Abstract
NKCC1 and KCC2, related cation-chloride cotransporters (CCC), regulate cell volume and γ-aminobutyric acid (GABA)-ergic neurotranmission by modulating the intracellular concentration of chloride [Cl−]. These CCCs are oppositely regulated by serine-threonine phosphorylation, which activates NKCC1 but inhibits KCC2. The kinase(s) that performs this function in the nervous system are not known with certainty. WNK1 and WNK4, members of the WNK (with no lysine [K]) kinase family, either directly or via the downstream SPAK/OSR1 Ste20-type kinases, regulate the furosemide-sensitive NKCC2 and the thiazide-sensitive NCC, kidney-specific CCCs. What role the novel WNK2 kinase plays in this regulatory cascade, if any, is unknown. Here, we show that WNK2, unlike other WNKs, is not expressed in kidney; rather, it is a neuron-enriched kinase primarily expressed in neocortical pyramidal cells, thalamic relay cells, and cerebellar granule and Purkinje cells in both the developing and adult brain. Bumetanide-sensitive and Cl−-dependent 86Rb+ uptake assays in Xenopus laevis oocytes revealed that WNK2 promotes Cl− accumulation by reciprocally activating NKCC1 and inhibiting KCC2 in a kinase-dependent manner, effectively bypassing normal tonicity requirements for cotransporter regulation. TiO2 enrichment and tandem mass spectrometry studies demonstrate WNK2 forms a protein complex in the mammalian brain with SPAK, a known phosphoregulator of NKCC1. In this complex, SPAK is phosphorylated at Ser-383, a consensus WNK recognition site. These findings suggest a role for WNK2 in the regulation of CCCs in the mammalian brain, with implications for both cell volume regulation and/or GABAergic signaling.
Keywords: Brain, Chloride Transport, Neurons, Serine Threonine Protein Kinase, Sodium Transport
Introduction
Homeostasis of the intracellular concentration of chloride [Cl−]i is important for the regulation of cell volume and γ-aminobutyric acid (GABA)5 neurotranmission in the mammalian nervous system. Cell volume regulation is important for neurons, which experience considerable channel-mediated influxes of cations and Cl− during synaptic activity (1). Because ionotropic GABAA receptors function as Cl− channels, [Cl−]i affects the strength and polarity (excitatory or inhibitory) of GABA neurotransmission (2).
[Cl−]i is established by cellular Cl− influx and efflux pathways. The SLC12a cation-chloride cotransporters (CCCs) contain the Cl−-importing Na-(K)-2Cl cotransporters (NCC, NKCC1, and NKCC2) and Cl−-exporting K-Cl cotransporters (KCC1-KCC4) (3). NKCC1 and KCC2 play essential roles in the function of the CNS and PNS (4, 5). NKCC1 is widely expressed in cortical and subcortical neuroglia, and in dorsal root ganglia (6). KCC2 expression is neuron-specific in the cortex, thalamus, cerebellum, and spinal cord (7). Early in development, cortical GABAergic neurons are excitatory due to an elevated [Cl−]i established in part by high NKCC1 activity and low KCC2 activity, but are subsequently inhibitory due to a developmental shift in the relative activity of KCC2 versus NKCC1 (8, 9). The co-expression of NKCC1 and KCC2 in specific neurons, and the rapid changes in the neuronal response to GABA that can occur due to shifts in [Cl−]i, suggest that NKCC1 and KCC2 can be tightly coordinated to generate precise neuronal Cl− gradients (10, 11). The mechanism underlying this regulation in the CNS is unknown.
Activities of the CCCs are regulated by serine-threonine phosphorylation, which has opposite effects on NKCC1 and KCC. Cell swelling and high [Cl−]i trigger net cotransporter dephosphorylation, activating the KCCs but inhibiting NKCC1, resulting in reduction of [Cl−]i; low [Cl−]i and cell shrinkage have the opposite effects (3). A regulatory pathway of [Cl−]i-sensing protein kinases has long been proposed to be the common mechanism that co-regulates both the NKCCs and KCCs (12, 13). Recent evidence suggests that members of the WNK (with no lysine = K) kinase family, directly or indirectly via the downstream SPAK/OSR1 Ste20-type kinases, serve this important function in the kidney (13, 14). Here, WNKs like WNK1 and WNK4 phosphorylate and activate SPAK/OSR1, which in turn phosphorylate and activate the furosemide-sensitive NKCC2 and the thiazide-sensitive NCC, kidney-specific CCCs. This pathway is necessary for proper blood pressure control and electrolyte homeostasis in humans (15). PRKWNK1 and PRKWNK4 are mutated in pseudohypoaldosteronism type II (PHAII), a Mendelian form of human hypertension (16).
In humans there are four WNK kinases (WNK1, WNK2, WNK3, and WNK4), encoded by genes on chromosomes 12, 9, X, and 17, respectively. WNK3, which is highly expressed in the brain, has robust effects in vitro upon the CCCs, simultaneously activating NKCC1, NKCC2, and NCC and inhibiting all four KCCs (17–19). The reciprocal actions of WNK3 on NKCC1 and the KCCs, and its co-expression with CCCs in GABAergic neuronal populations that undergo dynamic changes in [Cl−]i, suggest WNK3 might be involved in the regulation of neuronal CCCs (13, 18). Further evidence of a link between the WNKs and the CCCs in the brain includes the finding that mutations in the nervous system-specific HSN2 exon of WNK1 cause hereditary sensory and autonomic neuropathy type 2 (HSAN2) (20), and compound heterozygous mutations of the KCC3 gene causing Andermann syndrome can phenocopy HSAN2 (21). In addition, two recent studies have implicated the epigenetic silencing of WNK2 in the pathogenesis of multiple brain tumor types (22, 23).
WNK2 is the least characterized of all WNK kinases. Initial reports showed that WNK2 is most highly expressed in brain, and unlike other WNKs, is not expressed in the kidney (24). Here, we characterize the cellular and subcellular location and the function of WNK2.
MATERIALS AND METHODS
cDNA Constructs
A cDNA clone for human WNK2 was provided by Osamu Ohara (Kazusa DNA Research Institute, Chiba, Japan) (Nagase, T, 2000). A C-terminal hemagglutin A (HA) tag was added to WNK2 cDNA with PCR and subcloned into pCDNA3.1- (Invitrogen) for mammalian expression, and into pGH19 for studies in Xenopus oocytes (17). QuikChange (Stratagene) was used to introduce the D342A mutation into pGH19-WNK2-HA and pcDNA3.1-WNK2-HA. Constructs were verified by DNA sequencing.
Functional Assays in Oocytes
Xenopus oocytes were harvested and injected with the cRNAs of the indicated constructs, and bumetanide-sensitive and Cl−-dependent 86Rb+ uptakes were determined as described (18, 19). In brief, experiments with NKCC1 were performed in isotonic conditions (220 mm) in oocytes injected with water or with NKCC1 cRNA alone or together with WNK2 or WNK3 cRNA. 2 or 3 days after injections oocytes were incubated overnight in a Cl−-free frog Ringer (19). Next day oocytes were exposed to a 30 min incubation period in the same Ringer, with ouabain (1 mm) to prevent activity of the Na+:K+:ATPase, followed by 60 min. uptake in the presence of NaCl (86 mm) and KCl (10 mm) (19) with 2 μCi of 86Rb+/ml. Parallele groups of oocytes were exposed to similar uptake conditions but in the presence of 100 μm bumetanide. KCC experiments were performed in both isotonic and hypotonic conditions (110 mm), as described (18, 19). At the end of the uptake period oocytes were washed five times in cold solution to eliminate tracer 86Rb+ from extracellular fluid, dissolved in 10% SDS and tracer activity was determined for each oocyte by β-scintillation counting. Each experiment was performed at least three times, with a minimum of 10 oocytes per group.
Oocyte Cell Surface Biotinylation
For surface expression analysis oocytes proteins were biotinylated as described previously (25). In brief, oocytes injected with NKCC1 or FLAG-KCC4 cRNA were washed in ND-96 TEA buffer five times and incubated at 4 °C during 40 min in the same buffer supplemented with 1.5 mg/ml Sulfo-NHS-LC-Biotin (Thermo, Pierce). Then, oocytes were homogenized using a 25-gauge needle in a sucrose-based buffer (4 μl/oocyte). After 5 min centrifugation at 8,000 × g, the supernatants were collected and protein concentration assessed by the Bradford assay (Bio-Rad). Streptavidin precipitation was performed with 50 μl of streptavidin-agarose beads in 50% slurry (Upstate, Cell Signaling Solutions) per 500 μg of biotinylated proteins diluted in 1 ml of Tris-buffered saline. Samples were rolled overnight at 4 °C. Beads were then washed one time with Buffer 1, twice with Buffer 2, and once with Buffer 3. Then, buffer 3 was substituted with 30 μl of Laemmli Sample Buffer (Bio-Rad). Protein samples were heated to 65 °C for 15 min before separation on a 7.5% acrylamide gel for Western blotting using the monoclonal antibody T4 against NKCC1 (DSHB, University of Iowa) or anti-FLAG.
Transfections, Immunoprecipitation, SDS-PAGE, and Immunoblotting
Transfection, immunoprecipitation (IP), and Western blot analysis of WNK2 for MS analysis in HEK-293 cells, or from Xenopus oocytes, was performed as described previuosly (17). For Western blot analysis, lysates of HEK cells post-transfection or Xenopus oocytes (post-injection;5 oocytes per group) were solubilized in sample buffer with 1% Triton X-100, proteins were separated by SDS-PAGE, transferred to PVDF, blocked, and probed with anti-HA (each at 1:4000). Membranes were then washed, incubated with secondary antibody, and processed with ECL reagents (Amersham Biosciences) as described (17).
Whole mouse brains (2 adult brains per IP) were collected immediately after sacrifice, dissected on ice, and homogenized in lysis buffer, centrifuged, and the supernatant collected and used for IP. Immunoprecipitations of native WNK2 protein complexes was carried out using 10–20 μg of anti WNK2 antibodies (NOVUS, #25910002) conjugated to protein A-agarose (Sigma).
In Situ Hybridization and Immunostaining
Antisense probes were prepared from cDNA clones in TOPO 2.1 vectors (Invitrogen) from WNK1 (bases 3835–4953 of NM_198703), WNK2 (2033–3088 of NM_029361), and WNK4 (2501–3500 of NM_175638) complementary to a unique stretch 5′ of the kinase domain in each message. RNA was prepared with incorporation of digoxigenin-UTP (26). Brains were fixed by intracardiac perfusion with 4% paraformaldehyde and 36-μm-thick sections were obtained. In situ hybridization was performed and specific hybridization detected by using antidigoxin antibodies (27). Co-localization was carried out by additional immunohistochemistry steps with mouse anti-SMI 32 1:1000 (Covance Inc., Princeton, NJ), guinea pig anti-GFAP 1:500 (Advanced ImmunoChemical, Long Beach, CA), or mouse anti-calbindin d-28K 1:1000 (SWANT, Bellinzona, Switzerland) followed by diaminobenzidine (DAB) staining. Sections were treated with 1% H2O2, washed in PBS, preincubated in blocking solution (BS) containing 5% normal donkey serum (Jackson ImmunoResearch Laboratories), 1% bovine serum albumin, 0.1% glycine, 0.1% l-lysine, and 0.03% Triton-X-100. The sections were then incubated with the primary antibody for 36–48 h at 4ƒC. Following washes with PBS, they were incubated with biotinylated secondary antibodies (Jackson Laboratories, 1:250 dilution) for 2 h at room temperature, washed in PBS, and then incubated with Vectastain ABC Elite solution (Vector Laboratories, Burlingame, CA) for 2 h. Sections were developed using 0.05% DAB (pH 7.4), 0.2% glucose, 0.01% nickel ammonium sulfate, 0.04% ammonium chloride, and 8 mg/ml glucose oxidase, and then rinsed, mounted onto glass slides, allowed to dry, dehydrated, and coverslipped.
TiO2 Enrichment and Mass Spectrometry
TiO2 enrichment and mass spectrometry of WNK2 from HEK 293 cells and mouse brain was performed essentially similar to the protocol detailed in Ref. 28 (47). In brief, HA-tagged WNK2 expressed in mammalian cell culture were immunoprecipitated as described above. Immunoprecipitated proteins are then resolved on denaturing SDS-PAGE. Proteins are visualized with Coomassie Blue staining (R-250). Protein bands are excised from the gel and washed in 50% CH3CN, 50 mm NH4HCO3. The gel slices are then crushed and resuspended 1:1 (wt:vol) in a solution of 10 mm NH4HCO3 with 20 μg/ml sequence grade trypsin (Promega), and incubated overnight at 37 °C. Peptides extracted with 0.5% TFA/50% ACN, dried, resuspended in 0.5% TFA/50% ACN, and applied to a pre-equilibrated TiO2 TopTip (Glygen Corp.) micro-spin columns. The TiO2 columns are pre-equilibrated by washing with 100% ACN, followed by 0.2 m sodium phosphate buffer pH 7.0, and finally with 0.5% TFA/50% ACN. The peptides are bound to the column, unbound non-phospho peptides are washed off with 0.5% TFA/50% ACN, and the phosphopeptides are eluted with a 1:33 solution of high purity saturated (28%) ammonia. Eluate fractions containing phosphopeptides are dried and dissolved in 3 μl of 70% formic acid, and then diluted to 10 μl with 0.1% TFA. 5 μl of the sample is directly injected onto a 100 micron x 150 mm Atlantis HPLC column (Waters) running at 500 nl/min directly interfaced to an ESI-QTOF mass spectrometer (Waters/Micromass Q-Tof Ultima). Phosphopeptides are identified by searching with Mascot 2.1 database search engine with improved phosphopeptide scoring features with additional manual inspection of spectra.
RESULTS
WNK2 Is a Neuron-enriched WNK Kinase in the Developing and Adult Mammalian Brain
We first examined the expression of WNK2 in human tissues. Northern blot analysis showed WNK2 is most highly expressed in brain and heart, with no detectable expression in kidney (supplemental Fig. S1). These results are concordant with previous RT-PCR data (24). WNK2 expression was further studied in mouse brain using in situ hybridization (ISH; see “Materials and Methods”). At embryonic day 14.5 and 15.5, WNK2 is highly expressed in differentiating neurons of the forebrain, midbrain, and hindbrain (Fig. 1 and data not shown). WNK2 expression is low in the ventricular neuroepithelium and basal ganglia, but high in post-mitotic neurons. WNK2 is also expressed in the developing cortical plate, thalamus, dorsal midbrain, and cerebellum (Fig. 1).
FIGURE 1.
Analysis of WNK2 mRNA expression in mouse brain and kidney as compared with other WNK kinases. WNK2 localization (blue) by in situ hybridization shows a high level of expression in mouse brain both in embryonic development (E14.5) and in the adult (top two panels). A representative coronal section of the E14.5 brain (top left) is compared with sections of the adult brain (top right). Whole sections are shown in the inset on top of representative magnified region of the cortex. WNK2 is highly expressed in post-mitotic neurons in the cortical plate (CP) and intermediate zone (IZ) at E14.5. WNK2 is expressed in all cortical layers in the adult cortex (ctx) (top right). WNK2 expression in the adult cerebellum (bottom left). A whole section is shown in the inset on top of a magnified region of the cerebellum. Purkinje cells (pjk), the granular cell layer (gr), and the white matter (wm) are indicated. WNK2, WNK1, and WNK4 mRNA expression in the mouse kidney (bottom right). A whole sagittal section is shown in the insets below images of a magnified region of the cortex (ctx) for each WNK kinase indicated.
At post-natal stages and in the adult, WNK2 is expressed in the olfactory bulb, septum, neocortex (in all cortical layers, with strongest expression in pyramidal neurons), hippocampus, anterior dorsal thalamus (with lower levels in the posterior thalamus), and the cerebellum, with particularly strong expression in deep cerebellar nuclei. Low levels of WNK2 were detected in the hypothalamus. WNK2 was not detected in the basal ganglia (Fig. 1).
The expression of WNK2 in neocortical pyramidal neurons was demonstrated by combining ISH and immunostaining with SMI-32, a marker of pyramidal neurons with long axonal projections (29). Though WNK2 and SMI-32 co-localized in the neocortex, not all WNK2-expressing cells were SMI-32 positive (Fig. 2). However, WNK2-expressing cells did not express the astroglial marker GFAP, suggesting WNK2 is expressed primarily in neurons (Fig. 2). In the cerebellum, WNK2 co-localized with Calbindin, a marker of Purkinje cells (Fig. 2).
FIGURE 2.
Characterization of WNK2-expressing cells in mouse brain. SMI-32 co-localization (brown) highlights WNK2 expression in pyramidal neurons (pyr) in the developing neocortex (top panel). GFAP-expressing astrocytes do not express Wnk2 (middle). The arrowhead marks an astrocyte (ast). WNK2 mRNA is expressed in the Purkinje cells (pc) in the cerebellum, labeled with calbindin (bottom). The molecular layer (mol) and granular layer (gr) border the Purkinje cell layer in the cerebellum.
These results demonstrate that WNK2 is a neuron-enriched kinase in both the cerebral cortex and cerebellum in the developing and adult mouse brain, with a pattern of expression temporally and spatially distinct compared with other WNKs (supplemental Fig. S2).
WNK2 Activates NKCC1 and Inhibits KCC2, Promoting Cl− Influx
The effects of the WNKs on the CCCs are heterogeneous (17–19, 30, 31). Given its high expression in the brain, we tested whether WNK2 might affect the activities of the key neuronal CCCs NKCC1 and KCC2 by utilizing the Xenopus laevis oocyte expression system.
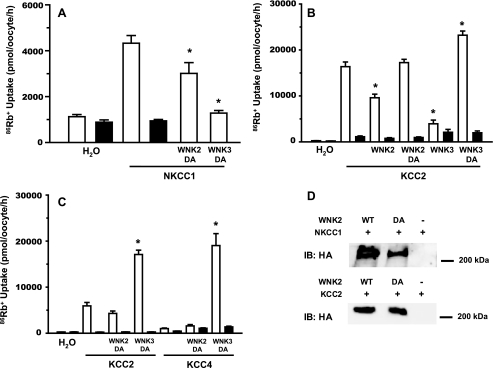
KCCs are activated by hypotonicity and inactivated by hypertonicity. KCC2 is unique among the KCCs in that it also exhibits significant activity in isotonic conditions, though less than in hypotonic conditions (3, 32). The effect of WNK2 on KCC2 activity was tested in both hypotonic (110 mOsm/kg) and isotonic (220 mOsm/kg) conditions. The related KCC4, which is virtually inactive in isotonic conditions, was tested in hypotonic conditions. WNK2 co-expression resulted in a significant reduction of the Cl−-dependent 86Rb+ uptake mediated by KCC2 in both isotonic (Fig. 3A) and hypotonic (Fig. 3B) conditions. WNK2 reduced the Cl−-dependent 86Rb+ uptake mediated by KCC4 in hypotonic conditions (Fig. 3B). These effects were similar to those observed in parallel experiments with WNK3 (Fig. 3).
FIGURE 3.
Modulation of KCC2, KCC4, and NKCC1 activity by WNK2. Xenopus laevis oocytes were injected with (A) water or 10 ng/oocyte of KCC2 cRNA, (B) water or 10 ng/oocyte of KCC2 or KCC4 cRNA, and (C) water, or 10 ng/oocyte of NKCC1 cRNA. In all figures, cotransporters cRNA were injected alone or together with 10 ng/oocyte of WNK2 or WNK3 cRNA, as stated. Four days later, 86Rb+ influx experiments were performed in isotonic (A and C) or hypotonic conditions (B) in the presence (open bars) or absence (closed bars) of extracellular chloride (A and B) or in the absence (open bars) or presence (closed bars) of 100 μm bumetanide (C). *, significantly different from the uptake observed in corresponding control. Each figure depicts combined data of at least three different experiments.
Conversely, NKCC1 is activated by hypertonicity and inactivated by hypotonicity. NKCC1 exhibits moderate activity in isotonic conditions (220 mOsm/kg). WNK2 co-expression resulted in a marked increase in bumetanide-sensitive 86Rb+ uptake induced by NKCC1 in isotonic conditions (Fig. 3C). These effects were qualitatively similar to, but quantitatively smaller, than effects observed in parallel experiments with WNK3 (Fig. 3).
We have previously shown that NCC activation by WNK3 is associated with increased expression of the cotransporter in the cell surface (17). In order to study the mechanism involved in NKCC1 or KCC4 modulation by both, WNK2 or WNK3, we performed functional assays experiments in oocytes injected with NKCC1 or FLAG-KCC4 cRNA alone or together with WNK2 or WNK3 cRNA, and oocytes from the same batch were used for extraction of total and biotinylated proteins. Western blots were performed using the anti-NKCC1 T4 monoclonal antibody developed by Forbush (33) or anti-FLAG monoclonal antibodies. A representative experiment is shown in Fig. 4 for each cotransporter. Functional expression in these experiments showed results that are similar to those discussed above. For NKCC1 there is a increase in both, the total amount of NKCC1 and the biotynilated fraction. Consistent with the functional data, the increase of NKCC1 induced by WNK3 is more apparent than the effect of WNK2. In the case of KCC4, no changes in total protein induced by WNK2 or WNK3 was observed. However, a decreased surface expression was induced by both kinases. Although the functional effect of WNK3 was higher than WNK2 (Fig. 4D), the decrease in surface expression of KCC4 by both kinases was similar (Fig. 4B).
FIGURE 4.
Effect of WNK2 and WNK3 on the protein level and surface expression of NKCC1 and KCC4. Western blot analysis of the total and biotinylated fraction of proteins extracted from oocytes injected with NKCC1 (A) or KCC4 (B) cRNA in the absence or presence of WNK2 or WNK3 cRNA. C and D depict the results of the functional expression performed the same day using oocytes from the same batch for NKCC1 or KCC4, respectively, expressed as bumetanide-sensitive (C) or chloride-dependent (D) 86Rb+ uptake.
All WNK kinases share a key aspartate residue in the kinase domain that is essential for catalytic activity (17, 31, 34–36). The effects of catalytically inactive WNK3 (WNK3D294A) on the CCCs are opposite to those of its wild type species, inhibiting NKCC1 and activating KCC2 (18, 19). We introduced the analogous mutation into the catalytic domain of WNK2 (WNK2D342A), and tested its effect on NKCC1 and KCC2 activity. In contrast with wild-type WNK2, WNK2D342A does not activate NKCC1 (Fig. 5A) or inhibit KCC2 (Fig. 5B), suggesting WNK2's effects on NKCC1 and KCC2 are dependent on its kinase activity. Similar to WNK3D294A, WNK2D342A inhibited NKCC1, though this effect was not as dramatic as with WNK3D294A (Fig. 5A). However, in contrast to WNK3D294A, WNK2D342A did not activate KCC2 (Fig. 5B).
FIGURE 5.
Effect of kinase-dead (DA) WNK2 upon NKCC1 and KCCs cotransporters activity. A, 86Rb+ uptake in oocytes injected with water, NKCC1 cRNA alone or together with WNK2-DA or WNK3-DA cRNA. Uptake was performed in isotonic conditions in the absence (open bars) or presence (closed bars) of 100 μm bumetanide. *, significantly different from the uptake observed in NKCC1 cRNA group. B, oocytes were injected with water, KCC2 cRNA alone or together with kinase-dead WNK2 or WNK3, as stated. C, oocytes were injected with water, KCC2, or KCC4 cRNA alone or together with kinase-dead WNK2 or WNK3, as stated. 86Rb+ Uptake was performed in in the presence (open bars) or absence (closed bars) of extracellular Cl−, in hypotonic (B) or isotonic (C) conditions. *, significantly different from the uptake observed in corresponding control. D, representative Western blot analysis of protein homogenates extracted from oocytes injected with wild type or kinase-dead HA-WNK2 cRNA, together with NKCC1 or KCC2 cRNA, as stated. Western blots were then performed using anti-HA monoclonal antibodies. Similar results were observed in the absence of NKCC1 cRNA injections.
We compared the activities of WNK2 on KCC2 (and KCC4) in isotonic conditions. In these conditions, KCC2 exhibits a small but significant activity, while KCC4 is inactive. In isotonicity, WNK3D294A induced a robust activation of both KCC2 and KCC4, while WNK2D342A imparted no effect (Fig. 5C). Similar expression of wild type WNK2 or WNK2D342A was observed by immunoblotting proteins from oocytes with anti-HA antibodies, demonstrating that any differences in effect were not due to protein expression (Fig. 5D). These results demonstrate functional differences between WNK2 and WNK3 on the CCCs in their active and inactive states.
WNK2 Forms a Phosphoprotein Complex with SPAK in Mouse Brain
Phosphorylation of the WNKs plays a key role in determining their kinase activities (34, 37), activation in response to osmotic stress (38), and regulation of ion transport proteins. Like many proteins, the comprehensive catalogue of sites that are phosphorylated in the WNKs has proved elusive given the typically low stochiometry of phosphopeptides in cells. Recent advances in the purification of phosphopeptides from complex mixtures have helped surmount this problem (39, 40).
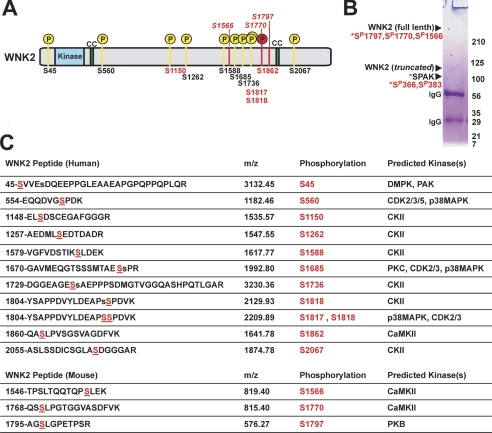
To identify phosphorylation sites in huaman WNK2 expressed in HEK cells, we used a simple and efficient phosphopeptide enrichment strategy coupled to liquid LC-MS (Fig. 6 and supplemental Fig. S3). This procedure resulted in a dramatic purification of phosphopeptides and enabled the identification of twelve phosphorylation sites in WNK2 (Fig. 6). These are distributed throughout the protein; none were identified in the kinase domain. These sites included potential phosphorylation sites mediated by Rho-associated protein kinase 1 (DMPK), cAMP-dependent protein kinase (PKA), casein kinase II (CKII), PKC protein kinase CΔ (PKC), p21-activated kinase (PAK), p38 mitogen-activated protein kinase (p38MAPK), and cyclin-dependent kinases (CDK2, 3, and 5).
FIGURE 6.
MS/MS analysis of WNK2 phosphorylation. A, phosphorylation sites are shown in the context of full-length human WNK2. The conserved kinase domain and two coiled-coiled regions are labeled. Phosphorylation sites identified in human WNK2 expressed in HEK cells are numbered on the bottom of the diagram. Sites unique to this study are in black, and sites in common with other studies are in red. Phosphorylation sites identified in native mouse WNK2 are numbered according to their position in the mouse sequence and mapped onto the corresponding position in human WNK2. Phosphorylation at Ser-1770 (mouse) and Ser-1862 (human) are homologous. B, SDS-PAGE analyisis of native WNK2 immunoprecipitates prepared from mouse brain for MS/MS. An example gel is shown with the results of the MS/MS analysis for each protein band shown to the left. Phosphorylation sites from full-length WNK2 and SPAK identified by MS/MS are listed (for additional peptide information see supplemental Tables S1 and S2. A brief Coomassie-staining method was used to reveal the proteins bands to the naked eye. Bands that were unique to brain IP and not visible in a control IP from mouse kidney (not shown) were cut and subjected to MS analysis. C, table of WNK2 phosphopeptides identified in MS/MS analysis of purified human and mouse WNK2. Mascot searches followed by manual MS/MS spectra interpretation places the possible phosphorylation site within each peptide. Unambiguous assignment of phosphorylation sites are indicated in underlined, red capital S, while ambiguous assignments have the second most likely site as a lowercase s. Kinase motifs corresponding to each phosphorylation site were identified with NetworKIN and NetPhorest analysis. Abbreviations are for Rho-associated protein kinase 1 (DMPK), cAMP-dependent protein kinase (PKA), casein kinase II (CKII), PKC protein kinase CΔ (PKC), p21-activated kinase (PAK), p38 mitogen-activated protein kinase (p38MAPK), and cyclin-dependent kinases (CDK2, 3, and 5).
Immunoprecipitates of native WNK2 from mouse brain were analyzed with tandem MS. SDS-PAGE analysis of the immunoprecipitated protein showed a full-length protein of the expected molecular weight of greater than 200 kDa. This full-length mouse WNK2 was unambiguously identified via tandem MS analysis (Fig. 5). Several sites of phosphorylation, Ser-1150, Ser-1817, Ser-1818, and Ser-1862, were identified that overlapped with those found for human WNK3 in vitro, suggesting sites of particular importance for WNK2's function in vivo (Fig. 6).
Interestingly, two proteins of sizes different from the expected full-length WNK2 were identified from the WNK2 immunoprecipitates from mouse brain. One of these, with an apparent molecular mass of 125 kDa, was also identified as WNK2, suggesting an alternative splice form of WNK2 exists in the mouse brain (Fig. 6). The other, with an apparent molecular weight of ∼100 kDa, was identified as SPAK (Fig. 6), a Ste20-type kinase known to interact with and phosphorylate NKCC1 (41). The apparent molecular mass of the protein was slightly higher than predicted 60 kDa; MS analysis revealed two phosphorylation sites within SPAK which may explain the gel shift. One of the SPAK phosphorylation sites detected in the WNK2 complex, S383 (supplemental Table S2), has been shown previously to be an important WNK-specific phosphorylation site (42). These data show that WNK2 exists in a phosphoprotein complex in vivo with SPAK, another kinase known to directly interact with NKCC1.
DISCUSSION
The WNK2 expression profile is unique among the WNK kinases. First, while WNK1, WNK3, and WNK4 are highly expressed in multiple Cl−-transporting epithelia (19, 43, 44), and most notably the nephron (16, 17, 19, 45), WNK2 is almost exclusively expressed in the brain, with no detectable expression in kidney. Second, in the CNS, while WNK1 is expressed throughout postnatal development, WNK3 is not significantly expressed until postnatal day 21 (19) and WNK2 is expressed since the embryonic life. Third, within the brain, WNK1 is predominantly expressed in non-neuronal cells and WNK3 localizes to both neurons and glia (19), but WNK2 is expressed primarily in cortical and thalamic neurons. Thus, WNK2 is more highly expressed in neurons and during early development than other WNKs. These differences might have important functional ramifications for the regulation of neuronal CCCs. In many other tissues in which WNKS and CCCs have been shown to co localize evidences show that WNKs are playing an important role in their regulation. In this regard, future development of WNK2 knock-out or knockin mice will be useful to analyze with detail the role that WNK2 might have on different parts of the brain function, during embryonic and in postnatal life.
It not clear at the moment how changes in osmolarity and/or intracellular chloride concentration modulates the activity of WNKs, but it has been observed that hypertonicity is clearly associated with activation of WNK1 by inducing autophosphorylation of the T-loop serine 328, known to be associated with activation of the kinase (37, 38, 46). However, similar to what we have observed for WNK3 (17–19, 47) WNK2 bypasses normal tonicity requirements to activate NKCC1 and inhibit KCC2 in the Xenopus oocytes. While caution must be exercised in extrapolating results to mammalian systems, oocytes have proven useful for studying the molecular mechanisms of CCC function and regulation (48). The opposing effects of WNK2 on cellular Cl− influx and efflux pathways would be expected to achieve a net increase in [Cl−]i, a property previously described for WNK3 (17–19). However, in contrast to WNK3, WNK2 is expressed during early development. As depicted in Fig. 7, after birth, the functional expression of the chloride-importing NKCC1 and chloride-exporting KCC2 cation-chloride cotransporters (CCC) is reciprocally related: NKCC1 is decreased, and KCC2 is increased. This shift is thought to underlie the polarity change in GABAergic signaling from excitatory in the immature brain to inhibitory in the mature brain. In the developing brain, WNK3 expression is low. In contrast, WNK2 is strongly expressed in both the developing and mature brain. The ontogeny of WNK2 expression, coupled with its ability to load Cl− into cells, suggests WNK2 might be important for establishing the elevated [Cl−]i seen early in development that is critical for the trophic functions of GABA excitatory signaling (2). In adulthood, both WNK2 and WNK3 are highly expressed. An appropriate mixture of WNK kinases might confer certain neuronal groups, like the suprachiasmatic nucleus, with the ability to rapidly change [Cl−]i, allowing for dynamic shifts in the response to GABA (Fig. 7).
FIGURE 7.
Relationship between GABA excitatory and inhibitory effect and the expression of WNK2, WNK3, NKCC1, and KCC2 in the CNS from embryonic to postnatal life.
Regulation of [Cl−]i by WNK2 might also be important for the role of the CCCs during regulatory volume increase (RVI) and regulatory volume decrease (RVD), key homeostatic counter-responses to maintain cell volume during shifts in extracellular osmolarity or intracellular solute content (Fig. 7). Acute activation of NKCC1 and other ion transport mechanisms (such as NHE1 functioning in parallel with a Cl−/HCO3− exchanger), are important in RVI and operate in neurons (49–51). KCCs like KCC3 are important for cell volume regulation via RVD in the nervous system (52). The WNK2 ability to activate NKCC1 and inhibit the KCCs suggests it may participate in RVI in neurons. Additional experiments will be required to investigate whether WNK2 activity is affected by changes in cell volume, or is altered in pathologic states like cerebral edema.
WNK kinases exhibit kinase-dependent and -independent effects on different ion transport pathways. WNK2 regulation of the CCCs is likely dependent on its catalytic activity, since its activation of NKCC1 or inhibition of KCCs is lost by an inactivating mutation in its kinase domain. Since serine-threonine phosphorylation is the primary mode of CCC regulation, we presume WNK2 activates NKCC1 and inhibits KCC2 by promoting net cotransporter phosphorylation. This could be due to: 1) direct phosphorylation of the CCCs by WNK2; 2) indirect phosphorylation via another downstream kinase; or 3) inhibition of protein phosphatases. The fact that WNK2 forms a complex with SPAK in vivo, that in this complex SPAK is phosphorylated at a consensus WNK-phospho site, and that SPAK has been shown to directly phsophorylate NKCC1 (35, 53), suggest WNK2 might activate NKCC1 indirectly via SPAK kinase. If so, this mechanism would be analogous to the WNK-SPAK-NCC pathway in kidney (46, 47, 54). We and others have previously shown that WNKs modulation of CCCs is associated with changes in protein expression at the cell surface (17, 55, 56). Here we shown that increased activity of NKCC1 induced by WNK2 or WNK3 is due to both, an increase in protein levels and also an increase in the surface expression of the cotransproter. In this regard, it has been shown that decreased activity of NCC induced by WNK4 is due to both, a reduction in the surface expression of the cotransporter (31, 55) and to a increased degradation of the NCC via lysosomes (57). In contrast, the down-regulation of KCC4 activity by WNK2 or WNK3 is more dependent on their effect upon the surface expression of the cotransporter.
WNK2 might utilize a similar mechanism to inhibit KCC2; however, SPAK does not appear to phosphorylate KCC2, and recent experiments demonstrate that WNKs and SPAK/OSR1 are essential for KCC phosphorylation (28). Because the regulation of KCCs by catalytically inactive WNK3 is prevented with protein phosphatase inhibitors such as calyculin or cyclosporin A, it is possible that a protein phosphatases are involved in the WNK2-KCC mechanism (18, 30).
SPAK and oxidative-stress response 1 (OSR1) kinase have been shown to bind, phosphorylate, and directly activate NKCC1 in multiple cell systems, including dorsal root ganglion neurons (14, 58). In this study, we isolated WNK2 in a phosphoprotein complex with SPAK from native mouse brain, suggesting these two kinases might work in concert to regulate neuronal CCCs. WNK1, WNK3, and WNK4 are known to interact with OSR1 and SPAK, and WNK1 and WNK4 phosphorylate Thr-233 and Ser-373 in SPAK, and Thr-185 and Ser-325 in OSR1 (35, 36, 46). SPAK activity requires phosphorylation at Thr-233 and Ser-373 (59). In our study, we have shown that SPAK is phosphorylated at Ser-366 and Ser-383 when complexed to WNK2 in vivo. These results suggest that a WNK2-SPAK-CCC pathway is operative in the brain. Further experiments will be required to examine the functional role of this pathway in both the CNS, including the spinal cord, and the PNS (e.g. in dorsal root ganglion cells).
WNK2 can be phosphorylated in at least 12 different sites. From the sequence flanking these phosphorylation sites, the Networkin prediction program (60) identified potential kinases that might phosphorylate WNK2. These include sites for Rho-associated protein kinase 1 (DMPK), cAMP-dependent protein kinase (PKA), casein kinase II (CKII), PKC protein kinase CΔ (PKC), and cyclin-dependent kinase 5 (CDK5) (Fig. 6). Among these putative upstream kinases, P38MAPK, PKA, and Cdk5 are particularly interesting owing to prior evidence of their role in neuronal function. Because WNK kinase regulation might be ultimately determined by the N- and C-terminal regions flanking the kinase domain, any of these kinases are candidates for WNK co-regulatory kinases.
At present, it is unclear if WNK2's regulation of CCCs is linked to its role in cell growth and proliferation (61). Hong et al. (22) recently identified WNK2 as a tumor suppressor gene in a large-scale genomic and epigenomic analysis of human infiltrative gliomas. Epigenetic silencing of WNK2 has also been shown in all grades of meningioma (23). Point mutations in other WNKs have also been associated with breast, lung and colonic cancer (62–64). In this context, it is interesting that NKCC1 has recently been shown to be the major pathway for Cl− accumulation in glioma cells, and genetic or pharmacologic inhibition of NKCC1 is associated with a marked reduction in glioma cell invasion in vivo (65). These issues will be topics of future experiments.
Supplementary Material
Acknowledgments
We thank Pedro San Cristobal MD for technical help in the early stages of the project and all members of the Molecular Physiology Unit for comments and suggestions.
This work was supported, in whole or in part, by NIDA, National Institutes of Health Grant 2P30DA018343 (to J. R., E. E. G., and R. P. L.), the Leducq Foundation Transatlantic Network on Hypertension (to G. G. and R. P. L.), and el Consejo Nacional de Ciencia y Tecnología (CONACYT-Mexico) Grant 59992 (to G. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S3.
- GABA
- γ-aminobutyric acid
- PHAII
- pseudohypoaldosteronism type II
- WNK
- with no lysine=K
- KCCs
- K-Cl cotransporters
- NKCC1
- Na-K-2Cl cotransporter.
REFERENCES
- 1. McManus M. L., Churchwell K. B., Strange K. (1995) N. Engl. J. Med. 333, 1260–1266 [DOI] [PubMed] [Google Scholar]
- 2. Ben-Ari Y., Gaiarsa J. L., Tyzio R., Khazipov R. (2007) Physiol. Rev. 87, 1215–1284 [DOI] [PubMed] [Google Scholar]
- 3. Gamba G. (2005) Physiol. Rev. 85, 423–493 [DOI] [PubMed] [Google Scholar]
- 4. Kahle K. T., Staley K. J., Nahed B. V., Gamba G., Hebert S. C., Lifton R. P., Mount D. B. (2008) Nat. Clin. Pract. Neurol. 4, 490–503 [DOI] [PubMed] [Google Scholar]
- 5. Blaesse P., Airaksinen M. S., Rivera C., Kaila K. (2009) Neuron 61, 820–838 [DOI] [PubMed] [Google Scholar]
- 6. Plotkin M. D., Kaplan M. R., Peterson L. N., Gullans S. R., Hebert S. C., Delpire E. (1997) Am. J. Physiol. 272, C173–C183 [DOI] [PubMed] [Google Scholar]
- 7. Williams J. R., Sharp J. W., Kumari V. G., Wilson M., Payne J. A. (1999) J. Biol. Chem. 274, 12656–12664 [DOI] [PubMed] [Google Scholar]
- 8. Plotkin M. D., Snyder E. Y., Hebert S. C., Delpire E. (1997) J. Neurobiol. 33, 781–795 [DOI] [PubMed] [Google Scholar]
- 9. Rivera C., Voipio J., Payne J. A., Ruusuvuori E., Lahtinen H., Lamsa K., Pirvola U., Saarma M., Kaila K. (1999) Nature 397, 251–255 [DOI] [PubMed] [Google Scholar]
- 10. Marty A., Llano I. (2005) Trends Neurosci. 28, 284–289 [DOI] [PubMed] [Google Scholar]
- 11. Duebel J., Haverkamp S., Schleich W., Feng G., Augustine G. J., Kuner T., Euler T. (2006) Neuron 49, 81–94 [DOI] [PubMed] [Google Scholar]
- 12. Lytle C., McManus T. (2002) Am. J. Physiol. Cell Physiol 283, C1422–C1431 [DOI] [PubMed] [Google Scholar]
- 13. Kahle K. T., Rinehart J., Ring A., Gimenez I., Gamba G., Hebert S. C., Lifton R. P. (2006) Physiology 21, 326–335 [DOI] [PubMed] [Google Scholar]
- 14. Delpire E., Gagnon K. B. (2008) Biochem. J. 409, 321–331 [DOI] [PubMed] [Google Scholar]
- 15. Kahle K. T., Ring A. M., Lifton R. P. (2008) Annu. Rev. Physiol. 70, 329–355 [DOI] [PubMed] [Google Scholar]
- 16. Wilson F. H., Disse-Nicodème S., Choate K. A., Ishikawa K., Nelson-Williams C., Desitter I., Gunel M., Milford D. V., Lipkin G. W., Achard J. M., Feely M. P., Dussol B., Berland Y., Unwin R. J., Mayan H., Simon D. B., Farfel Z., Jeunemaitre X., Lifton R. P. (2001) Science 293, 1107–1112 [DOI] [PubMed] [Google Scholar]
- 17. Rinehart J., Kahle K. T., De Los Heros P., Vazquez N., Meade P., Wilson F. H., Hebert S. C., Gimenez I., Gamba G., Lifton R. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16777–16782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Los Heros P., Kahle K. T., Rinehart J., Bobadilla N. A., Vézquez N., San Cristobal P., Mount D. B., Lifton R. P., Hebert S. C., Gamba G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1976–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kahle K. T., Rinehart J., De Los Heros P., Louvi A., Meade P., Vazquez N., Hebert S. C., Gamba G., Gimenez I., Lifton R. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16783–16788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shekarabi M., Girard N., Riviere J. B., Dion P., Houle M., Toulouse A., Lafreniere R. G., Vercauteren F., Hince P., Laganiere J., Rochefort D., Faivre L., Samuels M., Rouleau G. A. (2008) J. Clin. Invest. 118, 2496–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rudnik-Schöneborn S., Hehr U., von Kalle T., Bornemann A., Winkler J., Zerres K. (2009) Neuropediatrics 40, 129–133 [DOI] [PubMed] [Google Scholar]
- 22. Hong C., Moorefield K. S., Jun P., Aldape K. D., Kharbanda S., Phillips H. S., Costello J. F. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10974–10979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jun P., Hong C., Lal A., Wong J. M., McDermott M. W., Bollen A. W., Plass C., Held W. A., Smiraglia D. J., Costello J. F. (2009) Neuro. Oncol. 11, 414–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Verissimo F., Jordan P. (2001) Oncogene 20, 5562–5569 [DOI] [PubMed] [Google Scholar]
- 25. Arroyo J. P., Lagnaz D., Ronzaud C., Vazquez N., Ko B., Moddes L., Ruffieux-Hausel P., Koesters R., Yang B., Stokes J. B., Hoover R. S., Gamba G., Staub O. (2011) J. Am. Societ. Nephrol. 22, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Louvi A., Wassef M. (2000) Development 127, 4061–4071 [DOI] [PubMed] [Google Scholar]
- 27. Tole S., Patterson P. H. (1995) J. Neurosci. 15, 970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rinehart J., Maksimova Y. D., Tanis J. E., Stone K. L., Hodson C. A., Zhang J., Risinger M., Pan W., Wu D., Colangelo C. M., Forbush B., Joiner C. H., Gulcicek E. E., Gallagher P. G., Lifton R. P. (2009) Cell 138, 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Campbell M. J., Morrison J. H. (1989) J. Comp. Neurol. 282, 191–205 [DOI] [PubMed] [Google Scholar]
- 30. Garzón-Muvdi T., Pacheco-Alvarez D., Gagnon K. B., Vázquez N., Ponce-Coria J., Moreno E., Delpire E., Gamba G. (2007) Am. J. Physiol. Renal Physiol. 292, F1197–F1207 [DOI] [PubMed] [Google Scholar]
- 31. Wilson F. H., Kahle K. T., Sabath E., Lalioti M. D., Rapson A. K., Hoover R. S., Hebert S. C., Gamba G., Lifton R. P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 680–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Song L., Mercado A., Vázquez N., Xie Q., Desai R., George A. L., Jr., Gamba G., Mount D. B. (2002) Brain Res. Mol. Brain Res. 103, 91–105 [DOI] [PubMed] [Google Scholar]
- 33. Lytle C., Forbush B., 3rd (1992) J. Biol. Chem. 267, 25438–25443 [PubMed] [Google Scholar]
- 34. Xu B., English J. M., Wilsbacher J. L., Stippec S., Goldsmith E. J., Cobb M. H. (2000) J. Biol. Chem. 275, 16795–16801 [DOI] [PubMed] [Google Scholar]
- 35. Vitari A. C., Deak M., Morrice N. A., Alessi D. R. (2005) Biochem. J. 391, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moriguchi T., Urushiyama S., Hisamoto N., Iemura S., Uchida S., Natsume T., Matsumoto K., Shibuya H. (2005) J. Biol. Chem. 280, 42685–42693 [DOI] [PubMed] [Google Scholar]
- 37. Zagórska A., Pozo-Guisado E., Boudeau J., Vitari A. C., Rafiqi F. H., Thastrup J., Deak M., Campbell D. G., Morrice N. A., Prescott A. R., Alessi D. R. (2007) J. Cell Biol. 176, 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lenertz L. Y., Lee B. H., Min X., Xu B. E., Wedin K., Earnest S., Goldsmith E. J., Cobb M. H. (2005) J. Biol. Chem. 280, 26653–26658 [DOI] [PubMed] [Google Scholar]
- 39. Pinkse M. W., Uitto P. M., Hilhorst M. J., Ooms B., Heck A. J. (2004) Anal. Chem. 76, 3935–3943 [DOI] [PubMed] [Google Scholar]
- 40. Bodenmiller B., Mueller L. N., Mueller M., Domon B., Aebersold R. (2007) Nat. Methods 4, 231–237 [DOI] [PubMed] [Google Scholar]
- 41. Piechotta K., Garbarini N., England R., Delpire E. (2003) J. Biol. Chem. 278, 52848–52856 [DOI] [PubMed] [Google Scholar]
- 42. Richardson C., Alessi D. R. (2008) J. Cell Sci. 121, 3293–3304 [DOI] [PubMed] [Google Scholar]
- 43. Choate K. A., Kahle K. T., Wilson F. H., Nelson-Williams C., Lifton R. P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 663–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kahle K. T., Gimenez I., Hassan H., Wilson F. H., Wong R. D., Forbush B., Aronson P. S., Lifton R. P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2064–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. O'Reilly M., Marshall E., Macgillivray T., Mittal M., Xue W., Kenyon C. J., Brown R. W. (2006) J. Am. Soc. Nephrol. 17, 2402–2413 [DOI] [PubMed] [Google Scholar]
- 46. Richardson C., Rafiqi F. H., Karlsson H. K., Moleleki N., Vandewalle A., Campbell D. G., Morrice N. A., Alessi D. R. (2008) J. Cell Sci. 121, 675–684 [DOI] [PubMed] [Google Scholar]
- 47. Ponce-Coria J., San-Cristobal P., Kahle K. T., Vazquez N., Pacheco-Alvarez D., De Los Heros P., Juárez P., Muñoz E., Michel G., Bobadilla N. A., Gimenez I., Lifton R. P., Hebert S. C., Gamba G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8458–8463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gamba G. (2005) Am. J. Physiol. Renal Physiol. 288, F245–F252 [DOI] [PubMed] [Google Scholar]
- 49. Hoffmann E. K., Lambert I. H., Pedersen S. F. (2009) Physiol. Rev. 89, 193–277 [DOI] [PubMed] [Google Scholar]
- 50. Schomberg S. L., Bauer J., Kintner D. B., Su G., Flemmer A., Forbush B., Sun D. (2003) J. Neurophysiol. 89, 159–167 [DOI] [PubMed] [Google Scholar]
- 51. Breitwieser G. E., Altamirano A. A., Russell J. M. (1990) Am. J. Physiol. 258, C749–C753 [DOI] [PubMed] [Google Scholar]
- 52. Byun N., Delpire E. (2007) Neurobiol. Dis. 28, 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gagnon K. B., England R., Delpire E. (2006) Am. J. Physiol. Cell Physiol. 290, C134–C142 [DOI] [PubMed] [Google Scholar]
- 54. Rafiqi F. H., Zuber A. M., Glover M., Richardson C., Fleming S., Jovanović S., Jovanović A., O'Shaughnessy K. M., Alessi D. R. (2010) EMBO Mol. Med. 2, 63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cai H., Cebotaru V., Wang Y. H., Zhang X. M., Cebotaru L., Guggino S. E., Guggino W. B. (2006) Kidney Int. 69, 2162–2170 [DOI] [PubMed] [Google Scholar]
- 56. Golbang A. P., Cope G., Hamad A., Murthy M., Liu C. H., Cuthbert A. W., O'Shaughnessy K. M. (2006) Am. J. Physiol. Renal Physiol. 291, F1369–F1376 [DOI] [PubMed] [Google Scholar]
- 57. Subramanya A. R., Liu J., Ellison D. H., Wade J. B., Welling P. A. (2009) J. Biol. Chem. 284, 18471–18480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Geng Y., Hoke A., Delpire E. (2009) J. Biol. Chem. 284, 14020–14028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gagnon K. B., Delpire E. (2010) Am. J. Physiol. Cell Physiol. 299, C614–C620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Linding R., Jensen L. J., Ostheimer G. J., van Vugt M. A., Jørgensen C., Miron I. M., Diella F., Colwill K., Taylor L., Elder K., Metalnikov P., Nguyen V., Pasculescu A., Jin J., Park J. G., Samson L. D., Woodgett J. R., Russell R. B., Bork P., Yaffe M. B., Pawson T. (2007) Cell 129, 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Moniz S., Jordan P. (2010) Cell Mol. Life Sci. 67, 4481–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Davies H., Hunter C., Smith R., Stephens P., Greenman C., Bignell G., Teague J., Butler A., Edkins S., Stevens C., Parker A., O'Meara S., Avis T., Barthorpe S., Brackenbury L., Buck G., Clements J., Cole J., Dicks E., Edwards K., Forbes S., Gorton M., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Jones D., Kosmidou V., Laman R., Lugg R., Menzies A., Perry J., Petty R., Raine K., Shepherd R., Small A., Solomon H., Stephens Y., Tofts C., Varian J., Webb A., West S., Widaa S., Yates A., Brasseur F., Cooper C. S., Flanagan A. M., Green A., Knowles M., Leung S. Y., Looijenga L. H., Malkowicz B., Pierotti M. A., Teh B. T., Yuen S. T., Lakhani S. R., Easton D. F., Weber B. L., Goldstraw P., Nicholson A. G., Wooster R., Stratton M. R., Futreal P. A. (2005) Cancer Res. 65, 7591–7595 [DOI] [PubMed] [Google Scholar]
- 63. Sjöblom T., Jones S., Wood L. D., Parsons D. W., Lin J., Barber T. D., Mandelker D., Leary R. J., Ptak J., Silliman N., Szabo S., Buckhaults P., Farrell C., Meeh P., Markowitz S. D., Willis J., Dawson D., Willson J. K., Gazdar A. F., Hartigan J., Wu L., Liu C., Parmigiani G., Park B. H., Bachman K. E., Papadopoulos N., Vogelstein B., Kinzler K. W., Velculescu V. E. (2006) Science 314, 268–274 [DOI] [PubMed] [Google Scholar]
- 64. Greenman C., Stephens P., Smith R., Dalgliesh G. L., Hunter C., Bignell G., Davies H., Teague J., Butler A., Stevens C., Edkins S., O'Meara S., Vastrik I., Schmidt E. E., Avis T., Barthorpe S., Bhamra G., Buck G., Choudhury B., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Jenkinson A., Jones D., Menzies A., Mironenko T., Perry J., Raine K., Richardson D., Shepherd R., Small A., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Cahill D. P., Louis D. N., Goldstraw P., Nicholson A. G., Brasseur F., Looijenga L., Weber B. L., Chiew Y. E., DeFazio A., Greaves M. F., Green A. R., Campbell P., Birney E., Easton D. F., Chenevix-Trench G., Tan M. H., Khoo S. K., Teh B. T., Yuen S. T., Leung S. Y., Wooster R., Futreal P. A., Stratton M. R. (2007) Nature 446, 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Haas B. R., Sontheimer H. (2010) Cancer Res. 70, 5597–5606 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.