Abstract
Epigenetic marks (eg, DNA 5-methylcytosine [5mC] content or CpG methylation) within specific gene regulatory regions have been demonstrated to play diverse roles in stress adaptation and resulting health trajectories following early adversity. Yet the developmental programming of the vast majority of the epigenome has not yet been characterized, and its role in the impact of early stress largely unknown. In the present study, we investigated the relationships among early life stress, whole-epigenome and candidate stress pathway gene (serotonin transporter, 5-HTT) methylation patterns, and adult behavioral stress adaptation in a non-human primate model. Early in life, experimental variable foraging demand (VFD) stress or control conditions were administered to two groups each of 10 female bonnet macaques (Macaca radiata) and their mothers. As adults (3–13 years of age), these females were assessed for behavioral adaptation to stress across four conditions of increasing intensity. Blood DNA 5-HTT 5mC status was determined using sodium bisulfite pyrosequencing and total 5mC content was determined using ELISA. Neither stress reactivity nor DNA methylation differed based on early life stress. However, we found that both greater 5-HTT and whole-genome 5mC was associated with enhanced behavioral stress reactivity following early life stress, but not control conditions. Therefore, regardless of developmental origin, greater DNA methylation conferred a genomic background of “risk” in the context of early stress. We suggest that this may arise from constrained plasticity in gene expression needed for stress adaptation early in development. This risk may have wider implications for psychological and physical stress adaptation and health.
Keywords: Serotonin transporter, DNA methylation, genotype, variable foraging demand stress, development, Bonnet macaque
Early life stress has been linked with poor stress adaptation and related illnesses, such as cardiovascular disease, metabolic disorders, and psychiatric illness later in life (Wegman and Stetler, 2009). DNA methylation patterning, the addition of methyl groups to cytosine-guanine dinucleotides (CpG methylation; Bird, 1986) within gene regulatory regions, may moderate or even mediate the effects of early life stress on later stress adaptation. However, the developmental programming of the vast majority of the epigenome has not yet been characterized, and its role in the impact of early stress largely unknown. There are over 22 million genomic CpG sites that may be differentially methylated in each cell of the central (brain) and peripheral (heart, adrenal gland, blood) organs that regulate stress response. While methylation patterns in some gene regulatory regions may be sensitive to environmental cues during postnatal development (Weaver et al., 2004; Champagne et al., 2006; Roth et al., 2009; Murgatroyd et al., 2009), methylation patterns within other gene regulatory regions may be set before birth. They may be partially inherited (Jones and Takai, 2007; Schalkwyk et al., 2010), set during prenatal development, (Kaminsky et al., 2009) or dependent on DNA structure (Brandeis et al., 1994; Schalkwyk et al., 2010). The role of these marks in stress adaptation is largely unknown.
Epigenetic patterns of all developmental origins within stress pathway candidate genes may play a role in stress adaptation. Structural variation in the 5’ serotonin transporter (5-HTT) regulatory region has been reported by some investigators to moderate the effects of early life stress in humans and non-human primates (eg, 5-HTTLPR, Champoux et al., 2002; Bennett et al., 2002; Caspi et al., 2003; Barr et al., 2004; Coplan et al., 2011; although see Risch et al., 2009). We have recently shown that epigenetic variation in the 5-HTT regulatory region also moderates the effects of early stress. 5-HTT 5mC content was associated with enhanced behavioral reactivity to stress in infant rhesus macaques that had experienced early life maternal deprivation (Kinnally et al., 2010). Yet there was no association between early life stress and 5-HTT methylation patterns. We concluded that whatever the developmental origins of 5-HTT methylation patterns, taken as a whole, they compounded the risk of early life stress.
Thousands of genes are involved with stress response, however (Alter et al., 2008). We will better understand the role of candidate gene DNA methylation patterns if we compare them with individual differences in 5-methylcytosine across the genome. Genome-wide methylation is a potential confound of candidate gene methylation-stress associations because some important stress-related processes may occur at the epigenomic level. For example, pharmacological interventions that globally hypo- or hypermethylate the genome influence risk for stress susceptibility (Covington et al., 2009; Weaver et al., 2005). In the present study, we therefore tested whether candidate gene and global DNA methylation was associated with stress adaptation across the lifespan, even in tissue that is only secondarily involved in the stress response (e.g. blood, which includes cell subsets that have a lifespan of hours to years). We compared the roles of 5-HTT and/or whole-genome 5mC in the effects of experimental variable foraging demand (VFD) stress on behavioral stress adaptation in twenty adult (3–13 years of age) female bonnet macaques (Macaca radiata). Experimentally-induced variable foraging demand (VFD) conditions strain mother-infant relationships because dyads cannot predict whether food will be easily accessible on a given day (Coplan et al., 1996). Consequently, stress response is re-programmed in developing VFD infants and remains disrupted years after the manipulation, similar to maltreated humans. This experimental approach mimics early human adversity, while eliminating genome-environment correlations that can confound naturalistic human studies.
Materials and Methods
Subjects
Twenty female bonnet macaques (Macaca radiata), 3–13 years of age (mean 8.4 years), were included in this study. All were housed at SUNY Downstate Medical Center in indoor social enclosures (with 1–5 other females and juveniles) that measured approximately 2m x 3 m x 2.5m. Animals were maintained on 12:12 light dark cycle and fed monkey chow once per day at 1400 h. Ten subjects had been exposed to “variable foraging demand” (VFD) conditions in infancy (Coplan et al., 1996) and ten to “low foraging demand” (LFD) conditions. All were l/l rh5-HTTLPR homozygotes to control for potential allelic imbalance between methylation patterns and genotype (Kinnally et al., 2010).
Rearing Conditions
Variable foraging demand procedures have been described elsewhere (Coplan et al., 1996). All animals were reared from birth until three months of age under comparable conditions: housed with their mothers in small social groups comprised of 3–6 mother-infant dyads. From three to six months of age, all animals were required to forage for their food in a foraging device (a modified cart with wood chips to hide food). The mothers of control “LFD” animals were able to readily retrieve food from this device. VFD mothers were required to forage little or extensively for food, in randomized order. Subsequently, these females were housed in small social groups, usually including at least one other adult female and the offspring of all females.
Behavioral Testing
As adults, subjects were removed from social housing to temporary individual housing in the primate facility at SUNY Downstate Medical Center for all assessments. Assessments were conducted five days per week over the course of four weeks in the same testing room (6 m x 3 m x 4 m). Subjects’ anxiety-related and normal behavior (Kalin et al., 1991; Golub et al., 2008) were observed during four conditions, which were each repeated five times (once daily): following relocation, relocation with an unfamiliar human present, relocation with a familiar human making eye contact, and conditioned stimulus exposure following fear conditioning. Tasks were administered in a fixed order for all subjects. Prior to observation, observers reached 85% or better agreement on all ethogram behaviors (eg, locomotion, eating, scratching) for 10 consecutive 1-minute reliability trials. Inter-observer reliability was calculated as the number of behaviors the two observers agreed upon divided by the total number of observed behaviors (agree/[agree+disagree]).
Animals were tested in four cohorts of seven or eight animals each. No members from the same social groups were tested in cages in direct line of sight from other group members (i.e., not in the adjacent testing cage or the cage directly across). Order of testing was rotated each day such that cohorts were tested first, second, third, and fourth equal numbers of times. Each morning of testing, animals were removed from social housing and placed in transport boxes measuring 0.3 m x 0.5 m x 0.3 m. At approximately 0900 h, the first cohort of animals was placed in testing cages measuring 1 m x 0.8 m x 1 m. Testing cages were raised from the ground approximately 1.2 m in height (the top cages of quad housing), putting all subjects at about eye level with the observer. Immediately after the last animal was placed in the testing cage, and the animal care staff has vacated the testing room, the observer entered the room and observations commenced. For the unfamiliar human, human intruder and fear conditioning tests, the same second human entered with the observer. The order of testing within cohorts was rotated every day such that an animal was tested at a different time each day. Frequency of anxiety-related and normal behaviors was recorded for each subject during each condition. The same individual conducted all observations, and stood at least 1 m from the testing cage at all times during observation, averting eye contact.
Relocation Only
During the relocation only condition, behavioral responses were recorded for each animal in one-minute trials by a single observer as described above.
Relocation with Second Human Present
During the relocation with a second unfamiliar human present condition, a second human stood next to the observer, 1 m from the testing cage and averting eye contact throughout the 1-minute observation.
Relocation with Human Intruder
During the relocation with the second human making eye contact condition, baseline behavioral observations were conducted as in the relocation with unfamiliar human present condition. Observations were then repeated for each animal while the human initiated and maintained eye contact with the animal for the duration of the one-minute observation.
Fear Extinction Testing
Fear conditioning and extinction testing were conducted on the same day. The test included three parts: five consecutive 15-second presentations of a neutral stimulus (a light), five consecutive 15-second presentations of the light with simultaneous eye contact from the second human, and five consecutive 15-second presentations of the now-conditioned light stimulus without human eye contact. The second human operated the light apparatus and acted as human intruder. The conditioned stimulus was a round light, 5” diameter, fixed to a vertical rolling apparatus (metal pole, 1” diameter, 1.5 m height). The light was presented 1 m from the animal at the animal’s eye level. The observer recorded the frequency of anxiety-related and normal behaviors during five 15- second presentations of the conditioned stimulus (light). After a delay of 10 seconds, the conditioned stimulus was presented simultaneously with an unconditioned stimulus (second human making eye contact), and behavior frequencies recorded for five 15-second presentations by the observer. After a 10 second delay, the conditioned stimulus (light) was presented alone for five 15-second presentations and behavioral frequencies recorded.
Factor Analysis
Behavioral Reactivity scores were calculated to use in factor analysis. Frequencies of all behaviors were z-scored based on data from all trials within one test and summed for each individual within each trial within each test. Then, Cronbach’s alpha reliability for reactivity scores across trials for each test were computed. All showed Cronbach’s alpha above .76, indicating good internal consistency of scores within tests. To capture the time course of stress adaptation while minimizing the number of input variables for factor analysis, factor analysis was applied to behavioral reactivity composite scores from the first day of observation and the last day of the first week of observation (relocation only condition), the first and last observation with the presence of a second unfamiliar human (relocation with second human present), the first and last human intruder trial (relocation and human intruder), the first exposure to the conditioned stimulus following fear conditioning, and the last exposure to the conditioned stimulus following fear conditioning (fear extinction condition). Factor analysis of reactivity rates on the first and last exposure to each condition (to capture the time course of stress adaptation while minimizing the number of input variables) yielded a factor structure that accounted for 65% of the variance, and described reactivity to: 1.) first and final exposure to a second human present, first exposure to human intruder, and first and final fear conditioned stimulus exposure; 2.) first and final exposure to relocation and one human present, and first and final second human present condition, and 3.) final exposure of second human present, human intruder, and conditioned stimulus exposure. We have characterized these factors as reactivity to: 1.) high intensity stressors, 2.) low intensity stressors, and 3.) final exposure of stressors. Scores were calculated using the regression method based on factor loadings.
Sodium Bisulfite Pyrosequencing
Blood was collected from subjects via femoral venipuncture, decanted to EDTA-coated tubes and stored at 4°C. DNA was isolated from whole blood using commercially available DNA extraction kits (Qiagen, Inc., Valencia, CA). Peripheral blood DNA 5-HTT CpG island methylation status was determined using sodium bisulfite pyrosequencing as described previously (Kinnally et al., 2010). DNA was converted for methylation sequencing using a commercially available sodium bisulfite modification kit (Qiagen, Inc., Valencia, CA). Completion of sodium bisulfite conversion was confirmed by ensuring that known lone cytosines were determined to be thymines during sequencing. The target region was amplified using polymerase chain reaction. PCR amplification was conducted with three primer sets (Integrated DNA Technologies) targeting contiguous regions of the first 650 bp of the 5-HTT CpG island. Unfortunately, primers could not be designed for the 150 bp region of the CpG island that overlaps with the transcription initiation start site. Primers were as follows: F1 5’GGGAAGAAGTTTTGGAAAGA AA3’R1 5’CCACTATCTAAAAATCAAACCATATAA3’; F2 5’ GGTTGTAA AGTTATTGTAATTATAAAGG3’; R2 5’ TTTCTTTCCAAAACTTCTTCCC3’; F3 5’GGTTTTTTATATGGTTTGATTTTTAGATA3’; R3 5’ CCTACCCTACCCTACCT ACTACTCC3’. Reverse primers were tagged with biotin. Biotin-labeled amplicons were captured on streptavidin beads (Roche, Inc., Basel, Switzerland), washed sequentially with 70% ethanol, denaturing buffer (10mM Sodium hydroxide) and washing buffer (10 mM Tris, pH7.6). Beads with amplicon were incubated with binding buffer (10 mM Tris, 2 M NaCl, 1 mM EDTA, 0.1% Tween 20, pH 7.6) and 0.4 uM sequencing primer at 80°C to anneal primers to the template. Amplicons were subjected to pyrosequencing (Tost and Gut, 2007; Biotage, Inc., Uppsala, Sweden). The quantity of methylated residues was assessed using Q-CpG software (Biotage, Inc.). Each amplicon was sequenced in two to three separate reactions. Sequencing primers were as follows: SEQ1 5’AAGAAGTCTTGGAAAGAA A3’; SEQ2 5’TTGTAGGGTTGTGTTAGG3’; SEQ3 5’AAGTTATTGTAATTATAAA GGAAT3’; SEQ4 5’GGGYGTAGGGTTAGGAT3’; SEQ5 5’ATGGTTTGATTTTTAG ATAG3’; SEQ6 5’ TGAGGYGAATAAATTTAATG 3’; SEQ7 5’TAGGAGGGCAGG GAT3’. Sequencing was conducted using the PyroMark PSQ MA instrument and PyroGold reagents (Biotage, Inc., Uppsala, Sweden). Proportion of methylated residues in each reaction at each locus was assessed using Q-CpG software (Biotage, Inc. Uppsala, Sweden). Only samples with 100% sodium bisulfite conversion were included. Products were pyrosequenced using the PyroMark PSQ MA system (Biotage, Inc., Uppsala, Sweden). Data were analyzed for subjects with at least 70% coverage (mean = 88.81%) of the total 59 residues.
Assay reliability was ensured by conducting two separate sodium bisulfite conversions, PCR amplifications, and pyrosequencing reactions for each sample. Average C-methylation across 59 CpG sites were calculated from each separate run. Coefficients of variance of average C-methylation between the same samples between runs averaged 30% and individual values were significantly correlated (r = .52, p = .006). The high variance between runs likely arises from our conservative approach, to conduct two completely separate protocol runs to ensure replicability of the data. Taking the most conservative approach, the average of the two separate runs was therefore used for statistical analysis. A human 100% methylated DNA control (EpiGenDX, Waltham, MA) was included on every plate as a control, and C.V. among plates was 6.6%. This indicates good reliability of the pyrosequencing assay in the face of potential variation attributable to DNA extraction and sodium bisulfite conversion.
Whole-genome Methylation ELISA
Peripheral blood DNA 5-methylcytosine content was measured using a commercially available colorimetric ELISA kit (EpigenTek, Brooklyn, NY). Up to 100 nanograms of isolated whole blood DNA was added to pre-coated ELISA plates and incubated at 37° C. Plates were washed and incubated with secondary antibodies and developing reagents. Colorimetric intensity was imaged using a Labsystems Multiskan Plus (Fisher Scientific, Pittsburgh, PA) plate reader reading at 450 nm. Concentrations of unknown samples were interpolated from a standard curve of known 5-methylcytosine concentrations (EpiGentek, Brooklyn, NY). Regression fit of the standard curve, which was made up of linear standard dilutions, was good (R2 = .98). Samples were run on the same plate to facilitate comparison between samples. Samples were run in duplicate, and coefficients of variance averaged 18.1% (range 5–38%, standard deviation 9%). Analysis that included samples with a C.V. of 20% or less did not change the direction or significance of the results, and so all samples were retained in the analysis. Average 5mC between duplicates was used as an outcome measure.
Results
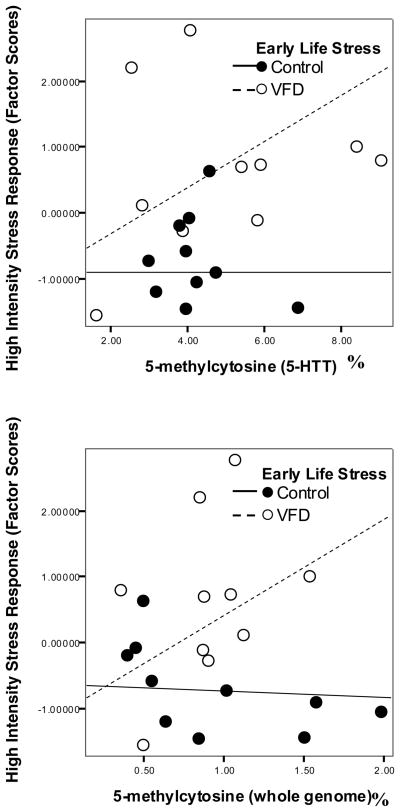
5-HTT and whole genome methylation were not correlated (Pearson’s correlation; r = .198, p = .403). Neither 5-HTT (Pearson’s correlation; r = .092, p = .699) nor whole genome methylation (Pearson’s correlation; r = −.409, p = .066) varied significantly by age, although there was a trend for older females to display lower whole blood whole genome 5-methylcytosine. Age was not significantly associated with behavioral stress reactivity (Pearson’s correlation; r = .312, p = .180). Neither 5-HTT nor whole genome methylation differed between VFD and control groups (independent samples t-test; t = −.847, df = 18, p = .408; t = .163, df=18, p = .862). Two separate backward multiple regression analyses were conducted for each DNA methylation measure. For the model including 5-HTT DNA methylation, rearing, and the interaction term, we found that 5-HTT 5mC (F (1, 19) = 4.756, p = .023, adjusted R2 = .283) interacted with early life stress to predict greater reactivity to high intensity stressors. The effect was such that higher methylation was associated with higher reactivity in VFD animals, but not controls (t = 2.9, p =.010). A trend level main effect of DNA methylation was also observed, such that 5-HTT DNA methylation (t = −1.966, p =.066) was negatively associated with stress reactivity. The same pattern of results was observed for whole genome DNA methylation (F (1,19) = 6.695, p = .008, adjusted R2 = .375), where an interaction between whole genome methylation and VFD predicted higher reactivity (t = 3.625, p = .002), but overall, methylation was inversely associated with reactivity scores (t = −2.98, p = .008).
Discussion
Previous studies have demonstrated a role for DNA methylation, or 5-methylcytosine (5mC), at specific genomic loci in the adverse effects of early life stress. We observed that both 5-HTT and genome-wide 5mC levels were linked with the same genomic background of “risk” in the context of early stress. VFD females that had higher average CpG methylation (in the 5-HTT regulatory region and across the epigenome) exhibited the greatest behavioral reactivity to stress. These findings suggest that both candidate gene and whole genome DNA methylation may confer a background of risk in individuals that experience early life stress. This is particularly notable because risk is present in carriers of the “protective” (long) 5-HTTLPR allele (Coplan et al., 2011). Importantly, our data suggest that this developmental risk influences stress adaptation across the lifespan, indicating that developmental epigenomic processes may permanently impact individual outcomes related to stress.
Our finding that DNA methylation across the epigenome is a risk factor for poor stress adaptation in individuals that experienced early life stress is novel. At-risk individuals showed enhanced reactivity to higher intensity stressors (such as human intruder exposure and fear conditioning), but not low intensity stress (such as relocation to temporary housing) or recovery from stress (during final exposures of each stressor). Reactivity to high intensity stressors is likely the most relevant factor to stress-related disorders in humans because these are the most fear-inducing conditions we presented our animals. We propose that reactivity to high intensity stress reflects either greater anxiety in response to stress or difficulty regulating its intensity under threatening conditions, both potentially adverse psychological and physical phenomena (Kalin et al., 1991). Future studies will examine whether poor stress adaptation is linked with stress-related disease outcomes in bonnet macaques, as is the case in humans (Wegman and Stetler, 2009).
The greatest risk posed by early life stress was observed in individuals with high 5-HTT and whole genome DNA methylation, however. Some studies have demonstrated a link between global hypermethylation and stress-related outcomes in humans (eg, coronary artery disease: Sharma et al., 2008; inflammation and cardiovascular mortality, Baccarelli et al., 2010). Our results suggest that risk is moderated by early life stress. We have recently shown that 5mC content within the 5-HTT regulatory region was associated with enhanced behavioral reactivity to stress in infant rhesus macaques that had experienced early life maternal deprivation (Kinnally et al., 2010). Our present data confirm and extend that finding. One implication of this result is that future candidate gene studies should consider the background of the entire epigenome to assess gene specificity, as some groups have done previously (Oberlander et al., 2008). We must also understand how global DNA methylation could confer generalized risk, when its effects on gene expression may differ across genes and tissue. Global DNA methylation includes 5mC of all developmental origins, including patterns that are inherited, set prenatally, and environmentally programmed (Jones and Takai, 2007; Schalkwyk et al., 2010; Kaminsky et al., 2009; Brandeis et al., 1994; Schalkwyk et al., 2010; Weaver et al., 2004). We did not observe a difference in global DNA methylation between VFD and controls, so individual differences in global 5mC likely arise from a combination of unique genetic, prenatal, and postnatal factors. Our data are consistent with the recent finding that susceptibility to post-traumatic stress disorder is linked with hypomethylation (and possibly higher expression) of the DNA methyl-transferase (DNMT) 3B gene, which is partly responsible for establishing methylation patterns across the genome (Uddin et al., 2010). Since DNA methylation of all developmental origins may limit genomic plasticity (Bird et al., 1996), it is possible that global 5mC constrains plasticity needed for stress adaptation across tissue, especially early in life. This hypothesis is consistent with rodent studies demonstrating that global genomic plasticity predicts behavioral adaptability to challenging circumstances (Alter et al., 2008) and also with studies demonstrating that relatively non-specific hypo-methylating drugs (eg, histone deacetylase inhibitors) are effective in treating stress-related disease outcomes in humans and rodent models (Covington et al., 2009). However, our data suggest that these processes are rooted in the developmental reorganization of stress response during exposure to early life stress, as an inverse relationship between 5mC level and stress reactivity was observed in control subjects.
A limitation to our study is the use of DNA from whole blood for epigenetic analyses. We cannot distinguish between methylation patterns in different hematocyte subtypes that were included our original sample. It is notable, however, that the present findings are paralleled by our previous report, that methylation patterns within a different leukocyte population (namely, mononuclear leukocytes) were linked to behavioral effects of early stress in macaques (Kinnally et al., 2010). The advantage of our approach is that if we can validate the use of peripheral cells to examine the relationship between methylation status and stress, we can track modification of risk in living individuals (eg, Uddin et al., 2010).
In conclusion, we have discovered that natural variation in global DNA methylation patterns is associated with stress responsiveness following early life stress. This finding highlights the need for in depth study of the global epigenomic processes that may put maltreated individuals at risk for poor stress adaptation not just at the time of early stress, but across the lifespan. Identifying the risk associated with DNA methylation patterns of all developmental origins will further clarify the role of epigenomics in stress-related health outcomes following early adversity.
Figure 1.
5-HTT CpG methylation moderates the effects of early life stress on behavioral stress reactivity. Higher 5-HTT CpG methylation was associated with higher reactivity scores in adults that experienced VFD stress as infants (F (1, 19) = 4.756, p = .023, adjusted R2 = .283). This effect is likely due to higher ratios of 5-methylcytosine across the epigenome, as they also confer risk for poorer stress adaptation (F (1,19) = 6.695, p = .008, adjusted R2 = .375).
Acknowledgments
We thank Yung-yu Huang, Charles LeDuc, Liyong Deng, Lutz Froenicke, Carol Novotney, and animal care staff at the SUNY Downstate Medical Center Primate Facility for assistance with this project. This study was funded by P50 MH062185-08 (to JJM), 1R01MH059990-01A1 (to JDC) and 5T32MH015174 and the Sackler Institute for Psychobiology at Columbia University provided fellowships to ELK. JDC disclosures; grants: Pfizer, GlaxoSmithKline; speaker: Pfizer, GlaxoSmithKline, Bristol Myers Squibb, Eli Lilly, Astra Zeneca. JJM received recent unrelated grants from GSK and Novartis.
Footnotes
Conflict of Interest Statement: JDC received grants from Pfizer, GlaxoSmithKline, Bristol Myers Squibb, Eli Lilly, Astra Zeneca. JJM received unrelated grants from GSK and Novartis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alter MD, Rubin DB, Ramsey K, Halpern R, Stephan DA, Abbott LF, Hen R. Variation in the large-scale organization of gene expression levels in the hippocampus relates to stable epigenetic variability in behavior. PLoS One. 2008;3:e3344. doi: 10.1371/journal.pone.0003344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Tarantini L, Wright RO, Bollati V, Litonjua AA, Zanobetti A, Sparrow D, Vokonas P, Schwartz J. Repetitive element DNA methylation and circulating endothelial and inflammation markers in the VA normative aging study. Epigenetics. 2010;5 doi: 10.4161/epi.5.3.11377. online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr CS, Newman TK, Lindell S, Shannon C, Champoux M, Lesch KP, Suomi SJ, Goldman D, Higley JD. Interaction between serotonin transporter gene variation and rearing condition in alcohol preference and consumption in female primates. Archives of General Psychiatry. 2004;61:1146–1152. doi: 10.1001/archpsyc.61.11.1146. [DOI] [PubMed] [Google Scholar]
- Bennett AJ, Lesch KP, Heils A, Long JC, Lorenz JG, Shoaf SE, Champoux M, Suomi SJ, Linnoila MV, Higley JD. Early experience and serotonin transporter gene variation interact to influence primate CNS function. Molecular Psychiatry. 2002;7:118–22. doi: 10.1038/sj.mp.4000949. [DOI] [PubMed] [Google Scholar]
- Bird A. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Champagne FA, Weaver IC, Diorio J, Dymov S, Szyf M, Meaney MJ. Maternal care associated with methylation of the estrogen receptor-alpha1b promoter and estrogen receptor-alpha expression in the medial preoptic area of female offspring. Endocrinology. 2006;147:2909–15. doi: 10.1210/en.2005-1119. [DOI] [PubMed] [Google Scholar]
- Champoux M, Bennett A, Shannon C, Higley JD, Lesch KP, Suomi SJ. Serotonin transporter gene polymorphism, differential early rearing, and behavior in rhesus monkey neonates. Molecular Psychiatry. 2002;7:1058–1063. doi: 10.1038/sj.mp.4001157. [DOI] [PubMed] [Google Scholar]
- Coplan JD, Abdallah CG, Kaufman J, Gelernter J, Smith EL, Perera TD, Dwork AJ, Kaffman A, Gorman JM, Rosenblum LA, Owens MJ, Nemeroff CB. Early-life stress, corticotropin-releasing factor, and serotonin transporter gene: a pilot study. Psychoneuroendocrinology. 2011;36:289–93. doi: 10.1016/j.psyneuen.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coplan JD, Andrews MW, Rosenblum LA, Owens MJ, Friedman S, Gorman JM, Nemeroff CB. Persistent elevations of cerebrospinal fluid concentrations of corticotropin-releasing factor in adult nonhuman primates exposed to early-life stressors: implications for the pathophysiology of mood and anxiety disorders. Proc Natl Acad Sci U S A. 1996;20:1619–23. doi: 10.1073/pnas.93.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington HE, 3rd, Maze I, LaPlant QC, Vialou VF, Ohnishi YN, Berton O, Fass DM, Renthal W, Rush AJ, 3rd, Wu EY, Ghose S, Krishnan V, Russo SJ, Tamminga C, Haggarty SJ, Nestler EJ. Antidepressant actions of histone deacetylase inhibitors. Journal of Neuroscience. 2009;29:11451–60. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott E, Ezra-Nevo G, Regev L, Neufeld-Cohen A, Chen A. Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nature Neuroscience. 2010;13:1351–1353. doi: 10.1038/nn.2642. [DOI] [PubMed] [Google Scholar]
- Golub MS, Hogrefe CE, Widaman KF, Capitanio JP. Iron deficiency anemia and affective response in rhesus monkey infants. Developmental Psychobiology. 2008;51:47–59. doi: 10.1002/dev.20345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- Kalin NH, Shelton SE, Takahashi LK. Defensive behaviors in infant rhesus monkeys: ontogeny and context-dependent selective expression. Child Development. 1991;62:1175–83. [PubMed] [Google Scholar]
- Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, Wong AH, Feldcamp LA, Virtanen C, Halfvarson J, Tysk C, McRae AF, Visscher PM, Montgomery GW, Gottesman, Martin NG, Petronis A. DNA methylation profiles in monozygotic and dizygotic twins. Nature Genetics. 2009;41:240–5. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- Kinnally EL, Capitanio JP, Leibel R, Deng L, LeDuc C, Haghighi F, Mann JJ. Epigenetic Regulation of Serotonin Transporter Expression and Behavior in Infant Rhesus Macaques. Genes, Brain and Behavior. 2010;9:575–82. doi: 10.1111/j.1601-183X.2010.00588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmühl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OF, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nature Neuroscience. 2009;12:1559–66. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Oberlander T, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin A. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang K, Eaves L, Hoh J, Griem A, Kovacs M, Ott J, Merikangas KR. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. Journal of the American Medical Association. 2009;301:2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth T, Lubin F, Funk A, Sweatt J. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biological Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalkwyk LC, Meaburn EL, Smith R, Dempster EL, Jeffries AR, Davies MN, Plomin R, Mill J. Allelic skewing of DNA methylation is widespread across the genome. American Journal of Human Genetics. 2010;86:196–212. doi: 10.1016/j.ajhg.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Kumar J, Garg G, Kumar A, Patowary A, Karthikeyan G, Ramakrishnan L, Brahmachari V, Sengupta S. Detection of altered global DNA methylation in coronary artery disease patients. DNA Cell Biology. 2008;27:357–65. doi: 10.1089/dna.2007.0694. [DOI] [PubMed] [Google Scholar]
- Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R, Goldmann E, Galea S. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proceeding of the National Academy of Sciences U S A. 2010;107:9470–5. doi: 10.1073/pnas.0910794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver I, Cervoni N, Champagne F, D'Alessio A, Sharma S, Seckl J, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nature Neuroscience. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, Szyf M. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. Journal of Neuroscience. 2005;25:11045–54. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegman HL, Stetler C. A meta-analytic review of the effects of childhood abuse on medical outcomes in adulthood. Psychosomatic Medicine. 2009;71:805–812. doi: 10.1097/PSY.0b013e3181bb2b46. [DOI] [PubMed] [Google Scholar]