Abstract
Collagen constitutes one third of the body protein in humans, reflecting its extraordinary role in health and disease. Of similar importance, therefore, are the idiosyncratic proteases that nature evolved for collagen remodeling. Intriguingly, the most efficient collagenases are those that enable clostridial bacteria to colonize their host tissues, but despite intense studies, the structural and mechanistic basis of these enzymes has remained elusive. Here we present the crystal structure of collagenase G from Clostridium histolyticum at 2.55 Å resolution. By combining the structural data with enzymatic and mutagenesis studies, we derive a conformational two-state model of bacterial collagenolysis, in which the recognition and unraveling of collagen microfibrils into triple helices as well as the unwinding of the latter go hand in hand with collagenase opening and closing.
By its mere quantity, collagen remodeling plays an outstanding physiological and pathological role in all vertebrates 1-3. Several independent lines of collagenases have evolved in nature to tackle the highly complex task of collagen hydrolysis, among them members of the mammalian matrix metalloproteases (MMPs) and bacterial collagenases. Interestingly, the most efficient collagenases are those found in clostridial bacteria which enable them to colonize and infiltrate the host tissue 4,5.
While considerable insight into the structure and mechanism of endogenous collagenases, belonging to the MMP family, has been obtained 6-14, exogenous bacterial collagenases so far have resisted a crystallographic analysis, partly due to the complexity of these multi-domain proteins. Therefore, much of the mechanistic basis for their enzymatic properties remained elusive despite their manifold medical and biotechnological use 15-17. Here we present the crystal structure and enzymatic analysis of a clostridial collagenase, resulting in a unified two-step model whereby recognized collagen microfibrils are mechanically unraveled and subsequently hydrolyzed.
RESULTS
Unexpected domain architecture of ColG
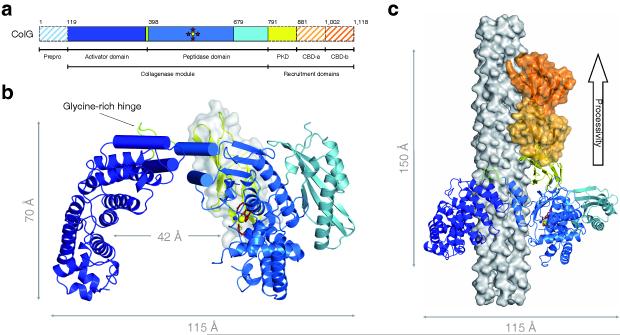
Clostridial, and other bacterial, collagenases have an approximate size of 120 kDa. Based on naturally occurring isoforms and bioinformatic analysis, their domain organization was expected to be composed of a pre-domain of variable length containing the export signal which is clipped in the mature protein; an N-terminal domain harboring the catalytic zinc (Tyr119 to Gly790; numbering corresponding to ColG from C. histolyticum); a variable number of polycystic kidney disease-like (PKD-like) domains (Ala791–Asn880); and one or more collagen binding domains (Glu881–Gly1001, Leu1002–Lys1118) 18, with structural information being available only on the latter domain 19 (Fig. 1a).
Figure 1. Domain organization and architecture of ColG.
(a) Schematic of the domain organization of ColG together with a functional annotation. The catalytic Zn2+ ion (yellow dot) and the catalytic residues (red stars) within the peptidase domain are indicated. The hatched domains (prepro-peptide and collagen binding domains) are not present in the crystal.
(b) Ribbon representation of the collagenase module, with identical color code as in a. The position of the PKD-like domain (yellow ribbon) at the rear of the peptidase domain is indicated in surface representation, reflecting a positional variance of up to 10 Å. The saddle-shaped collagenase is composed of an activator and a peptidase domain. The catalytic Zn2+ and the catalytic residues are highlighted by ball and stick representation. The seat of the saddle is formed by the distorted four-helix bundle, represented by four cylinders, and completed by the glycine-rich hinge, shown in green.
(c) Full-length model of ColG in complex with a collagen microfibril. The collagenase module (colored as in a) bound to a modeled collagen microfibril (in surface representation; grey) is shown in ribbon representation. The accessory domains are shown as surface representation. The two collagen binding domains (orange) were not present in the crystal, but oriented and positioned in order to satisfy the biochemically derived binding data, including the inferred binding epitopes. Direction of collagenase processivity is indicated at the right top.
Unexpectedly, the crystal structure discloses a distinct segmentation of the N-terminal collagenase module (Tyr119–Gly790), featuring a saddle-shaped two-domain architecture (Fig. 1b; Table 1). The smaller N-terminal saddle flap serves as an activator domain (Tyr119–Asp388), as to be discussed below, and comprises an array of 12 parallel α-helices. Starting with a distorted helix pair (Tyr119–Lys161), it continues with ten tandemly repeated HEAT motifs, ideally suited to generate a protein recognition interface 20. A solvent-exposed glycine-rich linker is positioned at the twist of the saddle seat (Gly389–Tyr397; Fig. 1b).
Table 1.
X-ray data collection and model refinement statistics.
| Zn-depleted (Selenomethionine MAD) |
Zn2+ bound |
Inhibitor complexed |
PKD-like domain |
||||
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | C121 |
| Cell dimensions | |||||||
| a, b, c (Å) | 57.8, 108.8, 182.0 |
57.3, 108.7, 181.5 |
58.1, 108.8, 181.0 |
68.2, 59.3, 55.3 |
|||
| α, β, γ (°) | 90, 90, 90 |
90, 90, 90 |
90, 90, 90 |
90, 125.6, 90 |
|||
| Peak | Inflection | High-Rem | Low-Rem | Peak | Remote | Remote | |
|
|
|||||||
| Wavelength | 0.97958 | 0.97969 | 0.97190 | 0.98783 | 1.28188 | 0.98793 | 0.98793 |
| Resolution (Å) | 50.3–2.75 | 50.3–2.75 | 36.6–2.75 | 48.3–2.55 | 35.6–2.80 | 35.7–3.25 | 40.5–1.18 |
| R merge | 0.116 (0.571) |
0.076 (0.639) |
0.084 (0.866) |
0.099 (0.695) |
0.094 (0.588) |
0.102 (0.300) |
0.066 (0.299) |
| I/σI | 10.1 (2.5) | 13.7 (2.3) | 12.5 (1.9) | 9.3 (1.8) | 12.4 (2.7) | 11.7 (4.7) | 13.9 (3.2) |
| Completeness (%) | 93.7 (70.5) | 93.7 (68.9) | 92.2 (67.9) | 88.1 (84.4) | 98.9 (98.1) | 99.5 (99.4) | 96.4 (78.4) |
| Redundancy | 6.4 (5.2) | 6.4 (5.1) | 6.4 (5.3) | 5.4 (5.0) | 6.9 (6.8) | 6.5 (6.2) | 6.0 (2.9) |
| Wilson B-factor | 76.7 | 79.9 | 81.3 | 62.2 | 72.6 | 69.0 | 9.7 |
| Refinement | |||||||
| Resolution (Å) | 38.9–2.55 | 34.4–2.80 | 34.8–3.25 | 39.61–1.18 | |||
| No. reflections | 33068 | 28436 | 18619 | 57689 | |||
| Rwork/ Rfree | 21.0 / 25.6 | 23.5 / 28.2 | 21.9 / 26.6 | 14.7 / 18.0 | |||
| No. atoms | |||||||
| Protein | 5434 | 5300 | 5341 | 2498 | |||
| Solvent | 181 | 158 | 137 | 364 | |||
| B-factors | |||||||
| Protein | 58.74 | 72.85 | 76.16 | 7.38 | |||
| Solvent | 73.44 | 67.75 | 75.20 | 38.77 | |||
| R.m.s deviations | |||||||
| Bond lengths (Å) | 0.011 | 0.0064 | 0.0068 | 0.0181 | |||
| Bond angles (°) | 1.152 | 0.908 | 0.915 | 1.805 | |||
| Ramachandran plot (%) | |||||||
| Most favorable | 93.1 | 91.2 | 91.9 | 97.2 | |||
| Allowed | 6.7 | 8.3 | 7.6 | 2.8 | |||
| Generously allowed | 0.0 | 0.2 | 0.3 | 0.0 | |||
| Disallowed | 0.2 | 0.3 | 0.2 | 0.0 | |||
Highest resolution shell is shown in parenthesis.
The peptidase domain resembles a thermolysin-like fold
The subsequent catalytic subdomain (Asp398–Gln669) adopts a thermolysin-like peptidase (TLP) fold of mixed α and β type, most closely resembling the structures of Leukotriene A4 hydrolase and F3 protease 21,22, and is accompanied by a catalytic helper subdomain. The flanking α-helix pairs of the activator domain and the catalytic subdomain combine to form a distorted four-helix bundle and thus comprise the seat of the saddle topology, which is completed by the glycine-rich linker. These structural elements latch the relative spatial arrangement of the peptidase domain and the N-terminal activator, which will turn out to be critical for collagen binding and triple-helix unraveling.
Topologically equivalent to thermolysin, the five-stranded β-pleated sheet of the catalytic subdomain presents the edge strand mediating the canonical substrate-recognition motif, as described below in more detail. Leu517–Leu534 form the “central helix” of the TLP fold 23 and contain the Zn2+-binding H523EXXH527 motif. The third Zn2+-ligand Glu555 resides on the neighboring helix, defining ColG as a gluzincin 24.
Another linker segment of eight amino acids (Gly670 to Asp678) connects the catalytic subdomain with a catalytic helper subdomain (Asp679–Asp788). The four-stranded antiparallel β-sheet partly connects to and extends the β-sheet of the catalytic subdomain, whereas the two antiparallel α-helices of the helper subdomain cover the β-sheet to the solvent.
Given the compact quaternary arrangement of the helper and catalytic subdomains, we refer to the combination of both subdomains as the peptidase domain. This interpretation is supported by the fact that the catalytic helper is indispensable for proper folding and full peptidase activity.
Position and hybrid structure of the PKD-like domain
Crystal packing as well as low-resolution contrast electron density analysis enabled us to position the PKD-like domain at the rear of the peptidase domain in Fig. 1b, with its exact orientation remaining flexible. We could further derive high resolution (1.18 Å) structural information from crystals of the isolated PKD-like domain. The PKD-like domain adopts a two-tiered β-barrel structure. Intriguingly, we found the conserved Trp833 buried inside the β-barrel (Supplementary Fig. 1). At first sight this orientation appears conflicting with literature data, reporting the critical role of Trp833 in collagen binding and swelling 25. However, our structure can reconcile this apparent conflict, as the elimination of the indole ring of Trp833 would destabilize and collapse the barrel structure and thus destroy the structural framework necessary for specific collagen binding (Supplementary Fig. 1).
The remaining C-terminal collagen binding domains (CBD) were not present in our crystal form; fortunately, crystal structure information on the isolated C-terminal CBD has been obtained previously 19, enabling us to construct a full-length structural model of ColG (Fig. 1c).
Enzymatic assessment of the collagenase domains
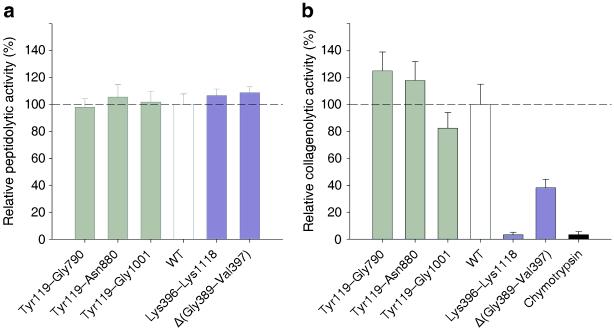
The unexpected domain architecture prompted us to re-investigate the functional assignment of the individual subdomains. Firstly, the structure analysis suggested that the enzymatic activity should be contained within the peptidase domain (Asp398 to Gly790). Indeed, an activator deletion construct, Lys396–Lys1118, showed 100% activity against small peptidic substrates as compared to the full-length ColG protease, thus confirming this domain to represent the peptidase domain of clostridial collagenases (Fig. 2a). Furthermore and remarkably, the peptidase domain was completely inactive towards collagen substrates. Full collagenolytic activity is, however, contained in the segment Tyr119–Gly790, comprising the activator and peptidase domains. This segment thus represents the collagenase module of the protein (Fig. 1a and Fig. 2b).
Fig. 2. The activator domain is necessary for the degradation of collagen.
(a) Histogram showing the relative peptidolytic activities of ColG variants, Tyr119–Gly790, Tyr119–Asn880, Tyr119–Gly1001, Lys395–Lys1118, and Δ(Gly389–Val397), normalized to the wild type (WT) protein, against FALGPA. Lys395–Lys1118 represents a variant lacking the N-terminal activator domain, whereas the glycine-rich hinge is short cut in Δ(Gly389–Val397). Data are graphed as mean ± s.d. (n=6).
(b) Histogram showing the collagenolytic activities given as relative initial velocities of Tyr119–Gly790, Tyr119–Asn880, Tyr119–Gly1001, Lys395–Lys1118, and Δ(Gly389–Val397), normalized to the WT, against fluorescein-labeled collagen type I. Chymotrypsin serves as a negative control, indicating that the collagen substrate was not denatured. Data are graphed as mean ± s.d. (n=36).
While these biochemical findings are consistent with, and were predicted by, our crystal structures, they are in seeming contrast with both earlier reports on clostridial collagenases 5 and the situation in mammalian collagenases, see also our discussion below. In the latter case, the analogous collagen binding domain, the hemopexin-like domain, is critical for collagen binding, unwinding of the triple helix, and catalytic turnover 26. The C-terminal accessory domains serve to recruit and partly to swell insoluble collagen without unwinding its triple-helical structure 19,25. These Ca2+-dependent recruitment domains tend to accelerate the hydrolysis of insoluble collagen substrate at low component concentrations.
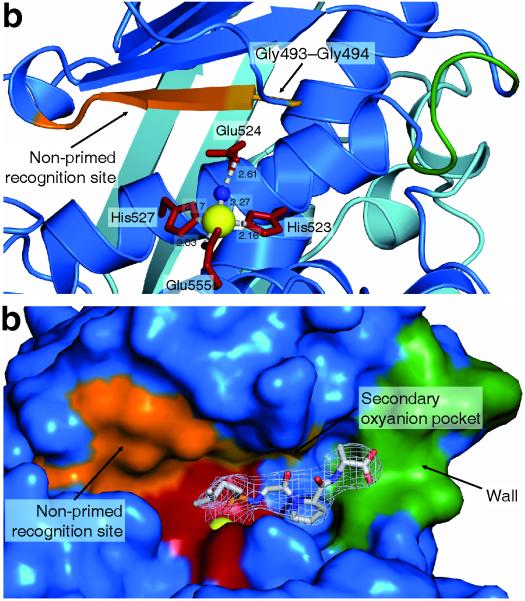
Catalytic Zn2+ site
We found the catalytic Zn2+ tetrahedrally coordinated by the side chains of His523, His527 and Glu555 as well as a water molecule which was further hydrogen-bonded to the general base Glu524 (Fig. 3a). Surprisingly, however, we also crystallized ColG in a zinc-depleted form, suggesting the zinc binding to be atypically weak 27. The ligating bond distances for the zinc (in brackets: the inhibitor complex) are 2.16 (2.08) Å, 2.17 (2.09) Å and 2.03 (2.09) Å to the three proteinacious ligands His523, His527 and E555, respectively. These distances tend to be somewhat larger than the analogous distances in thermolysin and related peptidases where the average ligation lengths are found to be at a distance of 2.00 Å ± 0.03 Å 21-23,27.
Figure 3. Mapping the peptidase active site.
(a) Ribbon representation of the active site. Three protein residues (His523, His527, Glu555; shown in red) and a water molecule (blue sphere) tetrahedrally coordinate the catalytic Zn2+ (yellow sphere). Glu524 hydrogen-bonds to the water molecule. The non-primed substrate recognition sites are formed by the antiparallel edge strand extending from Gly494 to Glu498 (orange). The S1′ oxyanion site is formed by the amides of Gly493–Gly494 (bright orange).
(b) Accessible surface representation of the ColG active site containing the inhibitor isoamyl-phosphonyl-Gly-Pro-Ala in a stick representation, with identical orientation and color code as in a. The inhibitor is overlaid with the experimental 2Fo-Fc omit electron density, contoured at the 1 σ over the mean (light grey). Substrate recognition is governed at the non-primed sites by the antiparallel edge strand; by the catalytic Zn2+ (dark red with yellow sphere); and by the secondary S1′ oxyanion pocket. A prominent wall delimits the S3′ site and explains the tripeptidyl-carboxypeptidase activity of ColG.
The partial zinc occupancy opens an unexpected route to tune enzyme activity; this regulation mechanism appears particularly sensible for a protease class that lacks a prodomain which would confer zymogen-like latency. In line with these observations, we found that His6-tagged protein variants show reduced activity, which can be explained by partial Zn2+ extraction 28. Importantly, the Zn2+ site was fully occupied in the presence of a substrate analogue inhibitor, consistent with the additional zinc coordination by the inhibitor. This finding suggests that substrates contribute to the stabilization, and correct positioning of the catalytic Zn2+, an effect which is expected to be kcat-dependent, consistent with our observations 28. Such substrate-assisted activation is indeed observed with substrates extending from P3 to P3′ residues, but not for the synthetic furylacryloyl-Leu+Gly-Pro-Ala (FALGPA) peptide lacking the non-primed edge strand stabilization 29.
Belonging to the metzincin family, MMPs contain a strictly conserved methionine residue that serves as a “hydrophobic basement” to the catalytic zinc. Positioned one helical turn downstream to the catalytic Glu555, Ala558 of ColG is located at a 3D position corresponding to the conserved metzincin methionine, with its Cβ side chain pointing towards the zinc at ~4.25 Å distance. Ala558 is conserved within clostridial collagenases. From the structure it seems unlikely that larger hydrophobic residues can be tolerated at this position.
Peptidic substrate recognition and processing
The comparison of apo, unliganded, and complexed active sites show a more than 3 Å contraction movement of the two TLP-half domains upon Zn2+ loading and inhibitor binding. The peptidomimetic inhibitor isoamyl-phosphonyl-Gly-Pro-Ala binds to the “edge strand” in an antiparallel orientation, consistent with TLP-like peptidase-inhibitor recognition. Preceding the edge strand (Leu495-Glu498) of the TLP β-sheet, two glycine residues form the entrance to the canonical antiparallel substrate recognition strand motif (Fig. 3a). This Gly493–Gly494 motif forms the S1′ recognition sites and thus serves as the shaping die for the conformation of the P1′ residue which can only be adopted by a glycine at P1′ position 29,30.
The complex structure of ColG with the peptidomimetic inhibitor revealed two additional noteworthy consequences resulting from the unique double glycine S1′ site in clostridial collagenases: First, the amides of both Gly493 and Gly494 point towards the P1′ carbonyl oxygen and thereby form a secondary S1′ oxyanion pocket (Fig. 3a). Second, the unusual topology of the strong S1′-P1′ oxyanion interaction dominates, yet misaligns the substrate binding geometry at the Zn2+ and the non-primed recognition sites. This will result in an unproductive binding of short substrates lacking P2 and P3 residues and explains why ColG starkly prefers substrates containing P2 and P3 residues 29,30. By virtue of the multi-valent interactions with the edge strand Gly493 to Glu498, N-terminally extended substrates can be bent and thus assist their hydrolysis.
The importance of the secondary oxyanion pocket (Gly493–Gly494) is witnessed by a serendipitous G494V mutation in the closely related collagenase H from C. histolyticum which completely inactivated the protease 28.
While the non-primed substrate recognition sites are defined by the backbone interaction with the edge strand (Gly494–Glu498), the wide cleft at the S3 to S1 sites allow for slightly alternative binding geometries, depending on the nature of the P3 to P1 substrate residues. In particular, prolines and hydroxyprolines, often found in collagen, cannot adopt an ideal antiparallel β-strand with the edge strand, but can still be accommodated, as indicated in figure 3b.
A loop segment Gln511–Phe515 preceding the central helix forms a prominent wall-like feature at the primed substrate recognition sites (Fig. 3b). By its geometric distance to the catalytic zinc, the wall enforces a major kink at the P4′ position of longer substrates, making its binding energetically less favorable. The wall thus serves as molecular ruler that explains the observed tripeptidyl-carboxypeptidase activity of clostridial collagenases 29.
Collagen recognition, unwinding, and processing
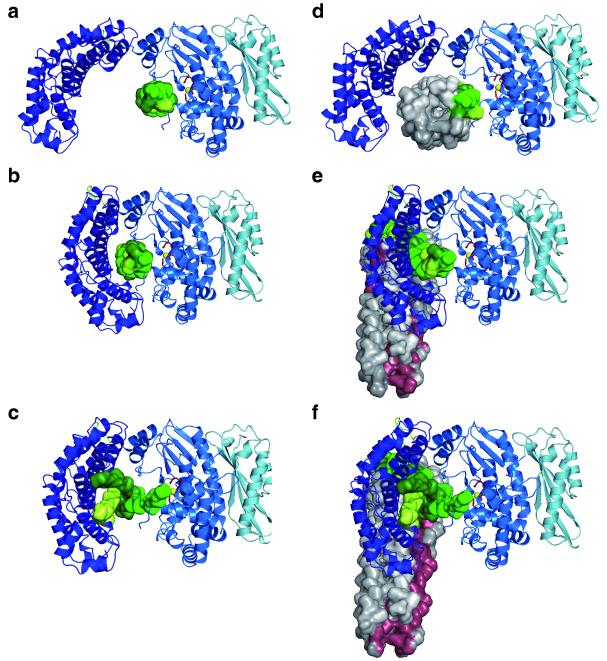
The integration of the crystal structure with the observed enzymatic properties of the different ColG variants (Fig. 2) suggests a two-step mechanism, whereby the N-terminal activator domain cooperates with the peptidase domain in both collagen triple helix and microfibril recognition and processing. The activator and peptidase domains, forming the two “saddle flaps”, have a distance of ~40 Å, whereas the diameter of the collagen triple helix is only approximately 15 Å 1 (Fig. 4a). Naively, one should therefore expect that the activator domain is neither involved nor required in triple helix recognition and processing – which is not the case (Fig. 2b). Consequently, one must conclude that the collagenase can adopt two preferred conformational states: In addition to the crystallized open state, there exists a closed state which allows the collagen triple helix to contact both the activator and peptidase domain (Fig. 4b). The closed state is latched by two major contacts at the bottom of the saddle and by an alternative 4-helix-bundle arrangement at the saddle seat. Only now are the activator HEAT-repeats able to interact with triple-helical collagen, and initiate the unwinding of the triple-helix α-chains which are processively cleaved 31,32. Consistent with these data we postulate that the activator and peptidase domains remain mostly closed during collagen cleavage, but relax to the open ground state, once the collagen is degraded, which constituted the source of the attractive interaction for collagenase closing (Fig. 4c).
Figure 4. Unified processing model of triple-helical and microfibrillar collagen.
(a) A collagen triple helix (green) initially docks to the peptidase domain of collagenase (colored as in Fig. 1). In the open state, the activator (dark blue) cannot interact with the substrate, with no hydrolysis occurring.
(b) Step 2, closed conformation, showing the activator HEAT repeats interacting with the triple helix, which is a prerequisite for collagen hydrolysis.
(c) Step 3, semi-opened conformation, allowing for exchange and processive degradation of all three α-chains 31,32. Once the triple helix is completely cleaved, the collagenase can relax back to the open ground state conformation, as found in our crystals, and thus complete the catalytic cycle.
(d) Collagenase with a docked collagen microfibril (grey; based on pdb entry 4clg). The micro-fibril typically consists of five triple-helical molecules 19; the triple helix analogous to a is indicated in green.
(e) Step 2, closed conformation with all triple helices but one (green) being expelled from the collagenase. The microfibril wound caused by the triple-helix stripping is indicated in red.
(f) Step 3, semi-opened conformation allowing for completely processing the triple helix, indicated in green. After that, the collagenase will relax back to the open state and only then allow the remaining part of the microfibril to enter the collagenase.
To test this chewing mechanism of collagen degradation, we constructed a variant where the glycine-rich hinge is short cut, Δ(Gly389–Val397). This variant should interfere with the closed conformation (Fig. 4b), and consequently with collagen chewing and digesting. While fully active towards small peptidic substrates (Fig. 2a), this variant exhibits only approximately one third of the wild type activity towards collagen substrates (Fig. 2b), in full agreement with our chew & digest mechanism.
Recognition, unwinding, and processing of microfibrils
One of the most glaring findings of the crystal structure analysis is the perfect match of the observed ground state conformation with the dimensions of collagen microfibrils, having a diameter of approximately 40 Å 33,34 (Fig. 4d). Albeit a bit more speculative, our model provides a strikingly simple mechanism how collagen microfibrils are proteolytically processed. Upon transition from the microfibril-loaded open state to the closed state, ColG will crimp the microfibril with its pliers, with only one triple helix remaining within the collagenase pliers (Fig. 4e). Analogous to the triple-helical processing, one triple helix remains embraced by the activator and peptidase domains until it is completely clipped, after which the collagenase will relax to the open conformation, allowing the remaining microfibril to enter the collagenase (Fig. 4f).
DISCUSSION
Collagenases are optimized to cleave glycine-rich substrates
One distinct difference between MMPs and ColG relates to the glycine residue which binds to the S1 and S1′ site, respectively. Consistent with transition state theory of enzyme catalysis, the substrate is bound to the active site in a highly distorted conformation (Fig. 3a). We found that Gly on P1′ versus P1. The distortion is counteracted by the flexibility of glycine residues, ubiquitously present in collagen. In ColG, this challenge is addressed by strong binding of the glycine P1′ carbonyl to the secondary oxyanion pocket.
Additionally, the position and orientation of the P1′ amid-nitrogen is fixed within the peptide plane by the P1 carbonyl via the classical oxyanion pocket formed by the Zn2+. The ColG solution to the flexible glycine-challenge may thus allow a more efficient substrate distortion as compared to the MMPs, because both dihedral angles (φ and ψ) are fixed upon substrate binding.
Entropically driven collagenolysis
Unraveling collagen (micro)fibrils and unwinding triple-helical collagen are prerequisites for collagen cleavage, both for mammalian MMPs and clostridial collagenases. There are two possible solutions how to store the required mechanical energy, namely either in the (i) protease or the (ii) substrate – or both. The protease-oriented view is supported by the crystal structure of the collagen binding domain of clostridial collagenase G. Here, a Ca2+-triggered conformational rearrangement 19 may implement the energy reservoir which could contribute to the triple-helicase activity of these enzymes.
However, our model suggests and emphasizes the substrate as the energy source powering the molecular machinery (Fig. 4). The required free energy is stored in the substrate itself: Triple-helical collagen is hydrated by at least two ordered water layers that are released upon its cleavage 35. With the accompanying entropy release the substrate thus assists its own catalytic turnover. Importantly, the release of water, and entropy, is independent of the exact mechanism of the triple-helicase and peptidase activity, and as such should hold true for all collagenases, including MMPs.
Transferring mechanistic insights from ColG to MMPs
Naturally, the question arises whether and to which extent the structural insights into the clostridial collagenase (Supplementary Fig. 2a) can be transferred to the mechanism of action in MMPs. It is particularly tempting to speculate which MMP domain may take the role of the activator domain in ColG. To us there are two interesting and alternative hypothetical models. In the first scenario, the MMP hemopexin domain takes the role of the activator domain (Supplementary Fig. 2b), reflecting their strict requirement for collagenase activity26,36.
The second scenario refers to the observation that MMPs form dimers 37-42. In this analogy, each the activator and peptidase domains of ColG correspond to a separate MMP catalytic domain, whereas the MMP hemopexin-like domains correspond to the PKD and collagen binding domains (Supplementary Fig. 2c). This model explains the necessity for hemopexin-like domain for collagenase activity by its critical role in MMP dimerisation 38,43,44. The latter model should be testable by chimeric MMP constructs, e.g. by replacing the hemopexin domain of an MMP collagenase with a suitable dimerisation motif such as GST.
Processivity of clostridial collagenases
Clostridial collagenases were traditionally divided into classes, class I and class II. Whereas the latter group is highly active towards peptidic substrates, class I enzymes such as ColG have a particular preference for collagen substrate degradation in a processive manner 4,17,28. For fibrillar collagen substrates, processivity includes two dimensions, namely the complete and the flush cut of a (micro-)fibril at one site of the substrate. For triple-helical substrates, processivity implies multiple cleavage events along the substrate, a process described as inch-worming by Overall and colleagues 45. The structure suggests how the accessory domains assist both aspects of processivity. When cleaving microfibrillar collagen, the activator and peptidase domains have to open to allow for the remaining microfibril to enter the collagenase active site (Fig. 3d-f). Here, the accessory domains avoid an inadvertant shift of the substrate. On triple helical collagen, but also on microfibrils, the accessory domains help to direct the collagenase module along the substrate. Importantly, this directed movement implies a polarity with the accessory domains leading and paving the way of processing (Fig. 1c). Furthermore, given the tri-carboxypeptidase activity of ColG, we suggest that the collagen processing occurs from the C-terminus of the collagen substrate to its N-terminus, consistent with the antiparallel substrate recognition by the edge strand (Fig. 2).
Potential impact
The presented structural and functional data provide, for the first time, an integrated mechanistic model of collagen recognition and processing by bacterial collagenases; this model may also prove as a valuable model for mechanistic studies on vertebrate collagenases. Given the use of collagenases in diverse biotechnological processes, including islet transplantation 16,46, the presented results open a structure-based strategy to optimize these processes, for instance by engineering substrate preferences or designing hyper-active collagenases, e.g. by combining key elements responsible for the catalytic properties of class I and class II collagenases. Another important opportunity resulting from this study is in the structure-based design of collagenase inhibitors 47. This approach is particularly attractive, as it is expected to enable a step change in “anti-biotics” development: Rather than attacking the pathogen directly, clostridial collagenase inhibitors block the colonization and infiltration of the host by the pathogens only, thereby escaping the Darwinian selection pressure which is encountered in classical antibiotics development.
METHODS
Protein preparation
Collagenase cloning, expression, purification, crystallization, and diffraction data collection, were performed as described previously 18,48. The PKD-like domain (Ile799–Asn880) was cloned, expressed, and purified analogously. Selenomethionine labeled ColG (Tyr119–Asn880) was expressed in E. coli B834 (DE3) cells overnight at 25 °C in ready-to-use selenomethionine medium (Molecular Dimensions), and purified as described for unlabelled protein.
Biochemical characterization
The peptidolytic assay was performed with FALGPA (Fluka, Sigma Aldrich) as substrate at a final concentration of 0.4 mM, as described previously 28. Collagenolytic activity was assayed with the EnzChek Gelatinase/Collagenase Assay Kit (Invitrogen), using fluorescein-labeled collagen type I from bovine skin according to the manufacturer’s protocol. In short, 0.4, 0.8, and 1.0 nM protein were incubated with 0.2 μg ml−1 substrate and fluorescence was measured in 30 s intervals using the microplate reader Infinite 200 (Tecan) (485 ± 9 nm; 530 ± 20 nm) at room temperature. For α-chymotrypsin from bovine pancreas (Fluka, Sigma Aldrich), 1.0, 10.0 and 100.0 mM were tested accordingly. Initial velocities were calculated using nonlinear regression 49.
Crystallization and data collection
Crystals of native, Zn2+ depleted, and inhibitor-complexed ColG (Tyr119–Asn880) were grown by sitting-drop vapor diffusion from 7.5 mg ml−1 protein solution, 20–23% PEG 3350, 175–225 mM Na3C6H5O7, pH 8.3, at 20 °C. Crystals of the PKD-like domain were obtained analogously from 70 mg ml−1 protein solution, 2.5 M C3H2O4Na2, 10 mM CaCl2, pH 7.5. Crystals were flash-frozen in a stream of nitrogen gas at 100 K, in which ColG crystals were supplemented with 30% glucose to the mother liquor for cryoprotection. Diffraction data were collected at beamline 14-1 at BESSY (Berlin), and 14-4 at ESRF (Grenoble), and processed within the CCP4 suite 50. Data collection and processing statistics are summarized in Table 1.
Structure determination
Using the Auto-Rickshaw four wavelength MAD protocol 51, heavy-atom positions were located by using the program SHELXD 52 and refined with MLPHARE 50, and the density improved by solvent flattening using DM 53. The dataset resulted in an initial model that was iteratively improved by a combination of refinement using REFMAC5 54 with the translation libration screw (TLS) description of the anisotropic rigid body motion 55, and manual model building in COOT 56. Electron densities at uninterpretable loop regions were sharpened by canonical electron densities, leading to effectively improved optical resolution, enabling their interpretation. The refinement was monitored throughout by using an Rfree calculated with 5% of the unique reflections. Refinement and Ramachandran statistics are summarized in Table 1. Molecular figures were created with PyMOL (http://pymol.sourceforge.net) 57.
Structure validation
The quality of all models was checked using the programs PROCHECK 58, WHATCHECK 59, and NQ-Flipper 60.
METHODS-ONLY-REFERENCES
- 48.Eckhard U, Nüss D, Ducka P, Schönauer E, Brandstetter H. Crystallization and preliminary X-ray characterization of the catalytic domain of collagenase G from Clostridium histolyticum. Acta Crystallograph. Sect. F Struct. Biol. Cryst. Commun. 2008;64:419–21. doi: 10.1107/S1744309108010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Briers Y, Lavigne R, Volckaert G, Hertveldt K. A standardized approach for accurate quantification of murein hydrolase activity in high-throughput assays. J. Biochem. Biophys. Methods. 2007;70:531–3. doi: 10.1016/j.jbbm.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 50.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 51.Panjikar S, Parthasarathy V, Lamzin VS, Weiss MS, Tucker PA. Auto-Rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr. D Biol. Crystallogr. 2005;61:449–57. doi: 10.1107/S0907444905001307. [DOI] [PubMed] [Google Scholar]
- 52.Sheldrick GM. A short history of SHELX. Acta Crystallogr. A. 2008;64:112–22. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 53.Cowtan KD, Zhang KY. Density modification for macromolecular phase improvement. Prog. Biophys. Mol. Biol. 1999;72:245–70. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 54.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 55.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439–50. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 56.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeLano WL. The case for open-source software in drug discovery. Drug Discov. Today. 2005;10:213–7. doi: 10.1016/S1359-6446(04)03363-X. [DOI] [PubMed] [Google Scholar]
- 58.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–67. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 59.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 60.Weichenberger CX, Sippl MJ. NQ-Flipper: recognition and correction of erroneous asparagine and glutamine side-chain rotamers in protein structures. Nucleic Acids Res. 2007;35:W403–6. doi: 10.1093/nar/gkm263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary Material
ACKNOWLEDGMENTS
We thank Roche (Penzberg) for providing plasmids; Paulina Klemm for cloning a Δ-activator construct; Simon Ginzinger and Manfred Sippl for help with canonical electron density expansion; Luis Moroder and Hideaki Nagase for valuable discussions; our anonymous reviewers for extraordinarily helpful suggestions and clarifications; staff at the synchrotrons ESRF (Grenoble), DESY (Hamburg), and BESSY (Berlin) for expert help in data collection; and the Austrian science foundation FWF for funding (H.B.; project P20582).
Footnotes
Accession codes. Protein Data Bank: Coordinates have been deposited with accession codes 2y3u, 2y50, 2y6i and 2y72.
Competing financial interests The authors declare no competing financial interests.
REFERENCES
- 1.Ottani V, Martini D, Franchi M, Ruggeri A, Raspanti M. Hierarchical structures in fibrillar collagens. Micron. 2002;33:587–96. doi: 10.1016/s0968-4328(02)00033-1. [DOI] [PubMed] [Google Scholar]
- 2.Brozek J, Grande F, Anderson JT, Keys A. Densitometric analysis of body composition: Revision of some quantitative assumptions. Ann. N. Y. Acad. Sci. 1963;110:113–40. doi: 10.1111/j.1749-6632.1963.tb17079.x. [DOI] [PubMed] [Google Scholar]
- 3.Sweeney SM, et al. Candidate cell and matrix interaction domains on the collagen fibril, the predominant protein of vertebrates. J. Biol. Chem. 2008;283:21187–97. doi: 10.1074/jbc.M709319200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bond MD, Van Wart HE. Purification and separation of individual collagenases of Clostridium histolyticum using red dye ligand chromatography. Biochemistry. 1984;23:3077–85. doi: 10.1021/bi00308a035. [DOI] [PubMed] [Google Scholar]
- 5.Matsushita O, et al. A study of the collagen-binding domain of a 116-kDa Clostridium histolyticum collagenase. J. Biol. Chem. 1998;273:3643–8. doi: 10.1074/jbc.273.6.3643. [DOI] [PubMed] [Google Scholar]
- 6.Borkakoti N, et al. Structure of the catalytic domain of human fibroblast collagenase complexed with an inhibitor. Nat. Struct. Biol. 1994;1:106–10. doi: 10.1038/nsb0294-106. [DOI] [PubMed] [Google Scholar]
- 7.Li J, et al. Structure of full-length porcine synovial collagenase reveals a C-terminal domain containing a calcium-linked, four-bladed beta-propeller. Structure. 1995;3:541–9. doi: 10.1016/s0969-2126(01)00188-5. [DOI] [PubMed] [Google Scholar]
- 8.Stams T, et al. Structure of human neutrophil collagenase reveals large S1′ specificity pocket. Nat. Struct. Biol. 1994;1:119–23. doi: 10.1038/nsb0294-119. [DOI] [PubMed] [Google Scholar]
- 9.Iyer S, Visse R, Nagase H, Acharya KR. Crystal structure of an active form of human MMP-1. J. Mol. Biol. 2006;362:78–88. doi: 10.1016/j.jmb.2006.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adhikari AS, Chai J, Dunn AR. Mechanical load induces a 100-fold increase in the rate of collagen proteolysis by MMP-1. J. Am. Chem. Soc. 2011 doi: 10.1021/ja109972p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han S, et al. Molecular mechanism of type I collagen homotrimer resistance to mammalian collagenases. J. Biol. Chem. 2010;285:22276–81. doi: 10.1074/jbc.M110.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minond D, Lauer-Fields JL, Nagase H, Fields GB. Matrix metalloproteinase triple-helical peptidase activities are differentially regulated by substrate stability. Biochemistry. 2004;43:11474–81. doi: 10.1021/bi048938i. [DOI] [PubMed] [Google Scholar]
- 13.Chung L, et al. Collagenase unwinds triple-helical collagen prior to peptide bond hydrolysis. EMBO J. 2004;23:3020–30. doi: 10.1038/sj.emboj.7600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tam EM, Moore TR, Butler GS, Overall CM. Characterization of the distinct collagen binding, helicase and cleavage mechanisms of matrix metalloproteinase 2 and 14 (gelatinase A and MT1-MMP): the differential roles of the MMP hemopexin C domains and the MMP-2 fibronectin type II modules in collagen triple helicase activities. J. Biol. Chem. 2004;279:43336–44. doi: 10.1074/jbc.M407186200. [DOI] [PubMed] [Google Scholar]
- 15.Fecteau KA, Haffner JC, Eiler H. The potential of collagenase as a new therapy for separation of human retained placenta: hydrolytic potency on human, equine and bovine placentae. Placenta. 1998;19:379–83. doi: 10.1016/s0143-4004(98)90077-7. [DOI] [PubMed] [Google Scholar]
- 16.Shi L, Carson D. Collagenase Santyl ointment: a selective agent for wound debridement. J. Wound Ostomy Continence Nurs. 2009;36:S12–6. doi: 10.1097/WON.0b013e3181bfdd1a. [DOI] [PubMed] [Google Scholar]
- 17.Hesse F, Burtscher H, Popp F, Ambrosius D. Recombinant enzymes for islet isolation: purification of a collagenase from Clostridium histolyticum and cloning/expression of the gene. Transplant. Proc. 1995;27:3287–9. [PubMed] [Google Scholar]
- 18.Ducka P, et al. A universal strategy for high-yield production of soluble and functional clostridial collagenases in E. coli. Appl. Microbiol. Biotechnol. 2009;83:1055–65. doi: 10.1007/s00253-009-1953-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson JJ, Matsushita O, Okabe A, Sakon J. A bacterial collagen-binding domain with novel calcium-binding motif controls domain orientation. EMBO J. 2003;22:1743–52. doi: 10.1093/emboj/cdg172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groves MR, Barford D. Topological characteristics of helical repeat proteins. Curr. Opin. Struct. Biol. 1999;9:383–9. doi: 10.1016/s0959-440x(99)80052-9. [DOI] [PubMed] [Google Scholar]
- 21.Kyrieleis OJ, Goettig P, Kiefersauer R, Huber R, Brandstetter H. Crystal structures of the tricorn interacting factor F3 from Thermoplasma acidophilum, a zinc aminopeptidase in three different conformations. J. Mol. Biol. 2005;349:787–800. doi: 10.1016/j.jmb.2005.03.070. [DOI] [PubMed] [Google Scholar]
- 22.Thunnissen MM, Nordlund P, Haeggstrom JZ. Crystal structure of human leukotriene A(4) hydrolase, a bifunctional enzyme in inflammation. Nat. Struct. Biol. 2001;8:131–5. doi: 10.1038/84117. [DOI] [PubMed] [Google Scholar]
- 23.Matthews BW, Jansonius JN, Colman PM, Schoenborn BP, Dupourque D. Three-dimensional structure of thermolysin. Nat New Biol. 1972;238:37–41. doi: 10.1038/newbio238037a0. [DOI] [PubMed] [Google Scholar]
- 24.Jung CM, et al. Identification of metal ligands in the Clostridium histolyticum ColH collagenase. J. Bacteriol. 1999;181:2816–22. doi: 10.1128/jb.181.9.2816-2822.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang YK, et al. Mechanistic insight into the function of the C-terminal PKD domain of the collagenolytic serine protease deseasin MCP-01 from deep sea Pseudoalteromonas sp. SM9913: binding of the PKD domain to collagen results in collagen swelling but does not unwind the collagen triple helix. J. Biol. Chem. 2010;285:14285–91. doi: 10.1074/jbc.M109.087023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez-Lopez R, Alexander CM, Behrendtsen O, Breathnach R, Werb Z. Role of zinc-binding- and hemopexin domain-encoded sequences in the substrate specificity of collagenase and stromelysin-2 as revealed by chimeric proteins. J. Biol. Chem. 1993;268:7238–47. [PubMed] [Google Scholar]
- 27.Vallee BL, Auld DS. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry. 1990;29:5647–59. doi: 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- 28.Eckhard U, et al. Biochemical characterization of the catalytic domains of three different Clostridial collagenases. Biol. Chem. 2009;390:11–8. doi: 10.1515/BC.2009.004. [DOI] [PubMed] [Google Scholar]
- 29.Mookhtiar KA, Steinbrink DR, Van Wart HE. Mode of hydrolysis of collagen-like peptides by class I and class II Clostridium histolyticum collagenases: evidence for both endopeptidase and tripeptidylcarboxypeptidase activities. Biochemistry. 1985;24:6527–33. doi: 10.1021/bi00344a033. [DOI] [PubMed] [Google Scholar]
- 30.Hu Y, et al. Rapid determination of substrate specificity of Clostridium histolyticum beta-collagenase using an immobilized peptide library. J. Biol. Chem. 2002;277:8366–71. doi: 10.1074/jbc.M111042200. [DOI] [PubMed] [Google Scholar]
- 31.French MF, Bhown A, Van Wart HE. Identification of Clostridium histolyticum collagenase hyperreactive sites in type I, II, and III collagens: lack of correlation with local triple helical stability. J. Protein Chem. 1992;11:83–97. doi: 10.1007/BF01025095. [DOI] [PubMed] [Google Scholar]
- 32.French MF, Mookhtiar KA, Van Wart HE. Limited proteolysis of type I collagen at hyperreactive sites by class I and II Clostridium histolyticum collagenases: complementary digestion patterns. Biochemistry. 1987;26:681–7. doi: 10.1021/bi00377a004. [DOI] [PubMed] [Google Scholar]
- 33.Hulmes DJ. Building collagen molecules, fibrils, and suprafibrillar structures. J. Struct. Biol. 2002;137:2–10. doi: 10.1006/jsbi.2002.4450. [DOI] [PubMed] [Google Scholar]
- 34.Wess TJ. Collagen fibril form and function. Adv. Protein Chem. 2005;70:341–74. doi: 10.1016/S0065-3233(05)70010-3. [DOI] [PubMed] [Google Scholar]
- 35.Berisio R, Vitagliano L, Mazzarella L, Zagari A. Crystal structure of a collagen-like polypeptide with repeating sequence Pro-Hyp-Gly at 1.4 A resolution: implications for collagen hydration. Biopolymers. 2000;56:8–13. doi: 10.1002/1097-0282(2000)56:1<8::AID-BIP1037>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 36.Chung L, et al. Identification of the (183)RWTNNFREY(191) region as a critical segment of matrix metalloproteinase 1 for the expression of collagenolytic activity. J. Biol. Chem. 2000;275:29610–7. doi: 10.1074/jbc.M004039200. [DOI] [PubMed] [Google Scholar]
- 37.Clegg PD, Burke RM, Coughlan AR, Riggs CM, Carter SD. Characterisation of equine matrix metalloproteinase 2 and 9; and identification of the cellular sources of these enzymes in joints. Equine Vet. J. 1997;29:335–42. doi: 10.1111/j.2042-3306.1997.tb03136.x. [DOI] [PubMed] [Google Scholar]
- 38.Itoh Y, et al. Cell surface collagenolysis requires homodimerization of the membrane-bound collagenase MT1-MMP. Mol. Biol. Cell. 2006;17:5390–9. doi: 10.1091/mbc.E06-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Itoh Y, Ito N, Nagase H, Seiki M. The second dimer interface of MT1-MMP, the transmembrane domain, is essential for ProMMP-2 activation on the cell surface. J Biol Chem. 2008;283:13053–62. doi: 10.1074/jbc.M709327200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malla N, Sjoli S, Winberg JO, Hadler-Olsen E, Uhlin-Hansen L. Biological and pathobiological functions of gelatinase dimers and complexes. Connect. Tissue Res. 2008;49:180–4. doi: 10.1080/03008200802151755. [DOI] [PubMed] [Google Scholar]
- 41.Olson MW, et al. Characterization of the monomeric and dimeric forms of latent and active matrix metalloproteinase-9. Differential rates for activation by stromelysin 1. J. Biol. Chem. 2000;275:2661–8. doi: 10.1074/jbc.275.4.2661. [DOI] [PubMed] [Google Scholar]
- 42.Saffarian S, Collier IE, Marmer BL, Elson EL, Goldberg G. Interstitial collagenase is a Brownian ratchet driven by proteolysis of collagen. Science. 2004;306:108–11. doi: 10.1126/science.1099179. [DOI] [PubMed] [Google Scholar]
- 43.Cha H, Kopetzki E, Huber R, Lanzendorfer M, Brandstetter H. Structural basis of the adaptive molecular recognition by MMP9. J. Mol. Biol. 2002;320:1065–79. doi: 10.1016/s0022-2836(02)00558-2. [DOI] [PubMed] [Google Scholar]
- 44.Tochowicz A, et al. The dimer interface of the membrane type 1 matrix metalloproteinase hemopexin domain: crystal structure and biological functions. J. Biol. Chem. 2010;286:7587–600. doi: 10.1074/jbc.M110.178434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Overall CM, Butler GS. Protease yoga: extreme flexibility of a matrix metalloproteinase. Structure. 2007;15:1159–61. doi: 10.1016/j.str.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Salamone M, et al. A new method to value efficiency of enzyme blends for pancreatic tissue digestion. Transplant. Proc. 2010;42:2043–8. doi: 10.1016/j.transproceed.2010.05.107. [DOI] [PubMed] [Google Scholar]
- 47.Supuran CT, Scozzafava A, Clare BW. Bacterial protease inhibitors. Med. Res. Rev. 2002;22:329–72. doi: 10.1002/med.10007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.