Abstract
We have attempted to use a familial genetics strategy to study mechanisms of Topoisomerase 1 (Top1) inhibition. Investigations have steadily been chipping away at the pathways involved in cellular response following Top1 inhibition for more than 20 years. Our system-wide approach, which phenotypes a collection of genotyped human cell lines for sensitivity to compounds, interrogates all genes and molecular pathways simultaneously. Previously, we characterized the in vitro sensitivity of fifteen families of CEPH cell lines (n = 142) to nine camptothecin analogues. Linkage analysis revealed a pattern of seven quantitative trait loci (QTLs) shared by all of the camptothecins. To identify which, if any, QTLs are related to the general mechanism of Top1 inhibition or should be considered camptothecin-specific, we characterized the in vitro sensitivity of the same panel of CEPH cell lines to the indenisoquinolones, a structurally distinct class of Top 1 inhibitors. Four QTLs on chromosomes 1, 5, 11, and 16 were shared by both the camptothecins and the indenoisoquinolines and are considered associated with the general mechanism of Top1 inhibition. The remaining three QTLs (chromosomes 6 and 20) are considered specific to camptothecin-induced cytotoxicity. Finally, eight QTLs were identified which were unique to the indenoisoquinolines.
INTRODUCTION
Since the discovery of Topoisomerase 1 (Top1) as a therapeutic target for the treatment of cancer, the design of Top1 inhibitors has been actively pursued. The camptothecins are the only Top1 inhibitors approved for clinical use by the Food and Compound Administration. In spite of activity in colon, lung, and ovarian cancers, their efficacy is compromised by a number of factors (1). The camptothecins are unstable and rapidly inactivated by ring opening of their lactone functionality at physiologic pH. Top1 cleavage complexes (Top1cc) reverse within minutes after compound withdrawal and require frequent and prolonged exposure (2, 3). Side effects are concentration-limiting and at times severe. These drawbacks prompted the search for additional novel Top1 inhibitors. The indenoisoquinolines (Ind) are one of the many non-camptothecin Top1 inhibitors in development. Their antiproliferative activity is comparable to the camptothecins in the NCI60 panel, a set of 59 human cancer cell lines derived from diverse tissues. However, differences in structure (4), interactions with Top1 and DNA (3, 5) and resistance mechanisms (3, 6) suggests that these compounds may have subtle distinctions in therapeutic activity and mechanisms of action.
Compound-to-compound comparative studies between structurally distinct compounds sharing a molecular target have been used to identify genes and pathways which are involved in a shared general mechanism and those which are class-specific (7, 8). Our own familial genetics strategy is one of the latest genomic tools for dissecting mechanisms of compound action and comparing genomic profiles across compound classes (Peters, EJ; Motsinger-Reif, A; Havener, TM; Everitt, L; Hardison, NE; Watson, VG; Richards, K; Province, MA; McLeod, HL. Pharmacogenetic dissection of FDA-approved cytotoxic drugs. Submitted. and (9)). In this model, genes influencing compound action are studied using the Centre d’Etude Polymorphisme Humain (CEPH) cell lines, a collection of densely genotyped, immortalized, human cell lines from multigenerational families. Compound-induced cytotoxicity is measured in these lymphoblastoid cell lines (LCLs) using a cell growth inhibition assay following treatment with increasing concentrations of compound. Linkage analysis is then used to identify regions of the genome, referred to as quantitative trait loci (QTLs), where genetic variation is correlated with observed variation in response.
We have recently used this model to establish a specific pattern of QTLs shared by the camptothecins, a group of Topoisomerase 1 (Top1) inhibitors, which is indicative of their shared mechanism of action (9). Seven QTLs were shared across multiple concentrations of six camptothecin analogues and then independently replicated in two separate validation experiments. The objective of this study was to determine which QTLs are specific to camptothecin-induced cytotoxicity and which are related to the general mechanism of Top1 inhibition. The CEPH LCLs were phenotyped for sensitivity to the indenoisoquinolines and the resulting genomic profiles compared with the camptothecins. While sharing the same molecular target, there are notable pharmacological distinctions between the two classes (4). By comparing the biological and genomic profiles of the indenoisoquinolines with the camptothecins we can demonstrate that this model can establish a pattern of QTLs (a) related to the general mechanism of Top1 inhibition, (b) specific to camptothecin-induced cytotoxicity, and (c) associated with indenoisoquinoline activity.
MATERIALS AND METHODS
Compounds
The following panel of camptothecins was purchased from LKT Labs (St Paul, MN): camptothecin (CPT), irinotecan (CPT11), 7-ethyl-10-hydroxycamptothecin (SN38), topotecan (TPT), 9-aminocamptothecin (9AC) and 9-nitrocamptothecin (9NC). The indenoisoquinolines, NSC 706744 (Ind1), NSC 725776 (Ind2) and NSC 724998 (Ind3), were generously supplied by Drs. Stephen Frye and Jian Jin of the Center for Integrative Chemical Biology and Compound Discovery (University of North Carolina, Chapel Hill, NC). All compounds were dissolved in DMSO. The chemical structures for all compounds studied are provided in Supplemental Table 1.
Cell lines
The CEPH cell lines are a set of immortalized lymphoblastoid cell lines (LCLs) collected from normal, healthy human volunteers which can be purchased from Corriell Cell Repositories (Camden, NJ). This collection is unique because the cell lines are established from large multigenerational families and every individual within the families has been genotyped, which enables investigators to perform genetics & pharmacogenomic analyses (10-12). For the purposes of this study, all CEPH cell lines from the following family identification numbers were used: 35, 45, 1334, 1340, 1341, 1345, 1350, 1362, 1408, 1420, 1447, 1451, 1454, 1459, 1463 (13). In total, cytotoxicity was studied in 142 CEPH cell lines from 15 families; the same families were used for study with the camptothecin analogues. The cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum at 37°C in humidified air containing 5% CO2. No cell line authentication was performed by the authors prior to initiating this study.
Measurements of Cell Growth Inhibition and Data Analysis
Wells in each black, clear-bottom 384 well plate (Corning, Corning, NY) contained either 1% DMSO (vehicle, 100% cell viability), 10% DMSO (100% cell death), or increasing concentrations of compound (8 concentrations per compound diluted in 1% DMSO) using a Tecan Evo 150 liquid handler (Tecan, San Jose, CA) and frozen at −20 C prior to experimentation. Each plate contained four replicates of each compound-concentration combination. On the day of experimentation, cells were plated at a density of 4000 cells/well (45 ul) in RMPI 1640 with 10% FBS. Alamar blue was added following 72 h compound exposure and fluorescence read at 96 h using a Tecan F200 multiwell plate reader (excitation 535 nm, Em 595 nm). Raw fluorescence values for each set of replicates were considered outliers if there was a ten-fold change in signal (in either directions) for a single replicate. Growth inhibition relative to untreated controls was determined using the manufacturer’s protocol. Final percent inhibition at each compound-concentration combination was averaged from 4 replicates of 2 independently plated experiments (n =8 replicates). Boxplots illustrating variation in cytotoxic response across the entire CEPH population for each compound are supplied in Supplemental Dataset 1. Boxplots illustrating intra- and inter-family variability in response are provided in Supplemental Dataset 2. The Pearson correlation coefficient (PCC) was used to analyze the degree of similarity between the biological activity profiles of the two classes of Top1 inhibitors. From the boxplot results in Supplemental Dataset 1, the concentration closest to yielding a population mean of 50% was selected for each compound. An activity profile or fingerprint was generated using the percent growth inhibition at this population mean GI50 across all cell lines (rather than constructing an activity profile using GI50 data across each cell line) for each compound. PCC was calculated using the formula:
where xi and yi are percent growth inhibition at the population mean GI50 for each compound A and B respectively, against each cell line, and xm and ym are the population mean values of xi and yi respectively.
Genomic Profiling and Peak Prioritization
The genotype data for each cell line were downloaded from V10 of the CEPH database using error checked markers (14, 15). The genetic map information was downloaded from the Marshfield database (16, 17). Error checking and nonparametric linkage analysis was performed as previously described (16). Linkage analysis tests for cosegregation of the phenotype of interest along with genetic markers at known locations. Phenotypes were defined as cytotoxic response at each compound-concentration combination. There were three indenoisoquinoline analogues and eight concentrations were investigated for each analogue (n = 24 phenotypes). For each phenotype, nonparametric linkage (NPL) analysis was used to construct Quantitative Trait Loci (QTL) maps by displaying the logarithm of odds (LOD) scores across each chromosome. The LOD score is a statistical estimate of linkage; it is the ratio of the likelihood that a chromosomal region is linked to the phenotype of interest over the likelihood that it is not. A LOD score of three indicates 1000 to 1 odds that the region is linked. A complete set of QTL maps for each compound, by chromosome can be found in Supplemental Dataset 3.
For each compound-concentration phenotype, gene-dropping permutations were conducted using Merlin to get a distribution of LOD scores which would occur under the null hypothesis of no linkage to the observed phenotypes. Such a permutation strategy has been evaluated statistically (18-20) and has been used for QTL and disease gene mapping in a wide range of human traits (21, 22). Three thousand genotype datasets were simulated for each compound-concentration phenotype, for a total of 72,000 simulated datasets. Results from the permutation testing were used to identify significant and suggestive linkage results. Using the permutation distributions, LOD score thresholds for significant linkage were defined as those LOD scores corresponding to p-values less than or equal to 0.002 (corrected for the entire genome: 0.05/ 22 chromosomes). This significance cut-off corresponds to a family-wise error rate of 5% for each phenotype and was used to define significant LOD score peaks. A region was considered significant if the highest LOD score in the region was greater than or equal to the permutation testing derived cut-offs. A complete list of LOD score significance cut-offs and significant peaks can be found in Supplemental Files 4 & 5 respectively. In addition to defining statistically significant peaks, suggestive QTLs were identified using the permutation distributions. LOD score cut-offs that define suggestive peaks were determined as the minimum LOD score needed to achieve a p-value less than or equal to 0.05 on each chromosome. Cut-offs that define suggestive linkage for each compound and concentration combination are also listed in Supplemental Dataset 4. QTLs observed for a compound-concentration phenotype were considered suggestive if the highest LOD score in that region surpassed the suggestive LOD score threshold for that compound-concentration phenotype on that chromosome. The start and end of a significant QTL region was defined as regions with LOD scores that were above either the suggestive LOD score threshold or peak LOD score minus one, whichever was greater. Understanding that the goal of this study is discovery, and very exploratory, these anti-conservative cut-offs were used for peak identification. The “significant” cut-offs are empirically derived to control the false positive rate to 0.05 across the genome for each drug tested, but do not control for the number of drugs tested, as each drug response phenotype was treated as an independent hypothesis. This is mitigated however, by looking for replication across drugs for significant peaks. The “suggestive” cut-offs are anticonservative even within a drug outcome, and even “replicated” suggestive peaks should be interpreted as hypothesis generating as opposed to strictly significant.
Identifying Class-Specific QTL profiles
To identify QTLs associated with the general mechanism of Top1 inhibition, the QTLs which have previously been identified as shared by all camptothecin analogues at multiple concentrations, were queried for replication in the indenoisoquinolines.2 A QTL was considered shared between the camptothecins and indenoisoquinolines only if it was present at the significant and/or suggestive level in multiple concentrations of all three indenoisoquinolines. QTLs which were not shared between the camptothecins and indenoisoquinolines were considered specific to camptothecin-induced cytotoxicity. Finally, QTLs which were significant in the indenoisoquinolines and shared across multiple concentrations of all three indenoisoquinolines were considered indenoisoquinoline-specific.
RESULTS
Variation in Biological Response Between Top1 Inhibitors
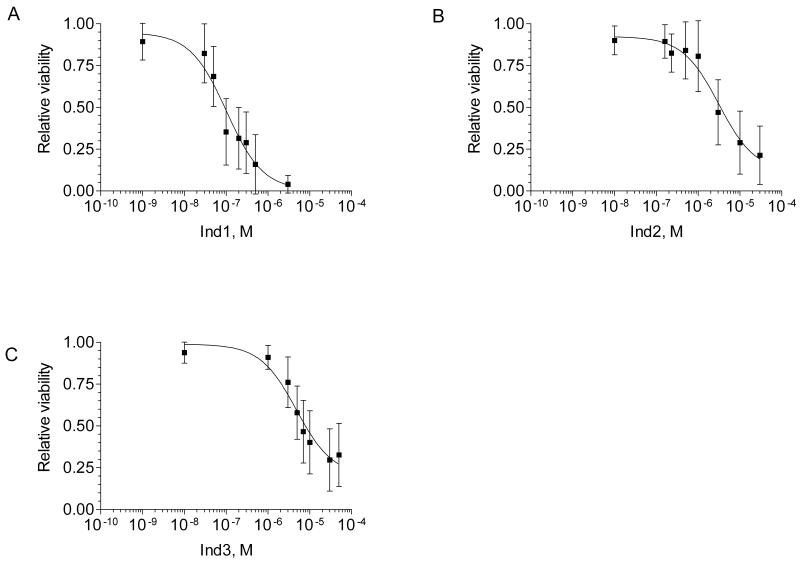
Some have argued that molecular & pharmacological distinctions between the camptothecins and the indenoisoquinolines could result in clinical differences (4). In this study, EBV-transformed LCLs derived from 15 CEPH pedigrees were used to evaluate differences in cytotoxic activity between the indenoisoquinolines and the camptothecins. Sensitivity to the indenoisoquinolines, Ind1 (NSC70644), Ind2 (NCS725776), and Ind3 (NSC724998), were evaluated in CEPH LCLs (n = 142) using the alamar blue assay. Ind1, Ind2, and Ind3 all showed concentration-dependent cytotoxicity (Figure 1). Considerable interindividual variation in sensitivity to the indenoisoquinolines was also observed (Figure 1, Supplemental Dataset 2).
Figure 1.
Concentration–response curve for (A) Ind1, (B) Ind2, and (C) Ind3. Data points represent the overall population mean (n=142) for growth inhibition relative to untreated controls at each concentration. Vertical bars represent the standard deviation for growth inhibition across the population.
Typically, for each compound, a finngerprint—its differential growth inhibitory activity against the cell lines—is compared using data for GI50, the concentration required for 50% growth inhibition. However, concentrations were insufficient to reach below the GI50, for 30% of the cell lines exposed to each of the idenoisoquinolines. As a result, for each compound fingerprint, we compared the percent growth inhibition at single concentration across all cell lines (rather than GI50 across each cell line). From the boxplot results in Supplemental Dataset 1, the concentration closest to yielding a population mean of 50% was selected for each compound. There was a wide range of sensitivity to the camptothecins and indenoisoquinolines at their respective population mean GI50s (Supplemental Figure 1). The Pearson correlation coefficient (PCC) was used to analyze the correlation between fingerprints for compounds based on percent growth inhibition at the population mean GI50. While both the indenoisoquinolines and camptothecins act by a similar mechanism, i.e. Top1 inhibition, a correlation matrix on percent inhibition at the population mean GI50 for each compound suggests distinctions between the two classes. The three indenoisoquinolines are highly correlated with each other (PCC > 0.8) and distinct from the camptothecins (PCC < 0) (Supplemental Table 2).
QTLs associated with Top1 inhibition
The next objective was to identify which peaks might be class-specific and which might be associated with the general mechanism of Top1 inhibition. Seven QTLs were shared among all camptothecin analogues investigated (Table 1).2 This pattern of QTLs was compared to the QTLs associated with indenoisoquinoline sensitivity. A peak was considered replicated if it was found in all three indenoisoquinolines at the significant or suggestive level. Four of the seven predefined QTLs were shared by all of the camptothecins and all of the indenoisoquinolines. These QTLs are summarized in Table 1. Peaks on chromosomes 11 and 16 were present at the significance level in multiple concentrations of all three indenoisoquinolines. The QTL on chromosomes 5 was significant at multiple concentrations of Ind2 and met the suggestive threshold for multiple concentrations of all three indenoisoquinolines. Finally, the QTL on chromosome 1 was shared by multiples concentrations of all three indenoisoquinolines at the suggestive level only. Most notably, the peak on chromosome 20, which contains Top1 and is shared by all of the camptothecins, was not present at the significant level in any indenoisoquinoline. However, it was present at the suggestive level in at least one concentration of Ind1 and Ind3. These results are illustrated in Supplemental Figure 2, a genome wide map of QTLs at the significant and suggestive level for both the camptothecins and indenoisoquinolines. The remaining QTLs on chromosomes 6 and 20, which were not replicated in the indenoisoquinoline, are considered camptothecin-specific (Table 1).
Table 1.
QTLs found in camptothecins and indenoisoquinolines
| Chromosome | Peak Start (cM) | Peak End (cM) | LOD† | LOD§ |
|---|---|---|---|---|
| 1* | 229 | 252 | 1.855 | 1.177 |
| 5* | 125 | 194 | 1.709 | 1.551 |
| 6 | 0 | 29 | 1.528 | - |
| 6 | 42 | 65 | 1.652 | - |
| 11* | 115 | 131 | 1.352 | 2.421 |
| 16* | 0 | 75 | 1.345 | 2.139 |
| 20 | 42 | 101 | 2.134 | - |
QTLs which were also shared by the indenoisoquinolines
Maximum LOD score observed in this region associated with the camptothecins
Maximum LOD score observed in this region associated with the indenoisoquinolines
QTLs associated with indenoisoquinoline-induced cytotoxicity
There are also QTLs which appear specific to the indenoisoquinolines alone. These QTLs are not replicated in the camptothecins and are summarized in Table 2. Of note, the QTLs on chromosomes 6 and 16 were identified in multiple concentrations of all three indenoisoquinolines with LOD scores that exceeded the significance thresholds (maximum LOD score 2.286 for Ind3 at 7uM and 2.139 for 3 uM Ind2). Similarly, the QTL on chromosome 13 was also present in multiple concentrations of all three indenoisoquinolines (maximum LOD score 1.44 for 10 uM Ind3). At least two indenoisoquinolines had significant peaks on chromosomes 4 and 10 at multiple concentrations which were replicated at the suggestive level in n ≥ 1 concentrations of the third indenoisoquinoline. There are also subtle distinctions between the indenoisoquinolines. Multiple concentrations of Ind1 and Ind3 had a QTL with significant LOD scores on chromosome 14 (98-134 cM); this QTL was not present in compound-concentration combinations of Ind2. A QTL on chromosome 19 (52-77 cM) surpasses the significance threshold for multiple concentrations of Ind1 but is not shared by the other members of that class. Regions with significant peak LOD scores for growth in vehicle did not overlap peaks with any of the significant QTLs associated with sensitivity to the indenoisoquinolines (Supplemental Figure 2) with the exception of a peak on chromosome 7 that is present in all three indenoisoquinolines. This suggests that the remaining QTLs are associated with cytotoxicity rather than the genetic effects from growth rate.
Table 2.
QTLs shared by all indenoisoquinolines
| Chromosome | Peak Start (cM) | Peak End (cM) | LOD* |
|---|---|---|---|
| 2 | 1 | 34 | 2.024 |
| 4 | 192 | 211 | 1.483 |
| 6 | 100 | 192 | 2.286 |
| 10 | 118 | 155 | 1.775 |
| 13 | 67 | 114 | 1.44 |
| 16 | 27 | 108 | 2.139 |
| 18 | 0 | 31 | 1.574 |
| 18 | 90 | 96 | 1.554 |
Maximum LOD score observed in this region
DISCUSSION
The similarities and differences in mechanisms between classes of compounds sharing the same molecular target have been characterized using genomic profiling (7). In the present study, we used pharmacologic and genomic profiling in the CEPH cell lines to investigate two classes of Top1 inhibitors, the camptothecins and the indenoisoquinolines. Reports indicate that while sharing the same molecular target, the indenoisoquinolines exhibit unique properties which may set them apart clinically from the camptothecins. We previously used our ex vivo familial genetics model to study mechanisms of camptothecin-induced cytotoxicity.2 The goal of this study was identify regions of the genome which were correlated to class-specific cytotoxicity and shared mechanisms of action.
The sensitivity of the CEPH cell lines to the indenoisoquinolines and the camptothecins was evaluated using a growth inhibitory assay. Percent growth inhibition at the population mean GI50 for each cell line was used to compare each compound. Interindividual variation in response to the indenoisoquinolines was found to be positively correlated (Supplemental Table 2). An inverse correlation was noted when comparing the two structural classes. A number of cell lines which were resistant to all camptothecins were sensitive to all indenoisoquinolines and vice versa. These differences in cytotoxic activity suggest that while they share the same molecular target, there may be subtle distinctions in the biochemical cascade required for compound action (e.g. uptake mechanisms, metabolism, and secondary molecular interactions). Pommier suggests that (a) the differential genomic targeting of Top1 cleavage complexes by the camptothecins and indenoisoquinolines, (b) the differences in chemical structure and chemical stability of the indenoisoquinolines compared with camptothecins, and (c) the low cross-resistance to camptothecins based on compound efflux and Top1 point mutations, make it likely that indenoisoquinolines will exhibit unique clinical and molecular properties which distinguish them from the camptothecins (4). Even with a shared mechanism of action, mutations may render a cell sensitive or resistant to these distinct structural classes. For example, Antony et al. reported that human leukemic cells which were resistant to the camptothecins as a result of a Top1 point mutation were sensitive to Ind1 (6). Not only can mutations in genes shared between these two classes change sensitivity, but this data suggest that there might be subtle distinctions in genes critical for action between the two classes (Table 2).
Nonparametric linkage analysis was used to identify regions of the genome which are specific to the general mechanism of Top1 inhibition, and distinct regions associated with the compound-induced cytotoxicity of camptothecins and indenoisoquinolines. Permutation testing was used to define empirical LOD score thresholds for significant and suggestive linkage for each compound-concentration combination on each chromosome. Seven of nine QTLs which had previously been identified as shared among camptothecin analogues were replicated in this study. Four of those QTLs were also replicated at the significant and/or suggestive level in all indenoisoquinolines (chr 1, 5, 11, 16; Table 1). These QTLs are considered specific to the general mechanism of Top1 inhibition, while QTLs in Table 2 are considered specific to the indenoisoquinolines. Different DNA cleavage patterns, biological activity within the CEPH, and unique QTLs patterns suggest some genes may be more selectively targeted by one compound class than the other (2, 3). While Ind1, Ind2, and Ind3 depend on Top1 for cytotoxic effect, siRNA knockdown of Top1 (at 80-90% efficiency) does not completely reverse growth inhibition which suggests additional targets (2, 6). Moreover, Ind2 and Ind3 have weak activity against Top2 (2). It’s too early to begin making hypotheses about what genes under those class-specific QTLs might be critical to the activity of the indenoisoquinolines. Since the discovery that this class of compounds acts on Top1 in 1998, our search of Pubmed reveals only 34 articles about the class. These articles predominately involve studies to optimize Top1 binding and potency.
The identification of QTLs on chromosomes 6 and 20, which are specific to the camptothecins, is also intriguing. Most exciting is the fact that maximum peak LOD scores observed for the QTL on chromosome 20 are centered over Top1 at 56 cM. This is the primary molecular target of both the camptothecins and the indenoisoquinolines. The absence of this QTL in the indenoisoquinolines does not mean that Top1 is not the molecular target of the indenoisoquinolines. Failure to find linkage in this region on chromosome 20 for the indenoisoquinolines could mean there was little or no genetic variation in the CEPH cell lines used in this study that could be correlated to the observed variation in phenotype. While the camptothecins and indenoisoquinolines share the same binding pocket on Top1, mutations in this pocket affect their activity differently. The camptothecins require hydrogen bonds to Top1 residues Arg364, Asn722, and Asp533 (4, 5). A mutation in any one of these residues confers resistance to the camptothecins. Conversely, the indenoisoquinolines hydrogen bond with Arg364 alone and are only sensitive to mutations in this Top1 amino acid residue (4). We have extensively reviewed other candidate genes under the QTL for chromosome 20 in another manuscript and refer the readers to that article (9).
Gene ontology terms which are enriched for genes within the QTLs on chromosome 6 were identified using the functional annotation clustering tool from (DAVID). The most overrepresented GO terms for genes under the QTL on chromosome 6 at 0-29 cM were cell differentiation (subcategory GO terms: apoptosis and cell death), protease inhibition, cell proliferation, and regulation of biological processes (subcategory GO terms: transcription, gene expression, DNA binding). Of the genes associated with these GO terms, there have been reports for the involvement of both WRN and FOXQ1 in camptothecin-induced cytotoxicity (1, 2). The QTL on chromosome 6 from 42-65 cM is enriched with genes associated with chromatin assembly, response to chemical stimulus, and NF-kappa binding. NF-kappa B interferes with the effect of most anti-cancer drugs through induction of anti-apoptotic genes. Blocking NF-kappa B activation sensitizes cells to camptothecin analogues (3). NF-kappa B tumor expression has been negatively correlated with response to irinotecan (4). A number of genes involved in the NF-kappa B signaling pathway are present in this QTL. This QTL also contains 22 members of the histone H1 family. The histones are proteins which package and order DNA into nucleosomes. Phosphorylation of histone H2AX is an extremely sensitive marker for double strand breaks induced by DNA damaging agents, such as the camptothecins and indenoisoquinolines (5, 6). It has been suggested that other histones are involved in DNA repair and cell proliferation (7). Studies indicate that histones are ADP-ribosylated in vivo in response to DNA damage (8). All core histones, including H1, can be mono-ADP ribosylated (7).
Although the CEPH LCLs have been shown to be a reasonable model for studying genes critical to drug action, the model is not without its limitations. This is an in vitro system was intended to simplify response to drug therapy by cancer patients in a clinical setting. As such, there are some pharmacokinetic and pharmacodynamic variables which cannot be captured by the model that impact patient response. Additionally, confounders specific to in vitro phenotyping, such as cellular growth rate, can affect cytotoxic response to anticancer agents in LCLs (23).
We have characterized a method for clarifying the mechanism of action for Top1 inhibition associated with the camptothecins and indenoisoquinolines. More than 25 years of research have been devoted to identifying the molecular pathways associated with camptothecin-induced cytotoxicity. Regions of the genome which contain known and potentially novel genes critical to their action have identified and validated. The QTL on chromosome 20 as well as those QTLs which are shared by both the indenoisoquinolines and camptothecins (Table 1) are considered the most important for followup. QTLs related to the general mechanism of Top1 inhibition may not only lead to predictions about camptothecin sensitivity and resistance in patient populations but could provide valuable insight into proposed functions of mammalian Top1 (24). We are currently performing ex vivo genome-wide association studies, using lymphoblastoid cell lines from large numbers of unrelated individuals. This will provide an approach to further investigate genetic influence on cytotoxic response to a large panel of anticancer agents including the Topoisomerase 1 inhibitors. This association approach will allow us to significantly narrow down the broad QTLs identified for the camptothecins and begin to identify genes critical to response. Finally, genes under these QTLs which are identified through association studies could be evaluated for their effect on camptothecin (and indenoisoquinoline) cytotoxicity using shRNA gene knockdown experiments. These steps would increase the knowledge base surrounding the molecular consequences of Top1 inhibition.
Supplementary Material
Acknowledgments
Financial Support: This work is supported in part by the Pharmacogenetics Research Network (U01 GM63340), a National Research Service Award Fellowship to V.G.W (F31 CA1199385) and a training grant to N.H. (NIEHS 2 T32 ES007329).
Abbreviations
- CEPH
Centre d’Etude Polymorphisme Humain (CEPH)
- cM
centimorgans
- DMSO
dimethyl sulfoxide
- EBV
Epstein-Barr Virus
- GI50
concentration required to inhibit growth by 50%
- NCI60
the 60 human cancer cell lines of the National Cancer Institute
- LOD
logarithm of odds
- LCLs
lymphoblastoid cell lines
- PCC
pearson correlation coefficient
- QTL
quantitative trait loci
- Top1
Topoisomerase 1
- Top1cc
Topoisomerase 1 cleavage complex
- CPT
camptothecin
- CPT11
irinotecan
- TPT
topotecan
- 9AC
9-aminocampothecin
- 9NC
9- nitrocamptothecin
- SN38
7-ethyl-10-hydroxycamptothecin
- Ind
indenoisoquinoline
Footnotes
Disclosure of Potential Conflict of Interest: HL McLeod serves as a consultant for Myriad Genetics, Medco Health Solutions, Affymetrix, Glaxo Smith Kline. HL McLeod has also received research support from Myriad Genetics.
REFERENCES
- 1.Jung LL, Zamboni WC. Cellular, pharmacokinetic, and pharmacodynamic aspects of response to camptothecins: can we improve it? Drug Resistance Updates. 2001;4:273–88. doi: 10.1054/drup.2001.0222. [DOI] [PubMed] [Google Scholar]
- 2.Antony S, Agama KK, Miao ZH, Takagi K, Wright MH, Robles AI, et al. Novel indenoisoquinolines NSC 725776 and NSC 724998 produce persistent topoisomerase I cleavage complexes and overcome multidrug resistance. Cancer Res. 2007;67:10397–405. doi: 10.1158/0008-5472.CAN-07-0938. [DOI] [PubMed] [Google Scholar]
- 3.Antony S, Jayaraman M, Laco G, Kohlhagen G, Kohn KW, Cushman M, et al. Differential Induction of Topoisomerase I-DNA Cleavage Complexes by the Indenoisoquinoline MJ-III-65 (NSC 706744) and Camptothecin: Base Sequence Analysis and Activity against Camptothecin-Resistant Topoisomerases I. Cancer Res. 2003;63:7428–35. [PubMed] [Google Scholar]
- 4.Pommier Y, Cushman M. The indenoisoquinoline noncamptothecin topoisomerase I inhibitors: update and perspectives. Mol Cancer Ther. 2009;8:1008–14. doi: 10.1158/1535-7163.MCT-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Staker BL, Feese MD, Cushman M, Pommier Y, Zembower D, Stewart L, et al. Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex. J Med Chem. 2005;48:2336–45. doi: 10.1021/jm049146p. [DOI] [PubMed] [Google Scholar]
- 6.Antony S, Kohlhagen G, Agama K, Jayaraman M, Cao S, Durrani FA, et al. Cellular topoisomerase I inhibition and antiproliferative activity by MJ-III-65 (NSC 706744), an indenoisoquinoline topoisomerase I poison. Mol Pharmacol. 2005;67:523–30. doi: 10.1124/mol.104.003889. [DOI] [PubMed] [Google Scholar]
- 7.Shimoyama T, Hamano T, Natsume T, Koizumi F, Kiura K, Tanimoto M, et al. Reference profiling of the genomic response induced by an antimicrotubule agent, TZT-1027 (Soblidotin), in vitro. The pharmacogenomics journal. 2006;6:388–96. doi: 10.1038/sj.tpj.6500386. [DOI] [PubMed] [Google Scholar]
- 8.Hamadeh HK, Bushel PR, Jayadev S, Martin K, DiSorbo O, Sieber S, et al. Gene Expression Analysis Reveals Chemical-Specific Profiles. Toxicol Sci. 2002;67:219–31. doi: 10.1093/toxsci/67.2.219. [DOI] [PubMed] [Google Scholar]
- 9.Watson VG, Motsinger-Reif A, Hardison NE, Peters EJ, Havener TM, Everitt L, et al. Identification and Replication of Loci Involved in Camptothecin-Induced Cytotoxicity Using CEPH Pedigrees. PLoS One. 2011;6:e17561. doi: 10.1371/journal.pone.0017561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dausset J, Cann H, Cohen D, Lathrop M, Lalouel JM, White R. Centre d’etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics. 1990;6:575–7. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- 11.Dolan ME, Newbold KG, Nagasubramanian R, Wu X, Ratain MJ, Cook EH, Jr., et al. Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity. Cancer Res. 2004;64:4353–6. doi: 10.1158/0008-5472.CAN-04-0340. [DOI] [PubMed] [Google Scholar]
- 12.Watters JW, Kraja A, Meucci MA, Province MA, McLeod HL. Genome-wide discovery of loci influencing chemotherapy cytotoxicity. Proc Natl Acad Sci U S A. 2004;101:11809–14. doi: 10.1073/pnas.0404580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coriell National Institutes for General Medicine (NIGMS) Human Genetic Cell Repository: CEPH Reference Families. 2010 cited 2010 March 11. Available from: http://ccr.coriell.org/sections/collections/NIGMS/CEPHFamilies.aspx?PgId=49&coll=GM.
- 14.Cohen D, Chumakov I, Weissenbach J. A first-generation physical map of the human genome. Nature. 1993;366:698–701. doi: 10.1038/366698a0. [DOI] [PubMed] [Google Scholar]
- 15.The Foundation Jean Dausset–Centre d’Etude du Polymorphisme Humain (CEPH) Genotyping Database Version 10. 2010 cited 2010 March 11. Available from: ftp://ftp.cephb.fr/ceph_genotype_db/ceph_db/Ver_10/mkr.
- 16.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–9. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Heart L. Blood Institute. Mammalian Genotyping Service (MGS) CEPH Family Genotyping Data. 2010 at Marshfield. cited 2010 March 11. Available from: http://research.marshfieldclinic.org/genetics/home/index.asp.
- 18.Medland SE, Schmitt JE, Webb BT, Kuo PH, Neale MC. Efficient calculation of empirical P-values for genome-wide linkage analysis through weighted permutation. Behav Genet. 2009;39:91–100. doi: 10.1007/s10519-008-9229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nature genetics. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 20.Jung J, Weeks DE, Feingold E. Gene-dropping vs. empirical variance estimation for allele-sharing linkage statistics. Genet Epidemiol. 2006;30:652–65. doi: 10.1002/gepi.20177. [DOI] [PubMed] [Google Scholar]
- 21.Fullerton JM, Donald JA, Mitchell PB, Schofield PR. Two-dimensional genome scan identifies multiple genetic interactions in bipolar affective disorder. Biol Psychiatry. 2010;67:478–86. doi: 10.1016/j.biopsych.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 22.Wojciechowski R, Moy C, Ciner E, Ibay G, Reider L, Bailey-Wilson JE, et al. Genomewide scan in Ashkenazi Jewish families demonstrates evidence of linkage of ocular refraction to a QTL on chromosome 1p36. Hum Genet. 2006;119:389–99. doi: 10.1007/s00439-006-0153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stark AL, Zhang W, Mi S, Duan S, O’Donnell PH, Huang RS, et al. Heritable and non-genetic factors as variables of pharmacologic phenotypes in lymphoblastoid cell lines. The pharmacogenomics journal. 2010 doi: 10.1038/tpj.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao ZH, Player A, Shankavaram U, Wang YH, Zimonjic DB, Lorenzi PL, et al. Nonclassic functions of human topoisomerase I: genome-wide and pharmacologic analyses. Cancer Res. 2007;67:8752–61. doi: 10.1158/0008-5472.CAN-06-4554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.