Abstract
Endothelial and mural cell interactions are vitally important for proper formation and function of blood vessels. These two cell types communicate to regulate multiple aspects of vessel function. In studying genes regulated by this interaction, we identified apolipoprotein D (APOD) as one gene that is downregulated in mural cells by coculture with endothelial cells. APOD is a secreted glycoprotein that has been implicated in governing stress response, lipid metabolism, and aging. Moreover, APOD is known to regulate smooth muscle cells and is found in abundance within atherosclerotic lesions. Our data show that the regulation of APOD in mural cells is bimodal. Paracrine secretion by endothelial cells causes partial downregulation of APOD expression. Additionally, cell contact-dependent Notch signaling plays a role. NOTCH3 on mural cells promotes the downregulation of APOD, possibly through interaction with the JAGGED-1 ligand on endothelial cells. Our results show that NOTCH3 contributes to the downregulation of APOD and by itself is sufficient to attenuate APOD transcript expression. In examining the consequence of decreased APOD expression in mural cells, we show that APOD negatively regulates cell adhesion. APOD attenuates adhesion by reducing focal contacts; however, it has no effect on stress fiber formation. These data reveal a novel mechanism in which endothelial cells control neighboring mural cells through the downregulation of APOD, which, in turn, influences mural cell function by modulating adhesion.
Keywords: vascular smooth muscle cells, coculture, NOTCH3
blood vessels are comprised of two major cell types, consisting of an inner layer of endothelial cells and an outer layer of mural cells (smooth muscle cells or pericytes). Numerous studies (2, 4, 9, 11) have highlighted the importance of heterotypic communication between these cells in normal vessel formation and function. More significantly, abnormal interactions between these two cell types have been linked to several pathological conditions, including tumor angiogenesis, vascular malformations, stroke, and the dementia syndrome CADASIL (7, 11, 23). Although it is well established that endothelial cells and mural cells communicate with one another to alter their functions, little is known about the complexities of these signaling mechanisms.
Using a microarray screen, we identified several genes that were regulated by endothelial/mural cell interactions (20), including NOTCH3 and apolipoprotein D (APOD). NOTCH3 belongs to an evolutionarily conserved family of membrane-bound receptors that have been implicated in an array of cellular functions ranging from lineage commitment, differentiation, and cancer progression (3). Recent studies (21) by our group have shown that NOTCH3 plays an important role in vascular cell communication, where endothelial cells promote NOTCH3 expression in mural cells, which facilitates smooth muscle maturation. The role of NOTCH3 has been extensively studied; however, there are relatively few studies pertaining to the function of APOD. APOD is a 29-kDa protein belonging to the lipocalin family of glycoproteins (29, 39). It is a secreted protein that is predicted to bind to small hydrophobic ligands and in plasma is found predominantly associated with high-density lipoprotein (29, 39). Recent studies (24) have highlighted a role for APOD in aging and stress resistance. Collectively, these data suggest that APOD plays a protective role in cellular and oxidative stress, and in fruit flies this effect has been linked to increased life span (12, 14, 31, 38). Furthermore, APOD has been associated with lipid metabolism and metabolic syndrome (26), with several studies (8, 18, 26, 37) demonstrating a correlation between APOD expression and healthy plasma lipid profiles. Additionally, in a mouse model of obesity, APOD was shown to regulate food intake and body fat accumulation through interaction with the leptin receptor (22). Whether APOD regulates these diverse processes through a common pathway or various mechanisms remains to be determined.
Within blood vessels, APOD has been shown to be prominently expressed in the perivascular cells surrounding the endothelium during development (28, 30, 33). In adults, expression has been localized to mature blood vessels and is also found at high levels in atherosclerotic lesions (26, 32). Studies (32) examining APOD function in vascular smooth muscle cells showed that APOD inhibited platelet-derived growth factor (PDGF)-BB-induced proliferation through inhibition of extracellular signal-regulated kinase 1/2. This same group also demonstrated that PDGF-BB and APOD synergized to mediate vascular smooth muscle migration (19). Since PDGF-BB acts as a signaling mediator between endothelial cells and smooth muscle cells, these data imply that APOD plays an important role in interpreting the signals between these vascular cell types. Our data presented here further strengthen this notion. In this study, we show that endothelial cell signaling regulates APOD expression. Endothelial cells cocultured with fibroblasts or smooth muscle cells cause a significant decrease in APOD expression within the mural cell subtypes. APOD expression within mural cells is modulated through both a secreted factor and the cell surface receptor NOTCH3. Modulation of APOD within mural cells likely has functional implications within the vasculature, since decreasing APOD levels within fibroblasts and smooth muscle cells causes an increase in cell adhesion and focal contacts, without affecting stress fiber formation. Thus the ability of endothelial cells to govern APOD expression in neighboring mural cells may facilitate binding of mural cells to the vascular basement membrane, leading to a more stable and mature blood vessel.
MATERIALS AND METHODS
Cell culture.
Primary cultures of human umbilical vein endothelial cells (HUVECs) were purchased from Lonza and grown in endothelial cell basal medium-2 (EBM-2) supplemented with the entire bullet kit as recommended. Human dermal neonatal fibroblasts (HDFNs) were purchased from Cascade Biological and cultured in DMEM (Mediatech) supplemented with 5% FBS (Hyclone), 2 mM glutamine, 1 mM sodium pyruvate, and 100 U/ml penicillin-streptomycin. Human aortic smooth muscle cells (HAoSMCs) and human coronary artery smooth muscle cells (HCASMCs) were purchased from Lonza and grown in DMEM supplemented with 10% FBS. Cells between passages 6–9 were used for all experiments. All cell culture experiments, unless indicated, were performed in media consisting of EBM-2 supplemented with all bullet kit components except FBS, VEGF, and basic fibroblast growth factor. These media were supplemented with 1% FBS and 30 ng/ml VEGF-A165 (Peprotech). For virus production, TN293 cells were purchased from Stratagene and cultured in 10% DMEM as above. All cultures were maintained in humidified 5% CO2 at 37°C. For coculture, 6 × 104 mural cells were plated in 12-well plates, and after adhesion, 6 × 104 HUVECs were added. To separate endothelial cells and fibroblasts, anti-platelet endothelial cell adhesion molecule 1-conjugated Dynabeads (Invitrogen) were used according to manufacturer's instructions. For conditioned media experiments, HDFNs were cultured for 24 h before addition of HDFN-conditioned media (as control) or HUVEC-conditioned media for indicated times. Media were conditioned for 24 h before addition to the HDFNs. The Transwell experiments were performed as previously described (21). Transwell inserts (12-well type; Corning Costar) with 0.4-μm pores were coated with 50 μg/ml collagen I. 2 × 104 HDFNs or HUVECs were first plated on the outside of the polycarbonate membrane of the Transwell inserts. After cell adherence, the Transwell inserts were inverted and reinserted onto 12-well plates, and 2 × 104 HDFNs or HUVECs were plated on the top surface of the insert and cultured in a final volume of 1.3 ml media (0.3 ml in the insert, 1 ml in the well). Following incubation for 48 h, cells grown on the top side of the inserts were harvested by trypsinization and processed for quantitative PCR (qPCR). For addition of the Jagged-1 peptide (JAG1) (CDD YYY GFG CNK FCR PR) (1), the peptide was added to the culture media for 48 h before cells were collected for analysis.
qPCR.
Total RNA was isolated using TRIzol reagent following manufacturer's instructions (Invitrogen), and reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Invitrogen) to generate cDNA. Real-time PCR was performed using a StepOne PCR system (Applied Biosystems) with SYBR Green and 50 ng cDNA template. The fold difference in various transcripts was calculated using ΔΔCT method with 18S as the internal control. Primer sequences were as follows: APOD For-5′-CTG GCC ACC GAC TAT GAG AA-3′, APOD Rev-5′-AAA ATC CAC GTG AAA AAG TTG GA 3′; NOTCH3 For-5′-CCT AGA CCT GGT GGA CAA G-3′, NOTCH3 Rev-5′-ACA CAG TCG TAG CGG TTG-3′; 18S For-5′-GTT GGT TTT CGG AAC TGA GGC-3′, 18S Rev-5′-GTC GGC ATC GTT TAT GGT CG-3′; and HES1 For-5′-TTG GAG GCT TCC AGG TGG TA-3′, HES1 Rev-5′-GGC CCC GTT GGG AAT G-3.′ Control primers to evaluate potential genomic DNA contamination were from SABiosciences (catalog no. PA-031).
ELISA.
ELISA kit (catalog no. E91968HU) for APOD expression was performed according to the manufacturer's instructions (USCN Life Sciences). Equal amounts of cell-conditioned media were collected and subjected to ELISA. Negligible amounts of APOD were found to be expressed by HUVECs. APOD concentration was determined based on a standard curve generated from known concentrations. Assays were performed in triplicate.
Immunoblotting.
Equivalent amounts of protein were run on 10% SDS-PAGE gels, transferred to immobilon-PVDF membranes (Millipore), and subjected to incubation using primary antibodies to NOTCH3 (Santa Cruz Biotechnology), vinculin (Sigma), and tubulin (Upstate), and secondary antibodies conjugated to horseradish peroxidase (Amersham). Protein was detected by enhanced chemiluminescence.
RNA interference.
HDFNs or HAoSMCs were plated in a 12-well plate at 6 × 104 cells/well. After 12 h, the cells were transfected with small interfering (si)RNA using Lipofectamine 2000 (Invitrogen). siRNA oligonucleotides for APOD were obtained from Ambion, and GFP control siRNA was obtained from Invitrogen. Efficiency of knockdown was assessed using qPCR and ELISA (Supplemental Fig. S1; Supplemental Material for this article is available online at the Am J Physiol Heart Circ Physiol website). NOTCH3 siRNA was synthesized by IDT as the following sequence: AAC UGC GAA GUG AAC AUU G; GUC AAU GUU CAC UUC GCA G, and used as previously described (21). All siRNA was used at 100 nM. Knockdown of APOD and NOTCH3 was assessed at 48 h by qPCR. For APOD knockdown experiments, after 5 h of transfection, the media were replaced with 0.25% FBS to minimize the presence of APOD in the serum having an effect on the cells. Cells collected 48 h following transfection were used for adhesion and immunostaining experiments. For NOTCH3 knockdown, cells were cocultured with HUVECs for 48 h, separated, and collected for qPCR analysis and Western blotting.
Lentivirus expression.
Human APOD cDNA from ATCC was cloned into pCDF1-GFP in front of the cytomegalovirus promoter using BamHI and BglII sites. The lentivirus plasmid containing GFP alone (lentiGFP) or APOD with GFP (lentiAPOD) were transfected into TN-293 cells using Lipofectamine 2000 (Invitrogen), and the viral particles were amplified and purified as described previously (21). For HDFNs and HAoSMCs infection, equal volumes of viral particles were diluted in 10% FBS DMEM and were incubated for 24 h. The efficiency of infection was evaluated using GFP expression and qPCR. LentiGFP and lentiAPOD viral particles were titrated to achieve 90 to 100% infection. Expression of APOD was confirmed using ELISA and qPCR (Supplemental Fig. S1). The NOTCH3 intracellular domain (NICD3) in plasmid, pCDF1-MCS2-EF1-copGFP-NICD3 (21), was overexpressed using lentivirus as described above.
Transfection and luciferase assays.
The human APOD promoter was cloned into the pGL3-basic-luciferase (Promega) construct by PCR. Primers containing APOD-specific sequences were used to amplify genomic DNA to clone the promoter region between −921 and +66 of the human APOD gene (10, 17). Primer sequences were 5′-atc taa atg tct cta aac ata cc-3′ and 5′-ctt ttc ttt cAA GAT GAA GGC AG-3′. To measure the transcriptional activity of the APOD promoter, HDFNs were transiently transfected at 80% confluency using Lipofectamine 2000 (Invitrogen) for 5 h. Cells were then cocultured with an equal number of HFDNs (control) or HUVECs for an additional 48 h. For conditioned media experiments, instead of the addition of cells, the media were replaced with conditioned media from HDFNs (control) or HUVECs. Cells were collected and promoter activity was measured by luciferase assays. To normalize the transfection efficiency, Hsp-β-galactosidase (LacZ) was cotransfected, and luciferase activities were normalized based on equivalent amount of LacZ activity. Luciferase and LacZ activity was measured as described previously (16) and quantified using a Turner Diagnostics luminometer. All experiments were performed in duplicate and repeated a minimum of three times. Statistical significance was determined using a Student's paired t-test.
Adhesion assay.
HDFNs and HAoSMCs were treated to overexpress or knockdown APOD as above. Forty-eight hours after treatment, the cells were trypsinized and counted for adhesion experiments (40). For addition of exogenous APOD, indicated concentrations of recombinant human APOD (PROSPEC) were added to the culture media 24 h before adhesion assay. The 96-well culture plates were coated overnight at 4°C with 10 μg/ml of either BSA, collagen type I, collagen type III, collagen type IV, laminin, or fibronectin (Sigma). Wells were then rinsed with PBS, and 100 μl of cells (2.0 × 105 cells/ml) in serum-free DMEM were placed in each well. The cells were allowed to adhere for 25 min. Plates were rinsed with PBS to remove nonadherent cells, and attached cells were fixed using 4% formaldehyde for 30 min. Following fixation, cells were rinsed in PBS and stained with 1% toluidine blue for 1 h. Plates were washed to remove the excess dye, and the remaining blue dye was solubilized in 2% SDS. Absorbance was measured at 620 nm. Cell adhesion to various matrices was normalized after subtracting the nonspecific cell adhesion to BSA. All experiments were performed in triplicate, and statistical analysis was done using GraphPad Prism software.
Immunofluorescence.
HDFNs were transfected with APOD siRNA as described above. Forty-eight hours posttransfection, cells were trypsinized, counted, and plated on coverslips coated with 10 μg/ml collagen I. Cells were allowed to attach for 30 min at 37C, fixed in 4% formaldehyde for 15 min, and permeabilized in 0.2% Triton X-100 for 3 min. Antibody incubations were carried out as described (35). Primary antibodies used: vinculin (1:300; Sigma), zyxin (25) (1:300), and phalloidin-Alexa-Fluor-594 (Molecular Probes; 1:100). Fluorescently tagged secondary antibodies were Alexa-Fluor-488 donkey anti-mouse (1:300) and Alexa-Fluor-594 donkey anti-rabbit (1:300; Molecular Probes). Coverslips were mounted in Prolong Gold antifade reagent (Molecular Probes), and cells were visualized using Zeiss LSM confocal microscope (×40 magnification). Data shown are representative of at least three independent experiments. From each experiment, five individual cells were randomly selected from the control and experimental group and vinculin intensity was measured using NIH ImageJ software.
Statistical analysis.
Data analyses were performed using GraphPad Prism, and comparisons between data sets were made using a Student's t-test. For addition of JAG1 peptide (see Fig. 5) and adhesion assays (see Fig. 6), data were analyzed by one-way ANOVA followed by post hoc t-tests between control and experimental groups. Differences were considered significant if P < 0.05, and data are presented as means ± SE. Data shown are representative of at least three independent experiments.
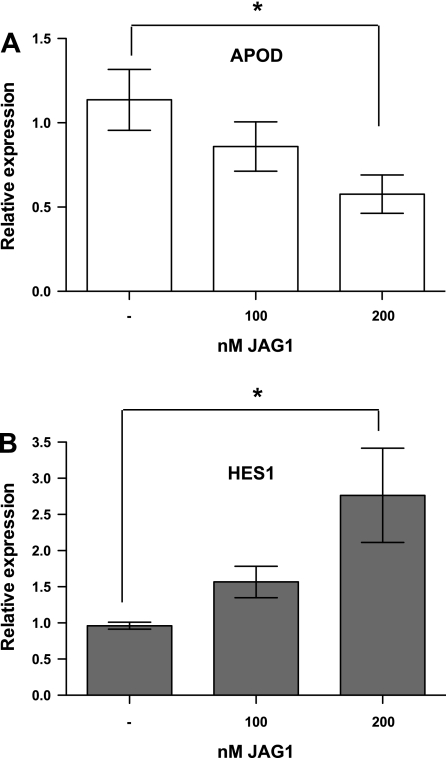
Fig. 5.
APOD expression is downregulated by JAG1 peptide. A: HDFNs were cultured in the presence or absence of indicated amounts of JAG1 peptide for 48 h, and harvested for RNA to measure APOD expression. B: expression of HES1 RNA was evaluated to verify Notch signaling was activated by addition of the JAG1 peptide. *P < 0.05.
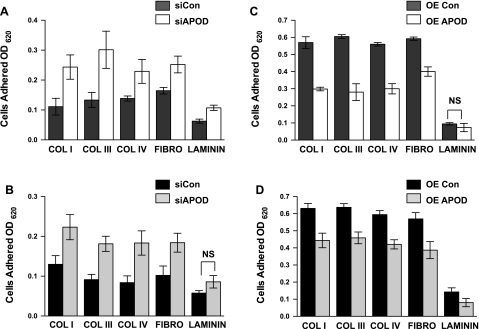
Fig. 6.
APOD regulates adhesion of mural cells to extracellular matrices. Cell attachment assays were performed to quantify cell adhesion of fibroblasts (A and C) and aortic smooth muscle cells (B and D) with altered levels of APOD expression. APOD expression was inhibited by knockdown with siRNA in HDFNs (A) and HAoSMCs (B). Cell adhesion was measured and compared with control (siCon). APOD was overexpressed (OE) by lentivirus and adhesion was measured in HDFNs (C) and HAoSMCs (D) and compared with control lentivirus-expressing GFP. Adhesion was quantified by normalizing each sample to binding of BSA, to account for nonspecific adhesion and differences in cell number. OD620, optical density at 620 nm; COL I, collagen I; COL III, collagen III; COL IV, collagen IV; FIBRO, fibronection. Differences between control and experimental were significantly different unless noted. P < 0.05, not significant (NS). Efficiency of APOD knockdown and overexpression was evaluated by qPCR and ELISA (Supplemental Fig. S1).
RESULTS
APOD is downregulated in mural cells by endothelial cells.
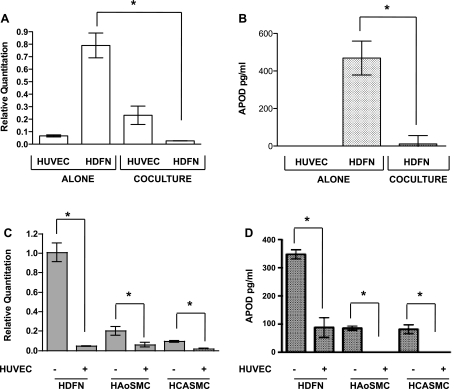
The interaction of endothelial cells and mural cells within the vasculature is important for proper formation and function of blood vessels. In investigating this interaction, we performed a microarray screen to identify genes, which were modulated by the communication between these two cell types (20). In doing so, we discovered that APOD is regulated by this interaction. We determined that APOD is highly expressed in HDFNs compared with HUVECs (Fig. 1A). Upon coculture of these two cells types, endothelial cells caused a dramatic decrease of APOD expression in mural cells. RNA transcripts for APOD showed a >20-fold decrease in expression when measured using qPCR (Fig. 1A). To determine if cocultured endothelial cells also decreased APOD protein, we performed ELISA. APOD is a secreted protein (29); therefore, we examined protein expression from the media of cultured cells. Consistent with the transcripts, expression of APOD protein was decreased upon coculturing with endothelial cells (Fig. 1B). Previous findings (21) from our laboratory revealed that HDFNs act as smooth muscle/mural cell precursors and can differentiate to acquire a smooth muscle phenotype upon coculture with endothelial cells. To determine if APOD downregulation occurred in other smooth muscle/mural cell subtypes, we examined RNA expression of APOD in HAoSMCs and HCASMCs and observed a decrease similar to HDFNs (Fig. 1C). A comparable decrease in APOD protein expression was also observed (Fig. 1D). Interestingly, the basal level of APOD was much lower in these smooth muscle cells, compared with the HDFNs, suggesting that more mature or differentiated mural cells may have reduced levels of APOD. These initial results demonstrate that APOD expression is modulated by the interaction of endothelial and mural cell interactions and suggest that it could play a role in how these two cells types cooperate to form a functional vessel.
Fig. 1.
Endothelial cells decrease apolipoprotein D (APOD) expression in mural cells. Endothelial cells [human umbilical vein endothelial cells (HUVECs)] and fibroblasts [human dermal neonatal fibroblasts (HDFN)] were cultured alone or cocultured with each other. After 48 h, cocultured cells were separated using anti-platelet endothelial cell adhesion molecule 1 (PECAM1)-conjugated Dynabeads for RNA expression analysis using quantitative (q)PCR (A). APOD protein quantification was performed using ELISA (B). HDFN, human aortic smooth muscle cells (HAoSMCs), and human coronary artery smooth muscle cells (HCASMCs) were cultured in the presence or absence of HUVECs for 48 h, separated, and subjected to qPCR analysis for APOD transcript expression (C). APOD protein expression was determined by ELISA (D). *P < 0.05.
Endothelial cells downregulate APOD through a secreted factor.
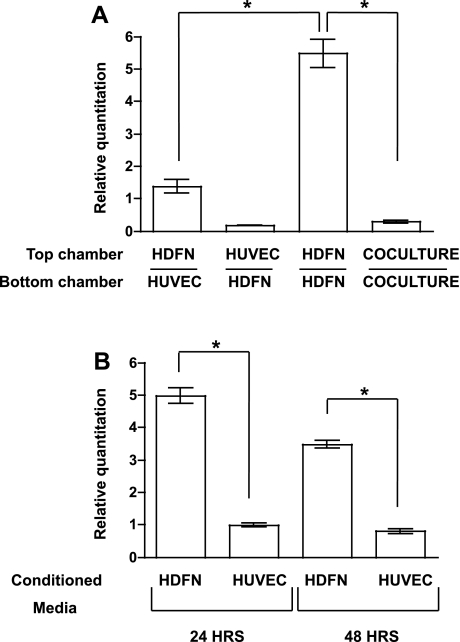
The ability of endothelial cells to decrease mural cell-expressed APOD prompted us to examine the mechanism of action. To determine if the decrease in APOD required cell-cell contact, we utilized a Transwell assay to coculture endothelial cells and fibroblasts in the absence of heterotypic cell contact. When HFDNs were cultured on the opposing side of a 0.4-μm porous membrane without physical contact with endothelial cells, expression of APOD RNA was significantly downregulated (Fig. 2A). However, unlike in coculture conditions, which exhibited a 20-fold reduction in expression, APOD expression in the absence of cell contact was decreased only fourfold. We further evaluated this using HUVEC-conditioned media. Similar to the Transwell assay, HUVEC-conditioned media significantly decreased APOD RNA expression in HDFNs by four- to fivefold at 24- and 48-h time points (Fig. 2B). These data indicate that while endothelial cells secrete a factor that attenuates APOD expression in mural cells, cell contact is likely required for the full effects of endothelial cell regulation of APOD. Alternatively, an unidentified secreted factor may be unable to diffuse to regulate APOD expression in mural cells.
Fig. 2.
APOD is partially downregulated in mural cells by an endothelial cell-derived secreted factor. A: HUVECs and HDFNs were plated on either side of a Transwell insert as indicated, cultured for 48 h, and the top chamber was harvested for qPCR to examine APOD mRNA expression. B: conditioned media from HDFNs or HUVECs were incubated with HDFNs for 24 or 48 h and cells were harvested to measure APOD RNA expression. *P < 0.05.
APOD promoter responds to endothelial cells.
The APOD promoter was previously shown to be active in fibroblasts and regulated by growth signals and cell density (10, 17). To determine if this promoter was responsive to signals emanating from endothelial cells, we cloned the proximal promoter (Fig. 3A) into the pGL3-luciferase vector and tested its response in coculture conditions using luciferase reporter assays (Fig. 3B). The promoter exhibited robust transcriptional activity in HDFNs, and upon coculturing with endothelial cells showed an approximate threefold reduction in activity. To determine if promoter attenuation was due to the secreted factor, we utilized endothelial cell-conditioned media and showed a similar, but slightly smaller, decrease in activity (2-fold compared with conditioned media control). These data indicate that the proximal promoter of APOD harbors response elements that contribute to endothelial cell-dependent downregulation; however, additional elements outside this region are likely required for the 20-fold reduction of APOD by endothelial cells.
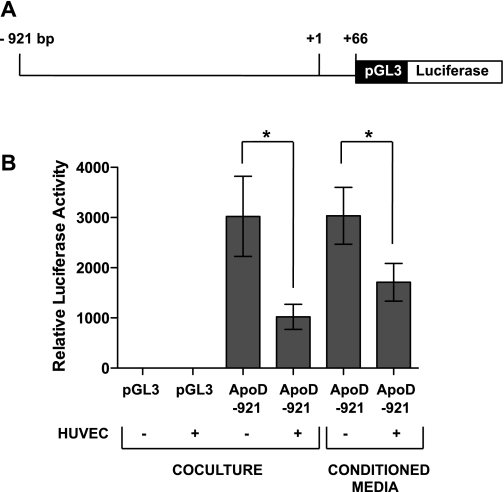
Fig. 3.
APOD promoter is responsive to signals from endothelial cells. A: diagram of APOD promoter depicting the region that was inserted upstream of the luciferase gene in pGL3-basic plasmid. B: luciferase reporter assays were performed to assess APOD promoter responsiveness to coculture and conditioned media. HDFNs were transiently transfected with the pGL3 empty vector or ApoD-921-pGL3-luciferase plasmid and cultured in the presence or absence of HUVECs, or supplemented with HUVEC-conditioned media. After 48 h, transcriptional activity was determined by luciferase activity. *P < 0.05.
NOTCH3 regulates APOD expression.
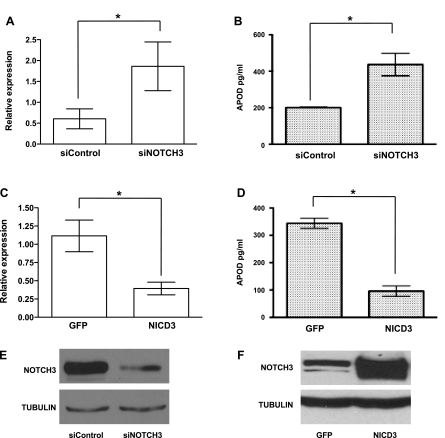
Previous findings (21) from our laboratory revealed that endothelial cells induce NOTCH3 and activate Notch signaling in mural cells. Given that APOD appeared to be partially regulated by cell-cell contact, we hypothesized that NOTCH3 might be regulating APOD. To test this hypothesis, we ablated NOTCH3 from mural cells using siRNA. Interestingly, knockdown of NOTCH3 under coculture conditions resulted in APOD levels remaining threefold higher compared with control siRNA-treated cells (Fig. 4, A and B). To determine if NOTCH3 was sufficient to cause a decrease in APOD levels, we overexpressed the NOTCH3 intracellular domain (NICD3). Overexpression of NICD3 caused a significant decrease in APOD RNA and protein levels (Fig. 4, C and D), indicating that NOTCH3 has a central role in the endothelial cell-dependent downregulation of APOD in mural cells. Previously, we demonstrated that JAGGED-1 (JAG1) present on endothelial cells promotes NOTCH3 expression and activates downstream Notch signaling (21). To determine if JAG1 could cause a decrease in APOD expression, we incubated HDFNs with a soluble JAG1 peptide (1). JAG1 caused a concentration-dependent decrease in APOD RNA (Fig. 5A). The 100 nM of peptide showed a slight, but not significant, decrease, while 200 nM of JAG1 caused a significant twofold reduction in APOD levels. The activity of Notch signaling was confirmed by analysis of the known downstream target gene HES1, which exhibited an expected increase in expression upon addition of JAG1 (Fig. 5B). Thus these data demonstrate that NOTCH3 contributes to the regulation of APOD in mural cells by endothelial cells, possibly through activation by JAG1.
Fig. 4.
APOD expression is dependent on NOTCH3. HDFNs were transiently transfected with control small interfering (si)RNA (siControl) or NOTCH3 siRNA (siNOTCH3) and cocultured with HUVECs. Forty-eight hours later, the cocultured cells were separated by anti-PECAM1-conjugated Dynabeads and total RNA was collected for qPCR to detect APOD RNA expression (A). APOD protein expression was determined by ELISA (B). NOTCH3 intracellular domain (NICD3) was overexpressed in mural cells by lentiviral infection. A lentivirus expressing GFP alone was used as control. Following infection, cells were analyzed for APOD expression by qPCR (C) and ELISA (D). *P < 0.05. Western blot analysis to demonstrate efficient knockdown of NOTCH3 by siRNA (E) and NICD3 overexpression (F). Tubulin expression was used as control.
APOD attenuates adhesion and focal contacts.
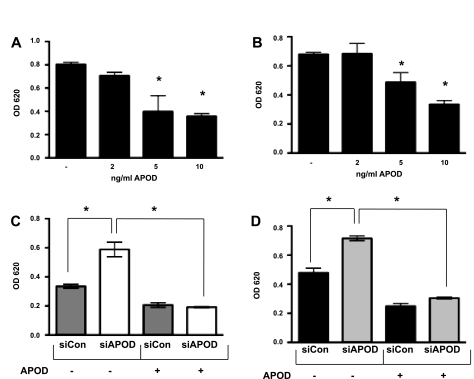
The decrease of APOD expression in mural cells caused by endothelial cells suggests that APOD acts to convey functional changes within the vessel. Previously, studies (19, 32) have linked APOD with the regulation of smooth muscle proliferation and migration. To further examine how APOD expression could affect vascular cell function, we measured adhesion of fibroblasts and smooth muscle cells with altered levels of APOD. Using siRNA directed against APOD transcripts, we performed adhesion assays with extracellular matrices (ECM) commonly found in the vascular wall (15). When APOD levels were decreased by siRNA (Supplemental Fig. S1), the adhesion to ECM was increased in both HDFNs and HAoSMCs (Fig. 6, A and B). The biggest increase in adherence was observed with collagen I and collagen III in HDFNs, and collagens III and IV in HAoSMCs, suggesting that APOD is potentially influencing specific collagen-binding integrins. Corollary to the knockdown results, when APOD levels were artificially increased using a lentivirus overexpressing APOD cDNA (Supplemental Fig. 1), we observed a decrease in adhesion in both cell subtypes compared with a lentivirus-expressing GFP control (Fig. 6, C and D).
To determine if exogenously added APOD would have a similar effect on adhesion, we added increasing amounts of recombinant APOD to the culture media and measured adhesion to collagen I. Similar to the APOD that was overexpressed by lentivirus, exogenous APOD regulated adhesion to collagen I in a concentration-dependent fashion (Fig. 7A and B). However, the amount of exogenous APOD that was required to see an effect on adhesion was much greater for reasons that are not clear. Moreover, we could rescue the knockdown affect of siAPOD by addition of exogenous APOD protein. In both HDFNs and HAoSMCs, knockdown of APOD by siRNA caused an increase in adhesion, which was blocked by addition of exogenous APOD (Fig. 7, C and D).
Fig. 7.
Exogenous APOD regulates adhesion. Adhesion assays were performed with HDFNs (A) and HAoSMCs (B) on collagen I in the presence OR absence of recombinant human APOD, APOD expression was inhibited by knockdown with siRNA (siAPOD) and compared with control siRNA (siCon) in HDFNs (C) and HAoSMCs (D). Exogenous APOD (10 ng/ml) was added to indicated samples 24 h before performing adhesion assays on collagen I. *P < 0.05.
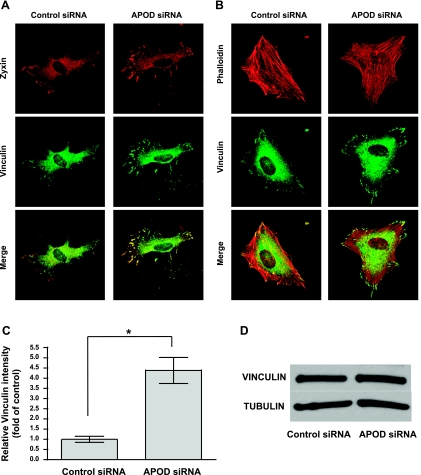
To investigate the mechanism by which APOD regulates adhesion, we evaluated the focal adhesions and stress fibers of APOD-deficient fibroblasts. Focal adhesions were identified by staining for vinculin and zyxin (6, 25). Cells that lacked APOD showed an increased presence of focal adhesions compared with control cells (Fig. 8A), suggesting that APOD attenuated the formation or stabilization of these structures. In contrast, we observed no difference in stress fibers using fluorescently labeled phalloidin for cell staining (Fig. 8B). vinculin staining in the focal contacts of APOD knockdown cells was measured and determined to be fourfold greater than in control cells (Fig. 8C). The difference in vinculin staining appeared due to a difference in cellular localization, as total vinculin protein expression in cells lacking APOD was comparable to control cells (Fig. 8D). Together, these results indicate that the expression of APOD in mural cells can modulate their ability to bind the ECM through regulating focal adhesion contacts, without affecting the gross structure of stress fibers. Thus the downregulation of APOD by neighboring endothelial cells likely has a role in regulating the recruitment and subsequent adherence of these cells within the blood vessel wall.
Fig. 8.
Focal adhesions are increased when APOD expression is attenuated. HDFNs were transfected with control siRNA or APOD siRNA to knockdown APOD expression. Forty-eight hours posttransfection, cells were plated on 10 μg/ml collagen I and allowed to attach for 30 min before fixing for immunostaining. A: cells were immunostained to visualize focal adhesions using zyxin and vinculin antibodies. B: cells were stained with fluorescently labeled phalloidin to visualize stress fibers, and vinculin to mark focal contacts. Pictures were captured at ×400 magnification. C: focal adhesions were quantified using vinculin staining intensity. *P < 0.0001. D: Western blot to detect vinculin expression in siRNA control and APOD-treated cells. Tubulin was used as a control to show equivalent amounts of total protein.
DISCUSSION
In an intact blood vessel, signaling between endothelial cells and smooth muscle cells is known to cause critical changes in vascular function. However, during the formation of the vasculature, little is known about how interactions between these cell-types act to build a blood vessel. Using a gene screening strategy, our laboratory identified genes, which were regulated by the direct interaction of vascular cells, with the goal of identifying factors that function in vessel assembly. One of these genes was APOD, which was intriguing since prior studies (19, 32) indicated that APOD plays a role in smooth muscle proliferation and migration. Additionally, APOD transcripts and protein were previously localized to atherosclerotic lesions (26, 32), suggesting a possible link to smooth muscle dysfunction.
Our findings initially indicated that APOD was regulated in mural cells by an endothelial cell-derived secreted factor. However, the comparative downregulation was significantly less than under coculture conditions. Indeed, we determined that endothelial cell contact was also involved in APOD downregulation. Why do endothelial cells utilize two modes of signaling control to regulate transcription of APOD in mural cells? One possibility is that the function of APOD in blood vessels is largely concentration dependent. Partial downregulation of APOD by an endothelial cell derived-secreted factor may alter or slow its activities. Once cell contact is established, this essentially blocks APOD expression and its subsequent function. Investigations to identify the nature of the secreted factor are ongoing. Alternatively, we identified NOTCH3 as a cell contact-dependent signal that decreases APOD expression. Our (21) previous analyses demonstrated an important role for contact-dependent Notch signaling in the regulation of endothelial/mural cell interactions. In particular, we showed that NOTCH3 promotes differentiation, which is generally associated with a quiescent state. The downregulation of APOD under these circumstances is inconsistent with previous reports that showed APOD possesses antiproliferative properties (27, 32). Yet, prior expression analyses localized APOD transcripts to both developing and mature vascular smooth muscle cells (28, 30, 33). Additionally, APOD has been associated with regeneration in the nervous system (5, 13, 36) and was found to be highly expressed in breast tumors (34). Thus, although under certain cellular conditions APOD might act as an antiproliferative agent, the decrease or absence of APOD expression might not be a direct indicator of a proliferative or undifferentiated state.
Our data show that APOD regulates adhesion through modulation of focal contacts. In our assays with both fibroblasts and smooth muscle cells, adhesion to collagen subtypes and fibronectin was attenuated with increasing APOD levels, while decreasing APOD levels promoted adhesion. Previous studies (19) performed in the Rabinovitch laboratory demonstrated that APOD enhanced migration in cooperation with PDGF-BB. The ability of APOD to modulate migration and adhesion offers a mechanism through which endothelial cells instruct neighboring mural cells. Endothelial cells are known to secrete PDGF-BB as a recruitment factor for surrounding mural cells (2). Possibly, high levels of APOD in undifferentiated mural cells promote their migration to endothelial cells in a PDGF-BB-dependent manner. Upon close contact with endothelial cells, the expression of APOD is strongly inhibited by both a secreted factor and cell contact. This results in increased adhesion, which serves to promote vessel maturation. Moreover, in atherosclerotic lesions where APOD is abundant (26, 32), endothelial dysfunction may lead to increased APOD, which, in turn, results in enhanced migration of smooth muscle cells within the vessel wall. Overall, these data demonstrate that gene expression changes caused by heterotypic interactions of vascular cells can provide critical molecular changes that impinge on the assembly and function of blood vessels.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute Grant R01-HL-076428 (to B. Lilly) and American Heart Association Predoctoral Fellowship 09PRE2220351 (to H. Liu).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We thank Andrea Rutherford and Pooja Joshi for contributing to the JAG1 results.
REFERENCES
- 1. Aho S. Soluble form of Jagged1: unique product of epithelial keratinocytes and a regulator of keratinocyte differentiation. J Cell Biochem 92: 1271–1281, 2004 [DOI] [PubMed] [Google Scholar]
- 2. Armulik A, Abramsson A, Betsholtz C. Endothelial/pericyte interactions. Circ Res 97: 512–523, 2005 [DOI] [PubMed] [Google Scholar]
- 3. Baron M. An overview of the Notch signalling pathway. Semin Cell Dev Biol 14: 113–119, 2003 [DOI] [PubMed] [Google Scholar]
- 4. Bergers G, Song S. The role of pericytes in blood-vessel formation and maintenance. Neuro Oncol 7: 452–464, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boyles JK, Notterpek LM, Anderson LJ. Accumulation of apolipoproteins in the regenerating and remyelinating mammalian peripheral nerve. Identification of apolipoprotein D, apolipoprotein A-IV, apolipoprotein E, and apolipoprotein A-I. J Biol Chem 265: 17805–17815, 1990 [PubMed] [Google Scholar]
- 6. Burridge K, Fath K. Focal contacts: transmembrane links between the extracellular matrix and the cytoskeleton. Bioessays 10: 104–108, 1989 [DOI] [PubMed] [Google Scholar]
- 7. Carmeliet P. Angiogenesis in life, disease and medicine. Nature 438: 932–936, 2005 [DOI] [PubMed] [Google Scholar]
- 8. Curry MD, McConathy WJ, Alaupovic P. Quantitative determination of human apolipoprotein D by electroimmunoassay and radial immunodiffusion. Biochim Biophys Acta 491: 232–241, 1977 [DOI] [PubMed] [Google Scholar]
- 9. Darland DC, D'Amore PA. Cell-cell interactions in vascular development. Curr Top Dev Biol 52: 107–149, 2001 [DOI] [PubMed] [Google Scholar]
- 10. Do Carmo S, Seguin D, Milne R, Rassart E. Modulation of apolipoprotein D and apolipoprotein E mRNA expression by growth arrest and identification of key elements in the promoter. J Biol Chem 277: 5514–5523, 2002 [DOI] [PubMed] [Google Scholar]
- 11. Gaengel K, Genove G, Armulik A, Betsholtz C. Endothelial-mural cell signaling in vascular development and angiogenesis. Arterioscler Thromb Vasc Biol 29: 630–638, 2009 [DOI] [PubMed] [Google Scholar]
- 12. Ganfornina MD, Do Carmo S, Lora JM, Torres-Schumann S, Vogel M, Allhorn M, Gonzalez C, Bastiani MJ, Rassart E, Sanchez D. Apolipoprotein D is involved in the mechanisms regulating protection from oxidative stress. Aging Cell 7: 506–515, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ganfornina MD, Do Carmo S, Martinez E, Tolivia J, Navarro A, Rassart E, Sanchez D. ApoD, a glia-derived apolipoprotein, is required for peripheral nerve functional integrity and a timely response to injury. Glia 58: 1320–1334, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hull-Thompson J, Muffat J, Sanchez D, Walker DW, Benzer S, Ganfornina MD, Jasper H. Control of metabolic homeostasis by stress signaling is mediated by the lipocalin NLaz. PLoS Genet 5: e1000460, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hynes RO. Cell-matrix adhesion in vascular development. J Thromb Haemost 5, Suppl 1: 32–40, 2007 [DOI] [PubMed] [Google Scholar]
- 16. Kennard S, Liu H, Lilly B. Transforming growth factor-beta (TGF- 1) down-regulates Notch3 in fibroblasts to promote smooth muscle gene expression. J Biol Chem 283: 1324–1333, 2008 [DOI] [PubMed] [Google Scholar]
- 17. Lambert J, Provost PR, Marcel YL, Rassart E. Structure of the human apolipoprotein D gene promoter region. Biochim Biophys Acta 1172: 190–192, 1993 [DOI] [PubMed] [Google Scholar]
- 18. Lane DM, McConathy WJ. Factors affecting the lipid and apolipoprotein levels of cord sera. Pediatr Res 17: 83–91, 1983 [DOI] [PubMed] [Google Scholar]
- 19. Leung WC, Lawrie A, Demaries S, Massaeli H, Burry A, Yablonsky S, Sarjeant JM, Fera E, Rassart E, Pickering JG, Rabinovitch M. Apolipoprotein D and platelet-derived growth factor-BB synergism mediates vascular smooth muscle cell migration. Circ Res 95: 179–186, 2004 [DOI] [PubMed] [Google Scholar]
- 20. Lilly B, Kennard S. Differential gene expression in a coculture model of angiogenesis reveals modulation of select pathways and a role for Notch signaling. Physiol Genomics 36: 69–78, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu H, Kennard S, Lilly B. NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires endothelial-expressed JAGGED1. Circ Res 104: 466–475, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Z, Chang GQ, Leibowitz SF. Apolipoprotein D interacts with the long-form leptin receptor: a hypothalamic function in the control of energy homeostasis. FASEB J 15: 1329–1331, 2001 [DOI] [PubMed] [Google Scholar]
- 23. Louvi A, Arboleda-Velasquez JF, Artavanis-Tsakonas S. CADASIL: a critical look at a Notch disease. Dev Neurosci 28: 5–12, 2006 [DOI] [PubMed] [Google Scholar]
- 24. Muffat J, Walker DW. Apolipoprotein D: an overview of its role in aging and age-related diseases. Cell Cycle 9: 269–273, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nix DA, Fradelizi J, Bockholt S, Menichi B, Louvard D, Friederich E, Beckerle MC. Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J Biol Chem 276: 34759–34767, 2001 [DOI] [PubMed] [Google Scholar]
- 26. Perdomo G, Dong H. Apolipoprotein D in lipid metabolism and its functional implication in atherosclerosis and aging. Aging (Albany NY) 1: 17–27, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Provost PR, Marcel YL, Milne RW, Weech PK, Rassart E. Apolipoprotein D transcription occurs specifically in nonproliferating quiescent and senescent fibroblast cultures. FEBS Lett 290: 139–141, 1991 [DOI] [PubMed] [Google Scholar]
- 28. Provost PR, Villeneuve L, Weech PK, Milne RW, Marcel YL, Rassart E. Localization of the major sites of rabbit apolipoprotein D gene transcription by in situ hybridization. J Lipid Res 32: 1959–1970, 1991 [PubMed] [Google Scholar]
- 29. Rassart E, Bedirian A, Do Carmo S, Guinard O, Sirois J, Terrisse L, Milne R. Apolipoprotein D. Biochim Biophys Acta 1482: 185–198, 2000 [DOI] [PubMed] [Google Scholar]
- 30. Sanchez D, Ganfornina MD, Martinez S. Expression pattern of the lipocalin apolipoprotein D during mouse embryogenesis. Mech Dev 110: 225–229, 2002 [DOI] [PubMed] [Google Scholar]
- 31. Sanchez D, Lopez-Arias B, Torroja L, Canal I, Wang X, Bastiani MJ, Ganfornina MD, Hull-Thompson J, Muffat J, Sanchez D, Walker DW, Benzer S, Ganfornina MD, Jasper H. Loss of glial lazarillo, a homolog of apolipoprotein D, reduces lifespan and stress resistance in Drosophila. Control of metabolic homeostasis by stress signaling is mediated by the lipocalin NLaz. Curr Biol 16: 680–686, 2006 [DOI] [PubMed] [Google Scholar]
- 32. Sarjeant JM, Lawrie A, Kinnear C, Yablonsky S, Leung W, Massaeli H, Prichett W, Veinot JP, Rassart E, Rabinovitch M. Apolipoprotein D inhibits platelet-derived growth factor-BB-induced vascular smooth muscle cell proliferated by preventing translocation of phosphorylated extracellular signal regulated kinase 1/2 to the nucleus. Arterioscler Thromb Vasc Biol 23: 2172–2177, 2003 [DOI] [PubMed] [Google Scholar]
- 33. Smith KM, Lawn RM, Wilcox JN. Cellular localization of apolipoprotein D and lecithin:cholesterol acyltransferase mRNA in rhesus monkey tissues by in situ hybridization. J Lipid Res 31: 995–1004, 1990 [PubMed] [Google Scholar]
- 34. Soiland H, Skaland I, Varhaug JE, Korner H, Janssen EA, Gudlaugsson E, Baak JP, Soreide JA. Co-expression of estrogen receptor alpha and Apolipoprotein D in node positive operable breast cancer–possible relevance for survival and effects of adjuvant tamoxifen in postmenopausal patients. Acta Oncol 48: 514–521, 2009 [DOI] [PubMed] [Google Scholar]
- 35. Spector D, Goldman R, Leinwand L. Cells: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, 1998, p. 98–110 [Google Scholar]
- 36. Spreyer P, Schaal H, Kuhn G, Rothe T, Unterbeck A, Olek K, Muller HW. Regeneration-associated high level expression of apolipoprotein D mRNA in endoneurial fibroblasts of peripheral nerve. EMBO J 9: 2479–2484, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vijayaraghavan S, Hitman GA, Kopelman PG. Apolipoprotein-D polymorphism: a genetic marker for obesity and hyperinsulinemia. J Clin Endocrinol Metab 79: 568–570, 1994 [DOI] [PubMed] [Google Scholar]
- 38. Walker DW, Muffat J, Rundel C, Benzer S. Overexpression of a Drosophila homolog of apolipoprotein D leads to increased stress resistance and extended lifespan. Curr Biol 16: 674–679, 2006 [DOI] [PubMed] [Google Scholar]
- 39. Weech PK, Provost P, Tremblay NM, Camato RN, Milne RW, Marcel YL, Rassart E. Apolipoprotein d–an atypical apolipoprotein. Prog Lipid Res 30: 259–266, 1991 [DOI] [PubMed] [Google Scholar]
- 40. Xu W, Baribault H, Adamson ED. Vinculin knockout results in heart and brain defects during embryonic development. Development 125: 327–337, 1998 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.