Abstract
The transcription factor interferon regulatory factor-1 (IRF-1) is induced by many tumor-suppressive stimuli and can mediate anti-proliferative and pro-apoptotic effects in cancer cells. Thus, identifying agents that enhance IRF-1 activity may be an effective approach to cancer therapy. A cell-based screening assay was developed to identify extracts and compounds that could enhance IRF-1 activity using an IRF-1-dependent luciferase reporter cell line. Through this approach, we identified a natural product extract and a known active component of this extract, baicalein, which causes a marked increase in IRF-1-dependent reporter gene expression and IRF-1 protein, with modulation of known IRF-1 targets PUMA and cyclin D1. Baicalein causes suppression of growth in vitro in multiple cancer cell lines in the low micromolar range. IRF-1 plays a role in this growth suppression as demonstrated by significant resistance to growth suppression in a breast cancer cell line stably transfected with shRNA against IRF-1. Finally, intraperitoneal baicalein by repeated injection causes inhibition of growth in both xenogeneic and syngeneic mouse models of cancer without toxicity to the animals. These findings indicate that identifying enhancers of IRF-1 activity may have utility in anticancer therapies, and that cell-based screening for activation of transcription factors can be a useful approach for drug discovery.
Keywords: IRF-1, baicalein
Introduction
The interferon (IFN) regulatory factors (IRFs) play a vital role in a variety of biological processes including growth regulation and immune activation (1). IRF-1 was the first member of the IRF family of transcription factors (TFs) to be identified, and was originally described as a TF induced by IFNs that up-regulates transcription of IFN-inducible genes. However, IRF-1 can also be effectively induced in most cell types after exposure to many other stimuli, including retinoic acid (2), ionizing radiation (3), and other cytokines (1). Interestingly, another IRF protein, IRF-2, binds the same sequence as IRF-1 but down-regulates or blocks up-regulation of the genes inducible by IRF-1. While IRF-1 has been found to act as a tumor suppressor, IRF-2 was determined to be pro-oncogenic (4).
The role of IRF-1 as a tumor suppressor is well established. IRF-1 −/− embryonic fibroblasts (EFs) can become transformed by introduction of just one oncogene, while wild-type EFs require at least two oncogenes for transformation (5). NIH3T3 cells transformed by IRF-2 over-expression can be reverted to the non-transformed phenotype by over-expression of IRF-1 (4). Furthermore, NIH3T3 cells transformed by the oncogenes c-myc and fosB can be reverted to their normal phenotype by ectopic expression of IRF-1(6). This is remarkable, since IRF-1 was able to suppress these unrelated oncogenes as opposed to replacing the same deficient tumor suppressor. This observation has been confirmed in vitro and in vivo using a conditionally activated IRF-1 system in transformed NIH3T3 cells (7). Using microarray analysis of inducibly transformed NIH3T3 cells in addition to inducible IRF-1 activity, cyclin D1 was found to be a key down-regulated element in the tumor suppression seen with IRF-1 expression (8).
IRF-1 has proven to be a mediator of apoptosis for novel and established agents against cancer. For example, IRF-1 has been found to mediate apoptosis of cancer cells through up-regulation of TRAIL by IRF-1 in retinoid- and IFN-induced apoptosis (9). Fulvestrant, an anti-estrogen that has successfully completed clinical trials, causes apoptosis in susceptible breast cancer cells, for which dominant negative IRF-1 cells were found to be resistant to this apoptosis (10). Also, IRF-1 has been determined to mediate the apoptotic effects of tamoxifen in ER-poor, acutely damaged, human mammary epithelial cells (HMECs) (11). While technically not breast cancer, these HMECs have been “acutely damaged” by HPV-E6, which inactivates p53 and is considered to be an oncoprotein.
In addition to inhibiting proliferation and survival of cancer cells, IRF-1 enhances the immunogenicity of tumor cells in part through enhancing IRF-1-dependent expression of MHC proteins. Previously, we demonstrated increased expression of MHC class I and II proteins in cancer cells transfected with an IRF-1 expression vector and found that these cells became immunogenic(12). This was confirmed in another study using an estradiol-regulated inducible IRF-1 system in a hepatic cancer cell line(13).
More recently, we demonstrated that over-expression of IRF-1 using Ad-IRF-1, a recombinant adenovirus expressing IRF-1, induced apoptosis of cancer cells in vitro and in vivo (14–16). IRF-1 expression resulted in apoptosis in mouse breast cancer cell lines in vitro and tumor growth suppression in vivo; this appeared to be caspase-mediated(14). Ad-IRF-1 caused apoptosis in human breast cancer cell lines, suppressed tumor growth in a xenogeneic model of breast cancer, and demonstrated up-regulation of p21 and down-regulation of survivin (15). We further demonstrated that apoptosis involves both the extrinsic (death-receptor) and intrinsic (mitochondrial) apoptotic pathways, but no death ligand appears to be involved(16). In the intrinsic pathway we demonstrated that IRF-1 transcriptionally up-regulates the mitochondrial pro-apoptotic protein PUMA in a p53-independent fashion(17).
Given the studies demonstrating the tumor-suppressive activity of IRF-1, an attractive therapeutic strategy may be to identify enhancers of IRF-1 activity based on screening for extracts and compounds that can enhance IRF-1-dependent gene expression. This strategy has been studied previously for the well-known TF, signal transducer and activator of transcription-1 (STAT-1), which is the classical upstream regulator of IRF-1 expression (18). However, STAT-1 appears to have pro-tumorigenic activities in certain contexts (19), which may be avoided by enhancement of a more downstream tumor-suppressive factor. Therefore, we developed a cell-based screening system using a luciferase reporter under the control of an IRF-1-dependent promoter sequence. We have successfully identified a natural compound, baicalein, capable of increasing IRF-1 activity that may have therapeutic or chemopreventative potential in humans. Baicalein is a flavonoid which is thought to be one of the active components of the Traditional Asian Medicine (TAM) extract Huang Qin, which comes from the root of Scutellaria baicalensis (SB). Baicalein has numerous purported activities, but we are the first to link it to IRF-1 activity. Baicalein causes tumor suppression of cancer cells in vitro and in vivo, further suggesting that identification of modulators of TF function may be a useful strategy in the treatment of cancer.
Material and Methods
Cell lines and culture
The human cancer cell lines AGS, MDA468, BT549, and SKBR3 were from ATCC (Manassas, VA), and passaged for fewer than 6 months after resuscitation. The murine carcinoma cell line C3L5 was described previously (14), and was tested for murine pathogens and mycoplasma and found to be negative. Cell line cross-contamination is unlikely since these murine cells formed tumors in syngeneic immunocompetent mice. Generation of MDA468/IRF-1-shRNA clone 23 is described in Supplemental Methods. All cell lines were cultured as described in Supplemental Methods.
Chemicals and reagents
Baicalein was from Cayman Chemical (Ann Arbor, MI). Baicalin, genistein, and retinoic acid were from Sigma (St. Louis, MO). Antibodies anti-IRF-1 (C-20) and anti-p21 (C-19) were from Santa Cruz (Santa Cruz, CA), anti-PUMA (P4743) from Sigma, and anti-β-actin from Millipore (Billerica, MA). Horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit-IgG were from Pierce Biotechnology (Rockford, IL). The enhanced chemiluminescence substrates were from Thermo Scientific (Waltham, CA). DMSO was from ATCC.
Construction of recombinant adenoviruses and transfection
Ad-IRF-3, which expresses the constitutively active form of IRF-3, was obtained from M.A. Rivieccio and previously described (20). Ad-Null (empty vector) and Ad-IRF-1 are previously described (14).
Construction and transfection of IRF-1-luciferase reporter plasmid; selection, characterization and screening of transfected cells
To construct a cell-based system by which IRF-1-dependent gene activation could be quantified, we used the pGL4.20 luciferase reporter vector from Promega (Madison, WI), which contains the firefly luciferase gene. The IRF-1-dependent promoter sequence which contains 3 tandem repeats of the IRF-1 binding sequence and a minimal promoter sequence, 5’-CAGTACTTTCAGTTTCATAGTACTTTCAGTTTCATAGTACTTTCAGTTTCATAGACACTAGAGGGTATATAATGGAAGCTCGACTTCCAGCTTG-3’, was cloned into the reporter vector pGL4.20. MDA468 and AGS cells were transfected with this plasmid using SuperFect (Qiagen). Cells were selected against 0.5μg/ml puromycin, and stable colonies were picked and expanded. For characterization, cells were seeded at 10,000 cells/well in 96-well plates and incubated overnight. Clones were then assessed for low basal luciferase activity and prominent induction following treatment with Ad-IRF-1 and retinoic acid. Assessment of luciferase activity is described in Supplemental Methods. The best clones were expanded and used for subsequent experiments as the reporter cell lines IRF-1-luc/AGS and IRF-1-luc/MDA468. For screening for enhancers of IRF-1 activity, cells were seeded and 1μl of 40 natural product extracts (Chinese University of Hong Kong, China) were added to each well in duplicate to a final concentration of 20μg/ml by using robotic transfer, with appropriate controls; or SB, active components, or appropriate controls were added as indicated, and luciferase activity assessed at 30h. Transient luciferase reporter assay using IRF-1 luciferase reporter and β-gal plasmid is described in Supplemental Methods.
Immunoblotting
Cells treated with SB extract, baicalein, baicalin, or wogonin as indicated were lysed, cleared by centrifugation, and immunoblotted as previously described (14–17).
Cell proliferation assay
Cell proliferation was measured by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide) assay as previously described. MTT conversion to formazan dye correlates with the number of living cells (14–17).
Animals and in vivo tumor growth suppression
Six-week-old female SCID-Bg mice (Charles River) had AGS tumor cells (5×106 cells/animal) implanted subcutaneously with Matrigel® in the flank. Mice were ranked by tumor volume and randomized into groups with equal numbers of sizes. Treatment was initiated on d7, when tumors were ~70mm3, with 20mg/kg of baicalein, genistein, or carrier administered by intraperitoneal (i.p.) injection 5 times/week. Tumor size was assessed by perpendicular caliper measurements performed weekly. For C3L5, six-week-old female C3H/HeJ mice (Jackson) were implanted with 5×105 C3L5 cells/animal in the mammary fat pad, and ranked and randomized as above. Baicalein or carrier control treatment was initiated on d5, administered by i.p. injection 5 times/week, at 20mg/kg. Tumor size was assessed by perpendicular caliper measurements every 2–3d. Tumor volumes were calculated using the formula π/6×(larger diameter)×(smaller diameter)2. n=5–7/group. Experimental protocols were approved by the Institutional Animal Care and Use Committee at City of Hope.
Statistical analyses
Experiments were performed in triplicate or more, with in vitro data presented as mean +/−SD, and in vivo data presented as mean +/−SEM. Statistical comparison of values were made using two-tailed Student t test and statistical significance was considered to be present when p<0.05.
Results
Characterization of reporter cell lines
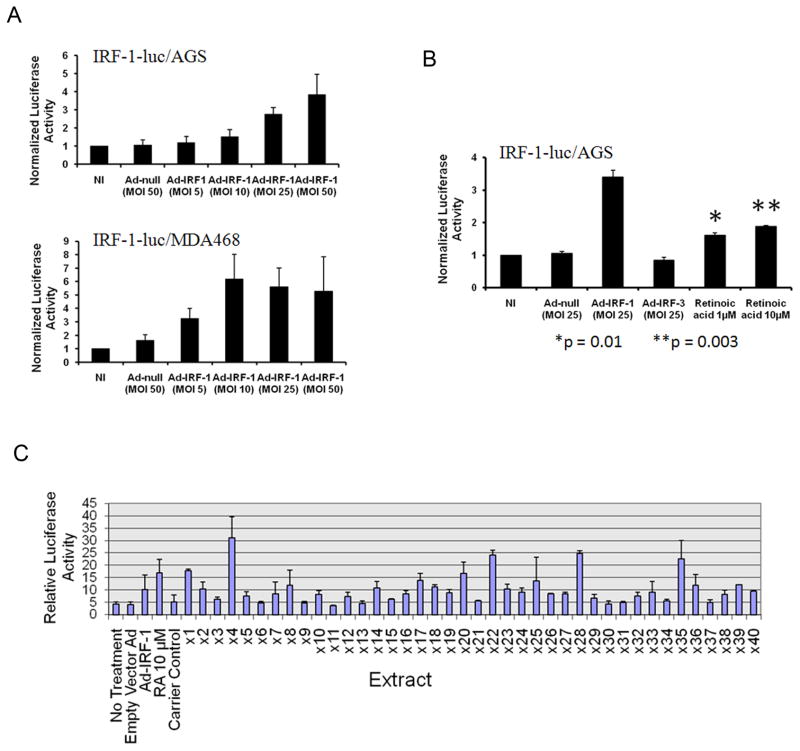
We first validated the sensitivity and specificity of the reporter cell lines. IRF-1-luc/AGS and IRF-1-luc/MDA468 were transfected with different MOI of Ad-IRF-1, and luciferase activity was determined after 24h. As shown in Figure 1A, luciferase activity was dramatically increased after Ad-IRF-1 transfection in a dose-dependent manner in both clones, validating the plasmid reporter construct. Since we are interested in compounds that enhance IRF-1 activity specifically, we chose IRF-1-luc/AGS for further characterization and screening because AGS cells do not express STAT-1 (17), which is a known upstream regulator of IRF-1. To further confirm the specificity of the assay, treatment of IRF-1-luc/AGS cells with recombinant Ad-IRF-3, which results in expression of a constitutively activated IRF-3 (20), caused no significant change in luciferase activity (Fig. 1B). To further test the sensitivity of the assay for cell permeable molecules, we treated IRF-1-luc/AGS with retinoic acid, which up-regulates IRF-1 by a non-STAT-1 mediated mechanism (2). Since IFN-γ up-regulates IRF-1 through STAT-1, this cytokine could not be used for characterization or as a control and is also not comparable to the cell permeable small molecules to be screened. Statistically significant differences in luciferase reporter activity were seen with retinoic acid at 1μM (p=0.01), and at 10μM, with ~2-fold induction (Fig. 1B, p=0.003). These assays were highly reproducible and showed little interwell variability. Thus, the IRF-1-luc/AGS cell line appears to be a sensitive and specific reporter for the activity of IRF-1.
Figure 1. Characterization of reporter cell lines and screening for enhancers of IRF-1 transcriptional activity.
A, IRF-1-luc/AGS and IRF-1-luc/MDA468 cells were plated and transfected with Ad-IRF-1. After 24h, luciferase activity normalized to cell viability was determined as described in Methods. Error bars are SD. B, IRF-1–Luc/AGS cells were plated and treated with no infection (NI), adenovirus, or retinoic acid for 24h, after which luciferase activities were quantitated. *p=0.01, **p=0.003. C, IRF1-Luc/AGS cells were plated and treated with 1μl/well natural extract, and after 30h, luciferase activity normalized to cell viability was determined as described in Methods. Positive controls: Ad-IRF-1, retinoic acid.
Screening for enhancers of IRF-1 transcriptional activity
Natural extracts have been long recognized as a potential source of anticancer drugs. Given the importance of IRF-1 as a tumor suppressor, the isolation of natural product extracts that enhance the activity of this protein would represent a novel approach to cancer therapy, and possibly chemoprevention. To identify such extracts, IRF-1-luc/AGS cells were seeded in 96-well plates and then treated with 40 TAM natural extracts at a final concentration of 20μg/ml. After 30h, luciferase activity was measured and normalized to cell viability. From this screen, we identified four herbal extracts (x4, x22, x28, and x35, Fig. 1C) that increased IRF-1-dependent luciferase activity by significantly more than 3-fold. Among these, the ethanol extract from x4, Scutellaria baicalensis (SB), was the greatest enhancer of IRF-1-dependent luciferase activity (Fig. 1C).
Scutellaria baicalensis enhances IRF-1 activity
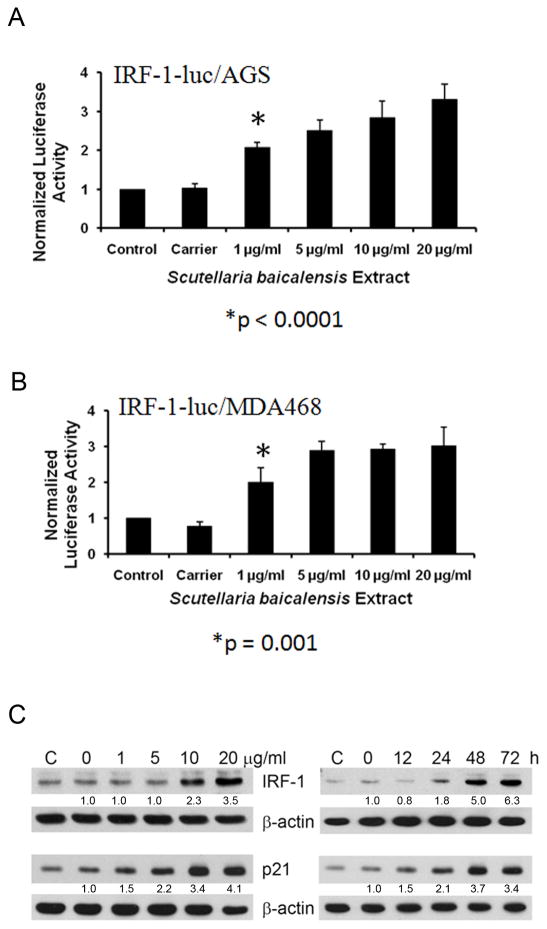
To further investigate and confirm the effect of SB on IRF-1 activity, IRF-1-luc/AGS cells were treated with SB extract and normalized luminescence was measured 30h later. As shown in Figure 2A, SB increased luciferase activity in IRF-1-luc/AGS cells in a dose-dependent manner. Luciferase activity was significantly increased at a dose of 1μg/ml of SB (p<0.0001). At 20μg/ml, SB showed maximal effects, enhancing IRF-1-dependent luciferase activity >3-fold.
Figure 2. Scutellaria baicalensis (SB) enhances IRF-1 activity.
A, IRF-1-luc/AGS cells were treated with indicated concentrations of SB extract. Luciferase activity was determined after 30h as described in Methods. B, IRF-1-luc/MDA468 cells were treated with SB extract for 30h, after which luciferase activity was measured. C, AGS cells were treated with SB extract for 24h or with 20μg/ml SB extract for the times indicated. Cell lysates were immunoblotted for IRF-1, p21, and β-actin. Densitometry ratios of indicated protein over β-actin, then normalized to 0 treatment are listed below each band as appropriate.
To determine whether SB enhances IRF-1 activity in other cell types, we also treated IRF-1-luc/MDA468 cells with SB. As shown in Figure 2B, in these cells SB also induced a significantly increased activity at 1μg/ml (p=0.001) and reached ~3-fold induction in normalized luciferase activity at 5μg/ml. This confirms that the SB activity on cancer cells can be generalizable to other cancer cell types, including breast cancer cells.
To further confirm whether SB enhances IRF-1 activity through up-regulating IRF-1, we examined IRF-1 protein levels in AGS cells by time and dose by immunoblotting. AGS cells were treated with SB extract for 24h and were clearly found to have increased protein at 10 and 20μg/ml at 24h in a dose-responsive fashion (Fig. 2C). Moreover, AGS cells incubated with 20μg/ml SB extract demonstrated increased IRF-1 protein at 24h post-treatment which further increased at 48 and 72h (Fig. 2C). We also performed immunoblot for p21, which is known to be increased by IRF-1 expression (15) and found that p21 increases in concert with IRF-1 (Fig. 2C).
Baicalein enhances IRF-1 activity and up-regulates IRF-1 expression
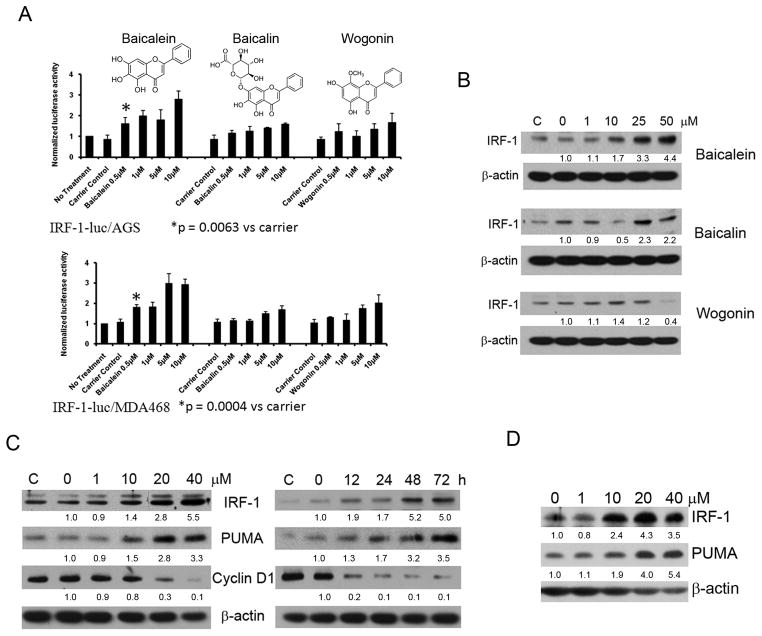
The active components of SB are purported to be flavones, including baicalein, baicalin, and wogonin (21, structures shown in Fig. 3A). We therefore screened these three flavones for IRF-1 activity using both IRF-1-luc/AGS and IRF-1-luc/MDA468. In both systems we found that baicalein demonstrated the most potent induction of IRF-1 activity. Baicalein at a concentration of 0.5μM resulted in a significant increase in IRF-1 dependent luciferase activity compared to control (p=0.0063 and p=0.0004 in AGS and MDA468 respectively), and increased normalized luciferase activity to ~3-fold over no treatment at 5–10μM, markedly higher than any concentration of the other flavones (Fig. 3A). Furthermore, when evaluating for IRF-1 protein by immunoblot, increased protein is identified at 10μM and is markedly increased at 25 and 50μM for baicalein, with no consistent increase with baicalin and perhaps even decrease of IRF-1 protein with wogonin in the AGS cell line (Fig. 3B). We then performed dose- and time-course studies of the induction of IRF-1 protein by immunoblot in baicalein treated AGS cells and found that once again there appears to be an increase in protein at 10μM at 24h which increases in a dose-related fashion and an increase in IRF-1 at 12h which increases significantly by 48 and 72h. Previously, we demonstrated that IRF-1 up-regulates PUMA, which mediates mitochondrial apoptosis in cancer cells (17), and in Figure 3C we demonstrate that PUMA does increase in concert with IRF-1 in baicalein treated AGS cells. Previously, IRF-1 has been shown to mediate its tumor-suppressive effect by down-regulation of cyclin D1 in transformed cells (8), and we found that cyclin D1 decreases in concert with IRF-1 in baicalein treated cells (Fig. 3C). Finally, since PUMA is also known to be induced by p53, we treated the p53-mutant human breast cancer cell line BT549 with baicalein and confirmed an increase in IRF-1 and an increase in PUMA independent of p53 (Fig. 3D), similar to results we had previously published with Ad-IRF-1 and tetracycline inducible IRF-1 cells in p53-mutant or deleted cells (17). We also examined cell lysates for mRNA levels of IRF-1 by quantitative real-time RT-PCR, and found significantly increased IRF-1 mRNA levels in AGS cells treated with baicalein in the ~50% to ~100% range (p=0.01 to 0.0008), implicating a transcriptional component to the enhanced IRF-1 protein and activity (Supplemental Data).
Figure 3. Baicalein enhances IRF-1 activity and up-regulates IRF-1 expression.
A, Evaluation of active components of SB in IRF-1-Luc cells. Upper panel, baicalein, baicalin, and wogonin structures are shown. IRF-1-Luc/AGS cells were plated, treated with indicated component, and assayed at 30h as described in Methods. *p=0.0063 versus control. Lower panel, IRF-1/MDA-468 cells were plated, treated, and assayed as in Methods. *p=0.0004 versus control. B, AGS cells were treated with the indicated concentrations of baicalein, baicalin and wogonin for 24h. Cell extracts were assessed for IRF-1 and β-actin by immunoblot. Densitometry ratios of indicated protein over β-actin, then normalized to 0 treatment are listed below each band as appropriate for B, C, and D. C, Immunoblot showing dose-dependence and time-course of IRF-1 and PUMA protein expression, as well as cyclin D1 suppression, in AGS cells treated with baicalein. AGS cells were either left untreated (C) or treated with baicalein at the indicated doses. Cells were collected after 24h. For time course, cells were treated at 20μM and collected at the times indicated. D, p53 mutant cell line BT549 was treated with baicalein and immunoblot performed of lysates after 24h as indicated.
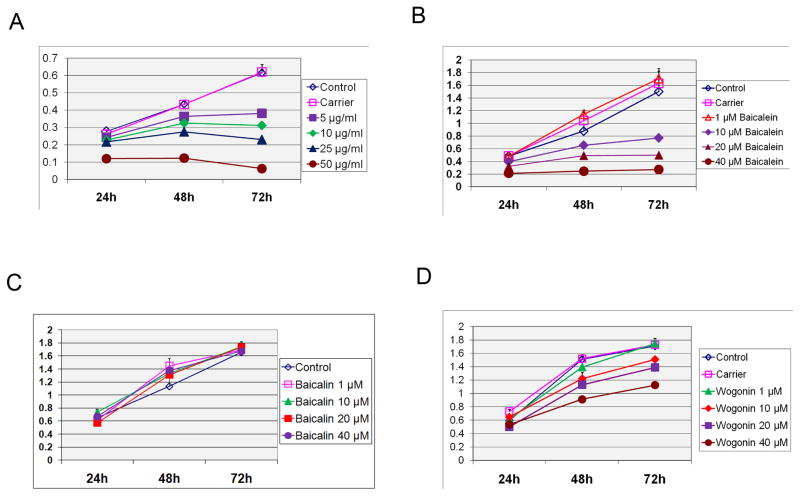
Baicalein inhibits growth of cancer cells in vitro
The rationale for screening for enhancers of IRF-1 activity was based on evidence that IRF-1 causes tumor suppression of cancer cells. This is further supported by data here demonstrating up-regulation of PUMA and down-regulation of cyclin D1 (Fig. 3D). We therefore examined the effect of SB extract and its active components on growth of AGS cells in vitro. As shown in Figure 4A, SB exerted a dose- and time-dependent inhibition on AGS cell growth. Among the active components of SB, MTT assay showed that baicalein inhibited the growth of AGS cells to the greatest extent (Fig. 4B). Baicalin had no inhibitory effect on cell growth (Fig. 4C) and wogonin had significantly less growth inhibition (Fig. 4D).
Figure 4. Growth inhibition of cancer cells by Scutellaria baicalensis and its active constituents.
MTT assay of AGS cells treated with SB (A), baicalein (B), baicalin (C) and wogonin (D). Cells were plated and treated as indicated over the time course as indicated and assessed by MTT assay. The y-axis represents absorbance at 520nm, which corresponds to the number of live cells.
Baicalein inhibits growth of cancer cells in vitro, which is in part mediated by IRF-1
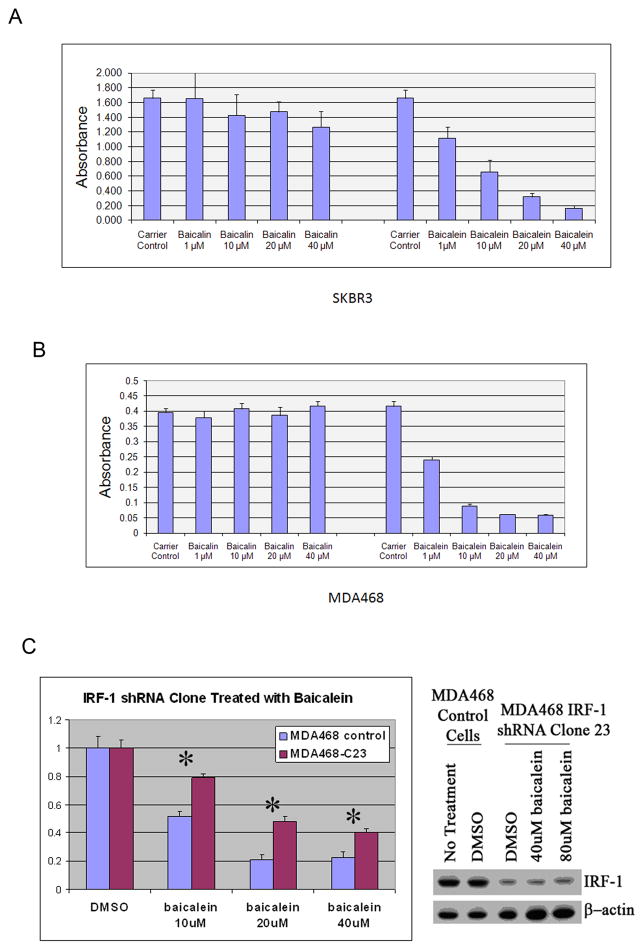
We also investigated the anti-proliferative effect of baicalein on the human breast cancer cell lines SKBR3 and MDA468. The MTT assays demonstrate that baicalein can inhibit cell growth significantly in both cell lines in a dose-dependent manner, with IC50’s between 1–10μM, while baicalin has minimal effect (Fig.5A and B). We have already demonstrated enhanced IRF-1 activity induced by baicalein in AGS and MDA468 cells (Fig. 3A), and have also confirmed this by luciferase assay in SKBR3 (Supplemental Data). We wished to use RNAi technology to see if knockdown of IRF-1 expression would decrease the growth inhibitory response by baicalein. We were unable to sufficiently inhibit the induction of IRF-1 protein by baicalein in AGS cells treated with siRNA transfection or clones that express shRNA (data not shown). We were successful with generation of MDA468 clones that express shRNA against IRF-1 that can suppress IRF-1 protein expression in the setting of baicalein induction. As seen in right panel of Fig.5C, shRNA efficiently knocked down the expression of IRF-1 protein in clone 23 of shRNA-IRF-1/MDA468 (MDA468-C23), even under the treatment of baicalein. MDA468-C23 was treated with the indicated dosage of baicalein for 72h, and as assessed by MTT assay, MDA468-C23 cells have significant resistance to the cell growth inhibition compared to that of the empty vector control MDA468 cells (Fig.5C). Taken together, these results suggest that the IRF-1 pathway is at least partly responsible for the baicalein–induced growth inhibition.
Figure 5. Baicalein inhibits growth of cancer cells in vitro, which is in part mediated by IRF-1.
A, MTT assay of SKBR3 cells treated with baicalin and baicalein. Cell viability was determined by MTT assay 72h post treatment as indicated. B, MTT assay of MDA468 cells treated with baicalin and baicalein. Cell viability was determined by MTT assay 72h post treatment as indicated. C, MDA468/IRF-1-shRNA clone 23 (MDA468-C23) was selected for knockdown of IRF-1 protein expression, both in the absence and presence of baicalein, using immunoblot (right panel). Cells were then compared to empty vector control cells in the presence of different concentrations of baicalein and assessed by MTT assay after normalization to DMSO treatment for each cell line (control, C23) at 72h. *p<0.0001 versus control.
Baicalein inhibits cancer cell growth in vivo
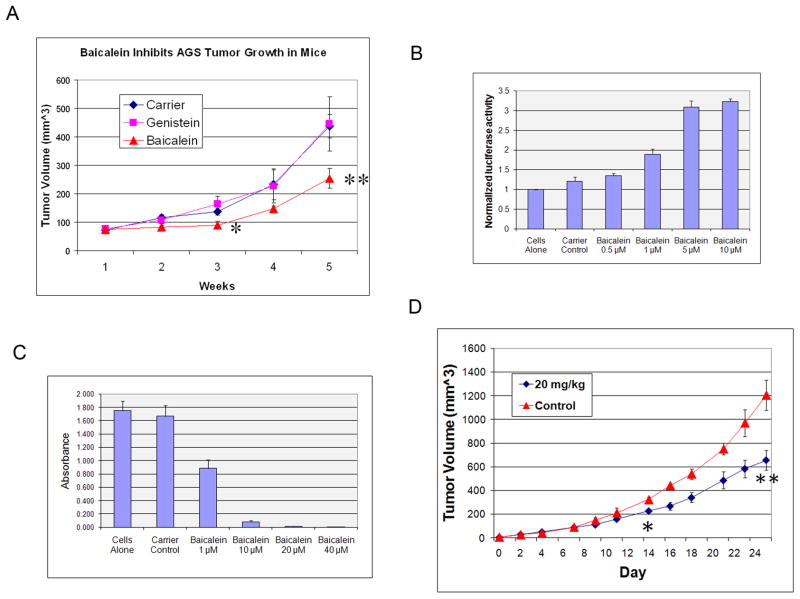
Baicalein can inhibit cell growth in different cancer cell lines in vitro, however, there has been little data on its effect on tumor growth in vivo. We performed studies in a xenogeneic model, using AGS cells. Tumor-bearing SCID-Bg mice were treated with carrier, 20mg/kg baicalein, or 20mg/kg genistein through i.p injection 5 times/week. Genistein was chosen as a comparison control because it is a flavonoid like baicalein and has the same chemical formula, but has a different structure. As shown in Fig. 6A, baicalein inhibits tumor growth in this model in comparison to either carrier or genistein, while genistein has no significant difference with carrier. There was no evidence of toxicity in any of the treated mice despite repeated doses. In order to prove that baicalein is able to inhibit tumor growth in a syngeneic mouse model, we used C3H/HeJ mice transplanted with the mouse breast cancer cell line C3L5, which we have shown previously to have marked tumor suppression by IRF-1 expression in vivo (14). We first confirmed that baicalein can enhance IRF-1 activity and inhibit cell growth in this cell line (Fig. 6B and C). Systemic administration of baicalein to C3L5 tumor bearing C3H/HeJ mice resulted in significant growth inhibition of these mouse breast tumors (Fig. 6D), with no evidence of toxicity in these animals. Taken together, these results provide strong evidence for the tumor growth inhibiting activity and tolerability of baicalein in vivo.
Figure 6. Baicalein inhibits cancer cell growth in vivo.
A, Baicalein suppresses growth of xenogeneic AGS tumors. AGS cells were implanted and treatment started as indicated on d7 as described in Methods. The x-axis represents weeks after AGS implantation. *p=0.03 for baicalein versus carrier or genistein; **p=0.006 for baicalein versus carrier. n=6–7/group. Error bars are SEM. B, Baicalein enhances IRF-1 activity in mouse C3L5 cells by luciferase reporter assay. C3L5 cells were treated with baicalein, and transient luciferase reporter assay was performed as described in Supplemental Methods. C, MTT assay of C3L5 cells treated with baicalein. Cell viability was determined by MTT assay at 72h post treatment as indicated. D, Intraperitoneal baicalein suppresses growth of syngeneic C3L5 tumors in C3H/HeJ female mice. Treatment was initiated on d5 after C3L5 implantation, when tumors were palpable, as described in Methods. n=5–6/group. *p=0.01, **p=0.001 versus control.
Discussion
A cell-based screening strategy was previously reported for identifying enhancers of STAT-1 activity, which identified a compound which enhanced IFN-γ induced STAT-1 dependent gene expression (18). This strategy had the advantage of rapidly excluding compounds that were not cell permeable and displaying non-specific toxicity. Furthermore, it allowed for the identification of active compounds independent of a predetermined mechanism of action, which in turn allowed for the identification of compounds that may function by a variety of mechanisms and may reveal levels of functional regulation that might not have been previously appreciated (18). Our screening strategy used a similar approach, making use of the same advantages. However, cancer cells may down-regulate STAT-1 as seen in AGS and other cancer cells (22). Furthermore, STAT-1 activity has been found to have pro-tumorigenic activities in certain contexts (19), and can also confer resistance to radiation and conventional cytotoxic agents (23). Focusing on a target farther downstream may circumvent signaling pathways that are pro-tumorigenic or confer treatment resistance. Notably, IRF-1 does not appear to be up-regulated in radio-resistant and other resistant cancer cells, despite significant expression of STAT-1 (19).
Significant IRF-1 enhancing activity of baicalein is seen at concentrations in the low micromolar range, with dose-dependent increase in IRF-1 protein and cell growth inhibition. We purposely limited our study to concentration levels of 50μM or lower, since these concentrations are achievable in vivo based on multiple studies of the pharmacokinetics of flavonoids, including baicalein. Studies of flavonoids in humans have demonstrated that dietary consumption of foods rich in flavonoids can result in plasma levels of individual flavonoids in the single-digit μM range. For example, women who consume varying amounts of flavonoids from soy milk have plasma genistein levels of 0.8–2.2 μM (24). After repeated injections of baicalein intraperitoneally into mice at 20mg/kg for five days/week, there was no evidence of toxicity. Furthermore, baicalein has been ingested by humans in human studies and in TAM, supporting the hypothesis that baicalein may be well tolerated in humans with plasma levels comparable to those used in our study (21).
The determination of the mechanism of enhancement of IRF-1 activity by baicalein requires further study. We evaluated mRNA transcript for IRF-1 by quantitative real time RT-PCR and found consistent increases in transcript in the AGS cell line when compared to retinoic acid as a positive control in the ~50% to ~100% range (Supplemental Data). However, the increases in protein appear to reach ~550% at the same doses although it is difficult to make such comparisons. We hypothesize that baicalein also affects the rate of protein turnover of IRF-1 in certain cancer cells which results in persistence of IRF-1 protein and increased transcriptional activity. The IRF-1 protein normally has a half-life of ~30 minutes. Treatments such as ionizing radiation have already been found to increase steady state levels of the IRF-1 protein through a concerted mechanism that includes increases in IRF-1 transcription and decreases in the rate of degradation (25). IRF-1 is known to be degraded by the ubiquitin-proteasome pathway (26). Baicalein may interfere with this process, perhaps through the same mechanism as ionizing radiation or DNA damaging agents. Indeed, numerous studies have demonstrated the potential role of reactive oxygen species in baicalein-induced cellular effects, including apoptosis (27).
It is notable that shRNA against IRF-1 attenuated, but did not abrogate, the tumor suppression caused by baicalein as assessed by MTT assay (Fig. 5C). This may be due to incomplete suppression of IRF-1 protein by the shRNA, since an IRF-1 band was still visible in our best shRNA clone, and small amounts of detected protein may have significant activity. Standard transient transfection techniques with siRNA to keep IRF-1 protein low in the presence of baicalein proved to be very difficult, since baicalein may affect IRF-1 protein expression at multiple levels. Regardless, there may be multiple off-target effects of baicalein not related to IRF-1 activity. Baicalein has been found to inhibit 12-LOX activity (28–30) as well as increase the TRAIL receptor DR5 (27). While baicalein was identified by screening for IRF-1 activity, IRF-1 certainly may not be the sole mediator of in vitro growth inhibition. This does not diminish the potential for this assay to identify useful compounds in cancer therapy.
In conclusion, we used a cell-based screening system of natural extracts and their active components to identify a compound that can enhance IRF-1 activity and cause tumor suppression of cancer cells in vitro and in vivo. Through further mechanistic and preclinical studies, we will be able to determine the potential clinical uses of this natural compound or its parent extract in cancer treatment.
Supplementary Material
Acknowledgments
Financial Support: JG was supported by the China Scholarship Council. JHY was supported by NIH K08CA098403 and Komen Foundation BCTR0708040.
The authors thank Drs. Richard Jove, David Horne, and Michael Weiss for their advice on the research plan and manuscript.
Abbreviations
- IRF-1
Interferon Regulatory Factor-1
- TNF
Tumor Necrosis Factor
- TRAIL
TNF Related Apoptosis Inducing Ligand
Footnotes
Disclosure of Potential Conflicts of Interests: No potential conflicts of interests were disclosed.
References
- 1.Savitsky D, Tamura T, Yanai H, Taniguchi T. Regulation of immunity and oncogenesis by the IRF transcription factor family. Cancer Immunol Immunother. 2010;59:489–510. doi: 10.1007/s00262-009-0804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Percario ZA, Giandomenico V, Fiorucci G, Chiantore MV, Vannucchi S, Hiscott J, et al. Retinoic acid is able to induce interferon regulatory factor 1 in squamous carcinoma cells via a STAT-1 independent signalling pathway. Cell Growth Differ. 1999;10:263–70. [PubMed] [Google Scholar]
- 3.Tamura T, Ishihara M, Lamphier MS, Tanaka N, Oishi I, Aizawa S, et al. An IRF-1-dependent pathway of DNA damage-induced apoptosis in mitogen activated T lymphocytes. Nature. 1995;376:596–599. doi: 10.1038/376596a0. [DOI] [PubMed] [Google Scholar]
- 4.Harada H, Kitagawa M, Tanaka N, Yamamoto H, Harada K, Ishihara M, et al. Antioncogenic and oncogenic potentials of interferon regulatory factors-1 and -2. Science. 1993;259:971–974. doi: 10.1126/science.8438157. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka N, Ishihara M, Kitagawa M, Harada H, Kimura T, Matsuyama T, et al. Cellular commitment to oncogene-induced transformation or apoptosis is dependent on the transcription factor IRF-1. Cell. 1994;77:829–839. doi: 10.1016/0092-8674(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka N, Ishihara M, Taniguchi T. Suppression of c-myc or fosB-induced cell transformation by the transcription factor IRF-1. Cancer Lett. 1994;83:191–196. doi: 10.1016/0304-3835(94)90318-2. [DOI] [PubMed] [Google Scholar]
- 7.Kroger A, Dallugge A, Kirchhoff S, Hauser H. IRF-1 reverts the transformed phenotype of oncogenically transformed cells in vitro and in vivo. Oncogene. 2003;22:1045–56. doi: 10.1038/sj.onc.1206260. [DOI] [PubMed] [Google Scholar]
- 8.Kröger A, Stirnweiss A, Pulverer JE, Klages K, Grashoff M, Reimann J, et al. Tumor suppression by IFN regulatory factor-1 is mediated by transcriptional down-regulation of cyclin D1. Cancer Res. 2007;67:2972–81. doi: 10.1158/0008-5472.CAN-06-3564. [DOI] [PubMed] [Google Scholar]
- 9.Clarke N, Jimenez-Lara AM, Voltz E, Gronemeyer H. Tumor suppressor IRF-1 mediates retinoid and interferon anticancer signaling to death ligand TRAIL. EMBO J. 2004;23:3051–60. doi: 10.1038/sj.emboj.7600302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouker KB, Skaar TC, Riggins RB, Harburger DS, Fernandez DR, Zwart A, et al. Interferon regulatory factor-1 mediates the proapoptotic but not cell cycle arrest effects of the steroidal antiestrogen ICI 182,780 (faslodex, fulvestrant) Cancer Res. 2004;64:4030–9. doi: 10.1158/0008-5472.CAN-03-3602. [DOI] [PubMed] [Google Scholar]
- 11.Bowie ML, Troch MM, Delrow J, Dietze EC, Bean GR, Ibarra C, et al. Interferon regulatory factor-1 is critical for tamoxifen-mediated apoptosis in human mammary epithelial cells. Oncogene. 2004;23:8743–55. doi: 10.1038/sj.onc.1208120. [DOI] [PubMed] [Google Scholar]
- 12.Yim JH, Wu SJ, Casey MJ, Norton JA, Doherty GM. IFN regulatory factor-1 gene transfer into an aggressive, nonimmunogenic sarcoma suppresses the malignant phenotype and enhances immunogenicity in syngeneic mice. J Immunol. 1997;158:1284–1292. [PubMed] [Google Scholar]
- 13.Kröger A, Ortmann D, Krohne TU, Mohr L, Blum HE, Hauser H, et al. Growth suppression of the hepatocellular carcinoma cell line Hepa1-6 by an activatable interferon regulatory factor-1 in mice. Cancer Res. 2001;61:2609–2617. [PubMed] [Google Scholar]
- 14.Kim PK, Armstrong M, Liu Y, Yan P, Bucher B, Zuckerbraun BS, et al. IRF-1 expression induces apoptosis and inhibits tumor growth in mouse mammary cancer cells in vitro and in vivo. Oncogene. 2004;23:1125–35. doi: 10.1038/sj.onc.1207023. [DOI] [PubMed] [Google Scholar]
- 15.Pizzoferrato E, Liu Y, Gambotto A, Armstrong MJ, Stang MT, Gooding WE, et al. Ectopic expression of interferon regulatory factor-1 promotes human breast cancer cell death and results in reduced expression of survivin. Cancer Res. 2004;64:8381–8. doi: 10.1158/0008-5472.CAN-04-2223. [DOI] [PubMed] [Google Scholar]
- 16.Stang MT, Armstrong MJ, Watson GA, Sung KY, Liu Y, Ren B, et al. Interferon regulatory factor-1-induced apoptosis mediated by a ligand-independent fas-associated death domain pathway in breast cancer cells. Oncogene. 2007;26:6420–30. doi: 10.1038/sj.onc.1210470. [DOI] [PubMed] [Google Scholar]
- 17.Gao J, Senthil M, Ren B, Yan J, Xing Q, Yu J, et al. IRF-1 transcriptionally upregulates PUMA, which mediates the mitochondrial apoptotic pathway in IRF-1-induced apoptosis in cancer cells. Cell Death Differ. 2010;17:699–709. doi: 10.1038/cdd.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch RA, Etchin J, Battle TE, Frank DA. A small-molecule enhancer of signal transducer and activator of transcription 1 transcriptional activity accentuates the antiproliferative effects of IFN-gamma in human cancer cells. Cancer Res. 2007;67:1254–61. doi: 10.1158/0008-5472.CAN-06-2439. [DOI] [PubMed] [Google Scholar]
- 19.Cheon H, Yang J, Stark GR. The functions of signal transducers and activators of transcriptions 1 and 3 as cytokine-inducible proteins. J Interferon Cytokine Res. 2011;31:33–40. doi: 10.1089/jir.2010.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivieccio MA, John GR, Song X, Suh HS, Zhao Y, Lee SC, et al. The cytokine IL-1beta activates IFN response factor 3 in human fetal astrocytes in culture. J Immunol. 2005;174:3719–26. doi: 10.4049/jimmunol.174.6.3719. [DOI] [PubMed] [Google Scholar]
- 21.Li-Weber M. New therapeutic aspects of flavones: the anticancer properties of Scutellaria and its main active constituents Wogonin, Baicalein and Baicalin. Cancer Treat Rev. 2009;35:57–68. doi: 10.1016/j.ctrv.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Abril E, Real LM, Serrano A, Jimenez P, García A, Canton J, et al. Unresponsiveness to interferon associated with STAT1 protein deficiency in a gastric adenocarcinoma cell line. Cancer Immunol Immunother. 1998;47:113–20. doi: 10.1007/s002620050511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khodarev NN, Beckett M, Labay E, Darga T, Roizman B, Weichselbaum RR. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proc Natl Acad Sci USA. 2004;101:1714–1719. doi: 10.1073/pnas.0308102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu X, Wang HJ, Murphy PA, Cook L, Hendrich S. Daidzein is a more bioavailable soymilk isoflavone than is genistein in adult women. J Nutr. 1994;124:825–32. doi: 10.1093/jn/124.6.825. [DOI] [PubMed] [Google Scholar]
- 25.Pamment J, Ramsay E, Kelleher M, Dornan D, Ball KL. Regulation of the IRF-1 tumour modifier during the response to genotoxic stress involves an ATM-dependent signalling pathway. Oncogene. 2002;21:7776–85. doi: 10.1038/sj.onc.1205981. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawa K, Yokosawa H. Degradation of transcription factor IRF-1 by the ubiquitin-proteasome pathway. The C-terminal region governs the protein stability. Eur J Biochem. 2000;267:1680–6. doi: 10.1046/j.1432-1327.2000.01163.x. [DOI] [PubMed] [Google Scholar]
- 27.Taniguchi H, Yoshida T, Horinaka M, Yasuda T, Goda AE, Konishi M, et al. Baicalein overcomes tumor necrosis factor-related apoptosis-inducing ligand resistance via two different cell-specific pathways in cancer cells but not in normal cells. Cancer Res. 2008;68:8918–27. doi: 10.1158/0008-5472.CAN-08-1120. [DOI] [PubMed] [Google Scholar]
- 28.Ding XZ, Kuszynski CA, El-Metwally TH, Adrian TE. Lipoxygenase inhibition induced apoptosis, morphological changes, and carbonic anhydrase expression in human pancreatic cancer cells. Biochem Biophys Res Commun. 1999;266:392–9. doi: 10.1006/bbrc.1999.1824. [DOI] [PubMed] [Google Scholar]
- 29.Tong WG, Ding XZ, Witt RC, Adrian TE. Lipoxygenase inhibitors attenuate growth of human pancreatic cancer xenografts and induce apoptosis through the mitochondrial pathway. Mol Cancer Ther. 2002;1:929–35. [PubMed] [Google Scholar]
- 30.Sekiya K, Okuda H. Selective inhibition of platelet lipoxygenase by baicalein. Biochem Biophys Res Commun. 1982;105:1090–5. doi: 10.1016/0006-291x(82)91081-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.