Abstract
Plant disease resistance can be triggered by specific recognition of microbial effectors by plant nucleotide binding-leucine rich repeat (NB-LRR) receptors. Over the last few years, many efforts have greatly improved the understanding of effector and NB-LRR function, but have left a lot of questions as to how effector perception activates NB-LRR induction of defense signaling. This review describes exciting new findings showing similarities and differences in function of diverse plant NB-LRR proteins in terms of pathogen recognition and where and how resistance proteins are activated. Localization studies have shown that some NB-LRRs can activate signaling from the cytosol while others act in the nucleus. Also, the structural determination of two NB-LRR signaling domains demonstrated that receptor oligomerization is fundamental for activation of resistance signaling.
INTRODUCTION
Understanding plant immunity mechanisms will provide a crucial input for improving disease control measures to protect agricultural production. Plant immunity relies on two major levels of resistance [1–3]. The first level is triggered by the recognition of conserved microbial molecules called Pathogen Associated Molecular Patterns (PAMPs) by cell surface located receptors PRRs (Pattern Recognition Receptors). This type of resistance is referred to as PTI for PAMP Triggered Immunity. PRRs generally consist of transmembrane proteins with an extracellular Leucine Rich Repeat (LRR) domain. Pathogens adapted to specific host plants avoid and/or suppress PTI through the action of virulence effectors. Host plants have evolved a second level of surveillance known as Effector-Triggered-Immunity (ETI) to counteract adapted pathogens. ETI is mediated by Resistance (R) proteins that directly or indirectly perceive pathogen effectors, then called Avirulence (Avr) proteins. This recognition is often characterized by a local cell death at the pathogen infection site termed the Hypersensitive Response (HR). Most ETI receptors (R proteins) are intracellular and belong to the conserved family of NB-LRR proteins containing a C-terminal LRR and a central nucleotide binding (NB) domain which is often referred to as the NB-ARC region (Nucleotide-Binding adaptor shared by Apaf-1, Resistance proteins and CED-4) [4]. Apaf-1 and CED-4 are part of the animal nucleotide-binding oligomerization domain (NOD)-like receptor (NLR) family, whose members also function as regulators of innate immune responses and apoptosis [5]. Evidence suggests that the NB-ARC domain can bind and hydrolyse nucleotides which may act as a molecular switch to regulate R protein activity upon pathogen perception [6]. Plant NB-LRRs can be divided in two subclasses depending on their N-terminal extremity [7]. The first class contains a Toll-Interleukin-1 Receptor (TIR) domain also found in the membrane-bound Toll-like receptor (TLR) family of animal immune receptors. The “non-TIR” class contains either a coiled-coil (CC) domain or a variable domain of unknown function [8].
Although our knowledge of plant immunity has improved considerably over the last ten years, the mechanisms by which effector perception is linked to NB-LRR activation remain elusive. What are the signals inducing receptor activation? Where and how do they trigger defense signaling? Here, we review the most recent findings providing new insights on NB-LRR function from pathogen perception to immune signaling activation.
1. Pathogen recognition: what is the signal leading to receptor activation and signaling?
Plant NB-LRR proteins can recognize pathogen effectors either by direct physical interaction [9–13], or indirectly by detecting modifications of host target proteins that are induced by the effector [14–17]. Recent advances described below, have demonstrated in two model-systems how effector enzymatic activity and/or effector-mediated modifications can be involved in both direct and indirect recognition events.
The canonical example of indirect recognition involves the Arabidopsis RIN4 protein which acts as an accessory protein to two NB-LRR immune receptors, RPS2 and RPM1. RPS2 activates resistance in response to loss of RIN4 due to cleavage by the bacterial protease avrRpt2 [14,15]. RPM1 activates resistance in response to either of two unrelated effectors, avrB and avrRpm1. RIN4 becomes phosphorylated in the presence of these effectors, leading to the hypothesis that RPM1 is activated by phosphorylated RIN4, although neither effector has a detectable kinase activity [16]. Two recent studies have now confirmed this hypothesis [18,19] and identified a host protein kinase that phosphorylates RIN4 [19] (Figure 1). Both studies identified the RIN4 Threonine residue in position 166 as a critical phosphorylation site required to activate RPM1. Antibodies raised against a RIN4 peptide containing phosphothreonine 166 could detect in vivo phosphorylation of this site in response to AvrB and AvrRpm1 induction. Alanine substitutions at this site prevented RPM1 activation, while aspartate or glutamate substitutions, which act as phosphomimics due to their negative charge, caused effector-independent activation of RPM1. Liu et al [19] identified the host protein kinase RIPK (RPM1-Induced Protein Kinase) in a pull down assay as a RIN4 interactor in the presence of AvrRpm1. The authors showed that RIPK is able to phosphorylate RIN4 in vitro on threonine 166, and also to bind AvrB, suggesting that AvrB binding to RIN4 and RIPK induces RIPK-mediated phosphorylation of RIN4, leading to activation of RPM1. However, a RIPK knock-out mutant only partially affected RIN4 phosphorylation and RPM1 mediated resistance, suggesting that other kinases may also contribute to these events.
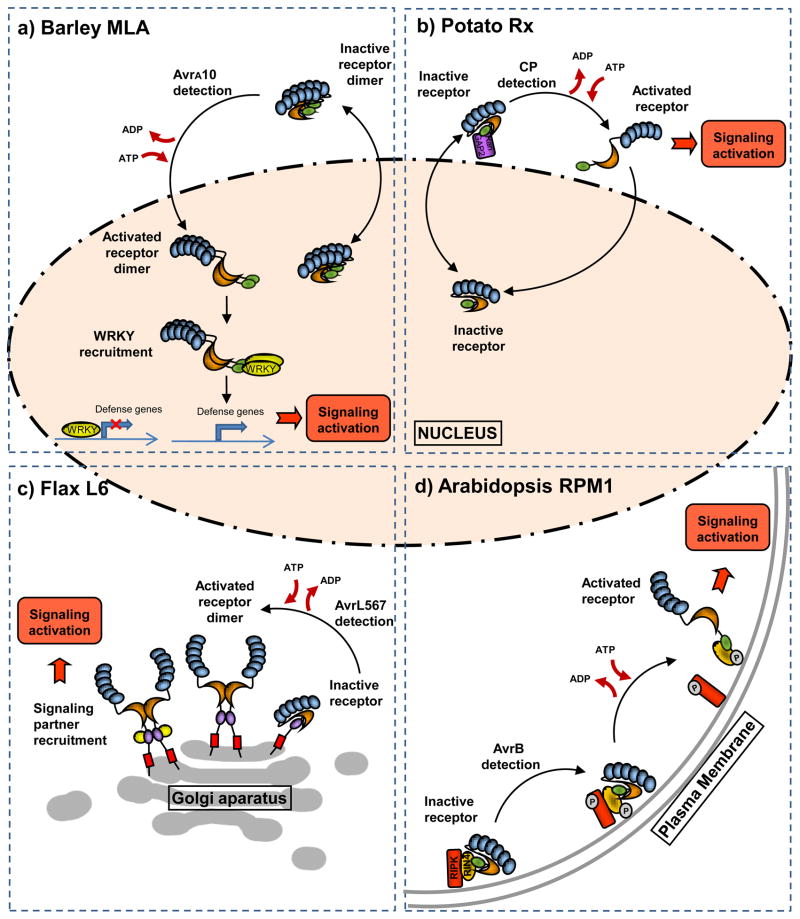
Figure 1. Different models for NB-LRR signaling activation.
Models for activation and signalling are presented for several R proteins to illustrate the potential variability in the mechanism and subcellular location of these events. The R protein N-terminal TIR and CC domains are represented by purple and green ovals respectively, the NB-ARC domain by an orange crescent and the Leucine Rich Repeats by a series of blue ovals.
a) In a resting state, Barley MLA navigates between the cytosol and the nucleus as an inactive homodimer interacting through the CC domain. The presence of its corresponding Barley mildew effector AvrA10 induces the accumulation of MLA in the nucleus, nucleotide exchange and conformational changes allowing the interaction of the CC domain with WRKY factors to derepress defense activation. b) The potato CC-NB-LRR Rx is also present in both the nucleus and cytosol. Its nucleocytoplasmic partitioning depends on the trafficking regulator RanGAP2 (purple), which acts as a cytoplasmic retention factor of Rx. The Potato Virus X Coat Protein (CP) is recognized in the cytosol and signaling is activated in this location. The Rx nuclear pool is required for correct regulation of resistance function. This protein is shown as a monomer as no direct evidence of its oligomerisation state is available. c) In the absence of pathogen, L6 is attached to the Golgi membrane through its N-terminal signal anchor (red rectangle). The protein is kept in an inactive state where the TIR domain dimerization interface is not exposed. Upon recognition of the flax rust effector AvrL567, nucleotide exchange and conformational change exposes the TIR domain for homodimerization and interaction with signaling proteins (yellow circles) to activate defense signalling. d) The Arabidopsis RPM1 protein is kept at the plasma membrane in a complex with the effector target RIN4 (yellow) and the protein kinase RIPK (red). The presence of the Pseudomonas effector AvrB induces RIPK and RIN4 phosphorylation. RIN4 modification leads to RPM1 activation and signaling at the plasma membrane. Again, in the absence of direct evidence otherwise, this protein is shown as a monomer..
The Arabidopsis RRS1-R resistance protein confers resistance to the causal agent of bacterial wilt Ralstonia solanacearum containing the effector PopP2 [12]. The two R/Avr partners physically interact in yeast and this interaction has been recently confirmed in the nucleus of living plant cells using Fluorescence Lifetime Imaging (FLIM) [20]. Mass spectrometric analysis revealed that PopP2 displays an autoacetyl-transferase activity in vitro that acetylates a conserved Lysine residue at position 383. This enzymatic activity depends on the PopP2 predicted catalytic core residues. Mutation of the K383 residue compromised RRS1-R mediated immunity without disrupting PopP2/RRS1 association in the nucleus, suggesting that RRS1 activation requires PopP2 enzymatic activity in addition to its physical contact. However no PopP2-mediated acetylation of its known interacting partners RRS1 and the Cysteine protease RD19 [21] could be detected in planta, so it is not yet clear how PopP2 acetylation activity contributes to its recognition and what are its substrate(s). A second resistance protein, RPS4, acts in concert with RRS1 to confer resistance to different plant pathogens [22,23], and may also be involved in recognition of enzymatically active PopP2.
2. Immune receptor localization: where does the action take place?
In the past five years, a number of studies have demonstrated the importance of nucleocytoplasmic trafficking of immune receptors and immune components for disease resistance activation [24–26]. Indeed, effector-triggered nuclear accumulation of nucleocytoplasmic NB-LRRs such as Barley MLA10, Tobacco N, and Arabidopsis RPS4 and SNC1 is required for efficient induction of defense responses [27–30] For instance, in the presence of the corresponding barley mildew effector AvrA10, MLA10 was found to accumulate in the nucleus and to associate with two WRKY family proteins that act as transcriptional repressors of PTI [28]. These observations led to a model where pathogen perception induces immune receptor accumulation in the nucleus where they activate immune signaling responses through transcriptional reprogramming [31].
However, two recent studies demonstrated that the nucleocytoplasmic Potato NB-LRR Rx protein activates immune responses from the cytosol even though it requires both cytosolic and nuclear pools for correct regulation of its activity [32**,33**] (Figure 1). Rx interacts with RanGAP2, a small cytosolic GTPase Ran required for Rx function [34,35], and this interaction appears to control Rx nucleocytoplasmic equilibrium. Overexpression of RanGAP2 sequesters Rx in the cytosol, while expression of a modified version of RanGAP2 fused to a nuclear localization signal (NLS) leads to Rx accumulation in the nucleus [33**]. Both versions lead to increased accumulation of Rx protein, but nuclear retention inhibits Rx function, while cytosolic accumulation leads to enhanced Rx function including a weak autoactivity. Similar results were observed for autoactive mutants of Rx, indicating that defense signaling is mediated by the cytoplasmic pool of Rx. Similarly, fusion of an NLS directly to Rx significantly compromised its activity, while a nuclear exclusion signal had a minimal effect [32**]. Furthermore, Rx is not activated when the Potato Virus X coat protein (CP), its corresponding elicitor, is forced to accumulate in the nucleus, suggesting that both pathogen recognition and resistance signaling have to take place in the cytoplasm [32**]. Thus, nuclear accumulation of Rx may be a negative regulatory mechanism to limit its activation in the absence of the CP.
Not all NB-LRRs show nuclear localization, such as the plasma membrane-associated RPM1. Interestingly, RPM1 remains membrane-associated in the presence of the autoactivating RIN4 T166E mutant [18], as does the autoactive RPM1 D505V mutant [36*]. Gao et al [36*] further showed that nuclear exclusion, or direct membrane tethering did not compromise RPM1 activity. These results strongly suggest that RPM1 activation and signaling occurs at the plasma membrane and initiates a cytosolic signaling pathway (Figure 1). Some NB-LRRs contain membrane anchors or palmitoylation or myristoylation sites, which direct the protein to specific intracellular membrane locations. For instance the flax rust resistance proteins L6, M and P2 are respectively targeted to Golgi membranes, tonoplast and the cytosol (Takemoto et al., unpublished). N-terminal domain swap experiments between L6, M and P2, showed that membrane attachment is important for L6 resistance protein function as well as for effector-independent signaling by the autoactive L6 TIR domain (M. Bernoux, unpublished). This suggests that L6 early signaling occurs at the Golgi membrane (Figure 1).. To date no common early R protein signaling partners have been identified and it is possible that different resistance signaling pathways may be activated from either cytosolic or nuclear locations.
One key component of the immune signaling is EDS1, which is required downstream of TIR-NB-LRR resistance proteins, including L6 [37,38]. Garcia et al [39**] showed that both cytosolic and nuclear pools of EDS1 are required for its function in coordinating immune responses. Upon pathogen perception, defense associated transcriptional reprogramming is dependent on EDS1 nuclear accumulation. However, fusion of a nuclear export signal or a cytoplasmic retention glucocorticoid receptor concentrated EDS1 in the cytosol but only partially compromised pathogen resistance compared to an eds1 mutant. This indicated that a cytoplasmic pool of EDS1 is maintained and required to allow complete resistance activation after pathogen perception. The role of the EDS1 cytoplasmic pool is not yet clear, but this observation also suggests that resistance signaling pathways may operate in both cellular compartments.
3. Receptor oligomerization: a common feature for signaling activation?
One of the key unanswered questions in plant immunity is how NB-LRR receptors are activated to initiate the resistance signal? Current models predict that effector recognition induces NB-LRR protein changes in intramolecular interactions and conformation associated with nucleotide exchange by the NB domain [40] thus exposing an N-terminal signaling domain to activate downstream defense response [7]. While there has been limited biochemical data available to confirm these general propositions, Williams et al [41*] recently showed that wildtype M resistance protein from flax binds preferentially to ADP, while an autoactive mutant preferentially binds to ATP. This is consistent with the proposition that the ADP-bound protein represents the signaling off state and the ATP-bound protein the on state. Activation and signaling of animal NLRs and TLRs have been characterized in detail and serve as a useful comparative model. Once activated, animal NLRs oligomerize through their central nucleotide binding domain generating a wheel-shaped oligomeric platform [42]. This scaffold allows proximity-induced association of their N-terminal effector domains which then recruit signaling partners to activate immune or apoptotic responses [43]. Similarly, structural and biochemical data from animal TLRs indicate that PAMP perception by the extracellular LRR region leads to the homodimerization of the cytosolic TIR domain [44]. This dimerisation provides a new scaffold that binds to adaptor proteins to initiate downstream immune signaling [45]. A few studies indicate that plant NB-LRR proteins can also oligomerise. The Arabidopsis RPS5 and Tomato Prf CC-NB-LRRs exist as oligomeric complexes prior to pathogen perception [46,47], while the Tobacco N TIR-NB-LRR protein forms TIR-dependent oligomers upon perception of the TMV p50 protein [48]. However, how these associations are linked to signaling activity is not well understood.
Protein structural studies of plant NB-LRRs are now coming to the front stage with the recent determination of the first crystal structures of two NB-LRR signaling domains, the CC domain of Barley MLA10 [49**] and the TIR domain of Flax L6 [50**]. These N-terminal domains are each sufficient to autonomously trigger defense signaling in planta and form homodimers in solution. Each protein fragment crystallized as a dimer and site directed mutations in the dimer interface disrupted both dimerization in vitro and signaling activity in planta, which strongly supports that oligomerization of the signaling domain is required to activate defense responses [49**,50**]. Similarly to Prf and RPS5, MLA1 (a closely related allelic variant of MLA10) self-associates in planta in the absence of the pathogen effector, and occurs in a 300–400kDa complex in mildew infected as well as in uninfected plants [49**]. According to size exclusion chromatography and BN-PAGE, the Prf complex fits with a Prf dimer associated with two molecules of the accessory Pto kinase protein [46,51], but it is not clear whether other proteins are present in the MLA1 complex. Interestingly purified full length MLA27 protein behaved as a monomer in vitro. Gel filtration chromatography coupled to multiangle laser light scattering (MALLS) together with Analytical Ultra Centrifugation demonstrated that the autoactive L6 TIR domain involves two molecules in vitro [50**]. However, non autoactive L6 protein fragments including the NB-ARC domain or the full length protein do not self-associate in yeast. This suggests that, like the Tobacco N protein, L6 needs to be activated to oligomerize, although there is no evidence for changes in L6 oligomerisation state in planta. Further mutational analyses of the L6 TIR domain identified a signaling region independent of the dimerization interface. Mutations in this region disrupt L6 TIR signaling activity in planta but not dimerization in yeast, suggesting that it may be involved in the recruitment of signaling partners subsequent to dimerization (figure 1).
Thus, oligomerization seems to be required for signaling activity for at least five different plant immune receptors. In the case of the three CC-NB-LRRs, an inactive dimer is present prior to activation, while dimerisation of the two TIR-NB-LRRs appears to be associated with activation (figure 1). This may reflect a basic mechanistic difference between these two classes of protein, but further analysis of additional examples of each will be required to test this. It is not yet clear whether higher order oligomeric complexes may also form after R protein activation, as is the case for the animal NLRs, although the MLA1 gel filtration profile remains unchanged after pathogen challenge suggesting no change in oligomerisation state [49**].
The steps following NB-LRR activation leading to signaling also remain obscure. So far very few studies have described signaling components interacting with plant immune receptors. Mutations in the MLA10 CC dimer interface not only affect signaling but also its interaction with the WRKY1 defense repressor [28,49**]. Although this interaction requires only the first 45 residues of MLA CC domain, which does not dimerize by itself, the whole MLA10 CC dimer unit may provide the right scaffold allowing WRKY factor recruitment to derepress defense genes transcription. The Arabidopsis Topless-related 1 (TPR1) protein has been identified in a genetic screen to suppress the snc1 mutation, an autoactive TIR-NB-LRR protein. Overexpression of TPR1 activates immune responses and this protein associates in vitro with SNC1 TIR domain [52]. However, knocking out TPR1 only partially affects SNC1 mediated resistance response, indicating that TPR1 is not the only component controlling defense signaling activation. However there is no evidence that WRKY1 and TPR1 are more generally involved in other R protein signaling pathways. Interestingly, although the CC or TIR domains from a number of different NB-LRRs show an autoactive phenotype when overexpressed, others do not [53,54]. Since these assays are mostly performed in Nicotiana benthamiana or N. tabacum, this may suggest the involvement of species-specific direct signaling partners. Purification of protein complexes containing activated receptors should provide further information on oligomerisation and early signaling partners initiating cascades downstream of R proteins.
Conclusion
The common structural and functional domain patterns of plant NB-LRRs suggest that these proteins rely on similar mechanisms for activation and signaling. However, although NB-LRRs clearly adopt some common strategies like recognition of effector-mediated enzymatic activity or receptor oligomerization for signaling activation, there are also significant functional differences related to their subcellular localization, mode of activation and interaction with specific signaling partners. The studies described here stand at the dawn of understanding resistance protein function and require further analyses to deepen these hypotheses and identify common and specific routes used by these receptors. For instance, studying and comparing the subcellular localization patterns of diverse NB-LRRs using real time microscopy techniques prior to and upon effector elicitation or pathogen infection will be crucial to understand the early stages of pathogen recognition and defense activation. Further detailed structural and biochemical approaches, such as solving the 3D structure of other plant NB-LRR domains, will be required to define structurally the intramolecular interactions and conformational changes that are involved in R protein activation. Identifying interacting partners of activated receptors will also be necessary to understand how activated resistance proteins induce defense signaling and whether common signaling routes are shared between NB-LRRs.
HIGHLIGHTS.
Effector-mediated enzymatic activity can be required in both direct and indirect R/Avr recognition systems.
Some NB-LRRs can activate defense signaling from the cytosolic compartment.
Crystal structures of two functional NB-LRRs signaling domains have been determined for the first time.
Receptor oligomerisation is a prerequisite for signaling.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Maud Bernoux, Email: Maud.Bernoux@csiro.au.
Jeffrey G. Ellis, Email: Jeffrey.Ellis@csiro.au.
Peter N. Dodds, Email: Peter.Dodds@csiro.au.
References
- 1.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. nature reviews genetics. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 3.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Van der Biezen EA, Jones JD. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- 5.Inohara, Chamaillard, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 6.Lukasik E, Takken FL. STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol. 2009;12:427–436. doi: 10.1016/j.pbi.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Rafiqi M, Bernoux M, Ellis JG, Dodds PN. In the trenches of plant pathogen recognition: Role of NB-LRR proteins. Seminars in Cell and Developmental Biology. 2009;20:1017–1024. doi: 10.1016/j.semcdb.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, Ayliffe MA, Kobe B, Ellis JG. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci U S A. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catanzariti AM, Dodds PN, Ve T, Kobe B, Ellis JG, Staskawicz BJ. The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. Mol Plant Microbe Interact. 2010;23:49–57. doi: 10.1094/MPMI-23-1-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. Embo J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci U S A. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krasileva KV, Dahlbeck D, Staskawicz BJ. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell. 2010;22:2444–2458. doi: 10.1105/tpc.110.075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 15.Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 16.Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 17.van der Hoorn RA, Kamoun S. From Guard to Decoy: a new model for perception of plant pathogen effectors. Plant Cell. 2008;20:2009–2017. doi: 10.1105/tpc.108.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18**.Chung E-H, da Cunha L, Wu A-J, Gao Z, Cherkis K, Afzal AJ, Mackey D, Dangl JL. Specific Threonine Phosphorylation of a Host Target by Two Unrelated Type III Effectors Activates a Host Innate Immune Receptor in Plants. Cell host & microbe. 2011;9:125–136. doi: 10.1016/j.chom.2011.01.009. This work shows that RIN4 is phosphorylated in vivo on residue Threonine at position 166 in presence of effectors AvrB and AvrRpm1. This phosphorylation event plays a central role in effector-triggered RPM1 activation as a RIN4 T166A mutant, that cannot be phosphorylated, affects RPM1 activation and a phosphomimic substitution induces effector independent RPM1 activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.Liu J, Elmore JM, Lin Z-JD, Coaker G. A Receptor-like Cytoplasmic Kinase Phosphorylates the Host Target RIN4, Leading to the Activation of a Plant Innate Immune Receptor. Cell host & microbe. 2011;9:137–146. doi: 10.1016/j.chom.2011.01.010. In this work, the authors describe a host protein kinase (RIPK) which is able to bind and phosphorylate RIN4 at different positions including the crucial Threonine at position 166 in response to bacterial effectors. RIPK also interacts with the effector AvrB suggesting that RIN4 phosphorylation is triggered indirectly by an effector-mediated RIPK activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Tasset C, Bernoux M, Jauneau A, Pouzet C, Briere C, Kieffer-Jacquinod S, Rivas S, Marco Y, Deslandes L. Autoacetylation of the Ralstonia solanacearum Effector PopP2 Targets a Lysine Residue Essential for RRS1-R Mediated Immunity in Arabidopsis. PLoS Pathog. 2010;6:e1001202. doi: 10.1371/journal.ppat.1001202. This paper describes RRS1 and PopP2 physical interaction in living plant nucleus using FLIM analysis. The authors also demonstrate that PopP2 displays an acetyl-transferase activity that is required for RRS1 mediated immunity but not for RRS1/PopP2 interaction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernoux M, Timmers T, Jauneau A, Briere C, de Wit PJ, Marco Y, Deslandes L. RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell. 2008;20:2252–2264. doi: 10.1105/tpc.108.058685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22*.Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 2009;60:218–226. doi: 10.1111/j.1365-313X.2009.03949.x. This and the following paper by Birker et al [20*], independently showed that a dominant locus RCH2 conferring resistance to the hemibiotrophic fungus Colletotrichum higginsianum contained two resistance genes, RRS1 and RPS4, previously characterised to confer resistance to the bacterial pathogens Ralstonia solanacearum and Pseudomonas syringae respectively. Phenotype analysis of T-DNA insertion mutants in RRS1 and RPS4 revealed that both proteins are required for resistance to several isolates of C. higginsianum but also to R. solanacearum and P. syringae suggesting that these genes act in concert for resistance to multiple pathogens. [DOI] [PubMed] [Google Scholar]
- 23*.Birker D, Heidrich K, Takahara H, Narusaka M, Deslandes L, Narusaka Y, Reymond M, Parker JE, O’Connell R. A locus conferring resistance to Colletotrichum higginsianum is shared by four geographically distinct Arabidopsis accessions. Plant J. 2009;60:602–613. doi: 10.1111/j.1365-313X.2009.03984.x. Together with Narusaka et al. [19*] this paper showed that the locus conferring resistance to C. higginsianum contains the RRS1 and RPS4 resistance genes and is conserved among different Arabidopsis accessions. Mutational analysis demonstrated that both proteins are required for recognition of multiple pathogens. Further analysis also showed that this resistance blocks the fungal invasion early at the penetration stage and that it is dependent on the immune regulator EDS1. [DOI] [PubMed] [Google Scholar]
- 24.Garcia AV, Parker JE. Heaven’s Gate: nuclear accessibility and activities of plant immune regulators. Trends Plant Sci. 2009;14:479–487. doi: 10.1016/j.tplants.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Deslandes L, Rivas S. The plant cell nucleus: A true arena for the fight between plants and pathogens. Plant Signal Behav. 2011;6:42–48. doi: 10.4161/psb.6.1.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 2007;5:e68. doi: 10.1371/journal.pbio.0050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science. 2007;315:1098–1103. doi: 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- 29.Wirthmueller L, Zhang Y, Jones JD, Parker JE. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol. 2007;17:2023–2029. doi: 10.1016/j.cub.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 30.Cheng YT, Germain H, Wiermer M, Bi D, Xu F, Garcia AV, Wirthmueller L, Despres C, Parker JE, Zhang Y, et al. Nuclear Pore Complex Component MOS7/Nup88 Is Required for Innate Immunity and Nuclear Accumulation of Defense Regulators in Arabidopsis. Plant Cell. 2009;21:2503–2516. doi: 10.1105/tpc.108.064519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiermer M, Palma K, Zhang Y, Li X. Should I stay or should I go? Nucleocytoplasmic trafficking in plant innate immunity. Cell Microbiol. 2007;9:1880–1890. doi: 10.1111/j.1462-5822.2007.00962.x. [DOI] [PubMed] [Google Scholar]
- 32**.Slootweg E, Roosien J, Spiridon LN, Petrescu AJ, Tameling W, Joosten M, Pomp R, van Schaik C, Dees R, Borst JW, et al. Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell. 2010;22:4195–4215. doi: 10.1105/tpc.110.077537. This paper shows that Rx mediated pathogen recognition and resistance activation is triggered from the cytosol as the nucleocytoplasmic Rx is not able to recognize its corresponding elicitor when the latter is sequestered in the nucleus. Relocating Rx to the nucleus by fusion of a nuclear localisation signal significantly inhibited resistance function, while nuclear exclusion had a minimal effect on resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33**.Tameling WI, Nooijen C, Ludwig N, Boter M, Slootweg E, Goverse A, Shirasu K, Joosten MH. RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell. 2010;22:4176–4194. doi: 10.1105/tpc.110.077461. This paper supports Slootweg et al [28**] and shows that Rx cytosolic and nuclear pools are both required for proper regulation of defense signaling. The authors demonstrated that Rx interacting protein RanGAP2 controls Rx nucleocytoplasmic partitioning and can act as a cytoplasmic retention factor of Rx. Concentrating Rx in the cytosol by overexpressing RanGAP2 enhanced resistance signaling, while sequestering Rx in the nucleus by interaction with a nuclear localized version of RanGAP2 inhibited resistance signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tameling WIL, Baulcombe DC. Physical association of the NB-LRR resistance protein Rx with a ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell. 2007;19:1682–1694. doi: 10.1105/tpc.107.050880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sacco MA, Mansoor S, Moffett P. A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J. 2007;52:82–93. doi: 10.1111/j.1365-313X.2007.03213.x. [DOI] [PubMed] [Google Scholar]
- 36*.Gao Z, Chung EH, Eitas TK, Dangl JL. Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc Natl Acad Sci U S A. 108:7619–7624. doi: 10.1073/pnas.1104410108. In this work, the authors demonstrate by microscopy analyses and membrane fractionation that RPM1 remains associated with the plasma membrane and is not relocated to the nucleus upon activation. Furthermore, its function is not affected by nuclear exclusion of direct physical tethering to the plasma membrane, suggesting that initiation of the defense signal occurs outside the nucleus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci U S A. 1998;95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howles P, Lawrence G, Finnegan J, McFadden H, Ayliffe M, Dodds P, Ellis J. Autoactive alleles of the flax L6 rust resistance gene induce non-race-specific rust resistance associated with the hypersensitive response. Mol Plant Microbe Interact. 2005;18:570–582. doi: 10.1094/MPMI-18-0570. [DOI] [PubMed] [Google Scholar]
- 39**.Garcia AV, Blanvillain-Baufume S, Huibers RP, Wiermer M, Li G, Gobbato E, Rietz S, Parker JE. Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog. 2010;6:e1000970. doi: 10.1371/journal.ppat.1000970. This paper shows that nucleo-cytoplasmic immune regulator EDS1 accumulates in the nucleus in response to pathogen infection where it activate defense gene reprogramming. However, the authors also showed that EDS1 cytoplasmic pool is important for full resistance activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takken FL, Tameling WI. To nibble at plant resistance proteins. Science. 2009;324:744–746. doi: 10.1126/science.1171666. [DOI] [PubMed] [Google Scholar]
- 41*.Williams SJ, Sornaraj P, deCourcy-Ireland E, Menz RI, Kobe B, Ellis J, Dodds P, Anderson PA. An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, while wild-type M protein has a preference for binding ADP. Molecular Plant-Microbe Interactions. 2011 doi: 10.1094/MPMI-03-11-0052. in press. In this paper the authors express a full length TIR-NB-LRR resistance protein, the flax M protein, in Pichia pastoris and show that the purified protein it is bound to ADP. Mutations in the NB domain P-loop abolish ADP binding, while an autoactive M protein (containing a D to V substitution in the MHD motif) is bound to ATP. This provides direct support for the ADP OFF/ATP ON switch model of plant NB-LRR protein activation. [DOI] [PubMed] [Google Scholar]
- 42.Danot O, Marquenet E, Vidal-Ingigliardi D, Richet E. Wheel of life, wheel of death: A mechanistic insight into signaling by STAND proteins. Structure. 2009;17:172–182. doi: 10.1016/j.str.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 43.Qi S, Pang Y, Hu Q, Liu Q, Li H, Zhou Y, He T, Liang Q, Liu Y, Yuan X, et al. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell. 2010;141:446–457. doi: 10.1016/j.cell.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 44.Botos I, Segal DM, Davies DR. The Structural Biology of Toll-like Receptors. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tapping RI. Innate immune sensing and activation of cell surface Toll-like receptors. Semin Immunol. 2009;21:175–184. doi: 10.1016/j.smim.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 46.Gutierrez JR, Balmuth AL, Ntoukakis V, Mucyn TS, Gimenez-Ibanez S, Jones AM, Rathjen JP. Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J. 2009 doi: 10.1111/j.1365-313X.2009.04078.x. [DOI] [PubMed] [Google Scholar]
- 47.Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci U S A. 2007;104:2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mestre P, Baulcombe DC. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell. 2006;18:491–501. doi: 10.1105/tpc.105.037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49**.Maekawa T, Cheng W, Spiridon LN, Töller A, Lukasik E, Saijo Y, Liu Shen Q-H, Micluta MA, Somssich IE, et al. Coiled-Coil Domain-Dependent Homodimerization of Intracellular Barley Immune Receptors Defines a Minimal Functional Module for Triggering Cell Death. Cell host & microbe. 2011;9:187–199. doi: 10.1016/j.chom.2011.02.008. This paper describes for the first time the crystal structure of the N-terminal Coiled coil domain of a functional plant NB-LRR, the barley MLA resistance protein. The authors demonstrated that MLA self-associates in a pathogen independent manner and that this association is directed by its CC domain. Mutations in the CC dimer interface compromised MLA resistance activity suggesting that MLA self-association is a prerequisite for receptor activation. [DOI] [PubMed] [Google Scholar]
- 50**.Bernoux M, Ve T, Williams S, Warren C, Hatters D, Valkov E, Zhang X, Ellis JG, Kobe B, Dodds PN. Structural and Functional Analysis of a Plant Resistance Protein TIR Domain Reveals Interfaces for Self-Association, Signaling, and Autoregulation. Cell host & microbe. 2011;9:200–211. doi: 10.1016/j.chom.2011.02.009. This paper describes for the first time the crystal structure of the N-terminal TIR domain of a functional plant NB-LRR, the flax L6 resistance protein. A fine structure function analysis demonstrated that L6 TIR signaling domain self-associates and that this association is required to trigger defense signaling. Further analyses identified a signaling interface which may be involved in downstream signaling partner recruitment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mucyn TS, Clemente A, Andriotis VM, Balmuth AL, Oldroyd GE, Staskawicz BJ, Rathjen JP. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell. 2006;18:2792–2806. doi: 10.1105/tpc.106.044016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, Li X. Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci U S A. 2010;107:13960–13965. doi: 10.1073/pnas.1002828107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swiderski MR, Birker D, Jones JD. The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol Plant Microbe Interact. 2009;22:157–165. doi: 10.1094/MPMI-22-2-0157. [DOI] [PubMed] [Google Scholar]
- 54.Collier SM, Hamel L-P, Moffett P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Molecular Plant-Microbe Interactions. 2011 doi: 10.1094/MPMI-03-11-0050. in press. [DOI] [PubMed] [Google Scholar]