Abstract
Replication protein A (RPA) is a single-strand DNA-binding protein with essential roles in DNA replication, recombination and repair. RPA is necessary for the formation of the preincision complex which is required for proper incision of damaged DNA nucleotides during DNA repair. We have previously identified small molecule inhibitors (SMIs) with the ability to disrupt RPA binding activity to ssDNA. Further characterization of these RPA inhibitors was assessed using both lung and ovarian cancer cell lines. Lung cancer cell lines demonstrated increased apoptotic cell death following treatment with the SMI MCI13E, with IC50 values of ~5 μM. The A2780 ovarian cancer cell line and the p53-null lung cancer cell line H1299 were particularly sensitive to MCI13E treatment with IC50 values below 3 μM. Furthermore, a cell cycle effect was observed in lung cancer cell lines which resulted in a lengthening of either G1 or S-phases of the cell cycle following single agent treatment. Sequential treatment with MCI13E and cisplatin resulted in synergism. Overall these data suggest that decreasing RPA’s DNA binding activity via a SMI may disrupt RPA’s role in cell cycle regulation. Thus, RPA SMIs hold the potential to be used as single agent chemotherapeutics or in combination with current chemotherapeutic regimens to increase efficacy.
Introduction
The nucleotide excision repair pathway (NER) is a highly versatile DNA repair pathway present in a number of organisms from bacteria to mammals which requires the contribution of over thirty proteins (1). The NER pathway repairs a wide array of bulky DNA damage from a variety of sources such as, reactive chemicals and exposure to UV light (1;2). Numerous non-enzymatic protein-DNA interactions are essential for the proper functioning of the NER machinery and play important roles in nearly every reaction in the pathway including lesion recognition (3;4). Damaged DNA is recognized by the trimeric complex consisting of Xeroderma Pigmentosum Group C (XPC), Rad23B and Centrin 2 during global genomic nucleotide excision repair (GG-NER) while the stalling of RNA polymerase during transcription is the method of damage recognition during transcription-coupled (TC) NER (5); (6). Following damage recognition the preincision NER complex is completed with the subsequent recruitment of Xeroderma Pigmentosum Group A (XPA) protein, Transcription Factor II H (TFIIH) protein and the human single-stranded DNA (ssDNA) binding protein, Replication protein A (RPA) to the site of DNA damage. RPA is one of the first proteins that functions in both the GG and TC-NER subpathways (2;7;8). RPA is a heterotrimeric DNA binding protein containing three subunits p70, p34, and p14 (kDa) and plays an important role in DNA replication and recombination in addition to repair (9;10). The p70 RPA subunit contains DNA binding domains A and B (DBD-A and DBD-B) and contributes most significantly to the RPA-ssDNA interaction (11). The RPA p34 subunit also contains an OB-fold and interacts with additional proteins including XPA while the 14 kDA subunit plays a role in protein stability (12;13). The RPA-DNA interaction is essential for the formation of the NER preincision complex and proper functioning of the NER pathway (14). Disruption of this essential protein-DNA interaction via small molecule inhibitors (SMIs) should reduce the NER efficiency. Previous reports have demonstrated that decreased expression levels of essential NER proteins, such as XPA result in decreased NER capacity and removal of cisplatin adducts (15-17). Furthermore, increased expression of ERCC1-XPF was demonstrated to correlate with cisplatin resistance in ovarian cancer cell lines (18). Taken together, these data suggest that expression level of essential NER proteins affects the efficiency of the NER machinery. Using SMIs to inhibit RPA-DNA interactions and consequently the function of the NER machinery may increase the efficacy of DNA-damaging chemotherapeutics, particularly in tissues where enhanced repair via NER is a resistance mechanism.
Cisplatin, (cis-diamminedichloroplatinum[III]), is a front-line treatment for a variety of neoplasms, including ovarian, lung and testicular cancers (19). Innate and acquired resistance to cisplatin therapy is a recurring issue in the clinic and a wide spectrum of responses are observed in cancer patients, warranting the discovery of novel chemotherapeutic treatments (19-22). Cisplatin induces it toxic effects by interacting with DNA, typically by intrastrand linkage of adjacent guanines (GpG). This produces an N-Pt-N cross-link from the imidazole nitrogens (N7), resulting in a 12-28° kink in the DNA. This kink is then recognized and repaired by the NER machinery (23-25). Disruption of protein-DNA interactions, resulting in a decrease in NER efficiency and DNA repair, may be exploited to increase efficacy of cisplatin and related platinum chemotherapeutics. Previous work has demonstrated that a decrease in NER efficiency elicited by decreasing the expression of essential NER proteins, resulting in increased sensitivity to cisplatin (16;18). Therefore targeting the RPA-ssDNA interaction via SMIs holds the potential to sensitize cancer cells to Pt-based chemotherapy. Combination treatments involving SMIs may result in increased accumulation of cisplatin adducts and therefore increased efficacy of treatment, potentially decreasing the probability of recurrence/resistance.
Cisplatin damage results in a cell cycle response arresting cells in the G2 phase ultimately leading to cellular apoptosis (26;27). The P53 tumor suppressor is a key element in DNA damage response. It is post-translationally modified upon the induction of DNA damage resulting in activation (28). Activated p53 transactivates the p21 cyclin-dependent kinase inhibitor which in turn results in cell cycle arrest(29). In the absence of the ability to repair the lesions, ultimately apoptosis can be induced. Therefore, decreasing NER catalyzed removal, via inhibition of the RPA-DNA interaction, may result in persistent cisplatin-DNA adducts and increased cellular sensitivity to cisplatin treatment.
We present here characterization of the physiological effect of SMIs of the RPA-ssDNA interaction in a cell culture model. The core inhibitor isoborneol structure was identified in a screen of the NCI library (30) and subsequent SAR led to the development of haloester derivatives that act as irreversible inhibitors of RPA in vitro (31;32). Two of these isobornyl derivatives MCI13E and MCI13F demonstrated cellular activity and were chosen for further characterization. The data presented herein demonstrates that the disruption of RPA’s activity in lung and ovarian cancer cell models results in increased apoptosis and lengthening of cell cycle stages. The induction f apoptosis is independent of p53 and these SMIs synergize with cisplatin in combination treatments.
Methods and Materials
Chemicals
Dulbecco’s Modified Eagle Media (DMEM), RPMI, fetal bovine serum (FBS), penicillin/streptomycin and trypsin were purchased from CellGro (Manassas, VA). Annexin V-FITC/propidium idodine (PI) Vybrant Apoptosis Assay kit and the GAPDH primary antibody were purchased from Invitrogen (Carlsbad, CA). Dimethyl sulfoxide (DMSO) and sodium dodecyl sulfate (SDS) was purchased from Fisher Scientific (Pittsburgh, PA). Cell Counting Kit-8 solution was purchased from Dojindo Laboratories (Rockville, MD) and primary antibodies against p53 and ATM were supplied by Abcam (Cambridge, MD). Secondary antibodies were purchased from Bio-Rad (Hercules, CA) and Santa Cruz (Santa Cruz, CA). All other reagents and chemicals were purchased from Sigma Aldrich (Milwaukee, WI) or Fisher Scientific (Houston, TX).
Cell lines
Tumor cell lines A549 (lung, CCL-185), H1299 (p53-deficient lung, CRL-5803), and H460 (HTB-177) were obtained from the American Type Culture Collection, verified via STR testing (Manassas, VA) and were not passaged over 6 months following resuscitation. The A2780 line was obtained from Tom Hamilton at FCCC. The A549, H1299 and A2780 cells were maintained in Dulbecco’s modified Eagle medium supplemented with 1% penicillin/streptomycin and 10% fetal bovine serum. H460 cells were maintained in RPMI media containing 1% penicillin/streptomycin and 10% FBS. Cells were maintained in a humidified incubator at 37°C with 5% CO2 supplementation.
Cell Viability Assays
A crystal violet cell viability assay was performed to determine cytotoxicity of SMI. Lung cancer cell lines, A549 and H460, were plated in 24-well dishes at 5×104 total cells. Forty-eight hours after plating, the cells were treated for 24 hours with variable concentrations of SMIs. The media was aspirated, cells washed with 500 μl of PBS/EDTA and stained for 10 minutes with crystal violet solution (50% Ethanol, 0.75% crystal violet). Stained cells were resuspended in 500 μl DMSO containing 1% SDS and absorbance was measured at 590 nm in a SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA) and analyzed using SoftMax Pro 5.2. Percent viability is taken as an absorbance percentage against control untreated cells. Values plotted represent four separate plate replicates and each plate replicate contained three assay replicates per plate.
Ovarian cancer cell line, A2780, cells are difficult to use for crystal violet analysis due to their enhanced sensitivity to treatment. Therefore, the cell counting kit-8 (CCK) was used to determine the viability of A2780 cells in addition to the p53 null lung cancer cell line H1299, following a 24 hour treatment with log dosing of SMI. Cells were plated (1×105) in 24-well dishes and treated with inhibitor after 24 hours. After treatment media was aspirated and 1 ml of DMEM was added in addition to 100 μL of CCK-8 solution. Absorbance at 450 nm was measured over 4 hours in a SpectraMax M5 plate reader and values are presented as the percent untreated control at the two hour time point for three separate plate replicates each containing three assays per plate.
Flow Cytometry
After determining the SMI cytotoxicity, H460, A549 and H1299 lung cancer cell lines were analyzed for apoptosis using the Annexin V-FITC/Propidium Iodide (PI) Vybrant® Apoptosis Assay kit (Invitrogen, Gaithersburg, MD). Cells were plated in 6-well dishes (2×105 cells total), treated for 24 (H1299) or 48 (H460 and A549) hours with increasing concentrations of SMI. Following treatment, both adherent and non-adherent cells were collected, processed via the manufacturer’s protocol (Invitrogen) and analyzed on a FACScan flow cytometer (Becton Dickson, San Jose, CA). Data was analyzed using WinDMI software (The Scripps Research Institute, Can Diego, CA) and results are presented as a percentage of control (untreated cells).
Cell cycle analysis was also performed on the three lung cancer cell lines using PI staining to identify the potential arrest resulting from treatment with SMIs. Briefly, cells were plated in 6-well dishes (2×105 cells total) and treated for 6 or 12 hours with increasing concentration of inhibitor. Adherent and non-adherent cells were collected, washed twice with PBS containing 2% bovine serum albumin (BSA), resuspended in 70% cold EtOH and incubated overnight at -20°C. Cells were again collected and stained with PI (1 μg/mL) and RNaseA (25 μg/mL) for 30 min. at 37°C followed by 1.5 hours at 4°C in the dark. Flow cytometry was performed as above and data were analyzed on a histogram with events plotted against the FL2-A parameter. Cell cycle distribution was analyzed using the ModFit software and data presented represents three independent trials.
A549 cells were also treated sequentially or concurrently with combination treatment utilizing cisplatin and SMIs. Sequentially treated cells were plated (3×105 total cells) in 6-well dishes and treated 12 hours later with cisplatin (5 μM) for 12 hours at which time SMIs (7 μM) were added as indicated. Cells were then processed (as described above for cell cycle analysis) at 6 and 12 and 24 hours after the addition of SMI. Concurrently treated cells were plated in 6-well dishes, media was changed 12 hours after initial plating and cells were treated with cisplatin (5 μM) and SMIs (7 μM) after an additional 12 hours. Cells were then processed at 6, 12, and 24 hours post combination treatment for PI staining as described above.
In order to keep the cisplatin treatment constant and monitor cell cycle progression, a similar methodology was applied. Twelve hours after initial plating, A549 cells were treated with cisplatin (2.5 μM). Following either 12 or 24 hours of cisplatin treatment, MCI 13E was added to the media (5 μM) and cells were processed 12 or 24 hours later and analyzed for PI incorporation as discussed above (see Supplemental Figure 1).
Combination Index Studies
Combination index studies, performed with combination SMI and cisplatin treatment, were used to determine the level of synergy or antagonism between the two compounds. Briefly, A549 cells were plated as described above in 24-well dishes (5×104cells per well) and treated either concurrently or sequentially with single agent cisplatin or MCI treatment or with combination treatments. For sequential treatment, A549 cells were treated with cisplatin (0, 0.9, 1.8, 3.7, or 15 μM) for 24 hours and then the inhibitor was added to the media for an additional 24 hours (0, 0.75, 1.5, 3, 6 or 12 μM for E and 0, 0.25, 0.5, 1, 2, 4 μM for F). Concurrently treated cells were treated with both cisplatin and inhibitor for a full 48 hours. Following 48 hours of total treatment cells were analyzed for viability using the crystal violet assay described above and combination index values were calculated based on the Chou and Talalay method (33) as we have previously described (34;35).
Real-Time PCR
A549 cells were plated in 10 cm dishes at 1.5 × 106, allowed to adhere, and treated for 24-48 hours with cisplatin or SMIs (5μM). For sequential treatment, A549 cells were exposed to cisplatin for 24 hours followed by the addition of SMIs for another 24 hours as above. Concurrently treated cells were incubated simultaneously with both cisplatin and MCIs for 24 hours. RNA was extracted using RNeasy minikit (Qiagen, Germantown, MD) per manufacturer’s protocol. cDNA was generated using Applied Biosystem’s (ABI) High Capacity RNA-to-cDNA kit. Validated gene primer/probe sets, also from ABI, were used for Quantitative Real Time-PCR. ABI’s ΔΔCT Relative Quantification methodology was used for the analyses (applied Biosystems). Data were normalized to the housekeeping gene GAPDH and calibrated to the mock treated samples. Results presented are the average of two separate cell treatments/RNA isolations with duplicate Taqman Assays per RNA sample set.
Western Blot Analysis
Cells lines (A549, H460, or H1299) were plated in 10 cm dishes (3×106) and treated with the ~IC50 concentration of SMIs in the absence or presence of cisplatin (5 μM for A549 and H460 and 10 μM for H1299 cells). Twenty-four hours following treatment the media was aspirated and adherent cells were washed with 5 mL of PBS/EDTA. Adherent cells were scraped from the plates into 100 μL of RIPA buffer (10 mM Tris, pH 7.2, 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 1% Deoxycholate, 5 mM EDTA) and cellular debris was pelleted by centrifugation (10,000 × g, 10 min., 4C). The supernatant was collected into a 1.5 mL microfuge tube and total protein concentration was determined (Bio-Rad, Hercules, CA) standardized against BSA. Equal amounts of protein (40 μg) were loaded onto SDS-PAGE and following electrophoresis proteins were transferred to polyvinylidene difluoride membranes. Proteins were detected with various antibodies (1:2000 dilutions) and goat anti-mouse or goat anti-rabbit horseradish peroxidase secondary antibodies (1:2500 or 1:3000), respectively. Chemiluminescence and an Image Reader LAS3000 (Fujifilm) were employed to visualize bands while Multi Gauge V3.0 software was used for data analysis.
Results
SMIs of RPA’s DNA binding activity decrease cell viability, induce apoptosis and lengthen cell cycle stages
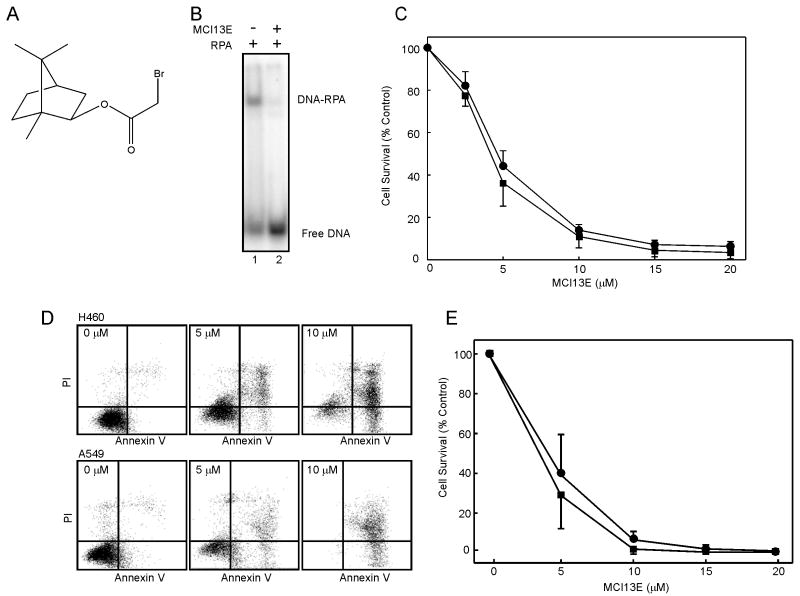
Previous work from our laboratory identified and characterized the in vitro activity of MCI13E (Figure 1A), a SMI of RPA which blocks the RPA-DNA interaction (31;34) (Figure 1B). Considering RPA in an essential protein which plays a central role in DNA replication we assessed the activity of the SMI MCI13E in assays measuring cellular viability. Experiments were performed in non-small cell lung cancer (NSCLC) cell lines A549, an adenocarcinoma, and the large cell carcinoma H460 cell line. Both lung cancer cell lines displayed a decrease in viability with increasing concentrations of SMI (Figure 1C), with similar sensitivity and IC50 values of approximately 5 μM. We also did not detet any change in the levels of endogenous RPA as a function of MCI13E treatment (Supplemental Figure 2).
Figure 1.
A) Chemical structure of MCI13E. B) MCI13E inhibits RPA-DNA binding in vitro. RPA was mock treated (lane 1) or treated with MCI13E (50 μM) (lane 2) and DNA binding to a single stranded 30 bp DNA (12.5 nM) assessed by EMSA analysis as previously described by Anciano et al. C) MCI13E displays single agent cytotoxic activity against A549 and H460 NSCLC cells. Cytotoxicity was analyzed via crystal violet analysis as described in “methods” 48 hours following MCI13E treatment. Data represent the average and standard deviation of four replicate experiments with each replicate containing triplicate determinations. D) MCI13E induced death via apoptosis in NSCLC cell lines. Cell were treated with the indicated concentrations of MCI13E for 48 hours and analyzed for the induction of apoptosis via Annexin V/PI staining. E) Percent survival was calculated for each cell line using untreated cells as a control. Data presented are the average and standard deviation from four replicate experiments.
Decreased viability in the staining assay can be a function of numerous factors. These include induction of cell death, decreased rates of proliferation or induction of cell cycle arrest. To assess induction of cell death, we assayed phosphatidylserine flipping, a measure of early apoptosis, in conjunction with PI staining, a measure of membrane integrity. Treatment of H460 and A549 cells produced a significant increase in Annexin V reactivity and PI staining (Figure 1D) demonstrating the induction of a classical apoptotic pathway following treatment with MCI13E. Quantification of the extent of cell death resulted in IC50 values of approximately 5 μM (Figure 1E).
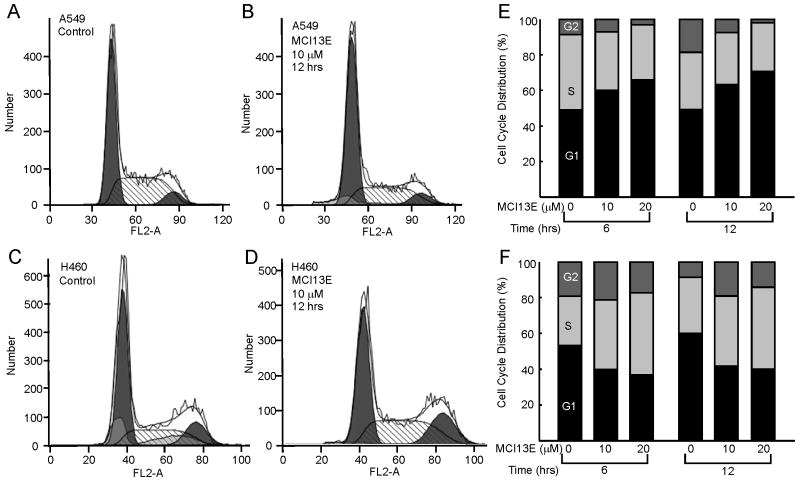
To determine the cellular mechanism by which apoptosis is induced by SMI treatment, we sought to assess effects on cell cycle progression. Interestingly treatment of A549 cells with increasing doses of MCI13E produced a greater proportion of cells in the G1 phase of the cell cycle whereas H460 cells demonstrate an increase in the proportion of cells in S-phase (Figure 2). Cell cycle progression was analyzed at 6, 12, and 24 hours of treatment with increasing concentrations of SMI and the 6 and 12 hour time points are presented in Figure 2. While cell cycle stages lengthened by 6 hours, there is no significant further increase after an additional 6 or 12 hours of treatment (Figure 2 and data not shown). This suggests that the effect of MCI13E occurs early and does not persist. This may also be due to chemical instability of the α-haloester moiety of MCI13E, resulting in reduced activity.
Figure 2.
Cell Cycle Analysis of Lung Cancer Cell Lines Following Treatment with MCI13E. A549 (Panels A and B) or H460 (C and D) were treated as indicated and analyzed for the cell cycle distribution using PI staining and flow cytometry. Quantification of data following 6 hours and 12 hour treatment with 10 and 20 μM MCI13E was performed for both A549 (panel E) and H460 (panel F) Data presented represent the averages of three replicate experiments.
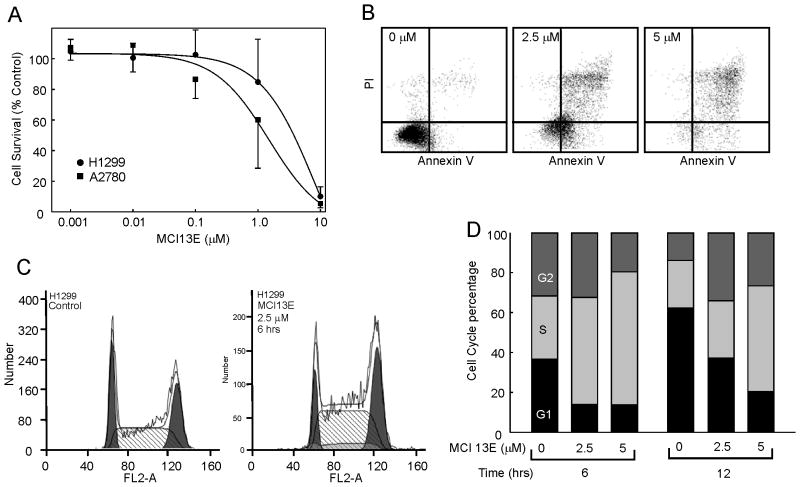
Considering this difference in cell type, we sought to determine the effects of MCI13E on other cell types. We selected the p53 null NSCLC cell line H1299 and the epithelial ovarian cancer cell line A2780. MCI13E was highly potent against A2780 cells with an IC50 of approximately 1 μM (Figure 3) while the H1299 cell line displayed a similar IC50 (~4 μM) to the other NSCLC cell lines despite the p53 status (Figure 3). Also note that this IC50 was determined following 24 hours of treatment, similar to that of the A2780 cell line, again demonstrating sensitivity to the SMI. Cell cycle progression was also measured in the H1299 cell line following 6, 12, and 24 hours of treatment with MCI13E and an increased percentage of cells accumulated in S-phase (Figure 3). As with the H460 cell line, an additional 6 or 12 hour of treatment did not result in a significant increase in the percentage of cells accumulating in S-phase.
Figure 3.
Cell Viability and Apoptosis of Ovarian and p53 null Lung Cancer Cell Lines. A) Ovarian cancer cell line A2780 and the p53 null lung cancer cell line, H1299, were treated with log dosing of MCI 13E and viability analyzed via CCK-8 metabolic assay as described in Methods. Data is presented as cell survival which was determined as a percent of the untreated control cells and represents the average and standard deviation of triplicate experiments. B) H1299 cells were analyzed for the induction of apoptosis via Annexin V and propidium iodine staining. A shift to the upper right quadrant was observed and is indicative of the induction of apoptosis. C) Cell cycle analysis was conducted in H1299 cells and following 6 hours of treatment cell accumulate in the S-phase of the cell cycle.
Synergy with cisplatin
Analyses thus far indicate that inhibition of RPA is effective in eliciting effects consistent with inhibition of RPAs role in cell cycle progression and DNA replication. To assess if SMI treatment impacts RPAs role in DNA repair, we assessed how MCI13E or MCI13F treatment influences cellular sensitivity to cisplatin. MCI13F is an isobornyl derivative MCI13F identified by Anciano et al. (2010) which demonstrates an IC50 similar to that of MCI13E in vitro (Anciano et al.) and in vivo (data not shown) but contains an iodide in place of the bromide. Cisplatin-induced DNA damage is primarily repaired via NER and homologous recombination. Considering that both cisplatin and MCI treatment possess single agent activity in the NSCLC cell lines, we employed combination index studies to analyze the effectiveness of combination treatments. We initially undertook a concurrent approach, similar to that we described for another RPA SMI, TDRL-505 (34). Briefly, A549 cells were treated for 48 hours with MCI13E, cisplatin, or combination treatment, with viability determined via crystal violet staining. The results from these experiments revealed an antagonistic effect (Table 1) suggesting that either cisplatin was rendering the SMI ineffective or vice versa. The analysis of cell cycle effects induced by MCI suggested that this compound may have a relatively short half-life eliciting its effects relatively quickly, while cisplatin typically requires at least 48 hours to produce effects. We therefore pursued a sequential treatment protocol, first treating cells with cisplatin for 24 hours, then adding the SMI and incubating for an additional 24 hours. The results from these experiments displayed synergy based on the Chou-Talaylay method (33). Interestingly, we observed slight differences with MCI13E, synergizing with cisplatin at higher dosing concentrations while MCI13F demonstrated synergy with cisplatin at all tested concentrations (Table 1).
Table 1.
Schedule dependent MCI-cisplatin synergy
| Treatment | % affected | CI
|
|
|---|---|---|---|
| Sequential | Concurrent | ||
| MCI 13E/Pt | 0.2 | 0.65 | 0.94 |
| 0.5 | 0.79 | 1.9 | |
| 0.75 | 0.86 | 3.0 | |
| MCI 13F/Pt | 0.2 | 0.38 | 2.1 |
| 0.5 | 0.50 | 3.7 | |
| 0.75 | 0.51 | 5.2 | |
A549 cells were treated sequentially or concurrently with cisplatin and MCI13E or MCI13F for a total of 48 hours and cell viability determined. Combination index values were calculated based on the Chou Talalay method.
Cell cycle analysis of combination treatments demonstrates a difference in cycle lengthening
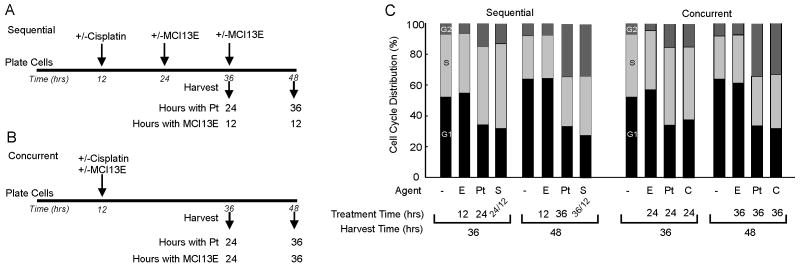
As discussed above MCI13E synergizes with cisplatin following sequential treatment while concurrent cisplatin/SMI treatment results in an antagonistic effect. Knowing that MCI13E induced a lengthening of the G1 stage of the cell cycle for A549 cells we wanted to determine if alteration in cycle progression with either sequential or concurrent cisplatin/SMI treatment could account for the differences in drug interactions. A549 cells were plated and treated sequentially or concurrently with cisplatin/SMI and processed for cell cycle analysis. The experiment was performed using two separate dosing schedules one keeping the time of SMI treatment consistent between the sequential and concurrent schedules and the other keeping cisplatin treatment time consistent. The data from the latter experiment is presented (Figure 4) using a reduced dose of SMI corresponding to the CI studies and demonstrates, under both schedules, the cisplatin induced S and G2 accumulation at 24 and 36 hours post treatment is modestly impacted by SMI treatment. These results suggest that differences in cellular response to the combined treatment schedule are not a function of differential cell cycle arrest between the concurrent and sequential dosing schedules.
Figure 4.
Cell Cycle Analysis with Combination Cisplatin and MCI Treatment. A) sequential treatment regimen and B) concurrent treatment regimen for maintaining constant cisplatin treatment time. C) A549 cells were treated sequentially or concurrently with single agent or combination treatments of cisplatin denoted “Pt” and/or MCI13E (denoted “E”) and analyzed for cell cycle distribution via PI and flow cytometry. Data represents the average percentage of cells in G1, S, or G2 phase of the cell cycle. This experiment was repeated twice with similar results.
Transcriptional and Post-Translational Modifications following combination SMI and cisplatin treatments
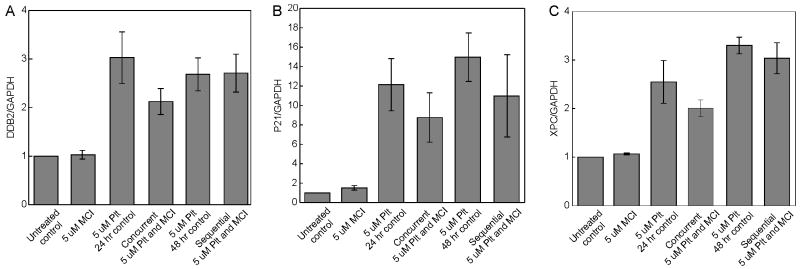
While no significant alteration in cell cycle progression was observed we sought to determine how activation of the DNA damage signaling pathways were impacted by the two treatment protocols. Using QPCR we analyzed the transcriptional activation of a series of genes encoding proteins involved in the DNA damage response (DDR). Three of these XPC, DDB2 and p21 showed consistent increases in expression in cisplatin treated cells as expected for p53 responsive genes. Interestingly in the concurrently treated cells, the expression of each gene was reduced compared to the cisplatin control while in the sequentially treated cells, only p21 showed a decrease in expression compared to the cisplatin control (Figure 5). This QPCR data was further confirmed by western blot analysis of p21 expression following the various treatment methods (Supplemental Figure 3). This suggests that p53 is differentially activated without a corresponding change in cell cycle distribution (Figure 5).
Figure 5.
Quantitative Real Time PCR analyses of mRNA expression changes of A549 cells treated with MCI13E, MCI13F and Cisplatin. Data represents the average of four separate Q-RT PCR Assays. Genes of interest were normalized to GAPDH transcript levels, and are presented as fold changes relative to untreated cell samples. Cisplatin treatment increases transcription of DDB2 (A), P21 (B), and XPC (C). Concurrent MCI/Cisplatin treated cells show a decrease in gene transcription when compared to cells treated with Cisplatin alone (24 hour).
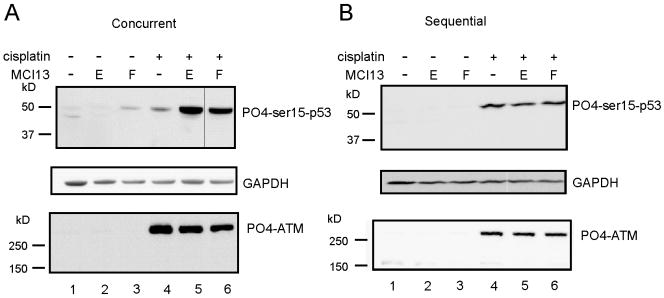
To assess p53 activation directly, we determined the phosphorylation level of serine-15 using the differing combination treatment protocols. Interestingly, bromide MCI13E and the iodide MCI13F had minimal single-agent effects on p53 phosphorylation while concurrent treatment with cisplatin produced a dramatic increase (Figure 6). This increase was not seen in the sequentially treated cells and is consistent with the transcriptional profiling analysis where differences were observed depending on treatment schedule. Analysis of total p53 was also analyzed, via western blot analysis, and a similar increase in total protein following concurrent treatment was observed. Following sequential treatment, however, total p53 was below the detection limit which we attribute to the difference in treatment times and the half life of p53 (Supplemental Figure 3). We also assessed a series of factors including DNA dependent protein kinase (DNA-PK), Ataxia-telangiectasia mutated and Rad3-related (ATR) kinase, ataxia-telangiectasia mutated (ATM) kinase, checkpoint kinase-1 (CHK1) and checkpoint kinase-2 (CHK2). Robust levels of phosphorylation and activation was observed only for ATM and in this case there was no significant difference between the concurrent sequentially treatments, suggesting that the differential activation of p53 was not applicable to ATM.
Figure 6.
Western Blot Analysis Following MCI 13E or F Treatment. A549 cells were treated sequentially or concurrently with cisplatin (5 μM) and MCI 13E or MCI13F for 24 total hours. Lysates were probed for p53-ser15-phosphorylation or phosphorylated-ATM. GAPDH was probed as a loading control.
Discussion
Targeting protein-DNA interactions with small molecules holds the potential to disrupt numerous essential cellular processes that could be used therapeutically. This approach holds much promise but presents significant challenges. Advances in screening technologies including high throughput screening and in silico screening of chemical libraries has aided in the identification of SMIs of a small number of protein-DNA interactions and previously our lab identified two different classes of SMIs capable of inhibiting the interaction of RPA with DNA (31;34). Here we describe the cellular effects of a novel isoborneol haloacetate SMI (MCI13E), which irreversibly inhibits RPA binding to ssDNA in vitro. We recently reported another RPA inhibitor, TDRL-505, a reversible inhibitor that disrupts RPA binding through the p70 central OB-fold-DNA interaction (31;34). However, MCI13E does not act via the p70 central DNA binding domain A/B. Considering that MCI13E and TDRL-505 target two distinct regions of RPA (31) it is not surprising that we observe some different cellular effects of treatment with these compounds. Both MCI13E and TDRL-505 possess single agent cytotoxic activity likely owing to RPA’s role in S-phase replication. However, MCI13E treatment induces death via classical apoptosis (Figure 2) while TDRL-505 is cytotoxic via a non-apoptotic pathway (34). Furthermore, the IC50 values associated with single agent treatment of A549 and H460 cells lines with MCI13E is lower than that following treatment with TDRL-505. Interestingly genetic knockdown of RPA p70 resulted in an increase in sub-G1 cells suggestive of apoptosis which was preceded by an accumulation of cells in S-phase (14) consistent with the MCI13E inhibitor data. Depending on cell type we also observe either an in increase in G1 or S-phase cells following MCI13E treatment (Figure 2, 3, 4). This difference in the cell cycle phase lengthening following treatment may be due to the difference in initial cell cycle distribution and may be magnified due to this initial difference. It is easy to attribute the differential effects of these two classes of compounds to the different mechanisms by which they inhibit RPA. However, we cannot rule out potential of off-target effects or differential activity as a function of compound stability and/or cellular uptake.
In addition to single agent cytotoxic activity, inhibition of RPA results in increased sensitivity to cisplatin (Figure 4 (34)). RPA’s role in NER is twofold. First RPA interacts with damaged duplex DNA and forms the preincision complex with XPA and TFIIH (36). Second RPA aids in the resynthesis and relegation in the final step of the NER pathway (37). More recently, mutational analysis of RPA has revealed differential effects on DNA repair activity, sometimes producing a dramatic reduction in ss-DNA binding and abrogating repair, but with less of an effect on replication (14). Consistent with the importance of ssDNA binding in repair, we observe a greater than additive effect when combining MCI13E treatment with cisplatin. Interestingly, this is a schedule-dependent phenomenon, and synergy was only observed in cells treated sequentially with cisplatin then MCI13E. Schedule dependent synergy was not observed for TDRL-505 cisplatin interactions (34). The schedule-dependent synergy of MCI13E could not be attributed to alterations in cell cycle, but could point to compound stability and reactivity, as the haloester may be relatively unstable in medium. The potential also exists for a direct MCI13E-cisplatin interaction which inactivates either or both compounds.
Recent studies have also demonstrated RPAs role in DNA damage induced signal transduction through the ATR-interacting protein (ATRIP) which in turn initiates the recruitment of ATR (38;39). The formation of this complex and the activation of ATR by TOPBP1 results in the phosphorylation of CHK1, which can phosphorylate p53 to block cell cycle progression to allow time for the repair of damaged DNA (38). While the low levels of endogenous CHK1 precluded definitive analysis, p53 activation was assessed and differential phosphorylation was observed in cells which were subject to concurrent versus sequential cisplatin-MCI13E treatment. These data suggest that the synergy observed in sequential treatment could be a function of reduced p53 activation compared to that observed with the concurrent treatment (Figure 6). Similarly, the dramatic increase in p53 activation in the concurrent treatment is likely in part responsible for the significant antagonism observed between cisplatin and MCI in the concurrent treatment protocol (Table 1).
Interestingly, single agent treatment with SMIs MCI13E or MCI13F only slightly increases the expression level of p21 as compared to the untreated control while single agent cisplatin treatment produced a dramatic increase in p21 mRNA expression. Combination studies, using concurrent treatment resulted in an increase in p21 mRNA level. However this increase was maintained at a reduced level compared with the appropriate cisplatin control. Sequential treatment resulted in a similar pattern; however the differences between single agent cisplatin treatment and sequential combination treatment were not as pronounced. Taken together, these data suggest that apoptosis and the lengthening of the cell-cycle may be primarily due to the increase in p21 mRNA expression level. Furthermore, the moderate differences in mRNA expression apparent with sequential treatment verses the concurrent control likely contribute to the synergy observed with sequential treatment. However, the slight decrease in p21 mRNA expression comparing single agent cisplatin and combination concurrent treatment does not correlate with the increase in the phosphorylation of p53 detected via western blot analysis. It is possible; however, that the increase in phosphorylated p53 may not translate directly into an increase in p21 expression or that this is require for p21 as p53 independent mechanisms may be involved, a possibility supported by results with H1299 p53 null cells. Further characterization of DNA repair proteins DDB2 and XPC demonstrate similar trends in mRNA expression levels as with p21. Taken together, the mRNA expression data is supportive of the synergistic and antagonistic results noted above for each treatment method. It also suggests that the inhibitors are altering not only the RPA-DNA binding activity but also the expression of other DNA repair proteins (XPC and DDB2) and tumor suppressor protein p21.
ATM, a protein kinase similar to ATR, initiates cell-cycle arrest in response to DNA double strand breaks (DSBs). ATM, which is recruited to the site of a DBS via the MRN complex (MRE11,Rad50 and NSB1) phosphorylates H2AX (γH2AX) and CHK2 resulting in cell cycle arrest and a lengthening of the S-phase of the cell cycle (38). Again, ATM phosphorylation increases moderately with cisplatin and combination MCI13E treatment in all cell lines analyzed and no detectable CHK2 or γH2AX signal was detected. Taken together, this data suggests that the cell cycle lengthening following single agent MCI or combination treatment is not due to an increase in ATM activation. Furthermore, combination treatment of A549 cell lines with MCI13E or F and ionizing radiation results in, at best, an additive increase in cell death (data not shown). This data, in addition to the ATM data, suggests that the SMIs do not elicit their cytotoxic effects by damaging both DNA strands resulting in a double strand break.
Although our data demonstrates an increase in cellular apoptosis and synergy between the SMIs and cisplatin, the direct interactions between the RPA protein and SMIs remains unclear. MCI13E is thought to react with cysteine residues, of which RPA contains 13. Four cysteine residues reside within the zinc-finger domain while the remaining residues are dispersed throughout the protein. Mutation analysis of each individual cysteine holds potential to reveal the direct interaction between the SMI and RPA. Moreover, demonstrating in vivo a direct SMI-RPA interaction would further confirm the specificity of the interaction already confirmed in vitro (31). Overall, these identified SMIs of RPA binding hold great potential for further characterizing the RPA-DNA interaction and delineating the specific role of individual RPA domains in DNA repair and replication. The synergy with chemotherapeutics like cisplatin suggest potential clinical benefits as well.
Supplementary Material
Acknowledgments
The authors would like to thank member of the Turchi and Jalal lab for the helpful, stimulating discussion. This work was supported by NIH Grant CA82741 to JJT.
References
- 1.Nouspikel T. DNA repair in mammalian cells : Nucleotide excision repair: variations on versatility. Cell Mol Life Sci. 2009;66:994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shuck SC, Short EA, Turchi JJ. Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology. Cell Res. 2008;18:64–72. doi: 10.1038/cr.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clement C, Kaczmarek N, Mathieu N, Tomas M, Leitenstorfer A, Ferrando-May E, et al. Dissection of the Xeroderma Pigmentosum Group C Protein Function by Site-directed Mutagenesis. Antioxid Redox Signal. 2010 Oct 7; doi: 10.1089/ars.2010.3399. [DOI] [PubMed] [Google Scholar]
- 4.Dip R, Camenisch U, Naegeli H. Mechanisms of DNA damage recognition and strand discrimination in human nucleotide excision repair. Dna Repair. 2004;3:1409–23. doi: 10.1016/j.dnarep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Nouspikel TP, Hyka-Nouspikel N, Hanawalt PC. Transcription domain-associated repair in human cells. Mol Cell Biol. 2006;26:8722–30. doi: 10.1128/MCB.01263-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tornaletti S, Patrick SM, Turchi JJ, Hanawalt PC. Behavior of T7 RNA polymerase and mammalian RNA polymerase II at site-specific cisplatin adducts in the template DNA. J Biol Chem. 2003;278:35791–7. doi: 10.1074/jbc.M305394200. [DOI] [PubMed] [Google Scholar]
- 7.Wakasugi M, Sancar A. Assembly, subunit composition, and footprint of human DNA r epair excision nuclease. Proc Natl Acad Sci USA. 1998;95:6669–74. doi: 10.1073/pnas.95.12.6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patrick SM, Turchi JJ. Replication Protein A (RPA) Binding to Duplex Cisplatin-damaged DNA Is Mediated through the Generation of Single-stranded DNA. J Biol Chem. 1999;274:14972–8. doi: 10.1074/jbc.274.21.14972. [DOI] [PubMed] [Google Scholar]
- 9.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–37. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA- binding protein required for eukaryotic DNA metabolism. Annual Review of Biochemistry. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 11.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–81. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 12.Bochkareva E, Frappier L, Edwards A, Bochkarev A. The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J Biol Chem. 1998;273:3932–6. doi: 10.1074/jbc.273.7.3932. [DOI] [PubMed] [Google Scholar]
- 13.Bochkarev A, Bochkareva E, Frappier L, Edwards AM. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single- stranded DNA binding. EMBO J. 1999;18:4498–504. doi: 10.1093/emboj/18.16.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haring SJ, Mason AC, Binz SK, Wold MS. Cellular functions of human RPA1. Multiple roles of domains in replication, repair, and checkpoints. J Biol Chem. 2008;283:19095–111. doi: 10.1074/jbc.M800881200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koberle B, Grimaldi KA, Sunters A, Hartley JA, Kelland LR, Masters JRW. DNA repair capacity and cisplatin sensitivity of human testis tumour cells. Int J Cancer. 1997;70:551–5. doi: 10.1002/(sici)1097-0215(19970304)70:5<551::aid-ijc10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 16.Koberle B, Masters JRW, Hartley JA, Wood RD. Defective repair of cisplatin-induced DNA damage caused by reduced XPA protein in testicular germ cell tumours. Current Biology. 1999;9:273–6. doi: 10.1016/s0960-9822(99)80118-3. [DOI] [PubMed] [Google Scholar]
- 17.Welsh C, Day R, McGurk C, Masters JRW, Wood RD, Koberle B. Reduced levels of XPA, ERCC1 and XPF DNA repair proteins in testis tumor cell lines. Int J Cancer. 2004;110:352–61. doi: 10.1002/ijc.20134. [DOI] [PubMed] [Google Scholar]
- 18.Ferry KV, Hamilton TC, Johnson SW. Increased nucleotide excision repair in cisplatin-resistant ovarian cancer cells - Role of ERCC1-XPF. Biochem Pharmacol. 2000;60:1305–13. doi: 10.1016/s0006-2952(00)00441-x. [DOI] [PubMed] [Google Scholar]
- 19.Einhorn LH. Curing metastatic testicular cancer. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4592–5. doi: 10.1073/pnas.072067999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armstrong DK, Bundy B, Wenzel L, Huang HQ, Baergen R, Lele S, et al. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. New England Journal of Medicine. 2006;354:34–43. doi: 10.1056/NEJMoa052985. [DOI] [PubMed] [Google Scholar]
- 21.Juergens RA, Brahmer JR. Adjuvant therapy for resected non-small-cell lung cancer: past, present, and future. Curr Oncol Rep. 2005;7:248–54. doi: 10.1007/s11912-005-0046-5. [DOI] [PubMed] [Google Scholar]
- 22.Rigas JR, Kelly K. Current treatment paradigms for locally advanced non-small cell lung cancer. J Thorac Oncol. 2007;(Suppl 2):S77–S85. doi: 10.1097/01.JTO.0000269735.21209.bc. [DOI] [PubMed] [Google Scholar]
- 23.Teuben JM, Bauer C, Wang AHJ, Reedijk J. Solution structure of a DNA duplex containing a cis- diammineplatinum(II) 1,3-d(GTG) intrastrand cross-link, a major adduct in cells treated with the anticancer drug carboplatin. Biochemistry. 1999;38:12305–12. doi: 10.1021/bi9904757. [DOI] [PubMed] [Google Scholar]
- 24.Liedert B, Pluim D, Schellens J, Thomale J. Adduct-specific monoclonal antibodies for the measurement of cisplatin-induced DNA lesions in individual cell nuclei. Nucleic Acids Res. 2006;34:e47. doi: 10.1093/nar/gkl051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zamble DB, Mu D, Reardon JT, Sancar A, Lippard SJ. Repair of cisplatin--DNA adducts by the mammalian excision nuclease. Biochemistry. 1996;35:10004–13. doi: 10.1021/bi960453+. [DOI] [PubMed] [Google Scholar]
- 26.Ormerod MG, Orr RM, Peacock JH. The role of apoptosis in cell killing by cisplatin: a flow cytometric study. British Journal of Cancer. 1994;69:93–100. doi: 10.1038/bjc.1994.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen HN, Sevin BU, Averette HE, Perras J, Ramos R, Donato D, et al. Cell cycle perturbations of platinum derivatives on two ovarian cancer cell lines. Cancer Invest. 1993;11:264–75. doi: 10.3109/07357909309024851. [DOI] [PubMed] [Google Scholar]
- 28.Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13:1027–36. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- 29.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–14. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews BJ, Turchi JJ. Development of a high-throughput screen for inhibitors of replication protein A and its role in nucleotide excision repair. Mol Cancer Ther. 2004;3:385–91. [PubMed] [Google Scholar]
- 31.Anciano Granadillo VJ, Earley JN, Shuck SC, Georgiadis MM, Fitch R, Turchi JJ. Targeting the OB-folds of Replication Protein A with Small Molecules. Journal of Nucleic Acids. 2010;304035 doi: 10.4061/2010/304035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turchi JJ, Shuck SC, Short EA, Andrews BJ. Targeting Nucleotide Excision Repair as a Mechanism to Increase Cisplatin Efficacy. In: Bonetti A, Leone R, Muggia FM, Howell SB, editors. Platinum and Other Heavy Metal Compounds in Cancer Chemotherapy. New York: Humana Press; 2009. pp. 177–88. [Google Scholar]
- 33.Chou TC, Talalay P. Quantitative-analysis of dose-effect relationships - the combined effects of multiple-drugs or enzyme-inhibitors. Advances in Enzyme Regulation. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 34.Shuck SC, Turchi JJ. Targeted inhibition of Replication Protein A reveals cytotoxic activity, synergy with chemotherapeutic DNA-damaging agents, and insight into cellular function. Cancer Res. 2010;70:3189–98. doi: 10.1158/0008-5472.CAN-09-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boeckman HJ, Trego KS, Turchi JJ. Cisplatin sensitizes cancer cells to ionizing radiation via inhibition of nonhomologous end joining. Mol Cancer Res. 2005;3:277–85. doi: 10.1158/1541-7786.MCR-04-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wakasugi M, Sancar A. Order of assembly of human DNA repair excision nuclease. J Biol Chem. 1999;274:18759–68. doi: 10.1074/jbc.274.26.18759. [DOI] [PubMed] [Google Scholar]
- 37.Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem. 1997;272:23465–8. doi: 10.1074/jbc.272.38.23465. Review 50 refs. [DOI] [PubMed] [Google Scholar]
- 38.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–27. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–8. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.