Abstract
The stress inducible transcription factor, NF-κB induces genes involved in proliferation and apoptosis. Aberrant NF-κB activity is common in cancer and contributes to therapeutic-resistance. Poly(ADP-ribose) polymerase-1 (PARP-1) is activated during DNA strand break repair and is a known transcriptional co-regulator. Here, we investigated the role of PARP-1 function during NF-κB activation using p65 siRNA, PARP siRNA or the potent PARP-1 inhibitor, AG-014699. Survival and apoptosis assays showed that NF-κB p65−/− cells were more sensitive to ionizing radiation (IR) than p65+/+ cells. Co-incubation with p65 siRNA, PARP siRNA or AG-014699 radio-sensitized p65+/+, but not p65−/− cells, demonstrating that PARP-1 mediates its effects on survival via NF-κB. Single strand break (SSB) repair kinetics, and the effect SSB repair inhibition by AG-014699 were similar in p65+/+ and p65−/− cells. Since preventing SSB repair did not radio-sensitize p65−/− cells, we conclude that radio-sensitization by AG-014699 is due to downstream inhibition of NF-κB activation, and independent of SSB repair inhibition. PARP-1 catalytic activity was essential for IR-induced p65 DNA binding and NF-κB-dependent gene transcription, whereas for TNF-α treated cells, PARP-1 protein alone was sufficient. We hypothesize that this stimulus-dependent differential is mediated via stimulation of the Poly(ADP-ribose) polymer, which was induced following IR, not TNF-α. Targeting DNA-damage activated NF-κB using AG-014699 may therefore overcome toxicity observed with classical NF-κB inhibitors without compromising other vital inflammatory functions. These data highlight the potential of PARP-1 inhibitors to overcome NF-κB-mediated therapeutic resistance and widens the spectrum of cancers in which these agents may be utilized.
Keywords: PARP-1, NF-κB, Radio-sensitization, AG-014699, PAR
INTRODUCTION
The stress-inducible transcription factor, NF-κB is known to regulate genes involved in apoptosis, cell proliferation, angiogenesis and metastasis. NF-κB is the heterodimeric complex of Rel family proteins (p50, p52, p65, c-Rel and Rel B) that is held inactive in the cytoplasm through its interaction with inhibitor κB (IκB) proteins. Upon stimulus, by DNA damage or inflammatory cytokines, the canonical pathway of NF-κB activation is induced. Briefly, signaling cascades converge at the inhibitor of kappa-B kinase (IKK) complex which phosphorylates IκBα, targeting this for degradation and promoting nuclear translocation of the p65/p50 heterodimer, activating transcription of many genes, including the anti-apoptotic Bcl-XL and XIAP. (Ghosh and Karin, 2002).
Constitutively active NF-κB is common in many cancers and leukemia (Basseres and Baldwin 2006) and has been shown to be associated with a poor prognosis in CLL (Hewamana et al. 2009). Aberrant NF-κB activation correlates with increased metastatic potential, more rapid disease progression, a higher proportion of tumor recurrence and therapeutic resistance (Prasad et al., 2009). Cellular NF-κB activity is known to be induced by varying doses of ionizing radiation (IR) and this differential is likely to be both cell type and tissue specific (Brach et al., 1991; Criswell et al., 2003). DNA damage-activated NF-κB induces anti-apoptotic genes, thereby inhibiting apoptosis (Barkett and Gilmore, 1999; Wang et al., 1998). Aberrant NF-κB contributes to radio- and chemo-resistance in breast tumors (Biswas et al., 2001; Wu and Kral, 2005) and loss or inhibition of NF-κB has been shown to chemo- or radio-sensitize in many different tumour types (Hewamana et al. 2008a and b, Criswell et al., 2003; Jung and Dritschilo, 2001; Russo et al., 2001). However, is it important to note that the dose of IR required to active NF-κB may be cell type or tissue-dependent. For example, Brach et al., (1991) showed that the clinically relevant dose of 2 Gy could induce NF-κB activation in the myeloid cell line, KG-1. Furthermore, in the EBV-transformed 244B human lymphoblastoid cell line, 0.5 Gy IR was shown to activate NF-κB activation (Sahijdak et al., 1994). Whereas, other studies, such as Stilmann et al. (2009), used hyper-lethal doses, of 80 Gy when inducing an NF-κB response.
Thus, inhibition of NF-κB represents a potential therapeutic strategy in cancer, and there are a number of inhibitors targeting various pathway components in development. These include inhibitors of the IKK complex, such as Parthenolide or proteosomal inhibitors like Bortezomib which may prevent degradation of IκB. (Gilmore and Herscovitch 2006). A novel strategy may be to inhibit DNA damage induced NF-κB activity to overcome toxic or off-target effects often encountered with IKK or proteosmal inhibitors. Here we describe a novel function for the Poly(ADP-ribose) polymerase (PARP) inhibitor, AG-014699, developed at Newcastle University in collaboration with Pfizer, in specifically inhibiting DNA damage induced NF-κB activity.
PARP-1 is a nuclear enzyme which modulates the cellular response to DNA damage (Smith, 2001) and we have previously shown that PARP-1 acts as a sensor for both single and double stranded DNA breaks in response to IR (Veuger et al,. 2003). More recently, there is evidence for a role of PARP-1 in the co-regulation of transcription factors. This may be via through local modification of chromatin structure (Althaus et al., 1994) and/or modulation of transcription factor activity via direct binding to gene regulating sequences, or physical interactions with proteins including transcription factors including Oct-1, NF-κB, AP-1 and HIF-1α (Kraus and Lis, 2003; Martin-Oliva et al., 2006). We have previously demonstrated that PARP-1 activity is vital for NF-κB activation following IR using panel of breast cancer cell lines and the small molecule inhibitor of PARP-1, AG-14361 (Veuger et al. 2009).
The role of PARP-1 catalytic activity in the activation of NF-κB is somewhat contentious, and is likely to be mainly dependent on stimulus, but also on cell type (Hassa et al., 2001; Hassa and Hottiger, 1999; Oliver et al., 1999). In this study, we have tested this by comparing two canonical activators of NF-κB, IR and TNF-α. TNF-α is a cytokine released during the immune response which is capable of initiating both death receptor-mediated cell death and canonical activation of the NF-κB (Clevers, 2004). We illustrate that PARP-1 is differentially required in the induction of NF-κB, and this is dependent on the ability of the stimuli to activate the Poly(ADP-ribose) (PAR) polymer. To this end, the recent work of Stilmann et al. describes the formation of a PARP-1 signaling scaffold via direct protein-protein interactions with NEMO, PIASγ and ATM in response to DNA damage. This signalosome formation requires binding with the PAR polymer and leads to pro-survival NF-κB activation (Stilmann et al., 2009).
The potent and specific PARP-1 inhibitor, AG-014699 has been used in Phase I and II clinical trials for solid tumours in combination with standard chemotherapeutics and also as a standalone treatment. (Plummer et al., 2008). Studies have also shown that AG-014699 enhances tumor regression in xenograft models (Daniels et al., 2009). However the effect of this agent upon NF-κB transcriptional activity remains undetermined. Here we show, by using AG-014699 and siRNA targeting NF-κB p65 and mouse embryonic fibroblasts (MEFs) proficient and deficient for NF-κB p65, that inhibition of PARP-1 positively modulates the apoptotic response to IR. Strikingly, we show that radio-sensitization by PARP-1 inhibition is exclusively due to inhibition of NF-κB and not due to the prevention of DNA repair. We also illustrate that PARP-1 is differentially required in the induction of NF-κB, and this is dependent on the activation stimulus and the ability of such stimuli to activate the PAR polymer.
RESULTS
Characterisation of cell lines
We have previously confirmed, using Western analyses the p50, p65, and PARP-1 status in all the cell lines. Briefly, the knock out MEFs lacked the relevant proteins, whilst showing bands of similar intensity for the other proteins (Veuger et al., 2009). PARP-1 activity is very similar in PARP+/+, p65+/+ and p65−/− MEFs and very low (< 5%) in the PARP-1−/− MEFs used here (Veuger et al., 2009). Transient transfection of 50nM p65 siRNA resulted in knockdown of p65 protein, and this was maximal (95% reduction) by 48 h (Figure 1A), and persisted up to 72 h. Similarly transfection of 50nM PARP-1 siRNA (Figure 1A) resulted in approximately 75% protein knockdown at 48 h, and this persisted up to 72 h. Non-specific (NS) siRNA had no effect on protein expression.
Figure 1. Characterisation of cell lines and siRNA knockdown.
(A) Western blots of whole cell extracts of p65+/+ and p65−/− MEFs at 48 h after transfection with vehicle alone (Mock), non-specific (NS) siRNA or p65 siRNA.
(B) Western blots of whole cell extracts of p65+/+ and p65−/− at 48 h after transfection with vehicle alone (Mock), non-specific (NS) siRNA or PARP-1 siRNA.
(C) Histogram showing PARP activity, measured using a validated immunoblot assay which quantifies PAR polymer formation, in p65+/+ MEFs after transfection with vehicle alone (mock), p65 siRNA, AG-014699, PARP-1 siRNA,a combination of p65 siRNA + AG-014699 or NS siRNA.
(D) PARP activity measured using a validated immunoblot assay which quantifies PAR polymer formation in p65−/− MEFs following transfection with vehicle alone (mock), p65 siRNA, AG-014699, PARP-1 siRNA,a combination of p65 siRNA + AG-014699 or NS siRNA
10H antibody was used measure PAR formation
Data are represented as the mean ±sem of three independent experiments.
*Significance relative to mock treated control was p<0.05 using unpaired Student’s t-test.
To confirm PARP knockdown and specificity of AG-104699, PARP activity was assessed in p65+/+ and p65−/− MEFs following transfection with PARP-1 or p65 siRNA, or following treatment with AG-014699 (Figures 1C and 1D). A combination of p65 siRNA and AG-014699 was also assessed. Cells treated with AG-014699, or a combination of AG-014699 and p65 siRNA exhibited very low (< 5%) PARP activity (p=0.03 and p=0.02 respectively, compared with NS siRNA controls). PARP-1 siRNA exhibited 25% PARP activity compared with controls (p=0.004), in both cell lines, which confirms Western analysis. siRNA targeting p65 does not have any significant effect on PARP-1 activity in either cell line, when compared with NS siRNA controls. It must be noted that there appears to be some effect when comparing NS siRNA with Mock treated controls. Other groups have documented effects on cellular processes including cell viability, proliferation, cell cycle distribution, apoptosis, and migration (Jackson and Linsley, 2004, Tschahatganeh, 2007). We suggest here that this could perhaps to due to the increased nucleic acid ratio in NS siRNA treated cells versus Mock transfected controls, causing the cells some stress.
Radio-sensitization by p65 knockdown, PARP-1 knockdown or AG-014699 is associated with the induction of apoptosis
We investigated the ability of p65 knockdown, PARP-1 knockdown or AG-014699 to sensitize p65+/+ and p65−/− MEFs, and PARP-1+/+ and PARP-1−/− MEFs to IR using colony forming assays. We also assessed a combination of p65 siRNA and AG-014699. In our previous study PF50 values illustrated that the p65−/− MEFs were 1.3-fold more sensitive to IR alone compared with p65+/+ MEFs (Veuger et al., 2009) (Figure 2A versus B). This is consistent with NF-κB conferring radio-resistance (Biswas et al., 2001; Wu and Kral, 2005). Here we show that, co-incubation with either AG-014699, p65 siRNA or PARP-1 siRNA radio-sensitized p65+/+ MEFs 1.4-fold compared to mock-transfected cells treated with IR. Significantly, a combination of p65 siRNA and AG-014699 showed no further sensitization (Figure 2A). Markedly, these treatments had no effect in p65−/− MEFs (Figure 2B). PARP-1−/− MEFs were 2.7-fold more sensitive to IR compared with PARP-1+/+ MEFs (Figure 2C versus D). PARP-1+/+ MEFs exhibited a 1.3-fold sensitization to IR by AG-014699, p65 knockdown or PARP-1 knockdown, compared with IR alone controls (Figure 2C).
Figure 2. Radio-sensitization by p65 knockdown, PARP-1 knockdown or AG-014699 is associated with the induction of apoptosis.
The effects of increasing doses of IR either alone, or co-incubated with p65 siRNA, AG-014699, PARP-1 siRNA or a combination of p65 siRNA and AG-014699, on cell survival were assessed using the clonogenic survival assay. Cells were treated with relevant siRNA, (or vehicle control), left for 48 h, pre-treated with AG-014699, or DMSO control 1 h prior to IR then re-plated after a further 24 h and allowed to form colonies for 7-21 days. (Survival curve (A) shows p65+/+ MEFs, (B) p65−/− MEFs, (C) PARP+/+ MEFs and (D) PARP−/− MEFs) E The effect of IR alone (mock, white bar) ± p65 siRNA (black bar) ± AG-104699 (AG alone, light grey bar, AG + p65 siRNA, dark grey bar) on the induction of apoptosis in p65+/+ MEFs. Cells were treated with relevant siRNA, or control left for 48 h, pre-treated with AG-014699, or control 1 h prior to IR then allowed to repair for a further 24 h before harvesting and assessment of the induction of apoptosis by Annexin V FACs analysis. Results shown are normalised to untreated controls.
F The effect of IR (mock, white bar) ± p65 siRNA (black bar) ± AG-104699 (AG alone, hatched bar, AG + p65 siRNA, striped bar) on the induction of apoptosis in p65−/− MEFs. Cells were treated with relevant siRNA, or control left for 48 h, pre-treated with AG-014699, or control 1 h prior to IR then allowed to repair for a further 24 h before harvesting and assessment of the induction of apoptosis by Annexin V FACs analysis. Results shown are normalised to untreated controls.
All data shown are represented as the mean ±sem of three independent experiments.
*Significance relative to mock treated control was p<0.05 using unpaired Student’s t-test.
We have also repeated these colony forming assays with IR alone, or in combination with AG-014699, in p65−/− cells that had been genetically complemented with wild-type (WT) p65, and in relevant control cells (Supplementary Figures 1B and 1C). Together with the data in Figures 1A and 1B, the results from the reconstituted cells confirm that AG-014699 radio-sensitized cells containing WT p65 but not the p65 null cells.
To assess the effects on apoptosis, we used FITC-conjugated Annexin V which has a high affinity for membrane phospholipid phosphatidylserine. Firstly, we found that p65−/− MEFs have a 3-fold higher intrinsic levels of apoptosis following IR, compared with p65+/+ (33.73 ± 3.1 for p65−/− versus 12.60 ± 2.7 for p65+/+, Data not shown). This is consistent with reports that NF-κB activation increases survival following DNA damage (Criswell et al., 2003, Russo et al., 2001, Wang et al., 1996). We then measured the levels of apoptosis following IR (2 Gy or 5 Gy) and either p65 knockdown or AG-014699 in p65+/+ and p65−/− MEFs (Figure 2E versus F). We also assessed a combination of p65 siRNA and AG-014699 (Figure 2E). Compared to IR alone (mock), both p65 knockdown and AG-014699 significantly increased apoptosis following IR (approx 2.5-fold) in the p65+/+ MEFs (p=0.03 and p=0.04 respectively). Importantly, when AG-014699 was used in conjunction with p65 knockdown, there was no change in the level of apoptosis in the p65+/+ MEFs, compared to either agent alone. No increase in apoptosis was seen in the p65−/− MEFs (Figure 2F) with any of the treatments, compared with IR alone. This data was confirmed using a caspase activity assay (Promega, Southampton, UK), (data not shown.)
AG-014699 inhibits Single strand break (SSB) repair to a similar extent regardless of cellular NF-κB status
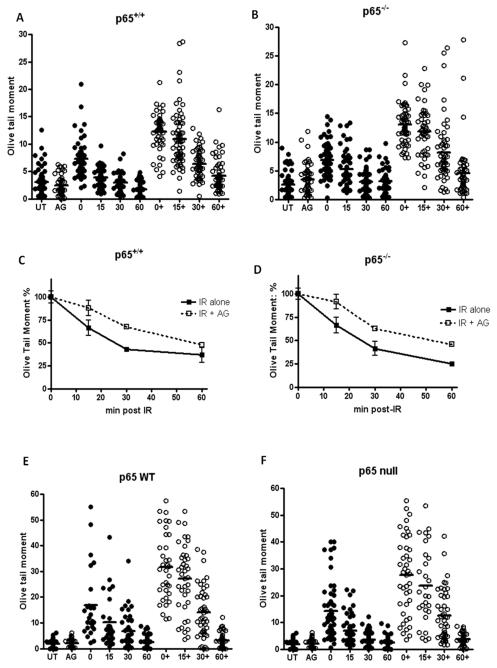
The alkaline Comet assay was used to examine IR-induced SSB formation and repair at the individual cell level. The kinetics of repair were assessed in the p65+/+ and p65−/− cell lines (Figures 3C and 3D). Figure 3 shows that immediately following 10 Gy IR, both cell lines had the same initial level of SSBs. There was no significant difference in the SSB levels remaining in the two cell lines 60 min post-IR, with > 85 % of breaks rejoined despite the difference in radio-sensitivity. Importantly, AG-014699 inhibited SSB repair (as demonstrated by increased residual SSB levels at all time-points post-IR) to the same extent in both cell lines (Figure 3A versus Figure 3B). Furthermore, we repeated these experiments in p65−/− cells that had been genetically complemented with WT p65, and also in p65 null controls. Importantly, we found that both the SSB repair kinetics and the extent of SSB inhibition by AG-014699 were very similar in both cell lines (Figure 3E versus Figure 3F). For all Comet assays, percentage tail DNA is shown in supplementary figure 2.
Figure 3. AG-014699 inhibits Single strand break (SSB) repair to a similar extent regardless of cellular NF-κB status.
Scatter diagrams showing the extent of single strand breaks (SSBs) in (A) p65+/+ MEFs and (B) p65−/− MEFs treated with 10 Gy IR ± AG-014699 (AG, denoted by +) and allowed to repair (0 min, 15 min, 30 min and 60 min). SSB repair was measured using the alkaline COMET assay and Olive Tail moment (shown here) is used as a measure of both the smallest detectable size of migrating DNA (reflected in the comet tail length) and the number of relaxed / broken pieces (represented by the intensity of DNA in the tail). All data were tested for Gaussian distribution and the lines shown indicate the mean of the data plotted.
Line graph showing the kinetics of single strand break repair in p65+/+ (C) and p65−/− MEFs (D). Data was normalised to relevant controls and in both cases solid black lines represent repair over time of cells treated with 10 Gy IR, and dotted lines represent repair over time of cells treated with 10 Gy IR + AG-014699.
Scatter diagrams showing the extent of single strand breaks (SSBs) in (E) p65−/− MEFs reconstituted with wild-type (WT) p65 and (F) p65−/− (control) MEFs treated with 10 Gy IR ± AG-014699 (AG, denoted by +) and allowed to repair (0 min, 15 min, 30 min and 60 min). SSB repair was measured using the alkaline COMET assay and Olive Tail moment is shown here. All data were tested for Gaussian distribution and the lines shown indicate the mean of the distribution.
All results shown are the mean of three independent experiments ± SEM
PARP activity is essential for NF-κB activation following IR, not TNF-α
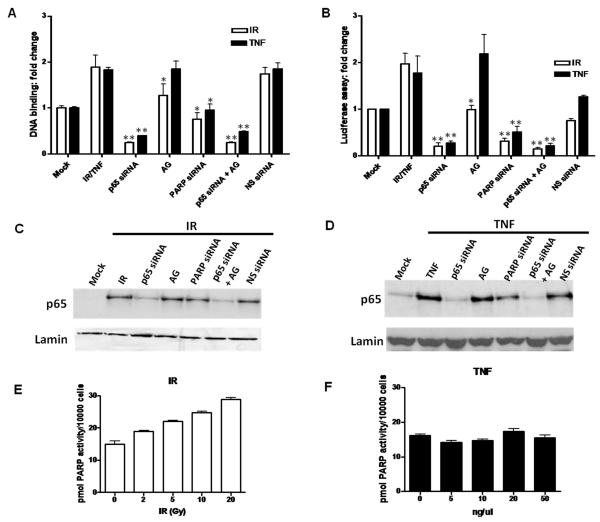
We investigated the ability of p65 to bind with its consensus sequence following either IR or TNF-α in p65+/+ or p65−/− MEFs. It was determined that 2 h post either stimuli p65 DNA binding activation was maximal (Supplementary Figures 3C and 3D). Binding was specifically competed out by excess wild type oligonucleotide, and unaffected by co-incubation with excess mutant oligonucleotide (data not shown). 10 Gy IR increased DNA binding activity 2-fold compared with Mock treated p65+/+ cells (Figure 4A). Also, siRNA targeting p65 or a combination of p65 siRNA and AG-014699 reduced DNA binding > 85% following IR in p65+/+ cells (p=0.008 and p=0.009, compared to IR alone, respectively). PARP-1 siRNA inhibited DNA binding by approximately 60% (p= 0.03, compared to IR). Significantly, AG-014699 also reduced IR-induced DNA binding to levels comparable with untreated controls (p=0.05, compared to IR). Non-specific siRNA had no effect on DNA-binding following IR. 10 ng/μl TNF-α increased p65 DNA binding activity 2.3-fold compared with Mock treated p65+/+ cells. p65 siRNA or a combination of p65 siRNA and AG-014699 reduced DNA binding to levels relative to Mock treated controls, following TNF-α (p= 0.009 and p=0.007, compared to TNF-α alone, respectively). Importantly, PARP-1 siRNA also reduced DNA binding 25% after TNF-α treatment (p=0.02, compared to TNF-α). In marked contrast, AG-014699 had no effect on p65 DNA binding following TNF-α. Negligible p65 DNA binding activity was seen after any treatments in the p65−/− cells with either IR or TNF-α (data not shown).
Figure 4. PARP activity is essential for NF-κB activation following IR, not TNF-α.
Histogram showing the effect of IR or TNF-α ± p65 siRNA ± AG-014699 (AG) ± PARP-1 siRNA ± (p65 siRNA + AG-014699) ±Non-specific (NS) siRNA control on NF-κB DNA binding activity, measured using an ELISA-based assay (A) and (B) NF-κB-dependent transcriptional activation, measured using a luciferase reporter assay, in p65+/+ MEFs. Open bars IR, black bars TNF-α. All results are the mean of three independent experiments with SEM. **Significance relative to mock treated control was p<0.01 using unpaired Student’s t-test. *Significance relative to mock treated control was p<0.05 using unpaired Student’s t-test.
Western blotting data showing nuclear translocation of p65 following (C) IR or TNF-α (D) ± p65 siRNA ± AG-014699 (AG) ± PARP-1 siRNA ± (p65 siRNA + AG-014699) ± NS siRNA control, in p65+/+ MEFs. Cells were treated with relevant siRNA, or control, left for 48 h, pre-treated with AG-014699, or control 1 h prior to IR and harvested 1 h post-IR. Loading was normalised to lamin nuclear loading control in all cases.
Histogram showing PARP activity, measured using a validated immunoblot assay which quantifies PAR polymer formation, in p65+/+ MEFs after increasing doses of IR (E) and TNF-α (F)
In order to assess the role of PARP-1 on NF-κB-dependent reporter gene transcription following IR or TNF-α, a luciferase reporter assay was used. Luciferase activity was found to be maximal 8 h post- IR or TNF-α treatment, and also to be dose-dependent in p65+/+ MEFs (data not shown). Consistent with the increased DNA binding, following either 10 Gy IR or 10 ng/μl TNF-α, luciferase activity was increased 2- and 2.5-fold respectively, compared to Mock treated controls (Figure 4B). Also, siRNA targeting p65 or a combination of p65 siRNA and AG-014699 reduced luciferase activity > 80% following IR (p=0.008 and p=0.007, respectively) or TNF-α (p=0.02 and p=0.03, respectively) in p65+/+ cells. PARP-1 siRNA inhibited luciferase activity by approximately 70%, following either stimulus (p=0.01 compared to IR, p=0.04 compared to TNF-α). Significantly, AG-014699 also reduced IR-induced luciferase reporter activity to levels comparable with untreated controls (p=0.02). However, AG-014699 had no effect on luciferase activity following TNF-α compared with TNF-α alone, consistent with other reports (Hassa et al., 2001). We have also previously shown that constitutive luciferase expression was about 10-fold lower in PARP-1−/− MEFs compared with PARP-1+/+ MEFs, and that IR failed to activate gene transcription in the PARP-1−/− MEFs, but clearly stimulated luciferase expression in the PARP-1+/+ MEFs (Veuger et al., 2009).
Furthermore, we show here that nuclear translocation of p65 in the p65+/+ MEFs observed 2 h after IR or TNF-α (Figures 4C and D), was unaffected by AG-014699 or PARP-1 siRNA.Cytoplasmic controls are shown in Supplementary Figures 3A and 3B.
IR, not TNF-α stimulates PAR polymer formation
The induction of PARP activity, by formation of the PAR polymer was determined using a validated assay (Plummer et al 2005, Zaremba et al., 2009). It is well documented that IR stimulates PARP activity, and consistent with that we show here that IR-induces PAR polymer formation in a dose-dependent manner in p65+/+ MEFs (Figure 4E). However, we found that, at a range of doses (5 - 50 ng/μl), TNF-α fails to stimulate PAR formation compared to basal levels in p65+/+ MEFs (Figure 4F).
Persistence of NF-κB DNA binding following PARG inhibition
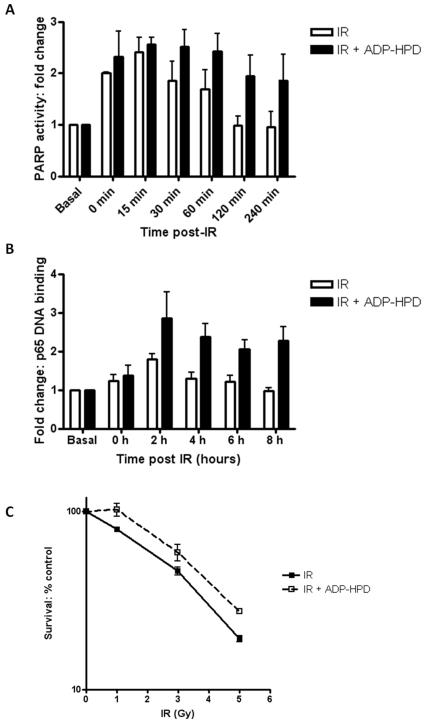
Poly (ADP-ribose) glycohydrolase (PARG) is the enzyme responsible for the catabolism of the PAR polymer. It is ubiquitously expressed at low levels in all cells and tissues (reviewed in Bonicalzi et al., 2005). Here, we have used the potent and specific inhibitor of PARG, ADP-HPD, (Slama et al., 1995) and the validated PARP assay (Zaremba et al., 2009) to show that co-incubation with ADP-HPD results in increased stability of the PAR polymer in IR-treated cells. Under normal conditions, polymer is degraded and returns to basal levels 1-2 hours following IR, whereas in ADP-HPD treated cells the polymer persists for upto 4-hours post-IR treatment.
Moreover, increased polymer stability correlates with a persistence in NF-κB DNA binding as shown in Figure 5B. DNA binding is maximal 2 hours post-IR before decreasing rapidly and returning to basal levels post-IR. Markedly, in cells treated with ADP-HPD DNA binding is maximal 2 hours post-IR and this persists up to 8 hours. Consistent with increased NF-κB DNA binding confers radio-resistance (Biswas et al., 2001; Wu and Kral, 2005), we show here, using the clonogenic survival assay (Figure 5C), that treatment with ADP-HPD protects against cell IR-induced cell kill in p65+/+ cells. Furthermore, there is no effect on survival in p65−/− cells (data not shown). In order to demonstrate that these findings were physiologically relevant we went on to determine whether PARG inhibition leads to an increase in the expression of some known NF-κB-mediated anti-apoptotic genes, in the p65+/+ cells. Using qRT-PCR, we show that IR significantly increases expression of Bcl-xL (p=0.04 compared with untreated controls) and this is further increased by co-incubation with ADP-HPD (p=0.0089 compared with ADP-HPD alone controls) (Supplementary Figure 4A). cFLAR expression (Supplementary Figure 4B) was also increased following IR (p=0.103) and this was significantly increased by co-incubation with ADP-HPD (p=0.0098). We also observed a similar trend with XIAP expression (Supplementary Figure 4C).
Figure 5. Persistence of NF-κB DNA binding following PARG inhibition.
(A) Histogram showing PARP activity timecourse, measured using a validated immunoblot assay which quantifies PAR polymer formation, in p65+/+ MEFs following 10 Gy IR ± ADP-HPD (open bars IR, black bars IR + ADP-HPD).
(B) Histogram showing NF-κB DNA binding timecourse measured using an ELISA-based method following IR ± ADP-HPD (open bars IR, black bars IR + ADP-HPD).
(C) The effects of increasing doses of IR on cell survival, either alone (solid line) or co-incubated with ADP-HPD (dashed line) were assessed using the clonogenic survival in p65+/+ MEFs. Cells were pre-treated with ADP-HPD, or control 1 h prior to IR then re-plated after a further 24 h and allowed to form colonies for 7-21 days.
All results are the mean of three independent experiments with SEM.
AG-014699 overcomes NF-κB-mediated therapeutic resistance in glioblastoma
Using Western blotting, we have confirmed the status of p65, and PARP-1 in U251 glioblastoma cells. We used siRNA to target either p65 or PARP-1 to knockdown these proteins (Figure 6A and 6B), and investigated the ability of p65 knockdown, PARP-1 knockdown or AG-014699 to sensitize U251 cells to IR using colony forming assays. We also assessed a combination of p65 siRNA and AG-014699. Similarly to the results observed in the p65+/+ MEFs, we show here that, co-incubation with either AG-014699, p65 siRNA or PARP-1 siRNA resulted in a 2.4-fold radio-sensitization compared to mock-transfected cells treated with IR. Again, a combination of p65 siRNA and AG-014699 showed no further sensitization (Figure 6D).
Figure 6. AG-014699 radio-sensitizes U251 glioblastoma cells via inhibition of NF-κB.
(A)Western blots of whole cell extracts of U251 cells 48 h post transfection with vehicle alone (Mock), non-specific (NS) siRNA or p65 siRNA.
(B) Western blots of whole cell extracts of U251 cells at 48 h after transfection with vehicle alone (Mock), non-specific (NS) siRNA or PARP-1 siRNA.
(C)Histogram showing the effect of IR ± p65 siRNA ± AG-014699 (AG) ± PARP-1 siRNA ± (p65 siRNA + AG-014699) ±Non-specific (NS) siRNA control on NF-κB DNA binding activity, measured using an ELISA-based assay. All results are the mean of three independent experiments with SEM. **Significance relative to mock treated control was p<0.01 using unpaired Student’s t-test.
(D)The effects of increasing doses of IR either alone, or co-incubated with p65 siRNA, AG-014699, PARP-1 siRNA or a combination of p65 siRNA and AG-014699, on U251 cell survival was assessed using the clonogenic survival assay. Cells were treated with relevant siRNA, (or vehicle control), left for 48 h, pre-treated with AG-014699, or DMSO control, 1 h prior to IR then re-plated after a further 24 h and allowed to form colonies for 7-21 days.
(E) Scatter diagrams showing the extent of single strand breaks (SSBs) in U251 cells treated with 10 Gy IR ± AG-014699 (denoted by +) and allowed to repair (0 min, 15 min and 30 min). SSB repair was measured using the alkaline COMET assay and Olive Tail moment (described above) is shown here. All data were tested for Gaussian distribution and the lines shown indicate the mean of the data plotted.
(F) Scatter diagrams showing the extent of single strand breaks (SSBs) in U251 cells treated with 10 Gy IR + p65 siRNA ± AG-014699 (denoted by +) and allowed to repair (0 min, 15 min and 30 min). SSB repair was measured using the alkaline COMET assay and Olive Tail moment (described above) is shown here. All data were tested for Gaussian distribution and the lines shown indicate the mean of the data plotted.
We then determined the effect of the treatments described above on NF-κB DNA binding using the ELISA-based method and found that our observations were consistent with those seen in the p65+/+ MEFs. 10 Gy IR increased DNA binding activity 2-fold compared with Mock treated cells (Figure 6C). Also, p65 siRNA alone or in combination with AG-014699 reduced DNA binding > 85% following IR in U251 cells (p=0.0003 and p=0.005, compared to IR alone, respectively). Importantly, both PARP-1 siRNA and AG-014699 reduced IR-induced DNA binding to levels comparable with untreated controls (p=0.0053 and p=0.0079, compared to IR, respectively). Non-specific siRNA had no effect on DNA-binding following IR.
The alkaline Comet assay was used to examine IR-induced SSB formation and repair in the U251 cells. Immediately following 10 Gy IR, cells treated with IR alone or IR in combination with p65 siRNA had the same initial level of SSBs (Figures 6E and F). There was no significant difference in the SSB levels remaining in U251 cells or U251 cells with p65 siRNA 30 min following IR, with > 95 % of breaks rejoined despite the observed differences in radio-sensitivity shown in Figure 6D. Importantly, AG-014699 inhibited SSB repair (as demonstrated by increased residual SSB levels at all time-points post-IR) to the same extent in cells regardless of the p65 status (Figure 6E versus Figure 6F).
DISCUSSION
In this study we demonstrate that PARP-1 knockdown and the PARP inhibitor, AG-014699 significantly decreased survival and induced apoptosis following IR in p65+/+ MEFs whilst having no effect in p65−/− MEFs. Sensitization was 1.3-fold and importantly occurred at clinically relevant doses of IR. A combination of AG-014699 and p65 knockdown did not further enhance this response, which is consistent with reports that PARP-1 and NF-κB are mechanistically linked in a common pathway (Chang and Alvarez-Gonzalez 2001, Hassa and Hottiger 1999, Stilmann et al., 2009, Veuger et al., 2009). The levels of SSBs in p65+/+ and p65−/− MEFs induced by IR were similar and both were equally proficient at their repair. AG-014699 was also able to increase the level of breaks to the same extent in both cell lines, consistent with the known role of PARP-1 in base excision repair (Durkacz et al., 1980). Thus, we can conclude that the potentiating effect of AG-104699 when used in conjunction with IR is not primarily due to the widely reported transient inhibition of SSB repair but a consequence of PARP-1 downstream signaling to NF-κB. This is consistent with the observation that cells severely compromised in DNA repair (e.g. PARP-1−/−) are able to divide and go on to survive despite having consistently higher levels of breaks compared to their wild type counterparts, even in the absence of exogenous damage (Trucco et al.,1998). These data, coupled with our previous findings that IR-induced DNA damage initiates signaling events in the nucleus and continues in the cytoplasm, resulting in the activation of NF-κB (Veuger et al.,2009), support the existence of an intermediary signaling protein/complex as described by Stilmann et al., (2009).
There is strong evidence that the effects we have observed using the Comet assay are likely to be due to SSBs, even though the alkaline Comet assay can detect a small amount of double strand breaks (DSBs) as well as SSBs. Firstly, Ogorek and Bryant (1985) showed that at the low doses of IR as used in our experiments, the ratio of SSB to DSB is approximately 20:1. Furthermore, we have previously shown that PARP-1 has a minor role in the repair of DSBs by non-homologous end joining (NHEJ) in the absence of the DNA-dependent protein kinase (DNA-PK) (Veuger et al., 2003). However the p65+/+ and p65−/− cells used here have functional DNA-PK, and thus any DSBs would be repaired by NHEJ, with AG-014699 being solely responsible for the inhibition of SSBs.
To investigate the effects on NF-κB activity induced by stimuli other than IR, we used TNF-α. While knockdown of the PARP-1 protein abrogated NF-κB activation following the inflammatory stimulus, PARP-1 enzymatic inhibition had no effect. In contrast, PARP protein and activity were necessary to inhibit the IR-induced NF-κB response. The role of PARP-1 protein rather its catalytic activity as a co-activator of NF-κB is contentious. Our data are consistent with some studies (Hassa et al., 2001) but others have shown that PARP-1 inhibition was able to alleviate TNF-α-stimulated NF-κB-dependent MMP-9-mediated neurotoxicity in microglia in a model of brain injury (Kauppinen and Swanson, 2005). This may be cell line or model system dependent and may also vary depending on the activation other NF-κB subunits. For example, we have recently shown that subunit activation is complex balance in CLL (Mulligan et al., manuscript submitted). However, is it also possible that in this case PARP-1 may facilitate a structural role in the NF-κB transcriptional complex, as Hassa et al., (2005) illustrated by demonstrating that the acetylation of PARP-1, by HDAC1 and p300, regulated NF-κB activity. A further explanation is that PARP inhibition may produce different physiological effects compared with protein knockdown. Although inhibition of other PARP family proteins, such as PARP-2 by AG-014699 cannot be rigorously excluded, the consistency in the data obtained from both the inhibitor studies and PARP-1 knockdown studies shown here indicate that this is not the case. We have also previously shown that the PARP-1−/− cells have negligible PAR formation using the PARP activity assay (Veuger et al., 2009), thus supporting our findings here that AG-014699 radio-sensitizes cells via inhibition of PARP-1, not PARP-2.
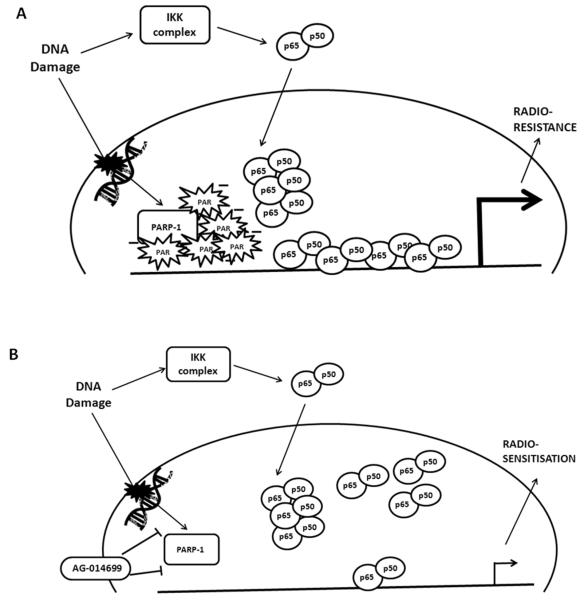
Since both stimuli resulted in similar amounts of p65 nuclear translocation, and this was unaffected by either AG-014699 or PARP-1 knockdown, then the differential requirements of PARP-1 for IR- and TNF-α-activated NF-κB must occur at the DNA binding and transcription stage. The requirement of the PAR polymer in DNA damage-activated NF-κB has been demonstrated (Stilmann et al., 2009) and this is consistent with our data that IR, not TNF-α stimulates polymer formation. Stilmann et al. (2009) clearly indentified a novel mechanism of pro-survival NF-κB activation mediated via the PAR polymer. Based on our observations we can propose a further mechanism (Figure 7). The DNA binding interface of p65 is buried until a conformational change is induced, usually when p65 is in close proximity to the negatively charged region of its DNA consensus sequence (Ghosh et al., 1999). The PAR polymer is known to have a large overall negative charge (Uedo and Hayaishi, 1985). Since we showed no change in levels of nuclear p65 following DNA damage or treatment with AG-014699, but yet that p65 DNA binding increased upon stimulation of polymer formation, we can postulate that the negative charge on the polymer induces this conformational change in p65, thus leading to increased binding and transcriptional activation (Figure 5). The data we present here using the PARG inhibitor, ADP-HPD, supports this hypothesis. We show that increased polymer stability, by virtue of inhibition of polymer degradation, leads to a persistence of NF-κB DNA binding, an increase in anti-apoptotic gene expression and a protection against IR-induced cell death. This confirms a role for the PAR polymer in the activation of NF-κB following DNA damage.
Figure 7. PAR is essential for DNA damage activated NF-κB.
(A) DNA damage, such as IR, activates PARP-1, recruiting it to the site of damaged DNA. This results in formation of the negatively charged PAR polymer. The NF-κB p65-p50 heterodimer is also translocated to the nucleus following DNA damage via activation of the canonical pathway of NF-κB activation. When in proximity to regions with overall negative charge, such as that of the PAR polymer, a conformational change in p65 can be induced, exposing the positively charged DNA binding interface of p65. Thus, we propose that it is this negative charge on the PAR polymer that is attracting p65 to DNA bind, (as PARP-1 and the polymer are recruited to sites of damaged DNA), up-regulating transcription of NF-κB-dependent genes, protecting against apoptosis and conferring radio-resistance.
(B) When the PARP inhibitor, AG-014699 is present, PARP-1 is no longer active and cannot form the polymer. Hence in this case, we see reduced DNA binding and transcriptional activation following IR, with induction of apoptosis and ultimately radio-sensitisation.
Inhibition of NF-κB is clearly an attractive target for cancer therapeutics (Gilmore and Herscovitch 2006). However, global inhibition may also have some negative aspects. For example, NF-κB plays a vital role in the inflammatory response, and thus complete inhibition of NF-κB, for example with an IKK inhibitor, may have adverse effects. The IKK complex has many known functions within the cell (Chariot, 2009, Criollo et al., 2010), and inhibition may be cytotoxic. Long-term NF-κB inhibition may also increase the likelihood of immunodeficiency, since it plays a pivotal role in the innate and adaptive immune response, and can possibly delay bone marrow recovery due to some reports of chemotherapeutic induced apoptosis of hematopoietic progenitors (Grossman et al., 1999, Turco, et al, 2004). The data here suggest that AG-014699 inhibits IR-induced NF-κB activation but not an inflammatory stimulus and thus may overcome these potential toxicities. Furthermore, ongoing gene expression analysis shows that AG-014699 has no effect on vital inflammatory response genes following TNF-α treatment (data to be published separately). Consequently, blockade of DNA-damage activated NF-κB by AG-014699 may therefore represent preferable therapeutic strategy, since we demonstrate here that the survival function of NF-κB can be directly inhibited without compromising other functions, such as the immune response.
AG-014699 has been shown to be efficacious both in vitro and in vivo (Daniels et al., 2009, Drew et al., 2010, Plummer et al., 2008, Zaremba et al., 2009) and is currently in phase II trials for BRCA defective breast and ovarian cancer as a single agent (www.clinicaltrials.gov). AG-014699 was the first PARP inhibitor to enter clinical trial for cancer therapy and Phase I trials showed that profound and sustained PARP inhibition could be achieved after a single intravenous infusion (Plummer et al., 2005). Phase II trials combining AG-014699 with temozolomide in metastatic melanoma patients showed an improvement in the response rate in comparison with temozolomide alone (Plummer et al., 2006). A number of pharmaceutical companies are now undertaking clinical trials of PARP inhibitors for the treatment of cancer (Drew and Plummer 2009); however, this is the first investigation of the clinically relevant PARP inhibitors in the context of their potential utility as inhibitors of DNA-damage induced NF-κB activation.
Radio-therapy is one of the main-stays of therapy for Glioblastoma multiforme, the most frequent and refractory adult brain tumour (Burger et al., 1985). Aberrant activation of NF-κB is known to play a role in the tumourigenesis of both diffuse and high-grade gliomas (Kawanza et al., 2003, Wang et al., 2004). Hence, here we have used the glioblastoma cell line, U251, to highlight the potential utility of AG-014699 in inhibiting DNA-damage activated NF-κB in tumor cells.
In summary, this is the first report to demonstrate that a very potent and specific PARP-1 inhibitor, AG-014699, can re-sensitise cells to irradiation and that this is mediated directly via NF-κB, rather than the inhibition of SSB repair. These data present a novel mechanism for PARP-1 in the regulation of NF-κB and highlight important new therapeutic avenues for the use of PARP inhibitors, which may prove useful in overcoming NF-κB-mediated therapeutic resistance.
METHODS
Cell lines and Reagents
The p65+/+ and p65−/− MEFs were kindly provided by Professor Ron Hay, Dundee University, UK. PARP-1+/+ and PARP-1−/− MEFs were a gift from Professor Gilbert de Murcia, École Superieure de Biotechnologie de Strasbourg, France. All of the above cell lines were cultured in RPMI-1640 medium (supplemented with 10 % (v/v) FCS and 2 mM glutamine). The p65−/− control MEFs, and the MEFs which have been genetically complemented with WT protein were a kind gift from Professor Neil Perkins, Newcastle University, UK. These were cultured in DMEM medium (supplemented with 10% (v/v) FCS, 2 mM glutamine and 4 μg/ml Puromycin). The U251 glioblastoma cell line was donated by Dr Gareth Veal, Newcastle University, UK, and was cultured in DMEM medium (supplemented with 10% (v/v) FCS and 2 mM glutamine).
PARP-1 Inhibitor
AG-014699 was synthesised by Pfizer GRD, CA. AG-014699 was dissolved in anhydrous DMSO (Sigma, UK) at a stock concentration of 10 mM and stored at −20 °C. AG-014699 was added in cell culture such that the final DMSO concentration was kept constant at 0.1 % (v/v), and was used at a final concentration of 0.1 μM in all experiments.
PARG Inhibitor
ADP-HPD was purchased from ENZO Life Sciences, Exeter, UK. It was dissolved in sterile H2O (Gibco, Paisley, Scotland) at a stock concentration of 5 mg/ml and stored at −20°C. ADP-HPD was added in cell culture at a final concentration of 1 μM in all experiments.
SiRNA Transfection
Cells were seeded in 6-well cell culture plates at a density of 5×104 in 2mls and incubated overnight. SiRNA targeting human p65 (GCCCUAUCCCUUUACGUCA); or PARP-1 (GGAGGAAGGUGUCAACAAA; both from Dharmacon, Cramlington, UK) was transfected at a final concentration of 50nM using Lipofectamine 2000 (Invitrogen, UK), according to the manufacturer’s instructions. Controls used included non-specific (NS) siRNA and mock/vehicle only transfected cells. p65 or PARP-1 knock down was confirmed by Western analysis (Figure 1).
PARP activity assay
PARP activity was determined in cells transfected with siRNA (or relevant controls, as described above), that had been treated with AG-014699 or DMSO, using a validated assay (Plummer et al., 2005, Zaremba et al., 2009). Briefly, basal PARP activity present in cells was measured in 1×104 permeabilized cells that had been incubated with NAD+ and oligonucleotide (to maximally stimulate PARP activity) (Sigma, UK). PARP activity was also assessed following treatment with IR or TNF-α, by omitting the oligonucleotide stimulation step. The reaction was terminated with excess AG-014699, and replicate samples (n=3) were blotted onto nitrocellulose membrane (Hybond N, Amersham, UK). A standard curve of purified PAR polymer (pmol of PAR) (Biomol, UK) was also blotted to aid with quantification. The membrane was probed with PAR-specific antibody (10H, a kind gift of Prof Burkle, University of Konstanz, Germany) and chemiluminescence was detected using a 5 min exposure on the Fuji-LAS 3000 CCD camera. Images were anaylzed using Aida Imaging Analyzer. Data was normalized to mock (vehicle alone) treated controls and results shown are the mean of 3 independent experiments.
Western Blotting
Proteins were resolved on 4-20 % (v/v) Tris-glycine gradient gels (Invitrogen Ltd, Paisley, UK) and electrotransferred onto nitrocellulose (Hybond C, Amersham, UK). Antibodies against PARP-1 (H-250), p50 (8414), p65 (8008) were purchased from Santa Cruz Biotechnology, (Santa Cruz, CA). As loading controls, actin antibody (CP01; Calbiochem) was used for whole cell extracts, lamin A/C (7293; Santa Cruz Biotechnology, Santa Cruz, CA) for nuclear extracts and -tubulin for cytoplasmic extracts (Sigma). This was followed by binding of peroxidase-conjugated goat anti-mouse/rabbit antibody (DAKO, Ely, UK) and detection by enhanced chemiluminescence (Amersham, UK). Cells were pre-treated with AG-014699 for 1 h and exposed to IR. Cells were harvested at different time points and whole cell extracts were taken using SDS lysis buffer (100 mM Tris-Hcl pH 6.8, 20 % v/v glycerol, 4 % w/v SDS). Nuclear and cytoplasmic extracts were prepared using the NE-PER extraction kit as per manufacturer’s instructions (Pierce-Perbio science, Cheshire, UK). α-tubulin was not detected in nuclear extracts and conversely, lamin was absent in all cytoplasmic extracts thus demonstrating the efficient separation of nuclear extracts. Bands were quantified and normalized to their relevant loading controls using densitometry (Fuji LAS 3000 Imaging).
Cytotoxicity Assay
Clonogenic assays were carried out as described previously (Veuger et al., 2003) and cloning efficiencies were normalized to untreated controls. PF50 (potentiation factor at 50% cell kill) values were calculated from the ratio of the individual LD50 (lethal dose producing 50% cell kill) values:- i.e., LD50 divided by LD50 in the presence of AG-014699.
Annexin-V Flow Cytometry
Apoptosis was measured using the FITC Annexin V Apoptosis Detection Kit I (BD Biosciences, Oxford, UK), as per manufacturers instructions. Briefly, cells were transfected with p65 siRNA (or relevant controls) 48 h prior to 1 h pre-treatment with AG-014699 and subsequent irradiation. Cells were harvested and resuspended in 100 μl Binding Buffer (0.01M Hepes/NaOH (pH 7.4), 0.14M NaCl, 0.25mM CaCl2). FITC Annexin V and Propidium Iodide Staining Solution were added and cells were incubated for 15 min at room temperature, in the dark. Cells were analyzed using the Becton Dickinson FACScan and Cell Quest Software. Results are the mean of 3 independent experiments.
COMET Assay
Following IR treatment, SSB levels were quantified using the alkaline Comet assay (Trevigen) following the manufacturer’s protocol. Briefly, cells were washed and resuspended in ice-cold PBS before mixing with proprietary low melting point agarose (37°C). This mixture was pipetted onto a CometSlide (Trevigen) and allowed to gel at 4°C. Following lysis, slides were subjected to electrophoresis under alkaline conditions at 20 V for 40 min and then fixed, dried, and stained with SYBR Green I. An Olympus BH2-RFCA fluorescence microscope (×10 objective) with Hamamatsu ORCAII BT-1024 cooled CCD camera was used to capture images. Image Pro Plus (Media Cybernetics) was used for image capture and analysis was performed using Komet software v 5.5 (Kinetic Imaging). Data for repair kinetics was expressed as a percentage, with 100% representing the mean number of breaks at time 0 post IR treatment, in the presence of absence of AG-014699. Subsequent time points are expressed as a percentage of the time 0 point.
NF-κB DNA binding Assay
NF-κB p65 DNA binding activity was determined using an ELISA-based EZ detect assay according to the manufacturer’s instructions (Promega Southampton, UK). Briefly, streptavidin-coated 96 well plates are bound with biotinylated κB consensus sequence oligonucleotides (5′GGGACTTTCC-3′). Cells were transfected with siRNA, or relevant controls 48 h prior to a 1 h pre-treatment with AG-014699 and subsequent irradiation. 5 μg of nuclear extracts (prepared 2 h after irradiation, as described above) were added to each well and incubated with primary antibody specific for p65. Binding was detected using a secondary HRP-conjugated antibody and chemiluminescence measured using a CCD camera (LAS-300; Fujifilm). Purity of cytoplasmic and nuclear extracts was confirmed as described above.
Reporter Gene Assay
Cells were seeded onto 24 well plates and incubated for 24 h. Cells were transiently transfected with 200 ng of an NF-κB-luciferase construct containing 3 tandemly repeated NF-κB consensus sequence binding sites in the promoter (Professor Ron Hay, Dundee University), together with 200 ng of a pCMB-β-galactosidase plasmid containing a minimal promoter element up-stream from the β-galactosidase gene, using Lipofecamine 2000 (Invitrogen, Paisley, Scotland) for 6 h. siRNA transfection was undertaken simultaneously, as described above. 48 h after transfection, cells were treated with AG-014699 for 1 h prior to IR. Following 8 h incubation, cells were lysed and assayed for luciferase activity according to the manufacturer’s instructions (Promega, Southampton, UK). Luciferase activity was corrected for β-galactosidase activity as described previously (Brady ME et al,.1999), and relative activities expressed as fold changes.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge Pfizer for the supply of AG-014699. This work was supported by Cancer Research UK, grant number C7369/A8048.
Footnotes
CONFLICT OF INTEREST ZH is an employee of Pfizer GRD. The other authors declare no conflict of interest.
REFERENCES
- Althaus FR, Hofferer L, Kleczkowska HE, Malanga M, Naegeli H, Panzeter PL, et al. Histone shuttling by poly ADP-ribosylation. Mol Cell Biochem. 1994;138:53–9. doi: 10.1007/BF00928443. [DOI] [PubMed] [Google Scholar]
- Barkett M, Gilmore T. Control of apoptosis by Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6910–24. doi: 10.1038/sj.onc.1203238. [DOI] [PubMed] [Google Scholar]
- Basseres DS, Baldwin AS. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25:6817–6830. doi: 10.1038/sj.onc.1209942. [DOI] [PubMed] [Google Scholar]
- Biswas DK, Shi Q, Baily S, Strickland I, Ghosh S, Pardee AB, et al. NF-kappa B activation in human breast cancer specimens and its role in cell proliferation and apoptosis. PNAS. 2004;101:10137–42. doi: 10.1073/pnas.0403621101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonicalzi ME, Haince JF, Droit A, Poirier GG. Regulation of poly(ADP-ribose) metabolism by poly(ADP-ribose) glycohydrolase: where and when? Cell Mol Life Sci. 2005;62:739–50. doi: 10.1007/s00018-004-4505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brach MA, Hass R, Sherman ML, Gunji H, Weichselbaum R, Kufe D. Ionizing radiation induces expression and binding activity of the nuclear factor kappa B. J Clin Invest. 1991;88:691–5. doi: 10.1172/JCI115354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady ME, Ozanne DM, Gaughan L, Waite I, Cook S, Neal DE, et al. Tip60 is a nuclear hormone receptor coactivator. J Biol Chem. 1999;274:17599–604. doi: 10.1074/jbc.274.25.17599. [DOI] [PubMed] [Google Scholar]
- Burger PC, Vogel FS, Green SB, Strike TA. Glioblsstoma multiforme and anaplastic astrocytoma pathologic criteria and prognostic implications. Cancer. 1985;56:1106–11. doi: 10.1002/1097-0142(19850901)56:5<1106::aid-cncr2820560525>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Chang WJ, Alvarez-Gonzalez R. The sequence-specific DNA binding of NF-kappa B is reversibly regulated by the automodification reaction of poly (ADP-ribose) polymerase 1. J Biol Chem. 2001;276:47664–70. doi: 10.1074/jbc.M104666200. [DOI] [PubMed] [Google Scholar]
- Chariot A. The NF-kappaB-independent functions of IKK subunits in immunity and cancer. Trends Cell Biol. 2009;19:404–413. doi: 10.1016/j.tcb.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Chiarugi A, Moskowitz MA. Poly(ADP-ribose) polymerase-1 activity promotes NF-kappaB-driven transcription and microglial activation: implication for neurodegenerative disorders. J Neurochem. 2003;85:306–17. doi: 10.1046/j.1471-4159.2003.01684.x. [DOI] [PubMed] [Google Scholar]
- Clevers H. At the crossroads of inflammation and cancer. Cell. 2004;118:671–4. doi: 10.1016/j.cell.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Criollo A, Senovilla L, Authier H, Maiuri M Chiara, Morselli E, Vitale I, et al. The IKK complex contributes to the induction of autophagy. EMBO J. 2010;29:619–31. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criswell T, Leskov K, Miyamoto S, Luo G, Boothman DA. Transcription factors activated in mammalian cells after clinically relevant doses of ionizing radiation. Oncogene. 2003;22:5813–27. doi: 10.1038/sj.onc.1206680. [DOI] [PubMed] [Google Scholar]
- Daniels RA, Rozanska AL, Thomas HD, Mulligan EA, Drew Y, Castelbuono DJ, et al. Inhibition of poly(ADP-ribose) polymerase-1 enhances temozolomide and topotecan activity against childhood neuroblastoma. Clin Cancer Res. 2009;15:1241–9. doi: 10.1158/1078-0432.CCR-08-1095. [DOI] [PubMed] [Google Scholar]
- Drew Y, Plummer R. PARP inhibitors in cancer therapy: two modes of attack on the cancer cell widening the clinical applications. Drug Resist Updat. 2009;12:153–6. doi: 10.1016/j.drup.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Drew Y, Mulligan EA, Vong WT, Thomas HD, Kahn S, Kyle S, et al. Therapeutic potential of Poly(ADP-ribose) Polymerase Inhibitor AG014699 in Human Cancers With Mutated or Methylated BRCA1 or BRCA2. JNCI. 2010. in press. [DOI] [PubMed]
- Durkacz BW, Omidiji O, Gray DA, Shall S. (ADP-ribose) participates in DNA excision repair. Nature. 1980;283:593–6. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- Ghosh G, Huang DB, Huxford T. Structural insights into NF-κB/IκB signaling. Gene Ther Mol Biol. 1999;4:75–82. [Google Scholar]
- Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Gilmore TD, Herscovitch M. Inhibitors of NF-kappaB signaling: 785 and counting. Oncogene. 2006;25:6887–99. doi: 10.1038/sj.onc.1209982. [DOI] [PubMed] [Google Scholar]
- Grossman M, Metcalf D, Merryfull J, Beg A, Baltimore D, Gerondakis S. The combined absence of the transcription factors Rel and RelA leads to multiple hemopoietic cell defects. Proc. Natl Acad Sci.USA. 1999;96:11848–11853. doi: 10.1073/pnas.96.21.11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO. A role of poly (ADP-ribose) polymerase in NF-kappaB transcriptional activation. Biol Chem. 1999;380:953–9. doi: 10.1515/BC.1999.118. [DOI] [PubMed] [Google Scholar]
- Hassa PO, Covic M, Hasan S, Imhof R, Hottiger MO. The enzymatic and DNA binding activity of PARP-1 are not required for NF-kappa B coactivator function. J Biol Chem. 2001;276:45588–97. doi: 10.1074/jbc.M106528200. [DOI] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, et al. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J Biol Chem. 2005;280:40450–64. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- Hewamana S, Alghazal S, Lin TT, Clement M, Jenkins C, Guzman ML, et al. The NF-kappaB subunit Rel A is associated with in vitro survival and clinical disease progression in chronic lymphocytic leukemia and represents a promising therapeutic target. Blood. 2008;111:4681–9. doi: 10.1182/blood-2007-11-125278. [DOI] [PubMed] [Google Scholar]
- Hewamana S, Lin TT, Jenkins C, Burnett AK, Jordan CT, Fegan C, et al. The novel nuclear factor-kappaB inhibitor LC-1 is equipotent in poor prognostic subsets of chronic lymphocytic leukemia and shows strong synergy with fludarabine. Clin Cancer Res. 2008;14:8102–11. doi: 10.1158/1078-0432.CCR-08-1673. [DOI] [PubMed] [Google Scholar]
- Hewamana S, Lin TT, Rowntree C, Karunanithi K, Pratt G, Hills R, et al. Rel a is an independent biomarker of clinical outcome in chronic lymphocytic leukemia. J Clin Oncol. 2009;27:763–9. doi: 10.1200/JCO.2008.19.1114. [DOI] [PubMed] [Google Scholar]
- Jackson A, Linsley P. Noise amidst the silence: off target effects of siRNAs? TRENDS in Genetics. 2004;20:521–524. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Jung M, Dritschilo A. NF-kappa B signaling pathway as a target for human tumor radiosensitization. Semin Radiat Oncol. 2001;11:346–51. doi: 10.1053/srao.2001.26034. [DOI] [PubMed] [Google Scholar]
- Kanzawa T, Ito H, Kondo Y, Kondo S. Current and future gene therapy for malignant gliomas. J Biomed Biotechnol. 2003;2003:25–34. doi: 10.1155/S1110724303209013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppinen TM, Swanson RA. Poly(ADP-ribose) ;polymerase-1 promotes microgial activation, proliferation and matrix metalloproteinase-9 mediated neuron death. J Immunol. 2005;174:2288–96. doi: 10.4049/jimmunol.174.4.2288. [DOI] [PubMed] [Google Scholar]
- Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113:677–83. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- Martin-Oliva D, Aguliar-Quesada R, O’valle F, Munoz-Gamez JA, Martinez-Romero R, Del-Moral R Garcia, et al. Inhibition of poly(ADP-ribose) polymerase modulates tumor-related gene expression, including hypoxia-inducible factor-1 activation, during skin carcinogenesis. Cancer Res. 2006;66:5744–56. doi: 10.1158/0008-5472.CAN-05-3050. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Nagaso H, Kakui N, Ishikawa M, Hiranuma T, Hoshiko S. Critical role of the automodification of poly(ADP-ribose) polymerase-1 in nuclear factor-kappaB-dependent gene expression in primary cultured mouse glial cells. J Biol Chem. 2004;279:42778–86. doi: 10.1074/jbc.M407923200. [DOI] [PubMed] [Google Scholar]
- Ogorek B, Bryant PE. Repair of DNA single-strand breaks in X-irradiated yeast.II. Kinetics of repair as measured by the DNA-unwinding method. Mutat Res. 1985;146:63–70. doi: 10.1016/0167-8817(85)90056-2. [DOI] [PubMed] [Google Scholar]
- Oliver FJ, de Murcia J Menissier, Nacci C, Decker P, Andriantsitohaina R, Muller S, et al. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. EMBO J. 1999;18:4446–54. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad S, Ravindran J, Aggarwal B. NF-κB and cancer: how intimate is this relationship. Mol Cell Biochem. 2009;336:25–37. doi: 10.1007/s11010-009-0267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer ER, Middleton MR, Jones C, Olsen A, Hickson I, McHugh P, et al. Temozolomide Pharmacodynamics in Patients with Metastatic Melanoma: DNA Damage and Activity of Repair Enzymes O6-Alkylguanine Alkyltransferase and Poly(ADP-Ribose) Polymerase-1. Clin Cancer Res. 2005;11:3402–3409. doi: 10.1158/1078-0432.CCR-04-2353. [DOI] [PubMed] [Google Scholar]
- Plummer R, Middleton M, Wilson R, Jones C, Evans J, Robson L, et al. First in human phase I trial of the PARP inhibitor AG-014699 with temozolomide (TMZ) in patients (pts) with advanced solid tumors. J Clin Oncol. 2005;23(No.16S,Part I):3065. [Google Scholar]
- Plummer R, Lorigan P, Evans J, Steven N, Middleton M, Wilson R, et al. First and final report of a phase II study of the poly(ADP-ribose) polymerase (PARP) inhibitor, AG014699, in combination with temozolomide (TMZ) in patients with metastatic malignantmelanoma (MM) J Clin Oncol. 2006;24(No.18S, Part I):8013. [Google Scholar]
- Plummer ER, Jones C, Middleton MR, Wilson R, Evans J, Olsen A, et al. Phase I Study Of The Poly(ADP-Ribose) Polymerase Inhibitor, AG014699, In Combination With Temozolomide in Patients with Advanced Solid Tumors. Clin Cancer Res. 2008;14:7917–23. doi: 10.1158/1078-0432.CCR-08-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo SM, Tepper JE, Baldwin AS, Jr, Lui R, Adams J, Elliott P, et al. Enhancement of radiosensitivity by proteasome inhibition: implications for a role of NF-kappaB. Int J Radiat Oncol Biol Phys. 2001;50:183–93. doi: 10.1016/s0360-3016(01)01446-8. [DOI] [PubMed] [Google Scholar]
- Sahijdak WM, Yang CR, Zuckerman JS, Meyers M, Boothman DA. Alterations in transcription factor binding in radioresistant human melanoma cells after ionizing radiation. Radiat. Res. 1994;138:S47–S51. [PubMed] [Google Scholar]
- Slama JT, Aboul-Ela N, Goli DM, Cheesman BV, Simmons AM, Jacobson MK. Specific inhibition of poly(ADP-ribose) glycohydrolase by adenosine diphosphate (hydroxymethyl)pyrrolidinediol. J Med Chem. 1995;38:389–93. doi: 10.1021/jm00002a021. [DOI] [PubMed] [Google Scholar]
- Smith S. The world according to PARP. Trends Biochem Sci. 2001;26:174–9. doi: 10.1016/s0968-0004(00)01780-1. [DOI] [PubMed] [Google Scholar]
- Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Schreiderit C. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Molecular Cell. 2009;36:365–78. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- Trucco C, Oliver F Javier, de Murcia G, de Murcia J Menissier. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acid Res. 1998;26:2644–9. doi: 10.1093/nar/26.11.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschaharganeh D, Ehemann V, Nussbaum T, Schrimacher P, Breuhahn K. Non-specific effects of siRNAs on tumor Cells with implications on therapeutic applicability using RNA Interference. Pathology Oncology Research. 2007;13:84–90. doi: 10.1007/BF02893482. [DOI] [PubMed] [Google Scholar]
- Turco MC, Romano MF, Petrella A, Bisgoni R, Tassone P, Ventuna S. NF-kappaB/Rel-mediated regulation of apoptosis in hematologic malignancies and normal hematopoietic progenitors. Leukemia. 2004;18:11–7. doi: 10.1038/sj.leu.2403171. [DOI] [PubMed] [Google Scholar]
- Ueda K, Hayaishi O. ADP-RIBOSYLATION. Ann. Rev. Biochem. 1985;54:73–100. doi: 10.1146/annurev.bi.54.070185.000445. [DOI] [PubMed] [Google Scholar]
- Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 2003;63:6008–15. [PubMed] [Google Scholar]
- Veuger SJ, Hunter JE, Durkacz BW. Ionizing radiation-induced NF-kappaB activation requires PARP-1 function to confer radioresistance. Oncogene. 2009;28:832–42. doi: 10.1038/onc.2008.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Mayo MW, Baldwin AS., Jr. TNF and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science. 1996;274:784–7. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS., Jr. NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281:1680–3. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang H, Zhang W, Huang HJ, Liao WS, Fuller GN. Analysis of the activation status of Akt, NFkappaB and Stat3 in human diffuse gliomas. Lab Invest. 2004;84:941–51. doi: 10.1038/labinvest.3700123. [DOI] [PubMed] [Google Scholar]
- Wu JT, Kral JG. The NF-kappaB/IkappaB signaling system: a molecular target in breast cancer therapy. J Surg Res. 2005;123:158–169. doi: 10.1016/j.jss.2004.06.006. [DOI] [PubMed] [Google Scholar]
- www.clinicaltrials.gov.
- Zaremba T, Ketzer P, Cole M, Coulthard S, Plummer ER, Curtin NJ. Poly(ADP-ribose) polymerase-1 polymorphisms, expression and activity in selected human tumour cell lines. Br J Cancer I. 2009;101:256–62. doi: 10.1038/sj.bjc.6605166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.