Abstract
Background
The cellular recycling process of autophagy is emerging as a key player in several longevity pathways in C. elegans; however, the underlying mechanism by which autophagy modulates aging is currently unknown. Here, we identify a role for autophagy in the extended lifespan induced by germline removal in C. elegans, and show that autophagy and lipid metabolism work interdependently to modulate aging in this longevity model.
Results
Germline ablation extends lifespan in C. elegans via genes such as the lipase LIPL-4, however less is known of the cellular basis by which longevity is achieved in these animals. Here, we show that germline loss induces autophagy gene expression via the FOXA transcription factor PHA-4, and that autophagy is required to extend longevity. We identify a novel link between autophagy and LIPL-4, as autophagy is required to maintain high lipase activity in germline-deficient animals. Reciprocally, lipl-4 is primarily expressed in autophagy-positive tissues and is required for autophagy induction. Coordination between autophagy and lipolysis is further supported by the finding that inhibition of TOR, a major negative regulator of autophagy, induces lipl-4 expression, and TOR levels are reduced in germline-less animals. TOR may therefore function as a common upstream regulator of both autophagy and of lipl-4 expression in germline-less animals. Importantly, we find that the link between autophagy and LIPL-4 is relevant to longevity, as autophagy is induced in animals overexpressing LIPL-4 and autophagy is required for the reported lifespan extension observed in these animals, recapitulating observations in germline-less animals.
Conclusions
Collectively, our data offer a novel mechanism by which autophagy and the lipase LIPL-4 interdependently modulates aging in germline-deficient C. elegans by maintaining lipid homeostasis to prolong lifespan.
Keywords: Autophagy, lipase, lipolysis, lipophagy, TOR, PHA-4, DAF-16, aging, C. elegans
INTRODUCTION
Reproductive capacity is closely linked to aging, and recent research has shown direct links between reproduction and longevity. For example, germ cell removal extends lifespan in the nematode Caenorhabditis elegans, where lifespan can be extended by up to 60% when the germline is ablated by a laser microbeam [1]. Interestingly, removal of the entire gonad results in a normal lifespan, suggesting that sterility per se is not necessary for lifespan extension and that signals from the germline and the somatic gonad likely cooperate to regulate lifespan [1]. Importantly, the effects of the reproductive system on longevity appear to be conserved between worms and flies, and are mediated via a forkhead box O (FOXO) transcription factor [2]. In germline-less C. elegans, DAF-16/FOXO translocates to the nucleus of intestinal cells [3, 4], suggesting the intestine as a central site of action for signals induced by the absence of germline cells. Such signals are likely to be mediated by lipophilic hormone signaling [2], but the cellular mechanisms underlying the extended longevity in germline-less animals remain relatively unexplored.
Macroautophagy (hereafter referred to as autophagy) is a major mechanism by which a cell degrades cytoplasmic components, including misfolded proteins and damaged organelles. During this multi-step process, double-membrane autophagosomes engulf cytosolic material and fuse to lysosomes, and the vesicular contents are then degraded and recycled [5]. In yeast, numerous conserved genes control autophagy [5], and homologs of many of these ATG genes have been identified in C. elegans [6]. Autophagy can be induced by multiple stress stimuli, including nutrient deprivation, through upstream regulators such as the nutrient sensor TOR, a key modulator of both metabolism and aging [7].
Autophagy was recently linked directly to aging in C. elegans, as autophagy was shown to play a critical role in several nutrient-sensing longevity processes, including the TOR and insulin-IGF-1 signaling pathways as well as in the dietary-restriction paradigm. Specifically, animals with reduced TOR or insulin/IGF-1 receptor daf-2 activity as well as dietary-restricted eat-2 worms show increased levels of autophagy, and autophagy-related genes are required for these animals to live long [8-12]. While these observations have suggested that autophagy may represent a potential common link between several conserved mechanisms that lead to an extended lifespan, it remains unknown how the autophagy process influences organismal aging in C. elegans.
Recent studies have suggested direct links between autophagy and lipid metabolism. Lipid droplet breakdown occurs in hepatocytes by a process termed lipophagy [13], possibly involving lysosomal lipases [14]. Notably, the long lifespan of germline-less glp-1 animals, as well as daf-2 mutants, was recently linked to increased expression of a predicted triglyceride lipase, LIPL-4/K04A8.5 [15]. Specifically, DAF-16 regulates the expression of lipl-4 in the intestine, a major site of fat storage in C. elegans. Moreover, lipl-4 is required for glp-1 animals to live long and LIPL-4 overexpression in the intestine is sufficient to extend lifespan [15]. However, the mechanism that links LIPL-4 activity to longevity in C. elegans remains elusive.
As autophagy appears to play a critical role in several longevity pathways in C. elegans, and the predicted lipase lipl-4 is important for lifespan extension by germline removal, we hypothesized that lipid metabolism could be linked to autophagy to provide a mechanism by which germline-less animals live long. In this study, we provide evidence for such a link between autophagy and LIPL-4 as we found that autophagy and LIPL-4 function interdependently in germline-less animals, possibly through the common upstream regulator TOR. Since germline-less animals and animals overexpressing LIPL-4 both display increased autophagy and require autophagy genes to prolong lifespan, we propose that this novel link plays a critical role in the extended longevity induced by germline removal.
RESULTS
Germline-Less glp-1 Animals Have Increased Autophagy Levels
To determine if autophagy is induced by germline removal in C. elegans, we used a genetic model of germline ablation, namely a temperature-sensitive glp-1 mutant, which lacks a germline and is long-lived at the non-permissive temperature [16]. To detect autophagy, we used electron microscopy (EM) to visualize and quantify autophagic events in several tissues of adult animals. We observed a dramatic increase in autophagosomes as well as in autolysosomes in intestinal and hypodermal seam cells in glp-1(e2141) mutants compared to N2 wild-type animals (Figs. 1A, B and S1A-D), consistent with increased autophagic flux in germline-less animals. To complement our EM studies, we also used a reporter strain expressing a GFP-tagged form of LGG-1 [7, 16], an ortholog of the mammalian LC3 protein that resides in pre-autophagosomal and autophagosomal membranes [8, 17]. In C. elegans, GFP::LGG-1 forms punctate structures or foci, reflecting LGG-1 sequestration to the membrane of nascent autophagosomes. We used this strain to quantify autophagic events in hypodermal seam cells (as described in [18]), and in the intestine. Consistent with our EM studies, we found a significant increase in GFP::LGG-1 foci in seam cells (Fig. 1C), as well as in the intestine (Figs. 1D and S2A) in 1-day old glp-1(e2141) mutants compared to wild-type animals. We also observed increased numbers of GFP-positive foci in another glp-1 loss-of-function mutant, glp-1(bn18) (Figs. S2B). Notably, a similar induction in GFP::LGG-1 positive foci was observed in glp-1 animals lacking the FOXO transcription factor DAF-16 (Fig. S2C). Collectively, these observations indicate that autophagy is induced in germline-less mutants, in a daf-16 independent fashion.
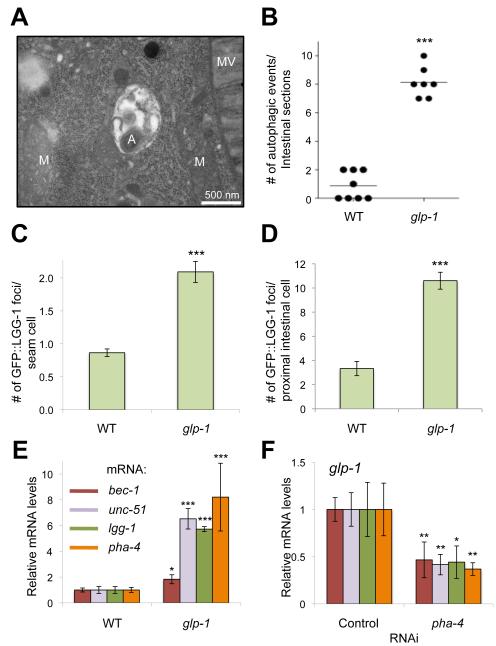
Fig. 1. Autophagy is Increased in glp-1 Animals Through a PHA-4-dependent Mechanism.
(A) Representative electron micrograph of an intestinal cell in a 1-day old adult glp-1(e2141) animal (A: autolysosome, M: mitochondria, MV: microvilli). See Fig. S1A for more information.
(B) Quantification of autophagic events detected by electron microscopy in the intestine of 1-day old N2 wild-type (WT) and glp-1(e2141) animals. The observed number of events per animal is shown as dots, and the mean is indicated by vertical line. ***, P<0.0001, t-test.
(C) Quantification of GFP::LGG-1-positive foci in seam cells of 1-day old WT and glp-1(e2141) animals. Mean number of foci ± SEM is shown. n=100-250 cells, ***, P < 0.001, ANOVA.
(D) Quantification of GFP::LGG-1-positive foci in intestinal cells of 1-day old adult WT and glp-1(e2141) animals. Cells from the proximal portion of the intestine were analyzed. Mean number of foci ± SEM is shown. n=15 worms, ***, P < 0.001, t-test.
(E) RT-PCR analysis of mRNA levels of autophagy genes (bec-1, unc-51, and lgg-1) and of pha-4 in 1-day old adult WT and glp-1(e2141) animals. Relative mean expression ± SD is shown. *, P < 0.05, ***, P < 0.001 vs. WT, ANOVA.
(F) RT-PCR analysis of mRNA levels of genes in (E) in glp-1(e2141) worms fed control bacteria or bacteria expressing pha-4 dsRNA from day 1 to day 3 of adulthood. Wild-type data is included in Fig. S2A. Relative mean expression ± SD is shown. *, P < 0.05, **, P < 0.01 vs. Control, ANOVA. In all experiments, animals were raised at the non-permissive temperature (25°C).
glp-1 Mutants Express Increased mRNA Levels for Autophagy Genes through the FOXA Transcription Factor PHA-4
To further investigate the mechanism by which germline removal induces autophagy, we used RT-PCR to measure mRNA levels of autophagy genes unc-51/ULK1, bec-1/Beclin 1, and lgg-1/LC3 in glp-1(e2141) mutants and in wild-type animals. Expression of all three genes was significantly upregulated between 2- to 8-fold in young glp-1(e2141) adults compared to wild-type animals (Fig. 1E). This upregulation was further increased during early adulthood (Fig. S2D), suggesting that autophagy is substantially induced in adult glp-1 animals. Notably, the same pattern of gene induction was observed in daf-16-deficient glp-1(e2141) animals (Fig. S2E), consistent with our results using the GFP::LGG-1 autophagy reporter (Fig. S2C). Thus, our data suggest that autophagy is transcriptionally upregulated in glp-1 animals in a daf-16-independent manner. However, we found that another FOXA transcription, PHA-4 was required for the induction of autophagy genes in glp-1 animals. As observed for autophagy genes, we found that pha-4 mRNA levels were significantly increased in glp-1(e2141) mutants compared to wild-type animals (Fig. 1E), and this was countered by feeding adult glp-1(e2141) animals with bacteria expressing double-stranded RNA (dsRNA) for pha-4 (Fig. 1E). In wild-type animals, pha-4 RNAi similarly decreased pha-4 mRNA levels but only had a modest effect on autophagy gene expression (Fig. S2F). Importantly, pha-4 RNAi significantly decreased the number of LGG-1-positive foci in seam cells of glp-1 animals expressing GFP::LGG-1 (Fig. S2G). Taken together, these data demonstrate that PHA-4 is a bona fide functional transcriptional regulator of autophagy genes in glp-1 animals.
pha-4 and Autophagy Genes are Required for glp-1 Animals to Live Long
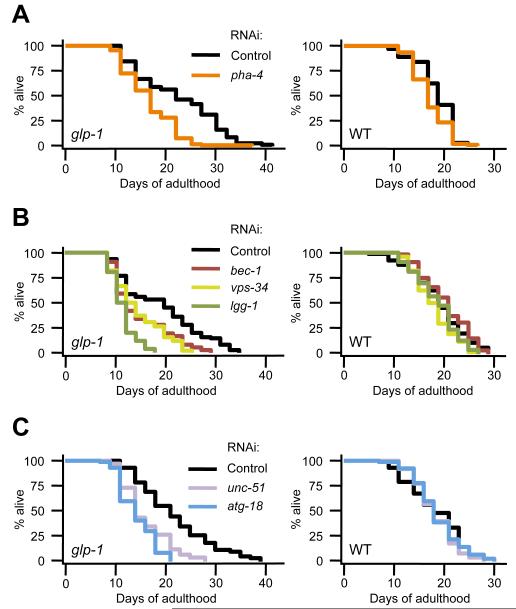
To explore further the role of PHA-4 in germline-less animals, we next determined its contribution to lifespan extension in glp-1 animals. Knockdown of pha-4 in adult glp-1(e2141) mutants significantly decreased their mean lifespan, but only modestly affected the mean lifespan of wild-type animals (Fig. 2A and Table S1), as reported previously [11, 19]. Thus, pha-4 is required for glp-1 mutants to live long, as was shown for dietary-restricted C. elegans [19], and animals with reduced TOR levels [20]. As glp-1 mutants also require daf-16 to live long [16], these data suggest that two forkhead transcription factors, DAF-16 and PHA-4, play critical roles in the germline-deficient longevity model.
Fig. 2. PHA-4 and Autophagy Genes are Required for glp-1 Animals to Live Long.
Lifespan analysis of wild-type N2 (WT) and glp-1(e2141) animals fed control bacteria (empty-vector) or (A) bacteria expressing pha-4 dsRNA, (B) bacteria expressing bec-1, vps-34, or lgg-1 dsRNA, or (C) bacteria expressing unc-51 or atg-18 dsRNA. See Table S1 for details and repeats. In all experiments, animals were raised at 25°C from hatching until the first day of adulthood and were then moved to 20°C for the remainder of their life.
To determine if autophagy genes were similarly required for glp-1 longevity, adult synchronized worms were fed bacteria expressing dsRNA against genes involved in different steps of the autophagy process [6]. We found that RNAi inhibition of unc-51, bec-1, vps-34, lgg-1, and atg-18 each significantly reduced the mean lifespan of glp-1(e2141) animals (Fig. 2B, 2C and Table S1), and we observed similar results with an additional germline-deficient mutant, mes-1(bn7) [16] (Fig. S2H, S2I). In contrast, RNAi inhibition of autophagy genes did not significantly affect the lifespan of adult wild-type animals (Fig. 2B, 2C and Table S1), consistent with previous findings [10, 11]. Taken together, these observations suggest that autophagy genes play critical roles in the extended lifespan of germline-less animals, as was shown for pha-4.
TOR is Downregulated in glp-1 Animals and Regulates LIPL-4 in Wild-type Animals
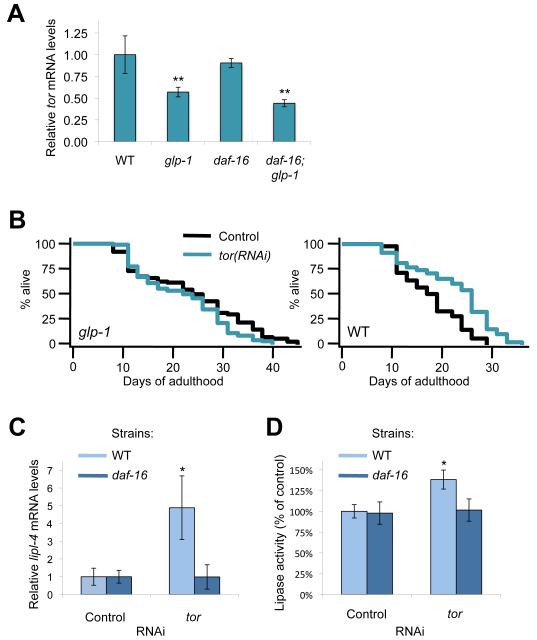
The nutrient sensor TOR is a conserved negative regulator of autophagy and aging. We considered whether TOR activity was decreased in glp-1 animals, providing a possible mechanism for increased autophagy in germline-less animals. We found that tor mRNA levels were reduced in a daf-16-independent manner in response to germline removal (Fig. 3A), and we similarly observed reduced TOR protein levels in glp-1(e2141) mutants compared to wild-type animals (Fig. S3A). Consistent with the reduced levels of TOR in glp-1 animals, we found that the lifespan of glp-1(e2141) animals was not significantly affected by tor RNAi (Fig. 3B, Table S1), whereas the lifespan of similarly treated wild-type worms was increased (Fig. 3B, Table S1), as reported previously [21]. Collectively, these experiments indicate that germline removal extends lifespan, at least in part, by reducing TOR signaling. This possibility is supported by the observations that (a) glp-1 animals require pha-4 to live long (Fig. 2A), (b) TOR inhibition induces pha-4 expression (Fig. S3B), (c) TOR inhibition induces unc-51 mRNA levels in a pha-4 dependent manner (Fig. S3B), and finally (d) TOR inhibition extends lifespan via pha-4 [20].
Fig. 3. Germline-less glp-1 Animals Have Reduced TOR mRNA levels and TOR Inhibition Results in Increased Levels of lipl-4.
(A) RT-PCR analysis of tor mRNA levels in wild-type (WT), glp-1(e2141), daf-16(mu86), and daf-16(mu86); glp-1(e2141) worms at day 1 of adulthood. Animals were raised at the non-permissive temperature (25°C) from hatching, and relative mean expression ± SD is shown. **, P < 0.01 vs. WT, ANOVA.
(B) Lifespan analysis of WT and glp-1(e2141) animals fed control bacteria or bacteria expressing tor dsRNA. Animals were raised as described in Fig. 2. See Table S1 for details and repeats.
(C) RT-PCR analysis of lipl-4 mRNA in 3-day old adult WT and daf-16(mu86) worms fed control bacteria or bacteria expressing tor dsRNA for 2 days starting from day 1. Animals were kept at 20°C throughout this experiment, and relative mean expression ± SD is shown. *, P < 0.05 vs. control, ANOVA.
(D) Lipase activity measurements in 3-day old adult WT and daf-16(mu86) worms fed control bacteria or bacteria expressing tor dsRNA for 2 days starting from day 1. Animals were kept at 20°C throughout this experiment, and relative mean expression ± SD is shown. *, P < 0.05 vs. control, ANOVA.
The predicted triglyceride lipase LIPL-4/K04A8.5 was recently shown to be upregulated in germline-less C. elegans in a daf-16-dependent mechanism [15], and we therefore asked if TOR could modulate lipl-4 gene expression in C. elegans. Interestingly, we found that RNAi inhibition of tor in wild-type worms significantly increased lipl-4 mRNA levels (Figs. 3C and S3B), which was dependent on daf-16 (Fig. 3C), but independent of pha-4 (Fig. S3B). Consistent with this, we observed a daf-16-dependent increase in lipase activity in tor RNAi-treated animals (Fig. 3D). Taken together, these data suggest that TOR may act as an upstream regulator of DAF-16 and PHA-4 to increase lipl-4 and autophagy, respectively, in germline-deficient C. elegans.
Autophagy and LIPL-4 Function Interdependently in glp-1 Animals
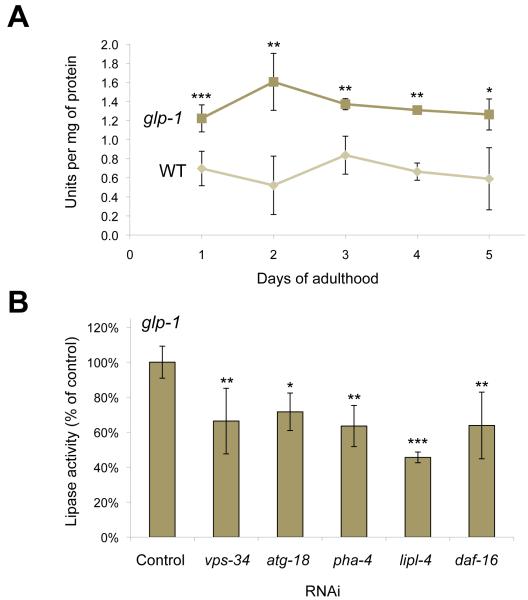
Since inhibition of both lipl-4 and of autophagy genes reduced the lifespan of glp-1 animals, we next considered that autophagy and LIPL-4 might converge to modulate longevity. Recent studies have demonstrated a link between autophagy and lipolysis in which lipid stores can be subjected to lysosomal-related hydrolysis through lipophagy [13]. To probe such a link between autophagy, lipolysis and longevity in C. elegans, we first measured lipase activity in adult glp-1 and wild-type animals. Lipase activity was significantly increased in adult glp-1(e2141) mutants compared to wild-type animals (Fig. 4A), consistent with the increased expression of lipl-4 in glp-1 animals [15]. Lipase activity was similarly increased in glp-1(bn18) mutants (Fig. S4A). To determine if autophagy was required for increased lipase activity, we used RNAi to inhibit expression of vps-34, atg-18, bec-1, and pha-4, and found that such reductions significantly decreased lipase activity in glp-1(e2141) animals (Fig. 4B). The increase in lipase activity, observed in glp-1 animals, was reduced significantly by lipl-4 RNAi, confirming that the measured lipase activity was at least a partial read-out for lipl-4 activity (Fig. 4B). Lipase activity was also dependent on daf-16 (Fig. 4B), consistent with lipl-4 being transcriptionally regulated by DAF-16 in glp-1 animals [15]. Notably, these RNAi treatments had only a modest effect on lipase activity in wild-type animals (Fig. S4B). These data suggest that autophagy is required for lipl-4 function in glp-1 animals. We also found that lipl-4 plays a role in autophagy induction, as lipl-4 RNAi significantly reduced the number of GFP::LGG-1-positive foci in seam cells of glp-1(e2141) animals (Fig. S5A), as observed when pha-4 and autophagy genes were inhibited (Figs. S2G and S5B). Collectively, these observations suggest that autophagy and and the putative lipase LIPL-4 are linked in an interdependent fashion in germline-less C. elegans.
Fig. 4. Increased Lipase Activity Observed in glp-1 Animals is Dependent on Autophagy Genes and PHA-4.
(A) Lipase activity was measured daily in wild-type N2 (WT, lighter line) and glp-1(e2141) animals (darker line) from day 1 to day 5 of adulthood. Animals were raised as in Fig. 2, and mean activity mean ± SD is shown. *, P < 0.05, **, P < 0.01, ***, P < 0.001 vs. WT, ANOVA.
(B) Lipase activity was measured in 4-day old adult glp-1(e2141) animals fed bacteria expressing dsRNA against several autophagy genes, pha-4, lipl-4 or daf-16 for 3 days starting from day 1. Animals were raised as in Fig. 2, and relative mean activity ± SD is shown. *, P < 0.05, **, P < 0.01, ***, P < 0.001 vs. Control, ANOVA.
Animals that overexpress LIPL-4 Display Increased Autophagy and Require Autophagy Genes and pha-4 to Live Long
To better understand the link between autophagy and LIPL-4, we analyzed animals overexpressing LIPL-4 from its endogenous promoter [15]. We confirmed that LIPL-4 overexpressing animals showed a significant increase in lipase activity (Fig. S5C), and we found that these animals were longer-lived (Table S2), as was observed in glp-1 mutants, and in animals overexpressing LIPL-4 from an intestinal-specific promoter [15]. Interestingly, we discovered that LIPL-4 overexpression increased GFP::LGG-1 positive foci as well as autophagy gene expression (Figs. S5C-D), suggesting that LIPL-4 overexpression was sufficient to induce autophagy. Consistent with a link between autophagy and LIPL-4, we found LIPL-4 was expressed not only in intestinal cells [15], but also in other tissues including the seam cells (Fig. S5E), where an increase in autophagy was detected in glp-1 mutants compared to wild-type animals (Fig. 1A-D). The presence of the glp-1 mutation further increased LIPL-4 expression levels, and also extended lifespan (data not shown), suggesting that the increased lipase activity in LIPL-4-overexpressing glp-1 mutants might be responsible for the extended lifespan.
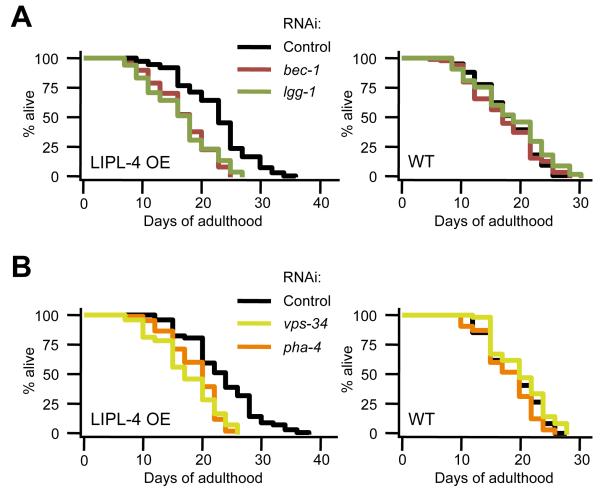
Because autophagy was increased in LIPL-4 overexpressing animals and in germline-less glp-1 mutants that require autophagy genes to live long, we next asked if autophagy genes were required for lifespan extension in LIPL-4 overexpressing animals. Interestingly, we found that RNAi-mediated inhibition of bec-1, lgg-1, vps-34, or the putative autophagy inducer pha-4 all significantly reduced the lifespan of LIPL-4 overexpressing animals, while having negligible effects on non-transgenic siblings (Fig. 5A, B, and Table S2). These data support our hypothesis that autophagy is required for the lifespan-extending effects of LIPL-4 overexpression. This is comparable to the effects of germline removal, and it is therefore possible that lipl-4 modulates longevity via an autophagy-related mechanism in germline-less animals. Consistent with this, we found that simultaneous knockdown of lipl-4 and vps-34 failed to further decrease the lifespan of glp-1(e2141) animals (Fig. S5F and Table S3). In summary, these experiments provide the first genetic evidence that lipolysis and autophagy are linked to positively modulate longevity in C. elegans.
Fig. 5. Autophagy Genes are Required for the Long Lifespan of Animals Overexpressing LIPL-4.
Lifespan analysis of animals overexpressing LIPL-4 (LIPL-4 OE) and non-transgenic siblings (WT) fed control bacteria or (A) bacteria expressing bec-1 or lgg-1 dsRNA, or (B) bacteria expressing vps-34 or pha-4 dsRNA. Animals were incubated at 20°C throughout their life. See Table S2 for additional information and repeats.
DISCUSSION
We investigated the role of autophagy in the extended lifespan induced by germline removal in C. elegans, and shown using multiple complimentary approaches that germline-less glp-1 animals display increased levels of autophagic events. We also detected that the increase in the expression of several autophagy genes was dependent on the FOXA transcription factor PHA-4, suggesting that autophagy is induced at the transcriptional level in response to germline removal. Accordingly, autophagy genes and pha-4 were required for glp-1 animals to live long. Taken together, these observations indicate that autophagy is induced in a beneficial manner in germline-less animals. While we detected autophagy by using steady-state methods, our data strongly argue for a functional role for autophagy turnover in germline-deficient animals because we obtain the same effects on long-lived glp-1 animals after RNAi against genes that act in multiple steps of the autophagy process. Consistent with this interpretation, we find that TOR, a known negative regulator of autophagy, is downregulated in glp-1 animals. As reduced TOR signaling plays an important role in other C. elegans longevity models that rely on autophagy genes, such as dietary restriction [10-12], these observations suggest a broader role for the TOR-regulated process of autophagy in preventing aging in C. elegans.
The FOXO transcription factor DAF-16 is essential for lifespan extension through germline removal [2], yet we find that autophagy gene expression and autophagy itself remained high in glp-1 animals lacking daf-16. Thus, in contrast to lifespan extension, induction of autophagy appears to be independent of daf-16 in these animals, similar to our previous observations in long-lived daf-2 insulin/IGF-1 receptor mutants [11]. These results suggest that daf-16 may act downstream of, or in parallel with, autophagy function in such long-lived animals. Nevertheless, DAF-16 is required to obtain the beneficial effects of autophagy on longevity in glp-1 animals. As discussed previously [11], we speculate that DAF-16 could play a regulatory role in the recycling of material from the autophagic process into new targets that have beneficial effects on longevity. This situation is different from overexpression of DAF-16, which is sufficient to induce autophagy in C. elegans [22], possibly because DAF-16 may be activated by different mechanisms in animals overexpressing DAF-16 than in glp-1 animals. Other proteins such as PHA-4 may induce autophagy in germline-less animals in the presence or absence of daf-16 activity. Specifically, we find that the expression of several autophagy genes (i.e., unc-51, lgg-1, and bec-1) is increased in glp-1 animals compared to wild type, and this induction requires pha-4. Consistent with these observations, we observed pha-4 to be required for autophagy induction in germline-less animals. Notably, we found that overexpression of PHA-4 significantly induced unc-51 but not lgg-1 and bec-1 levels (Fig. S6), suggesting that PHA-4 overexpression is sufficient to recapitulate some, but not all of the PHA-4 mediated effects observed in glp-1 animals. These experiments not only reveal a novel mechanism by which autophagy is induced in C. elegans, but also suggest that PHA-4 regulates the transcription of autophagy genes in metazoans. In support of this possibility, PHA-4 was recently shown to bind to the promoters of multiple autophagy genes, including unc-51, bec-1, and lgg-1 during development [23, 24] (see also Supplemental Information). Moreover, PHA-4 is required for the increase in autophagy observed in long-lived, dietary-restricted eat-2 mutants [11]. Taken together, these observations are consistent with PHA-4 modulating lifespan by directly inducing autophagy gene expression, but PHA-4 may also regulate additional targets to affect longevity in germline-less animals. It will be of interest to determine how DAF-16 and PHA-4 function in the same longevity model and whether they regulate certain shared targets [19], as well as to ask if PHA-4 also regulates autophagy in higher organisms.
In an effort to better understand how autophagy influences lifespan in germline-less animals, we have identified a new role for autophagy in modulating lipid metabolism in glp-1 animals through the predicted triacylglycerol lipase LIPL-4. We determined that LIPL-4 exhibits lipase activity, at least in vitro, and significantly contributes to the elevated lipase activity observed in glp-1 animals. Importantly, we found that autophagic activity as well as autophagy genes and pha-4 mRNA levels were increased in long-lived animals overexpressing LIPL-4 and that autophagy genes and pha-4 were required for the elevated lipase activity, as was observed in glp-1 mutants. As further support for a link between autophagy and LIPL-4, we observed that lipl-4 is required for the increased autophagy activity observed in glp-1 animals and thus an increase in LIPL-4 activity may promote autophagosome formation. In addition, we found that LIPL-4 was expressed in the same tissues in which we detected increased autophagy in germline-less animals, namely hypodermal seam cells and the intestine. Finally, the autophagy regulator TOR might function as a common upstream regulator of these two processes in germline-less animals, as we discovered that inhibition of TOR was sufficient to increase lipl-4 levels and lipase activity in a daf-16-dependent fashion, indicating that TOR has both daf-16-dependent as well as daf-16-independent functions [21, 25]. Collectively, these results strongly support the existence of a novel link between autophagy and LIPL-4 in germline-less C. elegans (see model in Fig. 6).
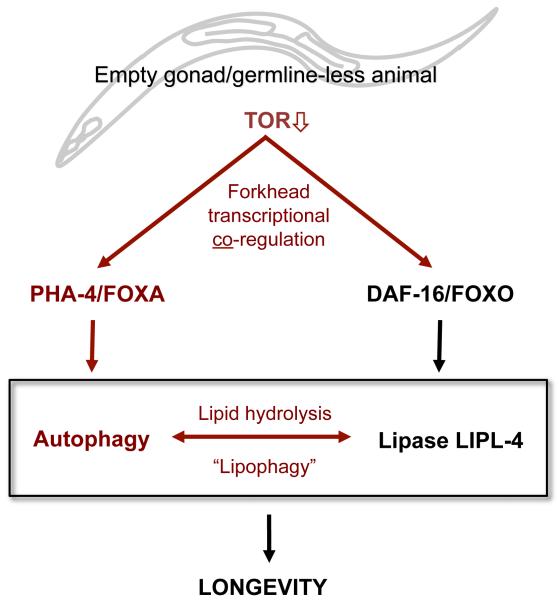
Fig. 6. Model for How Autophagy and LIPL-4 Coordinately Modulate Longevity in Germline-less C. elegans.
TOR levels are reduced in C. elegans with an empty gonad (germline-less), which show enhanced activity of two forkhead transcription factors that modulate longevity and lipid metabolism. PHA-4/FOXA stimulates autophagy, and DAF-16/FOXO upregulates LIPL-4, thereby possibly inducing lipid hydrolysis. In turn, LIPL-4 requires autophagy to modulate lifespan, possibly through a process involving lipophagy. Novel observations made in this study are shown in red, known links are shown in black. Possible feedback and cross-talk events are not included for simplicity; see text for details.
Our results further suggest that the connection between autophagy and LIPL-4 in germline-deficient C. elegans may be critical for lifespan extension in these animals. We found that LIPL-4 overexpressing animals are long-lived and both pha-4 and autophagy genes are required for this extended lifespan, as is the case for glp-1 animals. Although we do not yet know how LIPL-4 overexpression may induce autophagy to extend lifespan, it is possible that a lipase metabolite could trigger autophagy through regulation of TOR signaling (similarly to phosphatidic acid, a product of phospholipase D activity [26]) via an increase in PHA-4 activity. Such an explanation is consistent with the observation that directly reducing TOR levels by RNAi failed to extend lifespan in adult LIPL-4 overexpressing animals (Table S2), and LIPL-4 overexpressing animals have increased pha-4 mRNA levels. As further evidence for autophagy and LIPL-4 working by overlapping mechanisms, we observed that simultaneous inhibition of both lipase and autophagy functions did not further decrease the lifespan of glp-1 animals, compared to inhibiting each process separately. Taken together, our genetic and biochemical analyses are consistent with a model in which LIPL-4 and autophagy work in concert to extend the lifespan of glp-1 animals.
In this model (Fig. 6), the activity of the nutrient sensor TOR is reduced in response to germline removal, and this triggers the induction of two different pathways. One pathway involves activation of DAF-16 to induce lipl-4 expression, which again may increase lipid hydrolysis. In contrast, the other pathway causes an induction of PHA-4 and subsequent autophagy gene expression to ensure increased flux through the multi-step autophagy process. We note that feedback and crosstalk between components of these two pathways are possible. For example, we find that LIPL-4 overexpression causes a small (1.5 to 2-fold) increase in daf-16 mRNA levels, and inhibition of pha-4 by RNAi reduces lipl-4 mRNA levels to about 50% in glp-1 animals (data not shown). Consistent with the latter observation, PHA-4 can bind the lipl-4 promoter [23, 24]. Taken together, these data suggest that lipl-4 could be a common target of both DAF-16 and PHA-4. In turn, autophagy and LIPL-4 might work interdependently to ensure lifespan extension in germline-less animals.
What is the nature of the link between autophagy and LIPL-4, which may possess intracellular lipolytic activity, and how could this link lead to lifespan benefits? One possible mechanism may involve lipophagy, which is a large-scale hydrolysis of neutral lipid stores in the lysosome [13]. This scenario would predict lipases to be localized to the lysosome as seen for human lysosomal acid lipase, which we note shares a very high degree of sequence homology to LIPL-4. Alternatively, autophagy may be induced by a product of lipase activity, as is the case for autophagy induced by free fatty acids in pancreatic beta cells [27]. In this case, the lipase could be localized to the autophagosome, as has been observed for phospholipase D1 during starvation of mammalian cells [28]. It is also possible that enhanced lipolysis via autophagy prevents the accumulation of toxic byproducts or is critical for the partitioning of unused yolk, normally destined for oocytes. Lipolysis could process phospholipids to boost membrane formation for autophagosome maturation necessary to recycle components relevant to aging. Future experiments, including cytological and biochemical profiling of lipids in glp-1 and LIPL-4-overexpressing animals, should help clarify how autophagy is linked to lipid metabolism in germline-less animals, including determining the intracellular function of LIPL-4.
Taken together, this study proposes a potential mechanism by which autophagy affects lifespan: we suggest that autophagy and LIPL-4 modulate aging in germline-deficient C. elegans by maintaining lipid homeostasis to prolong lifespan. As such, our results advance our understanding of how autophagy affects organismal aging, and also offer new ideas as to how the regulation of lipid metabolism may be relevant to future treatments of metabolic disorders.
EXPERIMENTAL PROCEDURES
Strains
All strains were maintained as previously described [29]. See supplemental information for details on strains used.
Lifespan Analysis and RNAi Experiments
Lifespan assays were performed as described [30]. RNAi clones were obtained from the Ahringer and Vidal RNAi libraries. See supplemental information for details.
Autophagy Quantification
Autophagic events were quantified in C. elegans strains either by electron microscopy [31] or by use of a GFP::LGG-1 reporter [18]. See supplementary information for details.
Real-time Quantitative PCR
RNA was extracted from biological triplicates, reverse-transcribed and analyzed as previously described [32]. See supplementary information for details.
Lipase Activity Assay
Lipolytic activity was measured with a colorimetric assay kit (BioAssay Systems) and samples (biological triplicates) were prepared as previously described [33].
Statistical Analyses
For parametric analyses, Student’s t-test or one-way ANOVA was done using the Graph Pad Prism 5 Software. For lifespan assays, Kaplan-Meier survival curves and P values were obtained by analyzing data by the Log Rank (Mantel-Cox) test with the Stata 8.2 software.
Supplementary Material
Highlights.
-
-
Autophagy is required for germline-less C. elegans to live long.
-
-
Autophagy genes are under transcriptional regulation by PHA-4/FOXA.
-
-
Autophagy and the lipase LIPL-4 function interdependently to ensure prolonged lifespan.
ACKNOWLEDGMENTS
We thank all Hansen and Meléndez lab members for helpful discussions, and Drs. R. Bodmer, M. Van Gilst, C. Kumsta, E. Troemel, and A. O’Rourke for comments on the manuscript. We thank Drs. M. Wang and G. Ruvkun for sharing an unpublished transgenic strain expressing LIPL-4 from the endogenous promoter, and Drs. C. Kang and L. Avery for the integrated GFP::LGG-1 strain. We thank D. Hoffman in the Meléndez lab for creating new (non-Rol) LGG-1 reporter strains, Dr. M. Wood for technical assistance with electron microscopy analysis, and Dr. D. Hall for help with analyzing electron micrographs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: SG and MH created and analyzed new autophagy reporter strains, while LRL conducted all other experiments. LRL, AM, and MH designed the experiments, and LRL and MH wrote the manuscript with contributions from all authors. MH was supported by a Research Grant from American Federation for Aging Research, and by an R01 grant from the NIA, while AM was supported by a National Science Foundation Research Initiation Award. MH and AM are both Ellison Medical Foundation New Scholars in Aging.
REFERENCES
- 1.Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- 2.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 3.Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 4.Berman JR, Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124:1055–1068. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melendez A, Levine B. Autophagy in C. elegans. WormBook. 2009:1–26. doi: 10.1895/wormbook.1.147.1. [DOI] [PubMed] [Google Scholar]
- 7.Hansen M, Kapahi P. TOR Signaling and Aging. In: Hall MN, Tamanoi F, editors. The Enzymes. Vol. 28. Academic Press; Burlington: 2010. pp. 279–299. [Google Scholar]
- 8.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 9.Hars ES, Qi H, Ryazanov AG, Jin S, Cai L, Hu C, Liu LF. Autophagy regulates ageing in C. elegans. Autophagy. 2007;3:93–95. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- 10.Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- 11.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, Orosz L, Kovacs AL, Csikos G, Sass M, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 13.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czaja MJ, Cuervo AM. Lipases in lysosomes, what for? Autophagy. 2009;5:866–867. doi: 10.4161/auto.9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang MC, O’Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- 17.Kang C, You YJ, Avery L. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev. 2007;21:2161–2171. doi: 10.1101/gad.1573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melendez A, Hall DH, Hansen M. Monitoring the role of autophagy in C. elegans aging. Methods Enzymol. 2008;451:493–520. doi: 10.1016/S0076-6879(08)03229-1. [DOI] [PubMed] [Google Scholar]
- 19.Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 20.Sheaffer KL, Updike DL, Mango SE. The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr Biol. 2008;18:1355–1364. doi: 10.1016/j.cub.2008.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 22.Jia K, Thomas C, Akbar M, Sun Q, Adams-Huet B, Gilpin C, Levine B. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci U S A. 2009;106:14564–14569. doi: 10.1073/pnas.0813319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong M, Niu W, Lu ZJ, Sarov M, Murray JI, Janette J, Raha D, Sheaffer KL, Lam HY, Preston E, et al. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 2010;6:e1000848. doi: 10.1371/journal.pgen.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, Brdlik CM, Janette J, Chen C, Alves P, Preston E, et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 2010;21:245–254. doi: 10.1101/gr.114587.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 26.Foster DA. Regulation of mTOR by phosphatidic acid? Cancer Res. 2007;67:1–4. doi: 10.1158/0008-5472.CAN-06-3016. [DOI] [PubMed] [Google Scholar]
- 27.Komiya K, Uchida T, Ueno T, Koike M, Abe H, Hirose T, Kawamori R, Uchiyama Y, Kominami E, Fujitani Y, et al. Free fatty acids stimulate autophagy in pancreatic beta-cells via JNK pathway. Biochem Biophys Res Commun. 2010;401:561–567. doi: 10.1016/j.bbrc.2010.09.101. [DOI] [PubMed] [Google Scholar]
- 28.Dall’armi C, Hurtado-Lorenzo A, Tian H, Morel E, Nezu A, Chan RB, Yu WH, Robinson KS, Yeku O, Small SA, et al. The phospholipase D1 pathway modulates macroautophagy. Nat Commun. 2010;1:142. doi: 10.1038/ncomms1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Troemel ER, Felix MA, Whiteman NK, Barriere A, Ausubel FM. Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLoS Biol. 2008;6:2736–2752. doi: 10.1371/journal.pbio.0060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 2005;3:e53. doi: 10.1371/journal.pbio.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narbonne P, Roy R. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature. 2009;457:210–214. doi: 10.1038/nature07536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.