 units) of alkaline phosphatase (AlkPase). The incubation was finished by boiling the samples with SDS sample buffer, and proteins were resolved in a 14% SDS-PAGE followed by Western blot for FKBP51. D, content of FKBP51 in mitochondria from various rat organs. E, mitochondrial fractionation (31). Mito, mitoplasts; OM, outer membrane; IM, inner membrane; IMS, intermembrane space; MM, mitochondrial matrix. F, electron microscopy of mitochondria from 3T3-L1 cells. Preparations were incubated with mouse IgG anti-GR and rabbit IgG anti-FKBP51 (I) or non-immune IgGs (NI) followed by incubation with gold-conjugated counter-antibodies (average size of the gold particles: 6 nm for the anti-rabbit IgG and 15 nm for the anti-mouse IgG). The magnification is 120,000×. Some electrodense bodies of GR and FKBP51 are magnified next to the image. The Western blot is an immunoprecipitation of mitochondrial GR.
Fig. 2B shows that FKBP51 is also present in mitochondria isolated from rat liver. It is known that FKBP51 may show more than one band according to the resolution of the gel. However, we noted that this property was variable with different preparations, even when the same percentage of polyacrylamide gel was used (for example, compare Fig. 2, A and B). Because the presence of molybdate in the homogenization buffer always protected the higher molecular weight bands, we speculated that such variability could be due to the stability of phosphorylated forms of FKBP51. This was proven by treatment of 3T3-L1 cell extracts with alkaline phosphatase, and the pattern of FKBP51 bands was compared with controls incubated without the enzyme (Fig. 2C). Next, we used a protective buffer to test the expression of FKBP51 (and its isoforms) in mitochondria isolated from different organs. Fig. 2D shows that the immunophilin is present in the mitochondria of all assayed organs, the dephosphorylated isoforms being more abundant in liver and heart than in lung, brain, or kidney. Liver mitochondrial subfractionation showed that FKBP51 is located mostly in fractions that comprise the intermembrane space (Fig. 2E).
Fig. 2F shows immunogold labeling for FKBP51 and GR by electron microscopy (secondary antibodies conjugated to 6- and 15-nm gold particles, respectively). Interestingly, mitochondrial FKBP51 is associated with GR, as suggested by those “yeast-shaped” particles (see magnifications 1–3), as well as by the GR-free population of the immunophilin (see magnification 4). A quantification of the number of both types of particles from 37 mitochondria yielded 23% combined particles, suggesting that one of every four molecules of FKBP51 could be complexed with GR in the organelle. No isolated particles of free GR were observed. To confirm that GR and FKBP51 form similar complexes like those described for the cytosolic fraction of the immunophilin, milligrams of crude mitochondria were isolated from rat livers, a procedure that yielded at least 3 orders of magnitude higher amounts of mitochondrial protein than that obtained from cells in culture, and GR was immunoprecipitated from mitochondrial lysates. The Western blot shown in Fig. 2F, right panel, shows that Hsp90, Hsp70, and FKBP51 do coimmunoprecipitate with the mitochondrial receptor.
Studies on the FKBP51 Domain Responsible for Mitochondrial Localization
Fig. 3A shows confocal microscopy images for the distribution of FKBP51 and GR in 3T3-L1 fibroblasts grown in a medium containing charcoal-stripped serum. Although GR is primarily cytoplasmic in a steroid-free medium and rapidly translocates to the nucleus with dexamethasone, FKBP51 still shows the typical mitochondrial pattern observed in all previous figures. Interestingly, cytoplasmic GR also shows a pattern compatible with mitochondrial localization and colocalizes with FKBP51. In contrast to GR, FKBP51 does not abandon the mitochondria when cells are exposed to the steroid, although these cells always showed higher nuclear accumulation of FKBP51 than cells maintained in a steroid-free medium. As expected due to the slow nuclear export rate of the GR (36), GR still remains nuclear 1 h after washing the steroid, but it is almost entirely cytoplasmic in the presence of the Hsp90 inhibitor radicicol. This is in agreement with the putative participation of the chaperone in the export mechanism of steroid receptors (37). In contrast to the GR, FKBP51 translocates to the nucleus with radicicol, suggesting that the subcellular localization and trafficking of the immunophilin is independent of that of the GR. Also, this observation suggests a possible requirement of Hsp90 for the mitochondrial localization of FKBP51. To confirm that the mitochondrial localization of the immunophilin is independent of GR, the L929-derived cell line E82.A3 was used. This GR−/− cell line also shows FKBP51 in mitochondria by both methods, confocal microscopy and subcellular fractionation (Fig. 2B).
FIGURE 3.
The TPR domain of FKBP51 is required for mitochondrial localization. A, confocal images showing that GR and FKBP51 colocalize in mitochondria. Note that dexamethasone (DEX) promotes GR nuclear accumulation, whereas FKBP51 remains in mitochondria. Steroid washout does not change this distribution pattern after 1 h unless 1 μm radicicol (RAD) is present in the medium. DMSO, dimethyl sulfoxide. B, the presence of FKBP51 in mitochondria is independent of GR. Confocal images show immunofluorescence for FKBP51 (green) in both L929 (GR+/+) and E82.A3 (GR−/−) cells costained with MitoTracker (red). The Western blot shows the expression (or not) of GR in mitochondria as compared with the expression of the immunophilin. C, upper panel, FKBP51 shuttling. 3T3-L1 cells were treated with 1 μm radicicol for 30 min. The medium was replaced by fresh medium lacking radicicol, and cells were fixed 30 min after. Note the reversible nuclear accumulation of FKBP51 in the presence of the Hsp90 inhibitor. Lower panel, overexpression of the FLAG-TPR domain (blue cells) does not affect the mitochondrial colocalization of endogenous FKBP51 (green) and MitoTracker (red). D, upper panel, cells were transfected with FLAG-tagged FKBP51, with the K352A mutant unable to bind Hsp90, or with a TPR domain deletion mutant. Although FLAG-FKBP51 colocalizes with MitoTracker (red), both FKBP51 mutants (green) are excluded from mitochondria and concentrate in the nuclei. Lower panel, the photograph shows a wide field of transfected cells, and the bar graph quantifies the proportion of nuclear to mitochondrial accumulation of the immunophilin. E, overexpression of the PPIase domain or FKBP12 does not affect FKBP51 localization. Left panel, Western blot of purified mitochondria. Right panel, indirect immunofluorescence for FKBP51 (green) and FKBP12 (red). Bars in all panels = 10 μm.
Because radicicol promoted FKBP51 accumulation in the nucleus, it was tested whether nuclear FKBP51 can cycle back to mitochondria. Therefore, the cells were exposed to radicicol for 30 min and washed. As soon as 20–30 min after the removal of the Hsp90 inhibitor, FKBP51 abandoned the nucleus and cycled back to the mitochondria (Fig. 3C, upper panels). Inasmuch as Hsp90 seems to be involved in the nuclear-mitochondrial trafficking of FKBP51, we asked whether the TPR domain of the immunophilin is required for its subcellular localization. The overexpression of FLAG-TPR peptide does not affect the localization of endogenous FKBP51, which remains mitochondrial (Fig. 3C, lower panels). Nonetheless, the overexpression of the K352A-TPR domain mutant of FKBP51, which is unable to bind Hsp90, makes FKBP51 almost entirely nuclear (Fig. 3D). A similar result was also obtained with the deletion mutant of the domain (ΔTPR-FKBP51), so ∼90% of cells transfected with either mutant show nuclear rather than mitochondrial localization for FKBP51. This suggests that the TPR domain of the immunophilin is indeed involved in mitochondrial localization, but other domains of the molecule are also required.
When the cells were transfected with the PPIase domain, no immunofluorescence images could be taken because the antibody that recognizes the PPIase peptide also reacts with endogenous immunophilins, so the technical limitation was solved by analyzing the content of FKBP51 in mitochondria by subcellular fractionation. Fig. 3E shows that neither the PPIase domain nor the overexpression of FKBP12, which shares high homology with the PPIase domain of FKBP51 and also has enzymatic activity, modifies the subcellular localization of FKBP51. These results suggest that the functional presence of the TPR domain of the immunophilin rather than its isomerase activity may be responsible for its mitochondrial localization.
FKBP51 Protects the Cells against Oxidative Stress
Next, we investigated the possible role of FKBP51 in mitochondria. One tempting speculation was that the immunophilin could be involved in the mechanism of apoptosis. Cells were consequently exposed to hydrogen peroxide for 16 h, and the subcellular localization of FKBP51 and cell viability were analyzed. The cells shown in Fig. 4A demonstrate that FKBP51 translocates to the nucleus upon the onset of oxidative stress. This is also seen by subcellular fractionation of FKBP51. Scanning densitometry of these Western blots (n = 3) suggests that the mitochondrial pool of FKBP51 is the one that concentrates in nuclei because the percentage of total immunophilin in the cytosolic fraction remains approximately constant before and after treatment with H2O2 (33.3 ± 5.1% versus 36.0 ± 3.8%), whereas the mitochondrial pool is depleted (45.2 ± 4.9% versus 11.2 ± 3.3%) in favor of the nuclear pool (21.5 ± 2.6% versus 52.8 ± 5.6%). Paralleling the observation made in Fig. 3C with radicicol, FKBP51 also cycled back to mitochondria 30 min after washing the cells with regular medium (data not shown).
FIGURE 4.
FKBP51 protects cells from oxidative stress. A, 3T3-L1 fibroblasts were treated with 500 μm H2O2 for 30 min. Photographs show an indirect immunofluorescence for FKBP51 by confocal microscopy, and Western blots show a cell fractionation where Cox-IV was used as mitochondrial marker. T, total lysate; C, cytosolic fraction; M, mitochondrial fraction; N, nuclear fraction. Bar = 10 μm. B, cell viability is improved by FKBP51 expression. 293T cells were transfected with FLAG-FKBP51 or the empty vector. Then, cells were treated for 16 h with 500 μm H2O2. The bar graph shows the number of viable transfected cells, which were quantified by indirect immunofluorescence for FKBP51 (the average transfection efficiency was ∼50%). C, cell viability is reduced by FKBP51 knockdown. The experiment is similar to that of panel B except that cells were transfected with a specific siRNA for FKBP51, and the incubation was performed with 250 μm H2O2. Note the pyknotic nuclei in the most sensitive cells. The average efficiency of siRNA transfection was ∼80%. D, cell death activates caspases via Cyt c release. The Western blot shown on the top of the panel demonstrates the efficiency of the knockdown for FKBP51. The rate of Cyt c release from mitochondria to the cytoplasm during the first hour with peroxide is shown in the lower panel. E, caspase activity is increased in FKBP51-deficient cells. Cleavage of procaspase 3 (pro) for those cells shown in panel C incubated with 250 μm H2O2. Actin was used as loading control.
As expected, treatment with H2O2 diminished the number of attached cells to the dish to 30% of the control, and those cells that remained attached showed pyknotic nuclei (Fig. 4B). Overexpression of FKBP51 prevented the deleterious action of the peroxide. Accordingly, cell death was greatly increased after knocking down FKBP51 (Fig. 4C) to the point that the concentration of H2O2 used for the latter assay must be reduced to half to avoid massive cell detachment. Fig. 4D shows the impaired expression of FKBP51 in siRNA-treated cells (upper panel), whereas the lower panel shows the concomitant increased release of mitochondrial Cyt c to the cytosol. Consistent with this, Fig. 4E shows higher caspase activity in FKBP51-deficient cells. Taken together, these results suggest that the unexpected mitochondrial localization of FKB51 may be related to an antiapoptotic effect.
DISCUSSION
High molecular weight immunophilins are primarily regarded as regulators of the steroid receptor function because they were first described as members of those heterocomplexes. Nonetheless, this subfamily of proteins must have a number of still unknown functions, some of which could be related to nuclear receptor function, but others may be totally independent of them. In this work, we have reported a number of definitive evidences showing that FKBP51 is a mitochondrial protein. In view of the substantial proportion of mitochondrial FKBP51, it seems that this subcellular localization must be the major one when FKBP51 is not nuclear.
Our results demonstrate that the TPR domain of FKBP51 is required for its mitochondrial localization, and when its functional interaction with Hsp90 is disrupted by drugs such as radicicol or due to mutations, FKBP51 is no longer mitochondrial and concentrates in the nucleus. FKBP51 lacks a known mitochondrial localization signal and possesses a nuclear localization sequence that appears to be dominant in the overall balance for its subcellular localization when the immunophilin cannot be targeted to mitochondria.
Because TPR domain immunophilins were first described associated with steroid receptors via the C-terminal end sequence MEEVD of Hsp90, it has always been extrapolated that immunophilins are cytosolic or nuclear factors (38–40). This study demonstrates that this is not the case for FKBP51, which is also located in mitochondria in a TPR domain-dependent fashion. Whether FKBP51 may use the Hsp90/Hsp70-dependent mechanism for mitochondrial import is still uncertain, although it is possible in view of the requirement of the TPR domain of FKBP51. Recently, it was shown that neuronal FKBP51 also forms complexes with Hsc70 (28). Previous studies have already demonstrated that Hsp70 selectively targets the TPR domains in SGT1 (41) and heat-shock organizing protein (Hop) (42), whereas CyP40 (43) and carboxy terminus of Hsc-70 interacting protein (CHIP) (44) are less discriminatory and can bind either Hsp90 or Hsc70. The outer mitochondrial membrane transporter Tom-70 is also a TPR protein able to provide a docking site for both chaperones, favoring the mitochondrial import of proteins (45, 46). This makes Tom-70 a possible carrier for FKBP51. However, how FKBP51 is actually imported to mitochondria in a TPR domain-dependent manner remains to be elucidated.
The current functional models suggest that the TPR domains of Tom proteins recognize chaperones bound to the precursor protein rather than the targeted precursor protein itself, and there is also evidence that more than one cytosolic component could actually function as a chaperone-like protein (53). Moreover, it is clear that binding to the Tom complex may not be a prerequisite for mitochondrial import of proteins, which can occur even if the binding of the cargo protein to receptor components is poor. In short, although FKB51 does not show a known mitochondrial localization signal, there are alternative manners by which the immunophilin could be imported. It is noteworthy that evolutionary analyses showed that the TPR domains of FKBP51 are closely related to Tom-34 (54). Importantly, our data also suggest that when FKBP51 cannot be imported to mitochondria, it is mainly located in the nucleus, such that mitochondria may act as a class of reservoir for the immunophilin maintaining FKBP51 isolated from the nucleus.
The fact that the TPR domain of FKBP51 is essential is intriguing, but there is no competition by overexpression of the TPR peptide alone. This type of competition has been used before to analyze the relevance of the TPR protein·Hsp90 association (26, 27, 47–49). Clearly, the ineffective ability of the TPR peptide alone to prevent FKBP51 import to mitochondria indicates the need of additional sequences of the immunophilin involved in the effective mechanism of translocation. An alternative mechanism that cannot be ruled out includes interactions with other proteins.
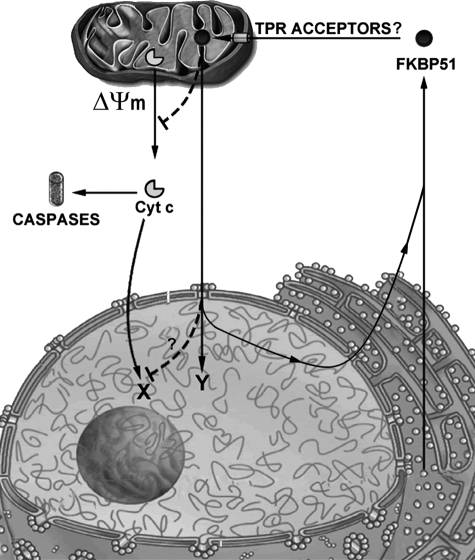
It is accepted that most soluble proteins are not confined to the cytoplasm or the nucleus in a static manner but are capable of shuttling dynamically through the nuclear pore (14, 38, 50). On the other hand, nuclear-mitochondrial shuttling is less frequently reported. Fig. 5 summarizes a model by which FKBP51 undergoes nuclear-mitochondrial shuttling. After translation, the FKBP51 population that is not in complexes with other soluble factors (for example, steroid receptors) can be translocated to mitochondria in a TPR domain-dependent manner. Changes in the membrane potential upon exposure to oxidants promotes the release of cytochrome c with the subsequent cleavage of procaspases (51). Our data show that although FKBP51 is also released from mitochondria (Fig. 4A), it has a protective role (Fig. 4, D and E), which could also be possible for the nuclear action of Cyt c on its sites of action (Fig. 5, shown as X) where it promotes chromatin condensation (52). In turn, FKBP51 also shows its own nuclear effects (Fig. 5, shown as Y), for example, modulation of the function of the steroid receptors. Actually, it should be noted that the protective mechanism of this immunophilin does not require its localization in mitochondria, but exactly the opposite. When the situation is back to normal, FKBP51 rapidly cycles back to mitochondria, closing its circular cycle.
FIGURE 5.
Integrated model for FKBP51 nuclear-mitochondrial shuttling. Soluble cytoplasmic FKBP51 can be translocated into mitochondria, perhaps via TPR domain acceptor factors. Mitochondrial membrane depolarization (ΔΨm) promotes Cyt c release from the organelle and caspase activation. FKBP51 impairs Cyt c release and, perhaps, Cyt c nuclear effects (question mark). When the stimulus ends, FKBP51 cycles back to mitochondria. X and Y refer to the nuclear sites of action for Cyt c and FKBP51 respectively.
The presence of the GR in mitochondria was first reported by Sekeris laboratory (55). It is known that the interaction of the receptors with regulatory elements of the mitochondrial genome and the activation of gene transcription underlie the hormonal stimulation of energy yield. Furthermore, the interaction of the receptors with apoptotic/antiapoptotic factors is possibly associated with the survival-death effects of the hormones (29). Glucocorticoids protect cells of epithelial origin against apoptotic stimuli (56). However, these hormones are also proapoptotic agents for cells of the hematopoietic system, such as monocytes, macrophages, thymocytes, and leukemic cells (57). Glucocorticoids induce a translocation of the cognate receptor from the cytoplasm to mitochondria, whereas in glucocorticoid-resistant cells (such as those used in our study), no such movement of GR is evident. Importantly, GR targeting to mitochondria in these cells by linking the receptor to a mitochondrial localization signal results in an apoptotic effect, irrespective of the presence of steroid. Perhaps the presence of GR·FKBP51 complexes in mitochondria may prevent the deleterious action of mitochondrial GR on cell survival due to the inhibitory effect of FKBP51 on the receptor action, just like it has been shown for the genomic effects.
Although our results demonstrate that the immunophilin forms complexes with the GR, FKBP51 also exists in forms not associated with this receptor (Fig. 2F), so it is possible that the latter population of FKBP51 may form complexes with other mitochondrial factors. In this regard, it has been shown that signaling cascades of p53 (11) and NF-κB (58) also involve immunophilins. Because both p53 and NF-κB are mitochondrial proteins, it could be possible that FKBP51 is an interacting factor with these proteins in the organelle. The presence in mitochondria of regulatory molecules regarded as solely involved in nuclear actions opens novel perspectives in deepening our knowledge on the physiology of mitochondria and on understanding the complexity of the interaction of this organelle with other cell compartments. The findings reported in this work open new prospects to unravel novel mitochondrial mechanisms that may regulate the antiapoptotic action and also create new perspectives for the development of selective agents able to regulate the antiapoptotic action by affecting the specific protective network of the organelle.
Acknowledgments
We are indebted to Drs. William Pratt, Jack M. Renoir, David Smith, Theo Rein, Marc Cox, Eddie Sanchez, and Michael Chinkers for providing essential reagents used in this study.
This work was supported by grants from the Guggenheim Foundation, Universidad de Buenos Aires (UBACyT Program), and Agencia Nacional de Promoción Científica y Tecnológica ProyectoS de Investigación Científica y Tecnológica (ANPCyT PICT) 2010-1170 (to M. D. G.) and by National Institutes of Health Fogarty International Research Collaboration Award (FIRCA) Award R03TW008143-01A1 and ANPCyT PICT 2007-00640 (to G. P. P.).
- FKBP
- FK506-binding protein
- TPR
- tetratricopeptide repeat
- PPIase
- peptidyl-(cis/trans)-isomerase
- Cyt c
- cytochrome c
- Tom
- translocase of outer mitochondrial membrane
- GR
- glucocorticoid receptor
- Tes
- 10 mm N-tris(hydroxymethyl)methyl-2-amino-ethane-sulfonic acid
- h
- human.
REFERENCES
- 1. Sinars C. R., Cheung-Flynn J., Rimerman R. A., Scammell J. G., Smith D. F., Clardy J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu B., Li P., Liu Y., Lou Z., Ding Y., Shu C., Ye S., Bartlam M., Shen B., Rao Z. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8348–8353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pirkl F., Buchner J. (2001) J. Mol. Biol. 308, 795–806 [DOI] [PubMed] [Google Scholar]
- 4. Riggs D. L., Roberts P. J., Chirillo S. C., Cheung-Flynn J., Prapapanich V., Ratajczak T., Gaber R., Picard D., Smith D. F. (2003) EMBO J. 22, 1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Das A. K., Cohen P. W., Barford D. (1998) EMBO J. 17, 1192–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lamb J. R., Tugendreich S., Hieter P. (1995) Trends Biochem. Sci. 20, 257–259 [DOI] [PubMed] [Google Scholar]
- 7. Pratt W. B., Toft D. O. (1997) Endocr. Rev. 18, 306–360 [DOI] [PubMed] [Google Scholar]
- 8. Galigniana M. D., Radanyi C., Renoir J. M., Housley P. R., Pratt W. B. (2001) J. Biol. Chem. 276, 14884–14889 [DOI] [PubMed] [Google Scholar]
- 9. Thomas M., Dadgar N., Aphale A., Harrell J. M., Kunkel R., Pratt W. B., Lieberman A. P. (2004) J. Biol. Chem. 279, 8389–8395 [DOI] [PubMed] [Google Scholar]
- 10. Gallo L. I., Ghini A. A., Piwien Pilipuk G., Galigniana M. D. (2007) Biochemistry 46, 14044–14057 [DOI] [PubMed] [Google Scholar]
- 11. Galigniana M. D., Harrell J. M., O'Hagen H. M., Ljungman M., Pratt W. B. (2004) J. Biol. Chem. 279, 22483–22489 [DOI] [PubMed] [Google Scholar]
- 12. Colo G. P., Rubio M. F., Nojek I. M., Werbajh S. E., Echeverría P. C., Alvarado C. V., Nahmod V. E., Galigniana M. D., Costas M. A. (2008) Oncogene 27, 2430–2444 [DOI] [PubMed] [Google Scholar]
- 13. Zhao W., Zhong L., Wu J., Chen L., Qing K., Weigel-Kelley K. A., Larsen S. H., Shou W., Warrington K. H., Jr., Srivastava A. (2006) Virology 353, 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Galigniana M. D., Echeverría P. C., Erlejman A. G., Piwien-Pilipuk G. (2010) Nucleus 1, 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kosano H., Stensgard B., Charlesworth M. C., McMahon N., Toft D. (1998) J. Biol. Chem. 273, 32973–32979 [DOI] [PubMed] [Google Scholar]
- 16. Ratajczak T., Carrello A. (1996) J. Biol. Chem. 271, 2961–2965 [DOI] [PubMed] [Google Scholar]
- 17. Gkika D., Topala C. N., Hoenderop J. G., Bindels R. J. (2006) Am. J. Physiol. Renal Physiol. 290, F1253–F1259 [DOI] [PubMed] [Google Scholar]
- 18. Sinkins W. G., Goel M., Estacion M., Schilling W. P. (2004) J. Biol. Chem. 279, 34521–34529 [DOI] [PubMed] [Google Scholar]
- 19. Sivils J. C., Storer C. L., Galigniana M. D., Cox M. B. (2011) Curr. Top. Pharmacol. 11, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cheung-Flynn J., Prapapanich V., Cox M. B., Riggs D. L., Suarez-Quian C., Smith D. F. (2005) Mol. Endocrinol. 19, 1654–1666 [DOI] [PubMed] [Google Scholar]
- 21. Warrier M., Hinds T. D., Jr., Ledford K. J., Cash H. A., Patel P. R., Bowman T. A., Stechschulte L. A., Yong W., Shou W., Najjar S. M., Sanchez E. R. (2010) Endocrinology 151, 3225–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Periyasamy S., Warrier M., Tillekeratne M. P., Shou W., Sanchez E. R. (2007) Endocrinology 148, 4716–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cox M. B., Smith D. F. (2006) in The Networking of Chaperones by Cochaperones (Blatch G. L. ed) pp. 13–25, Eurekah Bioscience, Austin, TX [Google Scholar]
- 24. Davies T. H., Ning Y. M., Sánchez E. R. (2002) J. Biol. Chem. 277, 4597–4600 [DOI] [PubMed] [Google Scholar]
- 25. Wochnik G. M., Rüegg J., Abel G. A., Schmidt U., Holsboer F., Rein T. (2005) J. Biol. Chem. 280, 4609–4616 [DOI] [PubMed] [Google Scholar]
- 26. Echeverría P. C., Mazaira G., Erlejman A., Gomez-Sanchez C., Piwien Pilipuk G., Galigniana M. D. (2009) Mol. Cell. Biol. 29, 4788–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galigniana M. D., Erlejman A. G., Monte M., Gomez-Sanchez C., Piwien-Pilipuk G. (2010) Mol. Cell. Biol. 30, 1285–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Quintá H. R., Maschi D., Gomez-Sanchez C., Piwien-Pilipuk G., Galigniana M. D. (2010) J. Neurochem. 115, 716–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Psarra A. M., Sekeris C. E. (2009) Biochim. Biophys. Acta 1787, 431–436 [DOI] [PubMed] [Google Scholar]
- 30. Fernández-Vizarra E., López-Pérez M. J., Enriquez J. A. (2002) Methods 26, 292–297 [DOI] [PubMed] [Google Scholar]
- 31. Gottlieb E., Armour S. M., Thompson C. B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12801–12806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kang B. H., Plescia J., Dohi T., Rosa J., Doxsey S. J., Altieri D. C. (2007) Cell 131, 257–270 [DOI] [PubMed] [Google Scholar]
- 33. Galigniana M. D. (1998) Biochem. J. 333, 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Galigniana M. D., Morishima Y., Gallay P. A., Pratt W. B. (2004) J. Biol. Chem. 279, 55754–55759 [DOI] [PubMed] [Google Scholar]
- 35. Galigniana M. D., Harrell J. M., Housley P. R., Patterson C., Fisher S. K., Pratt W. B. (2004) Brain Res. Mol. Brain Res. 123, 27–36 [DOI] [PubMed] [Google Scholar]
- 36. Galigniana M. D., Housley P. R., DeFranco D. B., Pratt W. B. (1999) J. Biol. Chem. 274, 16222–16227 [DOI] [PubMed] [Google Scholar]
- 37. Tago K., Tsukahara F., Naruse M., Yoshioka T., Takano K. (2004) Mol. Cell. Endocrinol. 213, 131–138 [DOI] [PubMed] [Google Scholar]
- 38. Pratt W. B., Galigniana M. D., Harrell J. M., DeFranco D. B. (2004) Cell. Signal. 16, 857–872 [DOI] [PubMed] [Google Scholar]
- 39. Ratajczak T., Ward B. K., Minchin R. F. (2003) Curr. Top. Med. Chem. 3, 1348–1357 [DOI] [PubMed] [Google Scholar]
- 40. Barik S. (2006) Cell. Mol. Life Sci. 63, 2889–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu F. H., Wu S. J., Hu S. M., Hsiao C. D., Wang C. (1999) J. Biol. Chem. 274, 34425–34432 [DOI] [PubMed] [Google Scholar]
- 42. Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. (2000) Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 43. Carrello A., Ingley E., Minchin R. F., Tsai S., Ratajczak T. (1999) J. Biol. Chem. 274, 2682–2689 [DOI] [PubMed] [Google Scholar]
- 44. King F. W., Wawrzynow A., Höhfeld J., Zylicz M. (2001) EMBO J. 20, 6297–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schmidt U., Wochnik G. M., Rosenhagen M. C., Young J. C., Hartl F. U., Holsboer F., Rein T. (2003) J. Biol. Chem. 278, 4926–4931 [DOI] [PubMed] [Google Scholar]
- 46. Fan A. C., Bhangoo M. K., Young J. C. (2006) J. Biol. Chem. 281, 33313–33324 [DOI] [PubMed] [Google Scholar]
- 47. Silverstein A. M., Galigniana M. D., Kanelakis K. C., Radanyi C., Renoir J. M., Pratt W. B. (1999) J. Biol. Chem. 274, 36980–36986 [DOI] [PubMed] [Google Scholar]
- 48. Harrell J. M., Murphy P. J., Morishima Y., Chen H., Mansfield J. F., Galigniana M. D., Pratt W. B. (2004) J. Biol. Chem. 279, 54647–54654 [DOI] [PubMed] [Google Scholar]
- 49. Aviezer-Hagai K., Skovorodnikova J., Galigniana M., Farchi-Pisanty O., Maayan E., Bocovza S., Efrat Y., von Koskull-Döring P., Ohad N., Breiman A. (2007) Plant Mol. Biol. 63, 237–255 [DOI] [PubMed] [Google Scholar]
- 50. DeFranco D. B. (2002) Mol. Endocrinol. 16, 1449–1455 [DOI] [PubMed] [Google Scholar]
- 51. Poyton R. O., Ball K. A., Castello P. R. (2009) Trends Endocrinol. Metab. 20, 332–340 [DOI] [PubMed] [Google Scholar]
- 52. Nur-E-Kamal A., Gross S. R., Pan Z., Balklava Z., Ma J., Liu L. F. (2004) J. Biol. Chem. 279, 24911–24914 [DOI] [PubMed] [Google Scholar]
- 53. Mukhopadhyay A., Avramova L. V., Weiner H. (2002) Arch. Biochem. Biophys. 400, 97–104 [DOI] [PubMed] [Google Scholar]
- 54. Schlegel T., Mirus O., von Haeseler A., Schleiff E. (2007) Mol. Biol. Evol. 24, 2763–2774 [DOI] [PubMed] [Google Scholar]
- 55. Demonacos C., Tsawdaroglou N. C., Djordjevic-Markovic R., Papalopoulou M., Galanopoulos V., Papadogeorgaki S., Sekeris C. E. (1993) J. Steroid Biochem. Mol. Biol. 46, 401–413 [DOI] [PubMed] [Google Scholar]
- 56. Evans-Storms R. B., Cidlowski J. A. (1995) J Steroid Biochem. Mol. Biol. 53, 1–8 [DOI] [PubMed] [Google Scholar]
- 57. Herr I., Gassler N., Friess H., Büchler M. W. (2007) Apoptosis 12, 271–291 [DOI] [PubMed] [Google Scholar]
- 58. Giordano A., Avellino R., Ferraro P., Romano S., Corcione N., Romano M. F. (2006) Am. J. Physiol. Heart Circ. Physiol. 290, H2459–H2465 [DOI] [PubMed] [Google Scholar]