Abstract
Prostate-derived sterile 20-like kinases (PSKs) 1-α, 1-β, and 2 are members of the germinal-center kinase-like sterile 20 family of kinases. Previous work has shown that PSK 1-α binds and stabilizes microtubules whereas PSK2 destabilizes microtubules. Here, we have investigated the activation and autophosphorylation of endogenous PSKs and show that their catalytic activity increases as cells accumulate in G2/M and declines as cells exit mitosis. PSKs are stimulated in synchronous HeLa cells as they progress through mitosis, and these proteins are activated catalytically during each stage of mitosis. During prophase and metaphase activated PSKs are located in the cytoplasm and at the spindle poles, and during telophase and cytokinesis stimulated PSKs are present in trans-Golgi compartments. In addition, small interfering RNA (siRNA) knockdown of PSK1-α/β or PSK2 expression inhibits mitotic cell rounding as well as spindle positioning and centralization. These results show that PSK catalytic activity increases during mitosis and suggest that these proteins can contribute functionally to mitotic cell rounding and spindle centralization during cell division.
Keywords: Cell Division, Cytoskeleton, Microtubules, Mitosis, Signal Transduction, PSK, Positioning, STE20, Spindle, TAOK
Introduction
The sterile 20 (STE20)3 group of mammalian protein kinases includes 28 proteins, which often act upstream of mitogen-activated protein kinase (MAPK) signaling pathways and regulate a diverse array of processes, including gene transcription, cell cycle progression, stress responses, cytoskeletal organization, and apoptosis (1–3). STE20s divide into two subfamilies according to their structure and regulation: six p21-activated kinases (PAKs), which have a C-terminal catalytic domain and an N-terminal Cdc42/Rac-interacting and binding domain (CRIB) and 22 germinal center kinase (GCK)-like kinases, which possess an N-terminal catalytic domain but no CRIB. PAKs can bind Rac or Cdc42 GTPases via their CRIB domain and act as downstream effectors to regulate the actin cytoskeleton (4–6), but much less is known about the GCKs and their upstream activators or downstream targets.
Prostate-derived STE20-like kinases (PSKs, also referred to as TAO kinases) have been classified as members of the GCK VIII subfamily of STE20 kinases and include PSK1-α and PSK1-β (splice variants with identical N termini, TAOK2 isoforms two and one, respectively), PSK2 (TAOK1) and PSK3 (TAOK3) (7–10). Each of these proteins appears to be expressed ubiquitously (7, 11, 12). PSK1-α and PSK2, but not PSK1-β or PSK3, can stimulate c-Jun N-terminal kinase (JNK) MAPK (7–10) and induce apoptotic morphological changes via their stimulation of JNK, caspases, and Rho kinase-1 (8, 9). PSK1-α and PSK2 also activate p38 MAPK (11, 13, 14). PSK1-α binds to microtubules (MTs) via its C terminus (amino acids 745–1235) and produces stabilized perinuclear MT cables that are nocodazole-resistant and contain increased levels of acetylated α-tubulin (lysine 40) (15). JNK and caspase-mediated cleavage of PSK1-α can remove the C-terminal MT-binding domain, permitting the N-terminal catalytic region of PSK1-α to relocate to the nucleus and induce apoptotic morphology, and PSK1-α can also down-regulate actin stress fibers (7, 9). In contrast, PSK2 (also referred to as MARK kinase) induces MT destabilization via activation of MT affinity-regulating kinase (MARK/PAR-1) and phosphorylation of MT-associated proteins (MAPs, e.g. tau), which dissociate from MTs resulting in their disassembly (16–18). Small interfering RNA (siRNA) knockdown of PSK2 shows that this protein is needed for neuritogenesis to occur (16). Much less is known about PSK1-β or PSK3 functions, but the opposing effects on MT stability and dynamics of PSK1-α and PSK2 suggest potential functional roles for this protein kinase family in regulating MT-dependent cellular processes.
Most of the work published previously on the PSK family of GCK-like kinases has focused predominantly on the analysis of transfected and overexpressed proteins, and few studies have examined the expression or activation of endogenous PSKs (7, 9). Here, we have used an antibody that recognizes catalytically active and phosphorylated PSKs 1-α, 1-β and 2 as well as additional PSK1-α/β- or PSK2-specific antibodies, to examine the endogenous proteins. PSKs are known to regulate the MT and actin cytoskeleton, and this study focuses on their expression and activation during mitosis, when dramatic alterations in the cytoskeleton occur and are required for cell division. We show that PSK1-α/β and PSK2 are activated catalytically during mitosis and that these proteins are needed for mitotic cell rounding and spindle positioning.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
pRK5-Myc PSK1-α, pRK5-Myc-PSK1-α (K57A), pRK5-Myc PSK1-β, pRK5-Myc-PSK1-β (K57A), pRK5-Myc-PSK2, and pRK5-Myc-PSK2 (K57A) were made using methods described previously (8, 9) and PSK1-α (K57A), subcloned into the pN-GFP-CB6 vector to express GFP-tagged protein. DAPI, propidium iodide, ribonuclease A, thymidine, nocodazole, paclitaxel, and mouse anti-α-tubulin or mouse anti-γ-tubulin were obtained from Sigma-Aldrich. Rabbit anti-ERK1, goat anti-PSK1-α/β, and mouse-anti-Myc antibodies were purchased from Santa Cruz Biotechnology, and mouse anti-PSK2 and rat anti-TGN38 antibodies were obtained from BD Biosciences. Mouse anti-cyclin A antibody was a gift from Dr. Tim Hunt. Affinity-purified rabbit PSK-Ser(P)-181 antibody and blocking peptide CPANS(P)FVGTC were made as described previously (Eurogentec) (9, 19). Recombinant PSK1-α/β and PSK2 proteins were obtained from SignalChem. PSK1 (TAOK2) and PSK2 (TAOK1) Smartpools (Sp) were purchased from Dharmacon. PSK1-Sp contained the four oligonucleotides TAOK2-si #1–4 with the respective sequences (5′-CUACAAACUUCGCAAGGAA-3′, 5′-GCAGUACGAUGGCAAAGUG-3′, 5′-GAGGUGCGGUUCUUACAGA-3′, 5′-GCUCUGACAACCUAUAUGA-3′), and PSK2-Sp contained the four oligonucleotides TAOK1-si #1–4 with the respective sequences (5′-CCAAGUAUCUCGUCACAAA-3′, 5′-UAAUAUGGUCCUUUCUAA-3′, 5′-CUAAAGUGAUGUCCAAUGA-3′, and 5′-GCUGUGAGUUGAUCAGAUU-3′). HeLa cells expressing mCherry-tubulin stably were a gift from Dr. Juan Martin-Serrano (20).
Cell Culture and siRNA Transfection
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS and antibiotics (10% CO2, 37 °C). For siRNA transfection 3 × 105 cells/3 ml of medium were seeded onto 60-mm Petri dishes containing 6 round glass coverslips (13 mm, VWR). After 16 h 22.5 μl of siRNA (20 μm stock) and 18 μl of HiPerFect (Qiagen) were added to 250 μl of OptiMEM (Invitrogen), mixed for 10 min at room temperature, and added to cultures in 3 ml of fresh medium without antibiotics, to provide a final siRNA concentration of 150 nm. For plasmid transfection HeLa cells (1.5 × 105) were seeded on 35-mm Petri dishes, and after 16 h the indicated plasmids (1 μg of DNA, 200 μl of OptiMEM, and 3 μl of Lipofectamine 2000 (Invitrogen)) were transfected into cells in medium without antibiotics for 4 h before replacement with normal growth medium.
Synchronous and Mitotic Cell Preparations
HeLa cells were used in the experiments described here as they can be synchronized using double thymidine blocks and provide a well characterized model system for cell cycle studies. HeLa cells were seeded on 60-mm dishes containing coverslips as described above. To prepare synchronous populations of HeLa cells, cultures were incubated in thymidine (2 mm) for 19 h, released into normal medium for 9 h, incubated in thymidine (2 mm) for 17 h, and released into normal medium. At the times indicated in the text, cells on coverslips were fixed in 4% paraformaldehyde/PBS (15 min, room temperature) for immunostaining and the remaining cells on dishes lysed in sample buffer for immunoblotting. To prepare semisynchronous HeLa cells, cultures were treated once with thymidine for 24 h, incubated in normal medium for 10 h, and then fixed in 4% paraformaldehyde/PBS for (15 min, room temperature) when a significant number of cells were in mitosis. For siRNA and PSK knockdown effects on mitosis, siRNA-transfected cells were incubated for 14 h before the addition of thymidine to the medium for 24 h and an additional incubation for 10 h in normal medium prior to fixation in 4% paraformaldehyde/PBS (15 min, room temperature), when a significant number of cells were in mitosis (48-h knockdown). To prepare semisynchronous cells in mitosis using nocodazole, cultures were treated with thymidine (2 mm) for 24 h, released into the cell cycle for 3 h, and then incubated with nocodazole (0.5 μm) for 12 h before analysis by immunoblotting or FACS.
Immunoblotting
Cultures were lysed in 200 μl of lysis buffer (1% Nonidet P-40, 130 mm NaCl, 1 mm dithiothreitol, 2 μg/ml leupeptin, 2 μg/ml aprotinin, 10 mm NaF, 0.1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, and 20 mm Tris, pH 7.4). 50 μg of total protein were separated using 8–15% SDS-PAGE and transferred to nitrocellulose. Immunoblotting was carried out as described previously (7).
Immunofluorescence and Confocal or Time Lapse Video Microscopy
For immunofluorescence experiments paraformaldehyde-fixed cells on coverslips were permeabilized with 0.2% Triton-X100 in PBS (5 min) and co-stained with the indicated primary antibodies in blocking buffer (PBS/20% goat serum) followed by appropriate secondary antibodies coupled to Alexa Fluor dyes (1:400, Invitrogen) and DAPI (3 μm). Cells were imaged with a Zeiss LSM 510 confocal laser-scanning microscope or Nikon TE-2000 microscope (time lapse).
Mitotic Cell Analysis
Metaphase cells with spindles parallel to the substratum were selected at random, and collected confocal images were exported to Adobe Photoshop (CS3), where each cell was overlaid with a 1-μm scale bar rotated to pass through both spindle poles. Distances between each pole and the cell membrane were measured and the data presented as the percentages of cells with decentralized spindles and the ratio of the longest over the shortest length for each cell (decentralization index). Cell rounding was examined by rotating the scale bar around the metaphase cell center to determine distances to the cell membrane, and changes in cell shape were scored as equal or unequal.
FACS Analysis
Cells were detached using Trypsin/EDTA (Sigma-Aldrich), pelleted by centrifugation (400 × g, 3 min), and resuspended in 300 μl of DMEM and 700 μl of ethanol. Fixed cells were pelleted again and resuspended in 0.2 ml of staining solution (40 μg/ml propidium iodide, 500 μg/ml ribonuclease A (boiled at 100 °C for 15 min)). Samples were analyzed using a BD Bioscience FACScan with filter sets to detect propidium iodide (585 nm). Doublets were removed by only including cells that were within linear gates for forward scatter versus forward scatter peak, for propidium iodide staining versus propidium iodide peak. For flow cytometry plots of DNA content, gates were set as 2n (G0/G1), 2n<x<4n (S), and 4n (G2/M).
RESULTS
Nocodazole Stimulates PSK Activity and Phosphorylation
PSK1-α and PSK1-β are isoforms produced by a single gene and the proteins are identical over their N-terminal kinase domains (amino acids 1–744) but possess different C termini (amino acids 745–1235 or 745–1049, respectively) (7–9). Previously we have used a phospho-antibody that only detects forms of PSK1-α or PSK1-β, which are catalytically active in in vitro kinase assays and undergo autophosphorylation on serine 181 as part of their conformational activation (9, 21). The PSK-Ser(P)-181 antibody was unable to detect point mutated PSK1-α (K57A) or PSK1-β (K57A), which are kinase-deficient and catalytically inactive in in vitro kinase assays, showing that this reagent specifically recognizes the catalytically active and autophosphorylated forms of each protein (9).
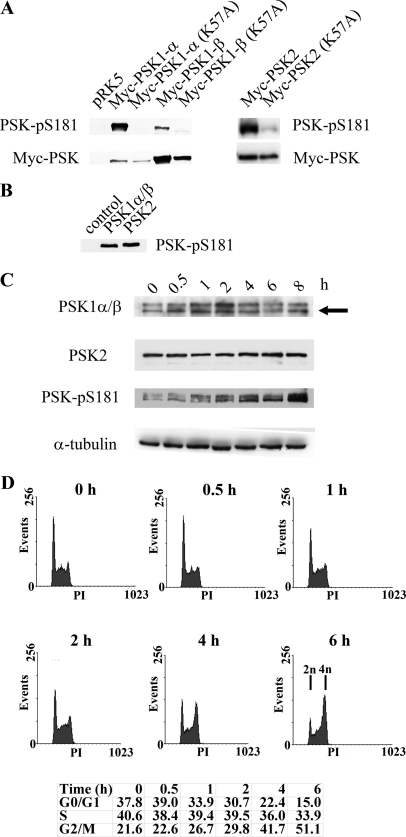
Although the antibody was originally made using data obtained from the PSK1-α crystal structure, the PANS181(P)FVGT sequence and epitope used to generate the PSK-Ser(P)-181 antibody are also present in PSK2. Immunoblotting experiments of kinase active and dead PSK transfected cell lysates show that this reagent also recognizes PSK2 and detects the activated form of the protein (Fig. 1A). In addition, the antibody recognizes purified, catalytically active, and phosphorylated recombinant PSK2, as well as PSK1-α and PSK1-β, which share identical N termini (Fig. 1B) (9). The ability of this antibody to recognize stimulated and phosphorylated forms of PSK1-α, PSK1-β, and PSK2 (PSK-Ser(P)-181) provides an opportunity to investigate the catalytic activity of these proteins at endogenous levels. PSK1-α, PSK1-β, and PSK2 do however have similar apparent molecular masses of 165, 160, and 150 kDa, respectively, and additional antibodies were obtained to detect endogenous PSK1-α/β (targeting their identical N termini) and PSK2 expression (8, 9). The antibodies were tested and able to detect transfected and overexpressed PSK1-α and PSK1-β or PSK2 by immunoblotting (data not shown). As PSK1-α binds and stabilizes MTs and PSK2 destabilizes MTs, these antibodies were used initially to investigate whether PSK expression and/or catalytic activity responds to drug-induced changes in MT dynamics (15, 16). HeLa cells were treated with nocodazole to depolymerize MTs and cell lysates taken at intervals and immunoblotted with antibodies to detect PSK1-α/β, PSK2, or activated and phosphorylated forms of these proteins. No significant changes in PSK1-α/β or PSK2 expression occur over the 8-h time course, and PSK-Ser(P)-181 levels are unchanged after 30 min of nocodazole treatment (Fig. 1C). However, PSK catalytic activity and phosphorylation (PSK-Ser(P)-181) increase thereafter throughout the 8-h time course (Fig. 1C). These results show that PSK activity does not change immediately in response to nocodazole-induced MT disruption but does increase with prolonged drug treatment. Analysis of DNA content by flow cytometry shows that PSK stimulation occurs as the cells accumulate in G2/M, with the percentage of cells with 4n DNA content increasing from 21.6% at time zero to 51.1% after 6 h of nocodazole treatment (Fig. 1D).
FIGURE 1.
A, PSK-Ser(P)-181 antibody detects catalytically active and phosphorylated PSK1-α, PSK1-β, and PSK2. Growing H1299 cells were transfected with pRK5-Myc vector, pRK5-Myc-PSK1-α, pRK5-Myc-PSK1-α (K57A), pRK5-Myc-PSK1-β, pRK5-Myc-PSK1-β (K57A), pRK5-Myc-PSK2, or pRK5-Myc-PSK2 (K57A). After 24 h, cell lysates were immunoblotted with anti-PSK-Ser(P)-181 antibody (upper panel) or anti-Myc antibody (lower panel), and proteins were detected by enhanced chemiluminescence. B, recombinant PSK1-α/β or PSK2 (amino acids 1–314, 30 ng) expressed and purified from Sf9 insect cells or controls without recombinant protein were subjected to in vitro kinase assays as described previously (9) and immunoblotted with the PSK-Ser(P)-181 antibody. C, nocodazole stimulates PSK activity and phosphorylation. Growing HeLa cells were treated with nocodazole (500 nm) for the times shown and then lysed and immunoblotted with antibodies to detect PSK1-α/β, PSK2, PSK-Ser(P)-181, or α-tubulin. PSK1-α/β proteins have similar mobilities, and the arrowed lower band represents both proteins (as confirmed using siRNA knockdown elsewhere in the manuscript) (“Results” and Fig. 7C). D, at the same times, some cells were fixed in 70% ethanol and stained with propidium iodide to determine cell cycle profiles using FACS and the percentages of cells in G0/G1, S, and G2/M are shown.
PSK Activity Is Reduced as Cells Exit Mitosis
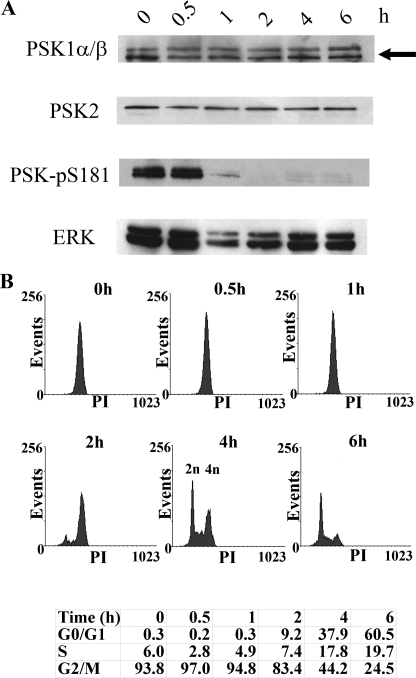
The stimulation of PSK activity that occurs as cells with 4n DNA content accumulate in G2/M, led us to investigate whether PSK activity could be reduced by removing nocodazole and permitting cells to pass through G2/M and exit mitosis. To do this, semisynchronous HeLa cell populations were prepared in S phase by incubating cultures in excess thymidine and then released into the cell cycle in the presence of nocodazole for 12 h to produce G2/M cells, with 4n DNA content. Flow cytometric analysis of these synchronized and nocodazole-treated cells shows that >90% of the population have 4n DNA content and are in G2/M, but these cells can recover between 1 and 2 h after drug removal and divide, thereby reducing their DNA content (2n) (Fig. 2B). Immunoblotting lysates taken from the same cell samples show that the expression of PSK1-α/β and PSK2 is unaltered, but that PSK activity is reduced significantly within 1 h of nocodazole removal (Fig. 2A). Analysis of the FACS data shows that this reduction in PSK activity occurs just before these cells begin to exit G2/M (Fig. 2B). Taken together, these nocodazole experiments show that PSK activity and phosphorylation (PSK-Ser(P)-181) increase as cells with 4n DNA content accumulate in G2/M, but decline rapidly after drug removal as the cells recover and complete and exit mitosis.
FIGURE 2.
PSK activity declines as cells exit mitosis after nocodazole removal. Growing HeLa cells were treated with thymidine (2 mm, 8 h) and then released into medium containing nocodazole (500 nm, 12 h). A, nocodazole was then removed, and at the times shown cells were lysed and immunoblotted with antibodies to detect PSK1-α/β, PSK2, PSK-Ser(P)-181, or ERK. PSK1-α/β proteins have similar mobilities and the arrowed lower band represents both proteins (as confirmed using siRNA knockdown elsewhere in the manuscript) (“Results” and Fig. 7C). B, at the same times, some cells were fixed in 70% ethanol and stained with propidium iodide to determine cell cycle profiles using FACS, and the percentages of cells in G0/G1, S, and G2/M are shown.
PSK Activity Increases in Mitotic Cells
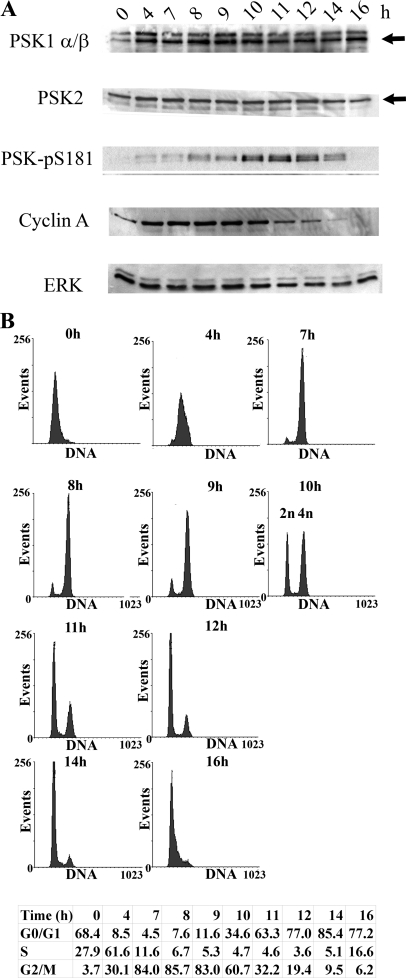
To investigate the apparent activation of PSKs during mitosis further, synchronous HeLa cell populations were prepared by introducing an additional thymidine cell cycle block to collect cells predominantly at the G1/S phase boundary. Synchronized HeLa cells were then released into the cell cycle by removing excess thymidine and lysates prepared at intervals over the following 16 h, as cells progress through S phase, G2/M, and G1. Immunoblotting these lysates for PSK1-α/β or PSK2 shows that their expression does not change dramatically over the 16-h time course; however, a marked but transient increase in PSK catalytic activity and phosphorylation is detected by the PSK-Ser(P)-181 antibody between 8 and 14 h after thymidine removal, and reaches a maximum between 10 and 12 h (Fig. 3A). FACS analysis of the same samples of cells shows that the percentages of cells in G2/M (4n DNA content) increases to 9 h and then begins to decline from 10 h and thereafter following thymidine removal. Immunoblotting shows that cyclin A levels are reduced from 10 h onward, as cells complete mitosis and divide (Fig. 3).
FIGURE 3.
PSKs are activated catalytically and phosphorylated during mitosis. Growing HeLa cells were synchronized using a double thymidine block and then released from G1/S into the cell cycle. A, at the times shown after thymidine removal cell cultures were lysed and immunoblotted with antibodies to detect PSK1-α/β, PSK2, PSK-Ser(P)-181, cyclin A, or ERK. Arrows indicate PSK1-α/β (lower band) or PSK2 (upper band) (as confirmed using siRNA knockdown elsewhere in the manuscript) (“Results” and Fig. 7C). B, alternatively, after thymidine removal, cell cultures were fixed in 70% ethanol and stained with propidium iodide to determine cell cycle profiles using FACS. The percentages of cells in G0/G1, S and G2/M are shown.
PSKs Are Activated Catalytically throughout Mitosis
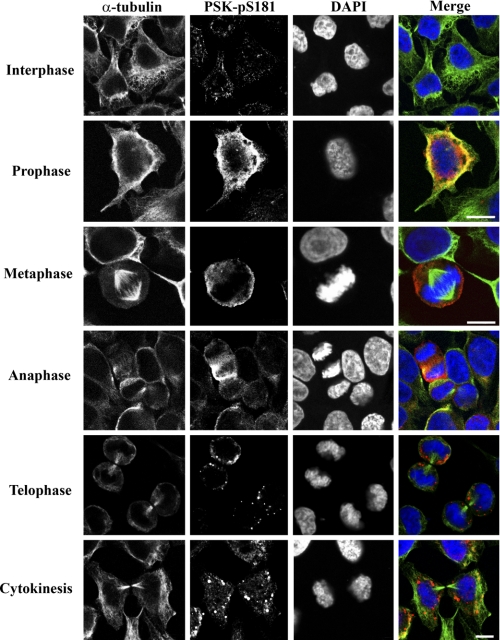
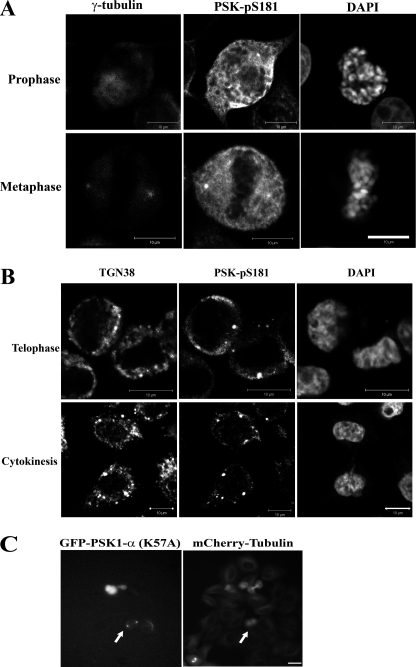
Immunoblotting and FACS analysis of synchronous populations of HeLa cells suggest that PSKs are activated as these cells progress through mitosis. To visualize PSK activation and phosphorylation in mitotic cells, synchronous cells on coverslips were fixed 10 h after the removal of excess thymidine and immunostained for analysis using confocal microscopy. Cells were co-stained for activated and phosphorylated PSKs (PSK-Ser(P)-181), α-tubulin (MTs) and also DAPI (DNA), to facilitate the identification of cells in prophase, metaphase, anaphase, telophase or cytokinesis, as well as interphase. Approximately 30% of the synchronized cell population were found to be in mitosis at these times. PSK-Ser(P)-181 staining increases significantly in ∼70% of cells in prophase compared with neighboring interphase cells, and intense staining for PSK-Ser(P)-181 is observed in all of the metaphase and anaphase cells analyzed (Fig. 4). The localization of activated PSKs was also examined. In interphase cells, PSK-Ser(P)-181 staining is weak and appears to localize to punctate spots, associated potentially with MTs, and increases in perinuclear and cytoplasmic PSK-Ser(P)-181 staining are observed in prophase cells (Fig. 4). In metaphase cells PSK-Ser(P)-181 staining localizes to the cytoplasm and toward the plasma membrane (Fig. 4) with additional staining detected at the spindle poles and co-localizing with an antibody that recognizes γ-tubulin (Fig. 5A). In telophase cells, and cells undergoing cytokinesis, PSK-Ser(P)-181 staining is reduced and less apparent in the cytoplasm and becomes more punctate (Fig. 4), co-localizing with trans-Golgi compartments identified using the trans-Golgi network protein 38 (TGN38) antibody (Fig. 5B). Additional analysis of PSK1-α localization using transfected and overexpressed kinase-defective protein fused to green fluorescent protein (GFP) shows that low levels of expression of transfected GFP-PSK1-α (K57A) resulted in punctate staining at both ends of the mCherry-tubulin-labeled spindle (Fig. 5C).
FIGURE 4.
PSK-Ser(P)-181 immunostaining increases during each stage of mitosis. Growing HeLa cells were synchronized using a double thymidine block and then released from G1/S into the cell cycle. 10 h after thymidine removal cells were fixed and co-stained with antibodies to detect catalytically active and phosphorylated PSK-Ser(P)-181, α-tubulin (MTs), or DAPI (DNA) to facilitate the identification of cells in each stage of mitosis. Pretreatment of the PSK-Ser(P)-181 antibody with the PSK-Ser(P)-181 epitope peptide abolishes its ability to immunostain mitotic cells (data not shown). Scale bars: 10 μm.
FIGURE 5.
PSK-Ser(P)-181 localizes to the spindle poles and trans-Golgi compartments. A and B, growing HeLa cells were synchronized using a double thymidine block, and 10 h after thymidine removal cultures were fixed and co-stained with antibodies to detect PSK-Ser(P)-181, γ-tubulin (spindle poles), or DAPI (DNA) (A) or antibodies to detect PSK-Ser(P)-181, trans-Golgi (TGN38), or DAPI (DNA) (B). C, growing HeLa cells stably expressing mCherry-tubulin were transfected with CB6-GFP-PSK1-α (K57A) for 4 h and incubated for an additional 10 h, and protein localization to both ends of the mCherry-tubulin labeled spindle (see arrows) was imaged using time lapse video microscopy. Scale bars: 10 μm.
siRNAs Targeting PSKs Inhibit Mitotic Cell Rounding and Spindle Positioning
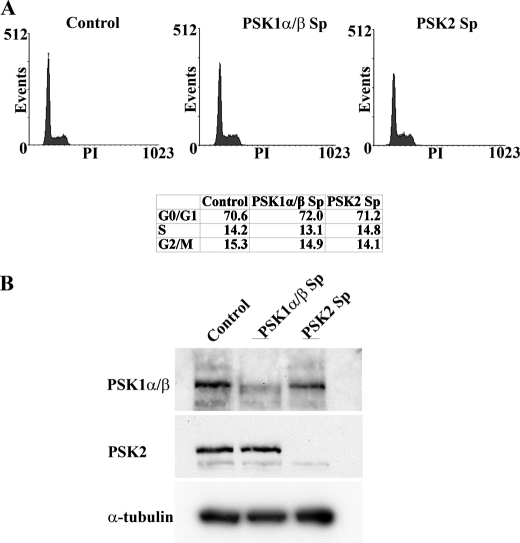
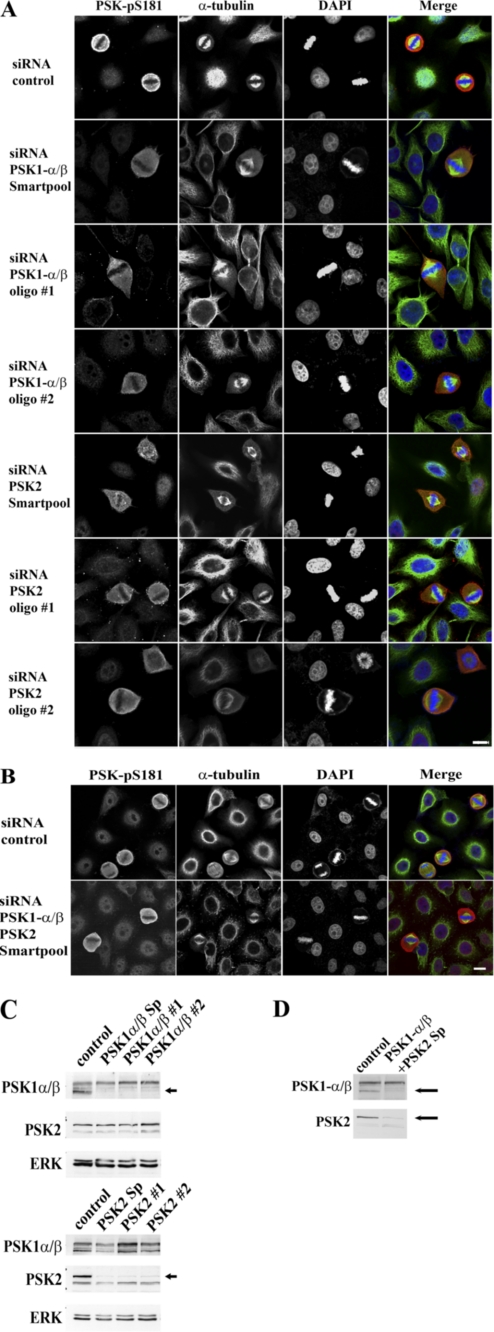
PSKs are therefore expressed, activated, and phosphorylated during each stage of mitosis, implicating potential functional roles for these proteins in regulating the cytoskeleton and cell division. To determine whether knockdown of PSK expression could alter cell cycle progression, small interfering RNAs (siRNAs, Smartpool) were used to target a common region of PSK1-α and PSK1-β or PSK2. siRNA-transfected and PSK1-α/β- or PSK2-depleted cells were examined initially using flow cytometry and showed that no significant changes in cell cycle kinetics or the percentages of cells in each stage of the cell cycle occurred in knockdown cultures compared with controls transfected with nontargeting siRNAs (Fig. 6). To investigate further the requirements for PSK expression and function during mitosis, HeLa cells were transfected with appropriate PSK1-α/β or PSK2 siRNAs (Smartpool mixture of four siRNAs or individual siRNAs #1 and #2) or control siRNAs for 14 h, then incubated with excess thymidine for 24 h and released from the thymidine cell cycle block for 10 h. Fixed cells were co-stained for PSK-Ser(P)-181, α-tubulin (MTs), and DAPI (DNA), and suitable metaphase cells with spindles parallel to the substratum were selected and analyzed by confocal microscopy (Fig. 7A). Interestingly, siRNA knockdown of either PSK1-α/β or PSK2 inhibits the ability of mitotic cells to round up and results in an irregular and more elongated and flattened morphology (Fig. 7A, rows 2–7). By contrast, cells transfected with control siRNAs and in mitosis retain their classic rounded morphology which is characteristic of mitotic HeLa cells (Fig. 7A, top row).
FIGURE 6.
siRNAs targeting PSK1-α/β or PSK2 do not alter cell cycle progression. A, growing HeLa cells were transfected with Smartpool (Sp) siRNAs targeting a common region of PSK1-α/β or PSK2 or control nontargeting siRNA, and after 48 h cultures were fixed in 70% ethanol and stained with propidium iodide (PI) to determine cell cycle profiles using FACS. The percentages of cells in G0/G1, S, and G2/M are shown. B, siRNA-transfected cells were also lysed, and knockdown of PSK target proteins was analyzed by immunoblotting.
FIGURE 7.
siRNAs targeting PSK1-α/β or PSK2 inhibit mitotic cell rounding and spindle centralization. A and B, growing HeLa cells were transfected for 14 h with control nontargeting siRNA or Smartpool (Sp, oligonucleotides 1–4) or individual (oligonucleotide 1 or 2) siRNAs targeting a common region of PSK1-α/β or PSK2 (alone (A) or Sp in combination (B)). Cultures were synchronized by incubation in 2 mm thymidine for 24 h and then incubated in the absence of thymidine for 10 h. A and B, siRNA-treated cells on coverslips were fixed and co-stained with antibodies to detect PSK-Ser(P)-181, α-tubulin (MTs), or DAPI (DNA) and analyzed by confocal microscopy. Scale bars: 10 μm. C and D, cells remaining on the Petri dish were lysed and siRNA knockdown of PSK target proteins by Smartpool or individual oligonucleotide 1 or 2 (as indicated) were analyzed by immunoblotting. Arrows indicate PSK1-α/β (lower band) or PSK2 (upper band) proteins that are knocked down by siRNAs targeting PSK1-α/β or PSK2.
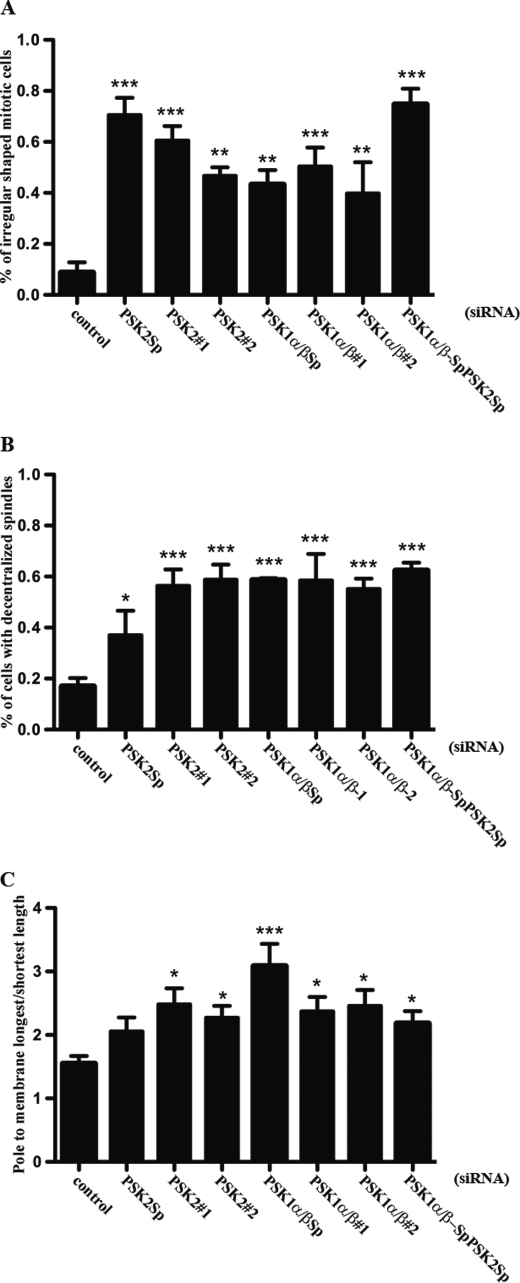
In addition, knockdown of either PSK1-α/β or PSK2 protein expression also affects the centralization of mitotic spindles, which localize to one side of the cell, compared with cells transfected with control siRNAs (Fig. 7A). Both inhibition of mitotic cell rounding and spindle positioning are also observed following knockdown of PSK1-α/β and PSK2 together, using a combination of siRNAs to target these proteins (Fig. 7B). The percentages of PSK1-α/β or PSK2 siRNA-transfected mitotic cells that display an irregular and nonrounded morphology or decentralized spindles are presented in Fig. 8, A and B. In addition, a spindle decentralization index is also shown where the distances between each spindle pole and the cell membrane have been measured and the data expressed as a ratio of the longest over the shortest length for each cell (Fig. 8C). Immunoblotting was used to confirm siRNA knockdown of PSK1-α/β and/or PSK2 protein expression, and the PSK1-α/β or PSK2 siRNAs were shown to be specific for their appropriate PSK targets (Fig. 7, C and D). Each of these siRNAs reduced but did not abolish the PSK-Ser(P)-181 fluorescence signal completely, suggesting that there is residual fluorescence from protein which is not knocked down (Fig. 7A). The results of the siRNA experiments show that both PSK1-α/β and PSK2 are required for mitotic cells to adopt a rounded morphology and to position the mitotic spindle at the cell center.
FIGURE 8.
SiRNAs targeting PSK1-α/β or PSK2 inhibit mitotic cell rounding and spindle centralization. The percentages of siRNA-transfected mitotic cells that showed nonrounded and irregular morphology (A) or decentralized spindles (B) are shown. C, decentralization index shows the distance between each spindle pole and the closest cell membrane as a ratio of the longest length over the shortest length. Mitotic cells were imaged and scored as described under “Experimental Procedures.” Graphs and error bars show mean ± S.E. (n = 3). Analysis of variance (ANOVA) was performed to assess the significance of the results shown in the graph. Subsequently, a Dunnet's multiple comparison test was used to determine whether the differences were significant between cell populations transfected with control siRNAs or the PSK siRNAs indicated in the figure. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
DISCUSSION
As with many GCK-like STE20s, not much is known about the upstream activation of PSKs or their downstream targets, apart from their regulation of MAPK signaling pathways, apoptosis and MTs (8, 9). Here we have used a phospho-PSK antibody that detects catalytically active and autophosphorylated endogenous PSK1-α/β and PSK2, to show that these kinases are activated catalytically in mitotic cells. PSK activity is stimulated significantly during prophase and peaks during metaphase and anaphase, and this elevation in PSK activity is also retained during telophase and cytokinesis. Activated PSKs are detected in the cytoplasm and at the spindle poles during prophase and metaphase, and our observations using transfected and overexpressed proteins, both here and previously, suggest that these cytoplasmic and polar localizations of activated PSKs are likely to be PSK2 and PSK1-α/β, respectively (8, 9). As cells progress into telophase and cytokinesis, activated PSKs localize predominantly to trans-Golgi compartments. These observations are not restricted to HeLa cells, and we have also detected enhanced PSK catalytic activity and phosphorylation during mitosis in other cell types, which include MCF7 breast cancer cells and Swiss 3T3 mouse fibroblasts (data not shown).
The stimulation of PSKs during mitosis and their ability to regulate MT dynamics are consistent with potential roles for these kinases in regulating the cytoskeleton during cell division. Highly organized changes in MTs, MT-associated proteins, and motor proteins are required to assemble, position, and organize the spindle, and MT-based forces and their regulating proteins are needed to align and segregate chromosomes correctly. Spindle tubulin is also a mixture of dynamic nonkinetochore spindle MTs and stable kinetochore MTs, which mediate spindle-chromosome attachment, and PSKs are likely to participate in some of these processes (22). siRNA knockdown of PSK1-α/β or PSK2 inhibited mitotic cell rounding and spindle centralization, and we also observed some additional rare mitotic phenotypes. The mitotic spindle appears to be more flattened than pyramidal, and there was also an increase in mitotic cells containing more than two spindle poles. Analysis of PSK1-α/β- or PSK2-depleted cells using time lapse video microscopy and flow cytometry, however, indicates that these cells do not undergo mitotic arrest but can exit or slip out of mitosis at least once. In support of this observation, others have used siRNAs targeting PSK2 to show that this protein is not a component of the spindle checkpoint (23, 24). PSK2 knockdown does not appear to delay mitotic progression significantly or override nocodazole-induced spindle assembly checkpoint arrest, nor does PSK2 interact with BubR1 (23, 24). Furthermore, PSK2 depletion does not alter Mad2 expression or displace Mad2 from kinetochores (23, 24).
The onset of mitosis is commonly accompanied by cell rounding, which involves the disassembly of focal adhesions, reduced cell attachment and actin reorganization to form rigid cortical actin (25, 26). The rounded cell morphology locates the spindle and centrosomes in close proximity to the actin cortex, which in turn contributes to the assembly of spindles as well as their orientation (26, 27). We found that PSK1-α/β and/or PSK2 siRNA knockdown inhibits mitotic cell rounding and spindle positioning, and these proteins may also influence the future cleavage plane and correct furrow location needed for symmetrical division (26, 28). The mechanisms involved in spindle positioning are poorly understood but do involve extension of the astral MTs outward to generate pushing forces between the astral MTs and cell cortex as well as cortical pulling forces, and Cdc42 and PAK2 have been linked to the regulation of spindle orientation (25, 29–32). Both PSK1-α/β and PSK2 can also contribute to mitotic cell rounding and spindle positioning, and these proteins are unable to compensate for each other following siRNA knockdown. Because MT growth and shrinkage are needed to generate the opposing forces required to position the spindle centrally, both proteins may be required to exert their opposing affects on MT stability to produce the normal phenotype. If this is the case, knockdown of either protein would be expected to result in spindle decentralization and this was observed (Fig. 7A).
In Drosophila melanogaster PSKs are represented by a single protein, dTao1/PSK, and recent work has shown that this kinase can regulate dynamic interactions between growing MT plus ends and the actin-rich cortex, and dTao1/PSK is required for cortical-induced MT catastrophe (19). dTao1/PSK can localize to the spindle but concentrates at the plus ends of MTs and limits their growth on contact with the actin-based cortex, and knockdown of dTao1/PSK results in the extension of MTs contacting cortical actin and the formation of long MT-based protrusions (19). PSK1-α/β and/or PSK2 could therefore regulate cortical actin-induced changes in MT dynamics directly, which in turn are needed for the control of cell shape and rounding and spindle centralization. Mechanistically, it has been suggested that dTao1 and PSKs may act in a manner similar to other MT-binding proteins, such as actin cross-linking family-7 (ACF7), cytoplasmic linker protein of 170 kDa (CLIP170), or CLIP-associated proteins (CLASPs), to bridge the interface between MTs and actin filaments and regulate MT dynamics (19, 33, 34). PSKs may also phosphorylate and alter the MT affinity of different microtubule plus end-binding proteins (+TIPs) and mediate their affects on MT dynamics in this way (33, 34). Other kinases that act in this manner include GSK3β, which phosphorylates and inhibits MT binding by CLASPs (37). Similarly, the budding yeast protein Aurora B/IPI 1p phosphorylates end-binding 1 protein (EB1) during anaphase and impairs MT binding (35), and FKBP12-rapamycin-associated protein (FRAP/mTOR) phosphorylates and regulates CLIP-170 and its interaction with MTs (36). Knockdown of dTao1/PSK induces changes in the growth rates of GFP-EB1 labeled MT plus ends, but our examination of MT dynamics in siRNA-treated and PSK-depleted mitotic cells using antibodies to detect acetylated (stable) or tyrosinated (dynamic) MTs did not detect significant changes in spindle staining with or without PSK expression (data not shown) (19). These results are consistent with the more subtle roles for PSKs described above. PSKs regulate MTs and influence actin, and these proteins may also provide additional links between these two cytoskeletal systems. PSK1-α stabilizes MTs and down-regulates actin stress fibers and is therefore similar to X-PAK5, which shuttles between MTs and actin-rich structures, depending on its activation status (7, 15, 38). By contrast, PSK2 destabilizes MTs but is inhibited by binding TESK1 (testis-specific protein kinase), which also stabilizes F-actin stress fibers via the phosphorylation of cofilin (16, 39).
Recent studies have identified a growing number of GCK-like STE20s that can regulate the MT and actin cytoskeleton and cell division, and some of these proteins share functional similarities with the PSKs. SLK not only binds and regulates MT organization but localizes to the mitotic spindle during metaphase and phosphorylates moesin to induce cortical actin rigidity and rounded cell morphology (40–42). SLK also phosphorylates and activates the mitotic polo-like kinase, and its catalytic activity is required for fibroblasts to complete the G2/M transition and exit mitosis (40, 43, 44). The catalytic activities of MST1 and MST2 are also elevated during mitosis, and knockdown of their expression impairs centrosome duplication, causes metaphase delay, and reduces cell proliferation (45–47). Another GCK PASK binds and stabilizes MTs, but little more is known about the functional roles for this protein (48). PAKs 1–3 have been investigated much more intensively than GCKs, as these kinases can act as downstream effectors for Cdc42 and Rac, and regulate the cytoskeleton. PAKs can control MTs and are activated during the G2/M transition and mitosis (49). PAK1 and PAK2 localize to the central spindle, mid body and poles, and overexpressed PAK1 induces multiple spindle orientations and centromere spots (49, 50). The localization of PAK1 is regulated by its phosphorylation on threonine 212 by Cdc2, which results in the movement of this protein to the spindle and MTOCs and the stimulation of astral MT elongation during metaphase (51, 52). Inhibition of PAK1 also delays G2/M transition and causes aberrant spindle formation (49). Downstream substrates that are phosphorylated and activated by PAK1 include the centrosomal kinase Aurora A as well as polo-like kinase, which regulates centrosome maturation, anaphase onset, spindle assembly, and cytokinesis (49, 53). It remains to be determined whether PSKs share some of these PAK substrates, but we have found that PSK1-α does not act like PAK1, which phosphorylates and inactivates the MT catastrophe promoting activity of stathmin to stabilize MTs (15, 54, 55). In addition, to date we have been unable to identify small GTP-binding proteins that activate PSKs, and in this respect these proteins are more similar to PAKs 4–6, whose catalytic activity appears to be regulated independently of GTPases (56). Much less is known about these PAKs, but PAK4 can phosphorylate and regulate Ran activity during mitosis and control the assembly of Ran-dependent complexes on the spindle. PAK5, unlike PSK2, can inhibit MARK activity and stabilize MTs (57, 58).
In conclusion, previous studies have shown that PSK1-α and PSK2 can stabilize or destabilize MTs, respectively, and regulate their organization. In this study we have used several different antibodies to investigate the expression and activation of the endogenous proteins during mitosis, when dramatic changes in the MT and actin cytoskeleton occur and are essential for cell division. We have demonstrated that PSKs are activated in dividing cells and are required for mitotic cell rounding and spindle positioning. Both of these processes are crucial to ensure correct partitioning and segregation of chromosomes into daughter cells, and it is likely that PSKs contribute by regulating interactions between MTs and actin. The ability of PSKs to regulate MTs suggests additional potential roles for these proteins in controlling spindle formation, positioning, and function, as well as the capture and segregation of chromosomes during cell division. Further work is now needed to test these possibilities and to identify the cellular mechanisms involved in the activation of these proteins as well as their potential downstream targets, which might include MT-associated proteins, molecular motors, or other mitotic kinases.
Acknowledgments
We thank Mark Shipman, Samantha King, Fariesha Hashim, Buzz Baum, Monica Agromayor, and Juan Martin-Serrano for many helpful discussions. We thank Maria Hernandez-Fuentes and Steve Orr for help and advice on cell cycle analyses.
This work was supported by the Association for International Cancer Research (St. Andrews, Scotland), King's Medical Research Trust, the Biotechnology and Biological Sciences Research Council United Kingdom Grant 29/C14086, and by a donation from Laura Price.
- STE20
- sterile 20
- CRIB domain
- Cdc42/Rac-interacting and binding domain
- GCK
- germinal center kinase
- MT
- microtubule
- PAK
- p21-activated kinase
- PSK
- prostate-derived sterile 20-like kinase
- PSK-Ser(P)-181
- catalytically active PSKs autophosphorylated on serine 181.
REFERENCES
- 1. Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 2. Dan I., Watanabe N. M., Kusumi A. (2001) Trends Cell Biol. 11, 220–230 [DOI] [PubMed] [Google Scholar]
- 3. Delpire E. (2009) Pflugers Arch. 458, 953–967 [DOI] [PubMed] [Google Scholar]
- 4. Bokoch G. M. (2003) Annu. Rev. Biochem. 72, 743–781 [DOI] [PubMed] [Google Scholar]
- 5. Arias-Romero L. E., Chernoff J. (2008) Biol. Cell 100, 97–108 [DOI] [PubMed] [Google Scholar]
- 6. Eswaran J., Soundararajan M., Kumar R., Knapp S. (2008) Trends Biochem. Sci. 33, 394–403 [DOI] [PubMed] [Google Scholar]
- 7. Moore T. M., Garg R., Johnson C., Coptcoat M. J., Ridley A. J., Morris J. D. (2000) J. Biol. Chem. 275, 4311–4322 [DOI] [PubMed] [Google Scholar]
- 8. Zihni C., Mitsopoulos C., Tavares I. A., Ridley A. J., Morris J. D. (2006) J. Biol. Chem. 281, 7317–7323 [DOI] [PubMed] [Google Scholar]
- 9. Zihni C., Mitsopoulos C., Tavares I. A., Baum B., Ridley A. J., Morris J. D. (2007) J. Biol. Chem. 282, 6484–6493 [DOI] [PubMed] [Google Scholar]
- 10. Tassi E., Biesova Z., Di Fiore P. P., Gutkind J. S., Wong W. T. (1999) J. Biol. Chem. 274, 33287–33295 [DOI] [PubMed] [Google Scholar]
- 11. Hutchison M., Berman K. S., Cobb M. H. (1998) J. Biol. Chem. 273, 28625–28632 [DOI] [PubMed] [Google Scholar]
- 12. Yustein J. T., Xia L., Kahlenburg J. M., Robinson D., Templeton D., Kung H. J. (2003) Oncogene 22, 6129–6141 [DOI] [PubMed] [Google Scholar]
- 13. Chen Z., Hutchison M., Cobb M. H. (1999) J. Biol. Chem. 274, 28803–28807 [DOI] [PubMed] [Google Scholar]
- 14. Raman M., Earnest S., Zhang K., Zhao Y., Cobb M. H. (2007) EMBO J. 26, 2005–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mitsopoulos C., Zihni C., Garg R., Ridley A. J., Morris J. D. (2003) J. Biol. Chem. 278, 18085–18091 [DOI] [PubMed] [Google Scholar]
- 16. Timm T., Li X. Y., Biernat J., Jiao J., Mandelkow E., Vandekerckhove J., Mandelkow E. M. (2003) EMBO J. 22, 5090–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Timm T., Balusamy K., Li X., Biernat J., Mandelkow E., Mandelkow E. M. (2008) J. Biol. Chem. 283, 18873–18882 [DOI] [PubMed] [Google Scholar]
- 18. Timm T., Matenia D., Li X. Y., Griesshaber B., Mandelkow E. M. (2006) Neurodegener. Dis. 3, 207–217 [DOI] [PubMed] [Google Scholar]
- 19. Liu T., Rohn J. L., Picone R., Kunda P., Baum B. (2010) J. Cell Sci. 123, 2708–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Agromayor M., Carlton J. G., Phelan J. P., Matthews D. R., Carlin L. M., Ameer-Beg S., Bowers K., Martin-Serrano J. (2009) Mol. Biol. Cell 20, 1374–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou T., Raman M., Gao Y., Earnest S., Chen Z., Machius M., Cobb M. H., Goldsmith E. J. (2004) Structure 12, 1891–1900 [DOI] [PubMed] [Google Scholar]
- 22. Zhai Y., Kronebusch P. J., Borisy G. G. (1995) J. Cell Biol. 131, 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Westhorpe F. G., Diez M. A., Gurden M. D., Tighe A., Taylor S. S. (2010) Chromosoma 119, 371–379 [DOI] [PubMed] [Google Scholar]
- 24. Hübner N. C., Wang L. H., Kaulich M., Descombes P., Poser I., Nigg E. A. (2010) Chromosoma 119, 149–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rosenblatt J., Cramer L. P., Baum B., McGee K. M. (2004) Cell 117, 361–372 [DOI] [PubMed] [Google Scholar]
- 26. Kunda P., Baum B. (2009) Trends Cell Biol. 19, 174–179 [DOI] [PubMed] [Google Scholar]
- 27. Heng Y. W., Koh C. G. (2010) Int. J. Biochem. Cell Biol. 42, 1622–1633 [DOI] [PubMed] [Google Scholar]
- 28. Théry M., Bornens M. (2006) Curr. Opin. Cell Biol. 18, 648–657 [DOI] [PubMed] [Google Scholar]
- 29. Mitsushima M., Toyoshima F., Nishida E. (2009) Mol. Cell. Biol. 29, 2816–2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grill S. W., Hyman A. A. (2005) Dev. Cell 8, 461–465 [DOI] [PubMed] [Google Scholar]
- 31. Pearson C. G., Bloom K. (2004) Nat. Rev. Mol. Cell Biol. 5, 481–492 [DOI] [PubMed] [Google Scholar]
- 32. Wühr M., Dumont S., Groen A. C., Needleman D. J., Mitchison T. J. (2009) Cell Cycle 8, 1115–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Akhmanova A., Steinmetz M. O. (2010) J. Cell Sci. 123, 3415–3419 [DOI] [PubMed] [Google Scholar]
- 34. Akhmanova A., Steinmetz M. O. (2008) Nat. Rev. Mol. Cell Biol. 9, 309–322 [DOI] [PubMed] [Google Scholar]
- 35. Zimniak T., Stengl K., Mechtler K., Westermann S. (2009) J. Cell Biol. 186, 379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Choi J. H., Bertram P. G., Drenan R., Carvalho J., Zhou H. H., Zheng X. F. (2002) EMBO Rep. 3, 988–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wittmann T., Waterman-Storer C. M. (2005) J. Cell Biol. 169, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cau J., Faure S., Comps M., Delsert C., Morin N. (2001) J. Cell Biol. 155, 1029–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johne C., Matenia D., Li X. Y., Timm T., Balusamy K., Mandelkow E. M. (2008) Mol. Biol. Cell 19, 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O'Reilly P. G., Wagner S., Franks D. J., Cailliau K., Browaeys E., Dissous C., Sabourin L. A. (2005) J. Biol. Chem. 280, 42383–42390 [DOI] [PubMed] [Google Scholar]
- 41. Burakov A. V., Zhapparova O. N., Kovalenko O. V., Zinovkina L. A., Potekhina E. S., Shanina N. A., Weiss D. G., Kuznetsov S. A., Nadezhdina E. S. (2008) Mol. Biol. Cell 19, 1952–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wagner S., Flood T. A., O'Reilly P., Hume K., Sabourin L. A. (2002) J. Biol. Chem. 277, 37685–37692 [DOI] [PubMed] [Google Scholar]
- 43. Ellinger-Ziegelbauer H., Karasuyama H., Yamada E., Tsujikawa K., Todokoro K., Nishida E. (2000) Genes Cells 5, 491–498 [DOI] [PubMed] [Google Scholar]
- 44. Johnson T. M., Antrobus R., Johnson L. N. (2008) Biochemistry 47, 3688–3696 [DOI] [PubMed] [Google Scholar]
- 45. Praskova M., Xia F., Avruch J. (2008) Curr. Biol. 18, 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hergovich A., Kohler R. S., Schmitz D., Vichalkovski A., Cornils H., Hemmings B. A. (2009) Curr. Biol. 19, 1692–1702 [DOI] [PubMed] [Google Scholar]
- 47. Dallol A., Hesson L. B., Matallanas D., Cooper W. N., O'Neill E., Maher E. R., Kolch W., Latif F. (2009) Curr. Biol. 19, 1227–1232 [DOI] [PubMed] [Google Scholar]
- 48. Tsutsumi T., Kosaka T., Ushiro H., Kimura K., Honda T., Kayahara T., Mizoguchi A. (2008) Arch Biochem. Biophys. 477, 267–278 [DOI] [PubMed] [Google Scholar]
- 49. Maroto B., Ye M. B., von Lohneysen K., Schnelzer A., Knaus U. G. (2008) Oncogene 27, 4900–4908 [DOI] [PubMed] [Google Scholar]
- 50. Vadlamudi R. K., Adam L., Wang R. A., Mandal M., Nguyen D., Sahin A., Chernoff J., Hung M. C., Kumar R. (2000) J. Biol. Chem. 275, 36238–36244 [DOI] [PubMed] [Google Scholar]
- 51. Banerjee M., Worth D., Prowse D. M., Nikolic M. (2002) Curr. Biol. 12, 1233–1239 [DOI] [PubMed] [Google Scholar]
- 52. Thiel D. A., Reeder M. K., Pfaff A., Coleman T. R., Sells M. A., Chernoff J. (2002) Curr. Biol. 12, 1227–1232 [DOI] [PubMed] [Google Scholar]
- 53. Zhao Z. S., Lim J. P., Ng Y. W., Lim L., Manser E. (2005) Mol. Cell 20, 237–249 [DOI] [PubMed] [Google Scholar]
- 54. Wittmann T., Bokoch G. M., Waterman-Storer C. M. (2004) J. Biol. Chem. 279, 6196–6203 [DOI] [PubMed] [Google Scholar]
- 55. Daub H., Gevaert K., Vandekerckhove J., Sobel A., Hall A. (2001) J. Biol. Chem. 276, 1677–1680 [DOI] [PubMed] [Google Scholar]
- 56. Molli P. R., Li D. Q., Murray B. W., Rayala S. K., Kumar R. (2009) Oncogene 28, 2545–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Matenia D., Griesshaber B., Li X. Y., Thiessen A., Johne C., Jiao J., Mandelkow E., Mandelkow E. M. (2005) Mol. Biol. Cell 16, 4410–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bompard G., Rabeharivelo G., Frank M., Cau J., Delsert C., Morin N. (2010) J. Cell Biol. 190, 807–822 [DOI] [PMC free article] [PubMed] [Google Scholar]