Abstract
Background
Anxiety disorders are common psychiatric conditions that are highly comorbid with each other and related phenotypes such as depression, likely due to a shared genetic basis. Fear-related behaviors in mice have long been investigated as potential models of anxiety disorders, making integration of information from both murine and human genetic data a powerful strategy for identifying potential susceptibility genes for these conditions.
Methods
We combined genome-wide association analysis of fear-related behaviours with strain distribution pattern analysis in heterogeneous stock mice to identify a preliminary list of 52 novel candidate genes. We ranked these according to three complementary sources of prior anxiety-related genetic data: (1) extant linkage and knock-out studies in mice, (2) a meta-analysis of human linkage scans, and (3) a preliminary human genomewide association study. We genotyped tagging SNPs covering the nine top-ranked regions in a two-stage association study of 1316 subjects from the Virginia Adult Twin Study of Psychiatric and Substance Use Disorders chosen for high or low genetic loading for anxiety-spectrum phenotypes (anxiety disorders, neuroticism, and major depression).
Results
Multiple SNPs in the PPARGC1A gene demonstrated association in both stages that survived gene-wise correction for multiple testing.
Conclusions
Integration of genetic data across human and murine studies suggests PPARGC1A as a potential susceptibility gene for anxiety-related disorders.
Keywords: anxiety disorder, depression, internalizing, candidate gene, genetic association, data integration
Introduction
Anxiety disorders (ADs), like generalized anxiety disorder (GAD), panic disorder, and phobias, are common, disabling conditions with substantial lifetime prevalence (1). They tend to persist throughout the life course and exhibit strong comorbidity with each other and with other internalizing disorders like major depressive disorder (MDD). Twin and family studies implicate genetic factors in their etiology, with moderate levels of familial aggregation and heritability (2). Neuroticism, a personality trait reflecting a tendency towards states of negative affect, not only increases risk of individual internalizing disorders but also their comorbidity (3–5). Twin studies suggest that genetic factors underlying neuroticism overlap substantially with those that increase susceptibility to anxiety and depressive symptoms and disorders (genetic pleiotropy) (6–8). Thus, studying these phenotypes in a coordinated manner may be an efficient and powerful approach for identifying susceptibility genes for ADs.
Identification of susceptibility genes for psychiatric and other complex medical conditions remains a challenge. Molecular genetic investigations of human ADs are somewhat behind that of other psychiatric conditions (9). The majority are candidate gene analyses targeting panic disorder, most either without consistent replication or with negative results (10). Two modest-sized genome-wide association studies (GWAS) for panic disorder have been recently published, the first in a Japanese sample (11) and the second in a German sample (12), the latter supporting a role for the gene TMEM132D.
Animal models of fear-related behaviors have been pursued as one strategy to explore the biologic and genetic basis of human ADs (13). One linkage study targeting regions implicated in rodent anxiety quantitative trait locus (QTL) studies (14) and several association studies (e.g., (15)) have taken this approach. Given the limited evidence supporting the set of candidate genes thus far examined for their putative role in AD susceptibility and the large number of potential candidates in the genome, integration across multiple, complementary sources of data are crucial to identifying the genes involved in these conditions. As yet, there have been no attempts to systematically combine genomewide rodent and human data to select a set of candidate susceptibility genes for testing in a powerful human sample.
The aims of this study are to (1) identify a set of candidate genes associated with fear-related behaviors in mice, (2) rank these using independent human and mouse data, and (3) confirm the association of the top candidates with human anxiety-spectrum disorders in a large twin sample.
Materials and Methods
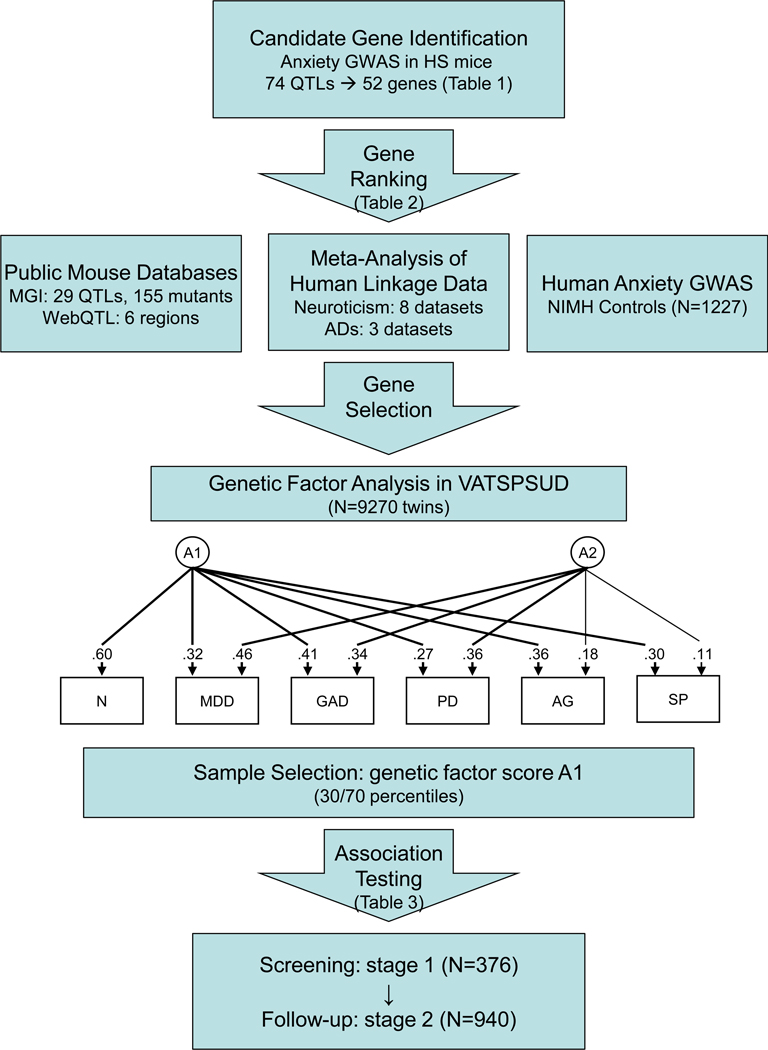
We employed gene identification, data integration, and data analysis procedures to accomplish the three proposed aims of the study, as illustrated in Figure 1 and summarized below.
Figure 1.
Gene identification, ranking, and analysis scheme. Path diagram depicts twin analysis and resulting genetic factor structure used for selecting VATSPSUD subjects high and low on common internalizing genetic risk A1 (adapted from (7)).
Abbreviations: GWAS – genomewide association study; QTL – quantitative trait locus; HS - heterogeneous stock; MGI - Mouse Genome Informatics; ADs – anxiety disorders; VATSPSUD - Virginia Adult Twin Study of Psychiatric and Substance Use Disorders; N – Neuroticism; MDD – major depressive disorder; GAD – generalized anxiety disorder; PD – panic disorder; AG – agoraphobia; SP – social phobia;
Candidate Gene Identification
We began with a set of genes derived from a GWAS of a large array of phenotypes measured in mice we conducted previously (16). That study exploited historical recombinants that have accumulated in genetically heterogeneous stock (HS) mice descended from eight inbred progenitor strains for high-resolution mapping of small-effect QTLs (17). The HS chromosomes are fine-grained mosaics of the progenitor haplotypes and capture most of the genetic variability in the founders. Anxiety-related phenotypic data of relevance for the current study included measures of open field activity, elevated plus maze activity, food hyponeophagia, startle, and contextual and cued freezing. The study obtained genotypes for 13,459 markers (mean interval 204.4 Kb) on 1,904 phenotyped mice and 298 parents. Using
Our primary analysis identified 74 QTLs that influence performance in the behavioural test battery, with 267 genes under their 95% confidence intervals. We further narrowed this list of genes by constraining the observed QTLs by data available in the progenitor strains (see Supplement for details). We were able to identify 52 genes most likely associated with the observed anxiety-related QTLs (Table 1).
Table 1.
Anxiety-related QTL Identified in Murine GWAS
| Mouse Gene | Mouse Chromosome |
Position (Mb) |
Human Gene |
Human Chromosomal Location |
Position (Mb) |
RMIP |
|---|---|---|---|---|---|---|
| Tnr | 1 | 159.8 | TNR | 1q25.1 | 173.6 | 0.29 |
| Lnp | 2 | 74.2 | KIAA1715 | 2q31.1 | 176.5 | 0.49 |
| Evx2 | 2 | 74.4 | EVX2 | 2q31.1 | 176.6 | 0.49 |
| Hoxd8 | 2 | 74.4 | HOXD8 | 2q31.1 | 176.7 | 0.49 |
| Hoxd3 | 2 | 74.5 | HOXD3 | 2q31.1 | 176.7 | 0.49 |
| Hoxd1 | 2 | 74.5 | HOXD1 | 2q31.1 | 176.7 | 0.49 |
| Flrt3 | 2 | 140.2 | FLRT3 | 20p12.1 | 14.2 | 0.86 |
| Kif16b | 2 | 142.1 | C20orf23 | 20p12.1 | 16.3 | 0.86 |
| Csrp2bp | 2 | 143.9 | CSRP2BP | 20p11.23 | 18.1 | 0.86 |
| Slc24a3 | 2 | 145.1 | SLC24A3 | 20p11.23 | 19.1 | 0.86 |
| Pygb | 2 | 150.3 | PYGB | 20p11.21 | 25.2 | 0.29 |
| BC057079 | 4 | 87.4 | KIAA1797 | 9p21.3 | 20.8 | 0.91 |
| Ppargc1a | 5 | 50.3 | PPARGC1A | 4p15.2 | 23.4 | 0.32 |
| Gats | 5 | 133.4 | GATS | 7q22.1 | 99.6 | 0.35 |
| Wbscr16 | 5 | 133.4 | WBSCR16 | 7q11.23 | 74.6 | 0.35 |
| Gtf2i | 5 | 133.5 | GTF2I | 7q11.23 | 73.7 | 0.35 |
| Pdk4 | 6 | 5.5 | PDK4 | 7q21.3 | 95.0 | 0.38 |
| Stk31 | 6 | 49.6 | STK31 | 7p15.3 | 23.7 | 0.34 |
| Eefsec | 6 | 88.7 | EEFSEC | 3q21.3 | 129.4 | 0.35 |
| Nek3 | 8 | 20.9 | NEK3 | 13q14.3 | 51.6 | 0.38 |
| Pdlim3 | 8 | 44.9 | PDLIM3 | 4q35.1 | 186.6 | 0.40 |
| 4933411K20Rik | 8 | 45.1 | KIAA1430 | 4q35.1 | 186.3 | 0.40 |
| Slc25a4 | 8 | 45.2 | SLC25A4 | 4q35.1 | 186.3 | 0.40 |
| Acsl1 | 8 | 45.5 | ACSL1 | 4q35.1 | 185.9 | 0.40 |
| Suhw4 | 9 | 72.5 | SUHW4 | 15q21.3 | 54.7 | 0.52 |
| Mns1 | 9 | 72.6 | MNS1 | 15q21.3 | 54.5 | 0.52 |
| Cyb561d2 | 9 | 107.6 | CYB561D2 | 3p21.31 | 50.3 | 0.49 |
| Tusc4 | 9 | 107.6 | TUSC4 | 3p21.31 | 50.3 | 0.49 |
| Zmynd10 | 9 | 107.6 | ZMYND10 | 3p21.31 | 50.3 | 0.49 |
| Rassf1 | 9 | 107.6 | RASSF1 | 3p21.31 | 50.3 | 0.49 |
| Hyal2 | 9 | 107.6 | HYAL2 | 3p21.31 | 50.3 | 0.49 |
| Usp19 | 9 | 108.6 | USP19 | 3p21.31 | 49.1 | 0.49 |
| Mtmr3 | 11 | 4.4 | MTMR3 | 22q12.2 | 28.6 | 0.81 |
| Ascc2 | 11 | 4.6 | ASCC2 | 22q12.2 | 28.5 | 0.81 |
| Nf2 | 11 | 4.7 | NF2 | 22q12.2 | 28.4 | 0.81 |
| BC018601 | 11 | 5.4 | CCDC117 | 22q12.1 | 27.5 | 0.81 |
| Pnpt1 | 11 | 29.1 | PNPT1 | 2p16.1 | 55.7 | 0.37 |
| AW011752 | 11 | 29.1 | SMEK2 | 2p16.1 | 55.7 | 0.37 |
| Cpeb4 | 11 | 31.8 | CPEB4 | 5q35.2 | 173.2 | 0.37 |
| Gria1 | 11 | 57.0 | GRIA1 | 5q33.2 | 152.9 | 0.28 |
| Gngt2 | 11 | 95.7 | GNGT2 | 17q21.32 | 44.6 | 0.45 |
| Slc10a1 | 12 | 77.8 | SLC10A1 | 14q24.2 | 69.3 | 0.33 |
| Kif21a | 15 | 91.0 | KIF21A | 12q12 | 38.0 | 1.00 |
| 4432406C05Rik | 16 | 8.3 | TMEM186 | 16p13.2 | 8.8 | 0.65 |
| Carhsp1 | 16 | 8.3 | CARHSP1 | 16p13.2 | 8.8 | 0.65 |
| Pmm2 | 16 | 8.3 | PMM2 | 16p13.2 | 8.8 | 0.65 |
| 1810013L24Rik | 16 | 8.5 | C16orf72 | 16p32.2 | 9.1 | 0.65 |
| Slc22a1 | 17 | 11.3 | SLC22A1 | 6q25.3 | 160.5 | 0.27 |
| Acat3 | 17 | 11.6 | ACAT2 | 6q25.3 | 160.1 | 0.27 |
| Mas1 | 17 | 11.7 | MAS1 | 6q25.3 | 160.2 | 0.27 |
| Crim1 | 17 | 76.2 | CRIM1 | 2p22.2 | 36.5 | 0.50 |
| Arl6ip2 | 17 | 77.7 | ARL6IP2 | 2p22.2 | 38.4 | 0.50 |
RMIP - resample inclusion model probability score (see text for explanation).
Data Integration
We obtained data from three independent and complementary sources of anxiety-related genetic data in order to prioritize these 52 genes for further study. For evaluating support from these other data, we grouped genes together that corresponded to the same original QTL signal (i.e., from the same murine chromosomal region). Due to their proximity and limited resolution of the independent data, most of these were assigned common p-values and final rankings (see Table 2). Some achieved differential ranking after applying all of the independent data and were separated in the final prioritization (e.g., Crim1 and Arl6ip2 on mouse chromosome 17).
Table 2.
Ranking of top 25 genes using data from public mouse databases, linkage meta-analysis, and human GWAS. Shaded genes were selected for further association testing.
| Human Gene | Chr | LOC (Mb) |
Mouse PMouse |
Linkage PLinkage |
GWAS Pmin |
GWAS PGwas |
GWAS QGwas |
Overall Rank |
|---|---|---|---|---|---|---|---|---|
| SLC10A1 | 14 | 69.3 | 0 | 0.0482 | 0.0133 | 0.066 | 0.066 | 1 |
| GATS | 7 | 99.6 | 0 | 0.0378 | 0.0805 | 0.161 | 0.16 | 2 |
| KIF21A | 12 | 38 | 0 | 0.1302 | 0.0551 | 0.441 | 0.16 | 3 |
| GTF2I | 7 | 73.7 | 0 | 0.2344 | 0.0005 | 0.003 | 0.001 | 4 |
| WBSCR16 | 7 | 74.1 | 0 | 0.2344 | 0.0005 | 0.003 | 0.001 | 4 |
| CPEB4 | 5 | 173.3 | 0 | 0.0102 | 0.0581 | 0.697 | 0.33 | 6 |
| TNR | 1 | 173.8 | 0 | 0.2172 | 0.0024 | 0.054 | 0.20 | 7 |
| PPARGC1A | 4 | 23.4 | 1 | 0.4504 | 0.0003 | 0.006 | 0.006 | 8 |
| HYAL2 | 3 | 50.3 | 0 | 0.2044 | 0.1518 | 0.455 | 0.31 | 9 |
| RASSF1 | 3 | 50.3 | 0 | 0.2044 | 0.1518 | 0.455 | 0.31 | 9 |
| TUSC4 | 3 | 50.3 | 0 | 0.2044 | 0.1518 | 0.455 | 0.31 | 9 |
| ZMYND10 | 3 | 50.3 | 0 | 0.2044 | 0.1518 | 0.455 | 0.31 | 9 |
| CYB561D2 | 3 | 50.4 | 0 | 0.2044 | 0.1518 | 0.455 | 0.31 | 9 |
| NF2 | 22 | 28.4 | 0 | 0.5021 | 0.0001 | 0.006 | 0.003 | 14 |
| CRIM1 | 2 | 36.5 | 1 | 0.0344 | 0.0266 | 1.0 | 0.90 | 14 |
| SMEK2 | 2 | 55.6 | 0 | 0.0644 | 0.0979 | 1.0 | 0.60 | 16 |
| PNPT1 | 2 | 55.7 | 0 | 0.0644 | 0.0979 | 1.0 | 0.60 | 16 |
| ARL6IP2 | 2 | 38.4 | 1 | 0.0244 | 0.3860 | 1.0 | 0.95 | 16 |
| CCDC117 | 22 | 27.5 | 0 | 0.5021 | 0.0329 | 0.165 | 0.16 | 19 |
| STK31 | 7 | 23.8 | 0 | 0.0677 | 0.1849 | 1.0 | 0.75 | 20 |
| SUHW4 | 15 | 54.8 | 0 | 0.1002 | 0.1752 | 1.0 | 0.95 | 21 |
| ACSL1 | 4 | 185.9 | 0 | 0.2039 | 0.0140 | 1.0 | 0.71 | 22 |
| KIAA1430 | 4 | 186.3 | 0 | 0.2039 | 0.0140 | 1.0 | 0.71 | 22 |
| SLC25A4 | 4 | 186.3 | 0 | 0.2039 | 0.0140 | 1.0 | 0.71 | 22 |
| MNS1 | 15 | 54.5 | 0 | 0.1132 | 0.1752 | 1.0 | 0.95 | 25 |
Chr – chromosome; loc – location; PMouse – whether homologous gene was found in “anxiety” keyword search of public mouse databases (1=”yes”, 0=”no”); PLinkage – Fisher’s combined p-value from human linkage meta-analysis; GWAS = human genomewide association study; Pmin – minimum p-value observed in gene; PGWAS – Bonferroni corrected minimum p-value; QGWAS – minimum q-value for the gene using conservative estimation (P0=1) based upon the set of genes in the study.
(1) Public Mouse Databases
We searched two public databases containing genetic data for anxiety phenotypes in mice. First, we searched for mouse QTLs using keyword “anxiety” in the Mouse Genome Informatics (MGI) database (http://www.informatics.jax.org/). We identified 29 anxiety-related QTLs (accessed 01/07/08). For these QTLs whose both start and end markers were known in MGI, we retrieved genes mapped to the corresponding genomic regions. Among the 52 candidate genes based on our mouse data, four appeared in the gene list within these QTLs. Also in the MGI database, we searched genes in the “mouse phenotypes & human disease” category using keyword “anxiety”. We found 155 gene mutations associated with anxiety-related phenotypes. Among them, two genetic knockouts appeared in the list of 52 candidate genes from the murine GWAS. We also searched anxiety-related records in WebQTL (http://www.genenetwork.org/). We found 6 anxiety-related records in mouse BXD group (accessed 01/07/08). Then, using the interval mapping function to identify genes under the significant peaks of these records, we found no correspondence with our candidate gene list. No genes appeared in more than one of these three searches. We categorically scored those with “hits” in these databases with a “1” vs. “0” for those not found in these lists.
(2) Linkage Meta-analysis
We applied the results of a genomewide linkage meta-analysis using primary data from eight independent genome-wide linkage studies of neuroticism and three of anxiety disorders, totalling 14,800 and 718 subjects, respectively (see Webb et al., in review, for details). We applied rank-based genome scan meta-analysis (GSMA) and Fisher’s method of combining p-values, with reasonable agreement between the methods genome-wide (r=0.87). The two phenotypes were initially analysed separately and rank-based p-values reported by chromosomal bin. Since these analyses also provided evidence of moderate correlation in genetic susceptibility between neuroticism and anxiety disorders in regions showing linkage, we then performed a joint meta-analysis across these phenotypes. Consistent with our strategy in the current study to maximally integrate information across anxiety-related phenotypes, we used Fisher’s combined p-value (designated PLinkage) calculated for all 11 datasets together to rank the candidate genes within each 1 Mb interval.
(3) Human GWAS
Finally, we performed a preliminary GWAS in the NIMH Center for Collaborative Genetics Studies on Mental Disorders dataset (www.nimhgenetics.org). We used early release data from the “control” sample originally part of a large schizophrenia study (Molecular Genetics of Schizophrenia (MGS): PI and Collaboration Coordinator, P.V.Gejman). The available sample consisted of unrelated subjects selected by random digit dialing from approximately 60,000 US households. The full MGS control sample is described in detail in a recent publication (18).
All control subjects completed an on-line psychiatric screening interview that included the lifetime version of the Composite International Diagnostic Interview, Short Form (CIDI-SF) (19). For those subjects with requisite response data (N=3775), we applied DSM-based algorithms to the CIDI-SF responses to obtain the following lifetime diagnoses (prevalence): MDD (28%), GAD (18%), panic (2%), agoraphobia (4%), social phobia (14%), and specific phobia (12%). The short form of the Eysenck Personality Questionnaire (EPQ-SF) (20) was used to calculate each subject’s total neuroticism score (range 0–12, mean 3.5).
Given prior evidence supporting shared genetic liability across these internalizing phenotypes, we performed factor analyses to estimate an overall internalizing score for each subject. We entered the six internalizing disorder phenotypes and the ordinal neuroticism score into factor analyses in Mplus (version 4) (21). Exploratory factor analyses (EFA) with one versus two latent factors imposed to explain correlation among these phenotypes each produced reasonable solutions that adequately fit the data. While the latter reproduced the familiar structure of correlated “misery” versus “fear” factors seen in other analyses of internalizing phenotypes (22), we chose to use the former solution representing a single overall “internalizing” factor for simplicity. Supporting this, EFA estimated one significant factor (eigenvalue=4.29) for the correlation matrix. The estimated factor loadings for each variable ranged from 0.64 (specific phobias) to 0.84 (GAD). We used confirmatory factor analysis to extract a factor score for each subject for use as the phenotype in association analyses.
We have previously provided details of another GWA analysis performed in this sample (23). Briefly, genotype data using the Affymetrix 500K "A" chipset (www.affymetrix.com/support/technical/datasheets/500k_datasheet.pdf) were generated at the Center for Genotyping & Analysis at the Broad Institute as part of a multi-institutional collaborative research study. In the early release of data from the NIMH repository used in the current study, genotypes were only available for about 1,700 subjects. Prior to performing association analyses, we removed individual samples with (1) call rates < 95%, (2) > 10% missing SNPs, or (3) unusual degrees of relatedness or heterozygosity, reducing the sample available for analysis to 1,227 Caucasian subjects. The total number of SNPs was reduced from 500,568 to 420,287 after deleting those with (1) > 10% missing genotypes, (2) minor allele frequencies < 0.005, and (3) HWE p-values < 1 e-05.
We performed regression analyses assuming additive genetic effects in PLINK (24) to test these 420,287 autosomal SNPs for association with the internalizing factor scores. As previously described, PLINK’s multi-dimensional scaling (MDS) module estimated that three dimensions adequately represented population substructure in the sample, included as covariates. Considering several different types of significance values as the optimal measure of gene effects, we calculated two statistical measures for ranking each of the 52 genes. First, we took the minimum p-value observed in the gene and conservatively corrected it by multiplying by the number of markers genotyped in that gene (PGWAS). Second, we used false discovery methodology to estimate the minimum q-value for each gene (QGWAS)(25). The higher the value of QGWAS, the more likely that the gene in question does not harbor a polymorphism with a true association.
Candidate Gene Ranking and Selection
Using these three sources of external data, we prioritized the 52 genes from the murine GWAS. Specifically, we calculated ranks separately for four measures: PMouse, PLinkage, PGWAS, QGWAS. We chose to use both PGWAS and QGWAS from the human GWAS analysis, since that dataset likely provides the most specific information for each gene regarding anxiety disorder susceptibility, and these two measures provide somewhat different types of evidence supporting association. We calculated an average rank across these four measures for each gene and rank-ordered the list of genes. We selected the top ranked genomic regions for replication testing in the following independent association sample.
Subjects
The subjects in this portion of the study derive from the longitudinal population-based Virginia Adult Twin Study of Psychiatric and Substance Use Disorders (VATSPSUD) (26;27) All subjects were Caucasian and born in Virginia by study design. Their age (mean, SD, range) at time of last interview was (37, 9, 20–58) for males and (36, 8, 21–62) for females. Approval of the local Institutional Review Board was obtained prior to the study and informed consent was obtained from all subjects.
Diagnostic Measures
We obtained lifetime psychiatric diagnoses via face-to-face or telephone structured psychiatric interview based on the Structured Clinical Interview for DSM-III-R (SCID) (28). Neuroticism was assessed using the 12 items from the EPQ-SF (29) via self-report questionnaire.
Sample Selection
As described previously (30), we have incorporated two novel strategies into our subject selection procedure (see lower portion of Figure 1). First, we have taken advantage of extant findings that suggest shared genetic susceptibility among neuroticism, ADs, and MDD (7;30–33) to estimate scores for common latent genetic factors across these phenotypes in the total twin sample (N=9270) [see (7) for details]. Briefly, an independent pathway model with two orthogonal additive genetic factors was found to best fit the data, with the first factor, A1, reflecting genetic risk shared most broadly across these phenotypes. Second, we used an extreme selection strategy (34–36) to choose one member from each twin pair for whom DNA was available as a case or control for genotyping based upon scoring above the 70th or below the 30th percentile, respectively, of the genetic factor A1 extracted from the above analysis. Selecting subjects from the extremes of their underlying genetic risk factor should provide a powerful method for detecting genes of small effect expected to contribute to complex genetic phenotypes. We obtained a total sample size N=1316 consisting of 713 cases (355 males, 358 females) and 603 controls (386 males, 217 females), of which 376 and 940 were used in stage 1 and stage 2 respectively. Overall, the cases had a mean neuroticism score of 5.9 out of 12 maximum (z-score= 0.69) and had the following frequencies of the target psychiatric illnesses: MDD (76.9%), GAD (50.7%), panic disorder (20.7%), agoraphobia (13.8%), and social phobia (17.0%). The controls were free of these five disorders and had a mean neuroticism score of 0.66 (z-score = −0.79).
Statistical Analysis
We used a 2-stage association design in which candidate loci were screened in stage 1, the positive results of which were tested for replication in stage 2. Selecting the most informative subjects from the extremes of the phenotypic distribution, the LGA972 program (37) indicated that we needed approximately 300 subjects in stage 1 and 1,000 in stage 2 to achieve 80% power to detect markers that explained 2% of the variance of the liability distribution while controlling the false discovery rate at 0.1 across the two stages (38). We used Pearson’s χ2 tests in PLINK to assess allelic differences by marker between cases and control subjects separately by stage. Any markers genotyped in stage 1 that met the screening p-value threshold of 0.1 were then tested in the stage 2 sample.
Given strong arguments for gene-based hypothesis testing (39), we examined the evidence for association across the SNPs in our most strongly implicated genes. Importantly, we perform gene-wise permutations as a means of describing the evidence at our most significant genes and not as our primary test statistic. Debate exists about the proper approach to calculating gene-based significance (see Supplement). Thus, we report multiple approaches to gene-based significance as a test of consistency across the methods.
Genotyping
DNA was extracted from buccal epithelial cells obtained via cytology brushes (40). SNPs were genotyped by the 5’ nuclease cleavage assay (TaqMan method) (41), as described in detail in prior studies in this sample (30). We selected SNP markers with MAF > 0.05 in each of the regions with the aim to tag the major haplotypes in the Caucasian panel of the HapMap project (42). Specifically, we used pair-wise tagging with r2>0.8 in the Tagger module of HAPLOVIEW 4.1 (43) with HapMap Phase II (release 22) data. We imposed quality control thresholds to exclude poorly performing assays: genotyping rate<90%, missingness>10%, HW <0.01.
Results
As illustrated in the upper portion of Figure 1, we used three complementary sources of prior anxiety-related genetic data to rank the 52 genes identified in the murine association study: (1) extant linkage and knock-out studies in mice, (2) a meta-analysis of human linkage scans, and (3) a preliminary human GWAS study. The results of this ranking procedure are depicted in Table 2 for the top 25 of the 52 candidate genes. Ties occurred in adjacent genes grouped within the same chromosomal region derived from the same murine QTL. We selected the top nine regions (14 genes – shaded in Table 2) to test for association in the human VATSPSUD sample. The genotyping and association results, by stage, are summarized in Table 3 for these regions. Details by region and stage are provided in Table S1 in the Supplement. As indicated, only 110 of the total 126 SNPs genotyped successfully. Seven of the 16 failed SNPs were located in the 7q11.23 region, six of these near the gene WBSCR16. We tried to replace the failed SNPs with others in LD with these, but we were unable to successfully genotype any available HapMap SNPs in that location, resulting in poor coverage of this region.
Table 3.
Stage 1 and stage 2 association summary results by gene region
| Region | Genes in Region | Region Size (Kb) / # Blocks |
# SNPs selected |
# SNPs P<0.1 of total genotyped (stage 1) |
# SNPs P<0.05 of total genotyped (stage 2) |
|---|---|---|---|---|---|
| 1q25.1 | TNR | 98.7 / 6 | 22 | 6 / 19 | 0 / 6 |
| 3p21.31 | HYAL2, RASSF1, TUSC4, ZMYND10, CYB561D2 | 34.5 / 1 | 5 | 0 / 3 | NA |
| 4p15.2 | PPARGC1A | 98.0 / 7 | 35 | 4 / 32 | 7 / 32 2 |
| 5q35.2 | CPEB4 | 74.5 / 2 | 13 | 1 / 13 | 0 / 1 |
| 7q11.23 | GTF2I, WBSCR16 | 477.2 / 3 | 15 | 0 / 8 1 | NA |
| 7q22.1 | GATS (STAG3) | 103.8 / 1 | 5 | 1 / 5 | 0 / 1 |
| 12q12 | KIF21A | 163.2 / 1 | 10 | 0 / 9 | NA |
| 14q24.2 | SLC10A1 | 38.4 / 1 | 6 | 0 / 6 | NA |
| 22q12.2 | NF2 | 99.3 / 3 | 15 | 3 / 15 | 0 / 3 |
| TOTAL | 9 regions, 14 genes | 1187.6 | 126 | 15 / 110 | 7 / 43 |
genotyping problematic in this region
genotyped all 32 markers in both stages
Of the 110 SNPs tested in the stage 1 sample, 15 showed preliminary association at our designated threshold (p<0.1) for genotyping in stage 2. These were in TNR, PPARGC1A, GATS, and NF2. None of the associations for SNPs from TNR, GATS, or NF2 brought forward in stage 2 were replicated. One (rs12640088) of the four stage 1 SNPs from PPARGC1A was associated in stage 2 and two others showed trend level association (p<0.1). The PPARGC1A gene is fairly large (98 Kb) and has a complex LD structure, with 7 haplotype blocks requiring 35 SNPs to tag the major allelic variation. Noting other trend-level p-values in stage 1 that were distributed across this gene, we decided to genotype all of the 32 successfully genotyped SNPs in the stage 2 sample. Seven of these showed association at p<0.05, and nine markers in total were significant when combining data across subjects from both stages (Table 4). One marker, rs3796407, with a nominal p-value of 4.2 × 10−4, remains marginally significant after correcting for the 110 SNPs tested in this study (corrected p=0.046). This SNP is part of a five marker LD block that contains other associated SNPs forming a common haplotype that is found in 31% of cases and 23% of controls (p=6 × 10−7). Using gene-based testing for PPARGC1A, we attained p-values after permutation of 0.003, 0.007, 0.006, 0.006 and 0.038 for Fisher's combined, minp, TPM (truncated at .05), TPM (at .01), and TPM (at .005), respectively.
Table 4.
PPARGC1A Association Results for Stage 1 + Stage 2 (677 cases, 592 controls)
| SNP | Allele Freq (Cases) |
Allele Freq (Controls) |
χ2 | P |
|---|---|---|---|---|
| rs2279525 | 0.285 | 0.307 | 1.404 | 0.24 |
| rs3821952 | 0.053 | 0.047 | 0.553 | 0.46 |
| rs7682765 | 0.070 | 0.078 | 0.728 | 0.39 |
| rs2932965 | 0.193 | 0.233 | 5.937 | 0.015 |
| rs3736265 | 0.060 | 0.055 | 0.378 | 0.54 |
| rs3755863 | 0.428 | 0.382 | 5.530 | 0.019 |
| rs8192678 | 0.366 | 0.337 | 2.290 | 0.13 |
| rs2970848 | 0.323 | 0.327 | 0.047 | 0.83 |
| rs16874194 | 0.082 | 0.072 | 0.956 | 0.33 |
| rs2305681 | 0.048 | 0.049 | 0.001 | 0.98 |
| rs4697046 | 0.321 | 0.354 | 2.860 | 0.091 |
| rs7665116 | 0.138 | 0.123 | 1.166 | 0.28 |
| rs12374310 | 0.412 | 0.401 | 0.336 | 0.56 |
| rs12374408 | 0.253 | 0.254 | 0.002 | 0.96 |
| rs7672915 | 0.428 | 0.431 | 0.032 | 0.86 |
| rs10002477 | 0.464 | 0.461 | 0.016 | 0.89 |
| rs10002590 | 0.119 | 0.150 | 5.109 | 0.024 |
| rs10002521 | 0.273 | 0.266 | 0.192 | 0.66 |
| rs12640088 | 0.155 | 0.119 | 6.692 | 0.0097 |
| rs6448228 | 0.420 | 0.389 | 2.430 | 0.12 |
| rs11724368 | 0.225 | 0.245 | 1.378 | 0.24 |
| rs13131226 | 0.298 | 0.304 | 0.108 | 0.74 |
| rs13117172 | 0.145 | 0.184 | 7.125 | 0.0076 |
| rs7674429 | 0.128 | 0.167 | 7.632 | 0.0057 |
| rs11734408 | 0.287 | 0.302 | 0.611 | 0.43 |
| rs4469064 | 0.077 | 0.072 | 0.224 | 0.64 |
| rs3796407 | 0.323 | 0.259 | 12.440 | 0.00042 |
| rs2946385 | 0.436 | 0.403 | 2.646 | 0.10 |
| rs2970873 | 0.127 | 0.163 | 6.631 | 0.010 |
| rs2970872 | 0.443 | 0.480 | 3.508 | 0.061 |
| rs2970871 | 0.421 | 0.463 | 4.480 | 0.034 |
| rs3774902 | 0.048 | 0.057 | 1.116 | 0.29 |
Discussion
We sought to identify candidate genes for human anxiety disorders starting with a genomewide association study of anxiety phenotypes in outbred mice. We identified the human homologues of the 52 associated murine genes and ranked them for further study using three independent and complementary sources of anxiety-related genetic data: (1) extant linkage and knock-out studies in mice, (2) a meta-analysis of human linkage scans, and (3) a preliminary human GWAS study. The top nine regions containing 14 genes were tested for association in a sample of twin subjects selected for high and low scores on a genetic factor shared across anxiety-related phenotypes. Of these novel candidates, one gene, PPARGC1A, had multiple associated SNPs and overall gene-based significant association.
Population stratification is a potential source of spurious associations for a genetic association study. While we did not have a set of ancestry informative markers genotyped in the twin sample to properly investigate this possibility, several prior analyses suggest that this was not a major source of Type I error (see Supplement). Type II error is likely more problematic, given the small effect sizes expected for complex genetic phenotypes. Our calculations predicted that the selected twin sample analyzed possessed the power to detect variants that explain ~2% of phenotypic variance, but this is only a rough estimate, as it did not take into account the complex nature of our study.
Another potential source of statistical confounding is the factor score distribution in the NIMH control sample used for GWAS. Like most psychiatric phenotypes, the distribution was quite skewed, with many of the subjects falling under a peak at the lower end of the scores (i.e., unaffected with low neuroticism). Plink’s regression routine makes the usual statistical assumption that the outcomes are normally distributed, which was not satisfied for our factor scores. Transformations provided little improvement. Post-hoc, we re-analyzed 100 randomly selected SNPs for association with the factor scores using permutation testing that does not depend upon such distributional properties, finding very similar p-values to those obtained by regression. This was reassuring, but p-values for some markers in the GWAS might have been biased by this effect.
One potential limitation of any study attempting to synthesize cross-species data is the homology of phenotypes; this is especially problematic for psychiatric phenotypes. While fear and anxiety are evident across mice and men, there are no clear isomorphisms. For this reason, we exploited a wide range of murine fear-related behaviors as well as a human phenotype that tapped into common genetic risk across anxiety-related disorders. However, as Smoller and colleagues have argued (44), mouse novelty-based tests, like some of the phenotypes utilized to identify the initial set of candidate genes, might find a better human homolog in behavioral inhibition in children rather than anxiety disorders in adults.
Since the time of the initial study planning and data analysis, genotype data for the other approximately half of the MGS control sample has been made publicly available via dbGaP (database of Genotypes and Phenotypes [http://www.ncbi.nlm.nih.gov/gap], Study Accession: phs000167.v1.p1 “nonGAIN sample”). This provided an opportunity for further replication of our finding (see Supplement for details). Only trend level association was observed for several SNPs in and around PPARGC1A (see Table S2 in the Supplement). We note that, while the nonGAIN sample was convenient due to the availability of phenotypes that overlapped with the twin sample, there are several importance differences between the two samples that affect the power to replicate our initial results. First, their ascertainment and assessment methods differed considerably. Second, the number of subjects in the nonGAIN sample available for analysis was somewhat smaller than for the twin sample. Third, while we utilized factor analysis applied to a similar compliment of internalizing phenotypes in both, the twin sample was selected for extremes of the common genetic factor. This results in two important potential differences in power to detect genetic association signals. First, utilizing a genetic versus a phenotypic factor score should provide a measure more directly associated with underlying allelic variants. Second, the twin sample was selected from factor score extremes and, therefore, contains much of the information contained in the larger sample from which it was derived, providing a substantially larger effective sample size.
We note that we were unable to successfully genotype markers in the 7q11.23 region near the gene WBSCR16. According to HapMap, this is a region with extensive copy number variation, possibly explaining why our SNP assays did not perform well. Looking back at the data in that region from the NIMH sample analysis, while 3 of the 5 SNPs had association p-values < 10-3, they also showed modest violation of HWE, supporting the complexity of that region. The WBSCR16 gene is a potentially interesting candidate for ADs, being one of a group of genes deleted in that region in Williams-Beuren Syndrome (WBS), a multisystem disorder with phenotype consisting of aortic stenosis, mental retardation, visiospatial impairment, and personality traits that include diminished social anxiety. Mice with disruptions of a neighboring, related gene (GTF2IRD1) have serotonergic alterations in several brain regions and exhibit reduced fear and increased social behaviors (45).
This study provides preliminary evidence for PPARGC1A as a novel candidate gene for anxiety-related psychiatric phenotypes. The PPARGC1A gene encodes the transcriptional coactivator, peroxisome proliferator-activated receptor gamma coactivator 1alpha (PGC-1α), which plays an important role in normal energy expenditure in peripheral tissue (46) and has been implicated in metabolic conditions like Type 2 diabetes mellitus (47). To our knowledge, our study is the first to suggest its association with psychiatric phenotypes, although a related gene, PPARG, appears in the list of SNPs with clusters of low p-values from the GAIN MDD GWAS (48). In the brain, PGC-1α is concentrated in GABAergic interneurons, and it may provide neuroprotection by activating genes involved in the metabolism of reactive oxygen species (49). As indicated in Table 2, this gene showed up using the public database search during the prioritization procedure from a knock-out model with anxiety phenotypes. Among various phenotypes described, PGC-1α −/− mice had increased thigmotaxis, a preference for staying at the perimeter walls of an open area or in an enclosed versus exposed area, which is an indicator of increased anxiety (50). PGC-1α −/− mice are also deficient in the interneuron-specific calcium binding protein, paralbumin, and exhibit signs of GABAergic dysfunction (51). However, these mice had adverse changes in multiple organ systems (50), so such a severe genetic change is unlikely to be a reliable predictor of effects from common variants. As with any novel genetic association finding, our identification of PPARGC1A as an anxiety candidate gene should be considered as tentative until adequate replication is demonstrated.
Supplementary Material
Acknowledgements
This work was supported by NIH grant R21MH79192 (JMH). We thank Alex Putnam, PhD, and Michael Miles, MD, PhD, for useful discussions regarding their mouse data (not included in the current study). We acknowledge the contribution of the Virginia Twin Registry, now part of the Mid-Atlantic Twin Registry (MATR), for the ascertainment of subjects for this study. The MATR is currently supported by UL1RR031990 from the National Center for Research Resources. Carol Prescott, PhD, provided critical help in the collection of the VATSPSUD sample. We also wish to thank Youfang Liu, PhD and Patrick Sullivan, MD, FRANZCP (University of North Carolina, Chapel Hill) for providing imputed genotypes for the nonGAIN sample used in our replication analysis.
NIMH Control Samples were from the National Institute of Mental Health Schizophrenia Genetics Initiative (NIMH-GI) and collected under the Molecular Genetics of Schizophrenia (MGS) collaboration. Funding support for these samples was provided by the Genomics Research Branch at NIMH, and the genotyping and analysis of samples was provided through the Genetic Association Information Network (GAIN) and under the MGS U01s: MH79469 and MH79470. Assistance with data cleaning was provided by the National Center for Biotechnology Information. Samples and associated phenotype data for the MGS GWAS study were collected under the following grants: NIMH Schizophrenia Genetics Initiative U01s: MH046276 (CR Cloninger), MH46289 (C Kaufmann), and MH46318 (MT Tsuang); and MGS Part 1 (MGS1) and Part 2 (MGS2) R01s: MH67257 (NG Buccola), MH59588 (BJ Mowry), MH59571 (PV Gejman), MH59565(Robert Freedman), MH59587 (F Amin), MH60870 (WF Byerley), MH59566 (DW Black), MH59586 (JM Silverman), MH61675 (DF Levinson), and MH60879 (CR Cloninger). Genotype data for the early release data used in our gene ranking scheme were generated at the Center for Genotyping & Analysis at the Broad Institute of Harvard and MIT as part of a multi-institutional collaborative research study (PIs: Pamela Sklar, M.D., Ph.D.; Jordan Smoller, M.D.,Sc.D.; Vishwajit Nimgaonkar, M.D., Ph.D.; and Edward Scolnick, M.D.). The MGS nonGAIN dataset used for the replication analysis described in the Supplement were obtained from the database of Genotype and Phenotype (dbGaP) found at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession numbers phs000167.v1.p1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Preliminary results from this study were presented at the XVIIth World Congress on Psychiatric Genetics, November 4–8, 2009 in San Diego, California, USA.
Financial Disclosures
The authors reported no biomedical financial interests or potential conflicts of interest.
Reference List
- 1.Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005 Jun;62(6):593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- 2.Hettema JM, Neale MC, Kendler KS. A review and meta-analysis of the genetic epidemiology of anxiety disorders. Am J Psychiatry. 2001 Oct;158(10):1568–1578. doi: 10.1176/appi.ajp.158.10.1568. [DOI] [PubMed] [Google Scholar]
- 3.Andrews G, Stewart G, Morris-Yates A, Holt P, Henderson S. Evidence for a general neurotic syndrome. Br J Psychiatry. 1990;157:6–12. doi: 10.1192/bjp.157.1.6. [DOI] [PubMed] [Google Scholar]
- 4.Bienvenu OJ, Brown C, Samuels JF, Liang KY, Costa PT, Eaton WW, et al. Normal personality traits and comorbidity among phobic, panic and major depressive disorders. Psychiatry Res. 2001 May 10;102(1):73–85. doi: 10.1016/s0165-1781(01)00228-1. [DOI] [PubMed] [Google Scholar]
- 5.Khan AA, Jacobson KC, Gardner CO, Prescott CA, Kendler KS. Personality and comorbidity of common psychiatric disorders. Br J Psychiatry. 2005 Mar;186:190–196. doi: 10.1192/bjp.186.3.190. [DOI] [PubMed] [Google Scholar]
- 6.Jardine R, Martin NG, Henderson AS. Genetic covariation between neuroticism and the symptoms of anxiety and depression. Genet Epidemiol. 1984;1(2):89–107. doi: 10.1002/gepi.1370010202. [DOI] [PubMed] [Google Scholar]
- 7.Hettema JM, Neale MC, Myers JM, Prescott CA, Kendler KS. A population-based twin study of the relationship between neuroticism and internalizing disorders. Am J Psychiatry. 2006 May;163(5):857–864. doi: 10.1176/ajp.2006.163.5.857. [DOI] [PubMed] [Google Scholar]
- 8.Kendler KS, Gardner CO, Gatz M, Pedersen NL. The sources of co-morbidity between major depression and generalized anxiety disorder in a Swedish national twin sample. Psychol Med. 2007 Mar;37(3):453–462. doi: 10.1017/S0033291706009135. [DOI] [PubMed] [Google Scholar]
- 9.Smoller JW. Who's afraid of anxiety genetics? Biol Psychiatry. 2011 Mar 15;69(6):506–507. doi: 10.1016/j.biopsych.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 10.Maron E, Hettema JM, Shlik J. Advances in molecular genetics of panic disorder. Mol Psychiatry. 2010 Jul;15(7):681–701. doi: 10.1038/mp.2009.145. [DOI] [PubMed] [Google Scholar]
- 11.Otowa T, Yoshida E, Sugaya N, Yasuda S, Nishimura Y, Inoue K, et al. Genome-wide association study of panic disorder in the Japanese population. J Hum Genet. 2009 Feb;54(2):122–126. doi: 10.1038/jhg.2008.17. [DOI] [PubMed] [Google Scholar]
- 12.Erhardt A, Czibere L, Roeske D, Lucae S, Unschuld PG, Ripke S, et al. TMEM132D, a new candidate for anxiety phenotypes: evidence from human and mouse studies. Mol Psychiatry. 2010 Apr 6; doi: 10.1038/mp.2010.41. [DOI] [PubMed] [Google Scholar]
- 13.Flint J. Animal models of anxiety and their molecular dissection. Semin Cell Dev Biol. 2003 Feb;14(1):37–42. doi: 10.1016/s1084-9521(02)00170-2. [DOI] [PubMed] [Google Scholar]
- 14.Smoller JW, Acierno JS, Jr, Rosenbaum JF, Biederman J, Pollack MH, Meminger S, et al. Targeted genome screen of panic disorder and anxiety disorder proneness using homology to murine QTL regions. Am J Med Genet. 2001 Mar 8;105(2):195–206. doi: 10.1002/ajmg.1209. [DOI] [PubMed] [Google Scholar]
- 15.Donner J, Pirkola S, Silander K, Kananen L, Terwilliger JD, Lonnqvist J, et al. An association analysis of murine anxiety genes in humans implicates novel candidate genes for anxiety disorders. Biol Psychiatry. 2008 Oct 15;64(8):672–680. doi: 10.1016/j.biopsych.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet. 2006 Aug;38(8):879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- 17.Mott R, Talbot CJ, Turri MG, Collins AC, Flint J. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci U S A. 2000 Nov 7;97(23):12649–12654. doi: 10.1073/pnas.230304397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders AR, Levinson DF, Duan J, Dennis JM, Li R, Kendler KS, et al. The Internet-based MGS2 control sample: self report of mental illness. Am J Psychiatry. 2010 Jul;167(7):854–865. doi: 10.1176/appi.ajp.2010.09071050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kessler RC, Andrews G, Mroczek DK, Ustun B, Wittchen H-U. The World Health Organization Composite International Diagnostic Interview Short Form (CIDI-SF) International Journal of Methods in Psych Res. 1998;7:171–185. [Google Scholar]
- 20.Eysenck H, Eysenck S, Barrett P. A revised version of the psychoticism scale. Personality and Individual Differences. 1985;6:21–29. [Google Scholar]
- 21.Muthen LK, Muthen BO. Mplus User's Guide. Fourth Edition. Los Angeles, CA: Muthen & Muthen; 2006. [Google Scholar]
- 22.Krueger RF. The structure of common mental disorders. Arch Gen Psychiatry. 1999 Oct;56(10):921–926. doi: 10.1001/archpsyc.56.10.921. [DOI] [PubMed] [Google Scholar]
- 23.van den Oord EJ, Kuo PH, Hartmann AM, Webb BT, Moller HJ, Hettema JM, et al. Genomewide association analysis followed by a replication study implicates a novel candidate gene for neuroticism. Arch Gen Psychiatry. 2008 Sep;65(9):1062–1071. doi: 10.1001/archpsyc.65.9.1062. [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007 Sep;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo PH, Bukszar J, van den Oord EJ. Estimating the number and size of the main effects in genome-wide case-control association studies. BMC Proc. 2007;1 Suppl 1:S143. doi: 10.1186/1753-6561-1-s1-s143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kendler KS, Prescott CA. A population-based twin study of lifetime major depression in men and women. Arch Gen Psychiatry. 1999 Jan;56(1):39–44. doi: 10.1001/archpsyc.56.1.39. [DOI] [PubMed] [Google Scholar]
- 27.Kendler KS, Prescott CA. Genes, Environment, and Psychopathology: Understanding the Causes of Psychiatric and Substance Use Disorders. New York: Guilford Press; 2006. [Google Scholar]
- 28.Spitzer RL, Williams JBW. Structured Clinical Interview for DSM-III-R (SCID) New York: Biometrics Research Department, New York State Psychiatric Institute; 1985. [Google Scholar]
- 29.Eysenck HJ&ESBG. Manual of the Eysenck Personality Questionnaire. London: Hodder and Stoughton; 1975. [Google Scholar]
- 30.Hettema JM, An SS, Bukszar J, van den Oord EJ, Neale MC, Kendler KS, et al. Catechol-O-methyltransferase contributes to genetic susceptibility shared among anxiety spectrum phenotypes. Biol Psychiatry. 2008 Aug 15;64(4):302–310. doi: 10.1016/j.biopsych.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Middeldorp CM, Cath DC, Van Dyck R, Boomsma DI. The co-morbidity of anxiety and depression in the perspective of genetic epidemiology. A review of twin and family studies. Psychol Med. 2005 May;35(5):611–624. doi: 10.1017/s003329170400412x. [DOI] [PubMed] [Google Scholar]
- 32.Jardine R, Martin NG, Henderson AS. Genetic covariation between neuroticism and the symptoms of anxiety and depression. Genet Epidemiol. 1984;1(2):89–107. doi: 10.1002/gepi.1370010202. [DOI] [PubMed] [Google Scholar]
- 33.Scherrer JF, True WR, Xian H, Lyons MJ, Eisen SA, Goldberg J, et al. Evidence for genetic influences common and specific to symptoms of generalized anxiety and panic. J Affect Disord. 2000 Jan;57(1–3):25–35. doi: 10.1016/s0165-0327(99)00031-2. [DOI] [PubMed] [Google Scholar]
- 34.Van Gestel S, Houwing-Duistermaat JJ, Adolfsson R, van Duijn CM, Van Broeckhoven C. Power of selective genotyping in genetic association analyses of quantitative traits. Behav Genet. 2000 Mar;30(2):141–146. doi: 10.1023/a:1001907321955. [DOI] [PubMed] [Google Scholar]
- 35.van den Oord EJCG. A comparison between different designs and tests to detect QTLs in association studies. Behav Genet. 1999 Jul;29(4):245–256. [Google Scholar]
- 36.Schork NJ, Nath SK, Fallin D, Chakravarti A. Linkage disequilibrium analysis of biallelic DNA markers, human quantitative trait loci, and threshold-defined case and control subjects. Am J Hum Genet. 2000 Nov;67(5):1208–1218. doi: 10.1086/321201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robles JR, van den Oord EJ. lga972: a cross-platform application for optimizing LD studies using a genetic algorithm. Bioinformatics. 2004 Nov 22;20(17):3244–3245. doi: 10.1093/bioinformatics/bth348. [DOI] [PubMed] [Google Scholar]
- 38.van den Oord EJ, Sullivan PF. False discoveries and models for gene discovery. Trends Genet. 2003 Oct;19(10):537–542. doi: 10.1016/j.tig.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 39.Neale BM, Sham PC. The future of association studies: gene-based analysis and replication. Am J Hum Genet. 2004 Sep;75(3):353–362. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Straub RE, Sullivan PF, Ma Y, Myakishev MV, Harris-Kerr C, Wormley B, et al. Susceptibility genes for nicotine dependence: a genome scan and followup in an independent sample suggest that regions on chromosomes 2, 4, 10, 16, 17 and 18 merit further study. Mol Psychiatry. 1999 Mar;4(2):129–144. doi: 10.1038/sj.mp.4000518. [DOI] [PubMed] [Google Scholar]
- 41.Livak KJ. Allelic discrimination using fluorogenic probes and the 5' nuclease assay. Genet Anal. 1999 Feb;14(5–6):143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 42.The International HapMap Project. Nature. 2003 Dec 18;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 43.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005 Jan 15;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 44.Smoller JW, Rosenbaum JF, Biederman J, Susswein LS, Kennedy J, Kagan J, et al. Genetic association analysis of behavioral inhibition using candidate loci from mouse models. Am J Med Genet. 2001 Apr 8;105(3):226–235. doi: 10.1002/ajmg.1328. [DOI] [PubMed] [Google Scholar]
- 45.Young EJ, Lipina T, Tam E, Mandel A, Clapcote SJ, Bechard AR, et al. Reduced fear and aggression and altered serotonin metabolism in Gtf2ird1-targeted mice. Genes Brain Behav. 2008 Mar;7(2):224–234. doi: 10.1111/j.1601-183X.2007.00343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005 Jun;1(6):361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Barroso I, Luan J, Sandhu MS, Franks PW, Crowley V, Schafer AJ, et al. Meta-analysis of the Gly482Ser variant in PPARGC1A in type 2 diabetes and related phenotypes. Diabetologia. 2006 Mar;49(3):501–505. doi: 10.1007/s00125-005-0130-2. [DOI] [PubMed] [Google Scholar]
- 48.Sullivan PF, de Geus EJ, Willemsen G, James MR, Smit JH, Zandbelt T, et al. Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol Psychiatry. 2008 Dec 9; doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006 Oct 20;127(2):397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 50.Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005 Apr;3(4):e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lucas EK, Markwardt SJ, Gupta S, Meador-Woodruff JH, Lin JD, Overstreet-Wadiche L, et al. Parvalbumin deficiency and GABAergic dysfunction in mice lacking PGC-1alpha. J Neurosci. 2010 May 26;30(21):7227–7235. doi: 10.1523/JNEUROSCI.0698-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.