Abstract
Background
Phlebotomus papatasi is a natural vector of Leishmania major, which causes cutaneous leishmaniasis in many countries. Simple sequence repeats (SSRs), or microsatellites, are common in eukaryotic genomes and are short, repeated nucleotide sequence elements arrayed in tandem and flanked by non-repetitive regions. The enrichment methods used previously for finding new microsatellite loci in sand flies remain laborious and time consuming; in silico mining, which includes retrieval and screening of microsatellites from large amounts of sequence data from sequence data bases using microsatellite search tools can yield many new candidate markers.
Results
Simple sequence repeats (SSRs) were characterized in P. papatasi expressed sequence tags (ESTs) derived from a public database, National Center for Biotechnology Information (NCBI). A total of 42,784 sequences were mined, and 1,499 SSRs were identified with a frequency of 3.5% and an average density of 15.55 kb per SSR. Dinucleotide motifs were the most common SSRs, accounting for 67% followed by tri-, tetra-, and penta-nucleotide repeats, accounting for 31.1%, 1.5%, and 0.1%, respectively. The length of microsatellites varied from 5 to 16 repeats. Dinucleotide types; AG and CT have the highest frequency. Dinucleotide SSR-ESTs are relatively biased toward an excess of (AX)n repeats and a low GC base content. Forty primer pairs were designed based on motif lengths for further experimental validation.
Conclusion
The first large-scale survey of SSRs derived from P. papatasi is presented; dinucleotide SSRs identified are more frequent than other types. EST data mining is an effective strategy to identify functional microsatellites in P. papatasi.
Background
The sand fly Phlebotomus (Phlebotomus) papatasi (Scopoli) is a natural vector of Leishmania major (Yakimov & Schokov), which is the causative agent of zoonotic cutaneous leishmaniasis in the Middle East and other countries [1,2]. Simple sequence repeats (SSRs) or microsatellites, are common components of eukaryotic genomes and are short, repeated nucleotide sequence elements arrayed in tandem and flanked by non-repetitive regions [3,4]. SSRs often harbour high levels of polymorphism, in terms of repeat number, and have been developed into one of the most common classes of genetic markers due to their high degree of ubiquity, co-dominance and variability in number among individuals. In recent years, microsatellites were extensively used to investigate genetic variability and the population structures of a wide range of organisms, including parasites and vectors of infectious diseases [5-13]. In the absence of genome sequences for sand flies, the isolation of microsatellite markers was carried out using various enrichment methods [14,15]. This approach has led to the development of a panel of five polymorphic and informative microsatellite markers for P. papatasi [16-18].
Parallel to the rapid increase in availability of diverse DNA sequence data, which resulted from the huge advancement of sequencing techniques, labour-intensive methods for the generation of microsatellite markers have been replaced gradually by in silico data mining of genomic and expressed sequence tag (EST) datasets [19-21]. Microsatellites are effectively randomly distributed throughout the genome and can represent transcribed elements. Although, SSRs derived from transcribed ESTs can still maintain allelic variability comparable with that in non-transcribed genomic DNA, they can serve as molecular markers for numerous applications [22,23]. EST databases have been a rich source of SSRs for the development of "genotyping" applications. Marker development from already existing sequence data is rapid, efficient and economical. Any type of SSR will be detected using an appropriate search program, whereas only SSRs with pre-defined motifs are captured by enrichment. In addition, SSRs are physically linked to expressed genes and thus represent functional markers.
The aims of this study were to expand the genomic resources for P. papatasi by analyzing 42,784 ESTs available in the GenBank database, increase the number of SSR markers by mining a previously developed ESTs, and evaluate specifically designed primer pairs for their abundance and motif type.
Results
Sequence analysis
The sequence analysis of the whole data set comprised ESTs of an average size of 469 bp. Sequence composition showed slight bias toward A and T; A+T = 13,235,131 (56.5%), whereas G+C = 10,149,530 (44.3%). The frequency of the main nucleotides (A), (C), (G) and (T) were comparable: 28.7, 21.8, 21.5, and 27.8%, respectively.
SSR types, distribution and frequency
Out of 42,784 ESTs analyzed; 1,499 (3.5%) SSRs were characterized. The number of repeats per SSR motif ranged from 5 to 16 repeats, with 5-9 being most frequent. On average, one SSR was found in every 15.55 kb of ESTs, and the total length of the regions containing repeats was 0.079% of the total ESTs size. A total of 93 ESTs were found to have more than one SSR motifs.
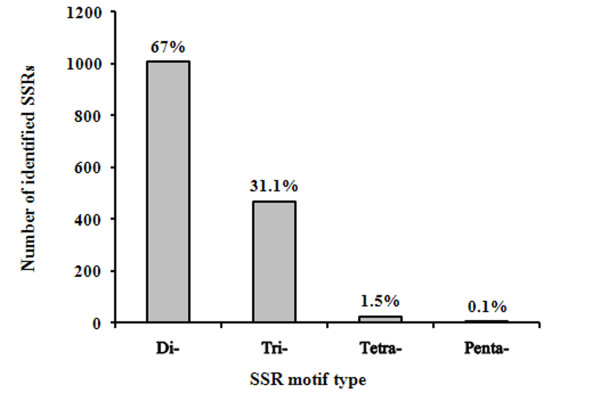
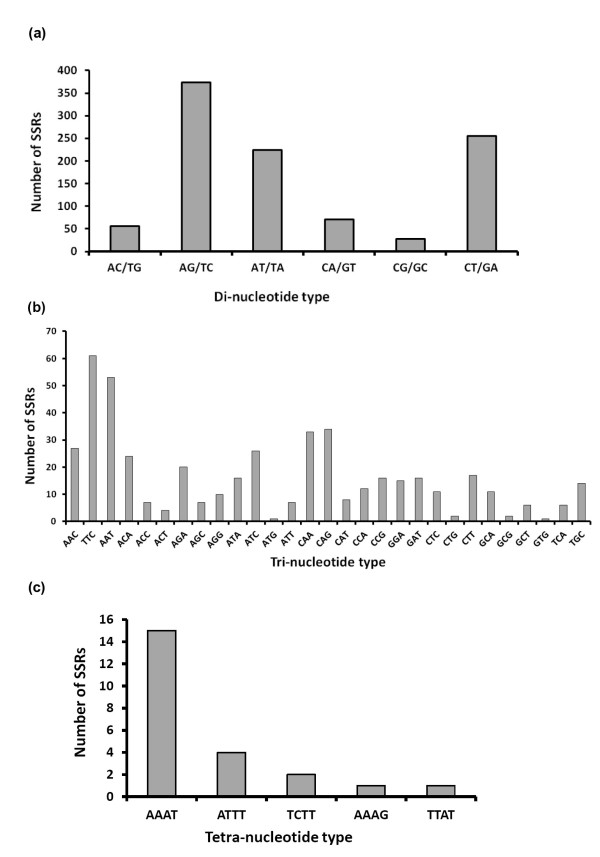
SSR loci were categorized by repeat type and structure; the dinucleotide repeat motifs were most abundant, accounting for 67% of the whole SSRs characterized followed by the trinucleotides (31.1%), tetranucleotides (1.5%), and pentanucleotides (0.1%), (Table 1). No hexanucleotide SSRs were detected in P. papatasi ESTs. Among the dinucleotide motifs, AG/TC type was more abundant (37%) than CT/GA (25.3%) and AT/TA types (22.2%); few CA/GT (7.1%), AC/TG (5.6%) and CG/GC (2.8%) types were characterized (Figure 1). For the trinucleotide SSRs, 467 motifs and 29 motif types were identified for P. papatasi; the TTC motif was the most abundant (13%), followed by AAT (11%), CAG, CAA (7%) each, AAC, ATC (6%) each, and ACA (5%) while the other motifs were at lower frequencies (Figure 2). Five types of tetranucleotide motifs were characterized; AAAT, ATTT, TCTT, AAAG, and TTAT, with frequencies of 15, 4, 2, 1, and 1%, respectively. Two identical pentanucleotide motifs of AATTG type (0.1%) were also identified.
Table 1.
Summary of in silico mining of EST sequences for P.papatasi sand fly
| Parameters | Number (%) |
|---|---|
| Total EST sequences | 42,784 |
| Total number of SSRs identified | 1,499 (3.5%) |
| Frequency of SSRs | One every 15.55 kb |
Numbers in parentheses is the percentage value of the repeat type.
Figure 1.
Frequency distribution of different repeat types (2-5 motif units) identified in ESTs from P. papatasi. The numbers on the bars indicate the percentage of each repeat type microsatellites in total number.
Figure 2.
Frequency distribution of (a) di-, (b) tri-, and (c) tetra-nucleotide repeat motifs of P. papatasi.
SSR marker development
Of 1,499 unique ESTs containing SSRs, 630 (42%) were suitable for primer design, comprising 425 dinucleotide, 271 trinucleotide and 9 tetranucleotide SSRs. The remaining sequences were inappropriate for primer design, mainly because of insufficient DNA sequence flanking the microsatellite core, or the sequences themselves not being suitable for primer design. Thus, overall SSR primers could be designed to amplify non-redundant loci from ~ 1.5% of the initial number of ESTs. Based on the size of repetitive motifs, a subset of 40 primer pairs were selected and designated as prime candidates to carry out polymorphism analysis using a minimum repeat length criterion of 5 repeats. This subset comprised 27 dinucleotide, 8 trinucleotide and 5 tetranucleotide (Table 2).
Table 2.
Primers designed and suggested to amplify repeat sequence (SSRs) including number of repeats product size in bp, forward (Fw-) and reverse (Rv-) primer sequences, and melting temperature (Tm).
| Name | Accession no. | SSR | Product size | Fw-Primer (5'-3') | Tm (°C) | Rv-Primer (5'-3') | Tm (°C) |
|---|---|---|---|---|---|---|---|
| PPEST1 | EY218895.1 | (CA)15 | 170 | AGTTCCGCCACATCCATTC | 60.9 | TTAGACAGCGGGAAAGAAGAAA | 60.4 |
| PPEST 2 | FG108562.1 | (GCA)13 | 141 | TGTCAATAGTGGCTCAATGCTC | 60.3 | ATTAGTCGTTTATCCTTCCCCG | 60.5 |
| PPEST 3 | EX474573.1 | (TA)13 | 188 | CAATTTTATGCGGTCTATGGGA | 61.0 | AGGTATGCAAAGTAATGGGTGG | 60.1 |
| PPEST 4 | FG116618.1 | (TC)13 | 197 | ACCCGACGCGAATTTACTTT | 60.8 | GGAGAGACAAGTTATGGGGTCA | 60.4 |
| PPEST 5 | FG107376.1 | (TGC)13 | 190 | GAGAGACATGGTGGATGGACTT | 60.4 | TGTCAATAGTGGCTCAATGCTC | 60.3 |
| PPEST 6 | FG108078.1 | (AT)12 | 291 | AAATCCACTATCCTCCTTTCCTC | 59.0 | TTTTGGGGTAAGATGGGG | 58.2 |
| PPEST 7 | EX473561.1 | (CA)12 | 245 | GTACCCTTTCCCTCCCTATGTC | 60.1 | GGGTTCACCAACATCCTCC | 60.2 |
| PPEST 8 | FG119248.1 | (CA)12 | 140 | CCACTGTAACTTGAGGAGGAGG | 60.2 | AGACTTGATGAGTGCGTCTCTG | 59.7 |
| PPEST 9 | FG117610.1 | (CA)12 | 175 | CGCACAAGAACAAAGTGGAAA | 61.2 | TCTTCTCGCTCCCTCGTTC | 60.6 |
| PPEST 10 | FG117371.1 | (TC)12 | 236 | ACTGAATCTTCTGCTTTCTCCATTC | 61.4 | TAAGGGAAGGGGCGGAAC | 62.2 |
| PPEST 11 | ES347986.1 | (GA)11 | 162 | GGTGGATACTTGTGACGACTGA | 60.0 | CCACTCAAACTAAACTGGAAAGC | 59.4 |
| PPEST 12 | FG113351.1 | (AT)10 | 233 | CTTTTCTGCCTTAGCTGCGTT | 61.0 | CGTGTCTCTTCCACCACTACAA | 60.2 |
| PPEST 13 | EY206382.1 | (TC)10 | 222 | AGCTGGAATCAGGAGCAAAT | 58.9 | CAGTATCAAGCGAAAGCCG | 59.6 |
| PPEST 14 | FK815057.1 | (GA)9 | 228 | ACGTGTTGTTTTCTGTGGAGTG | 60.1 | CTGGGTATTTTCTGCCTTGATT | 59.5 |
| PPEST 15 | FK812013.1 | (TGA)9 | 157 | AAGAAAGGTTTGGCTTCGTGT | 60.2 | AATGGTGCTTCATCTCCTCTTC | 59.7 |
| PPEST 16 | FK811085.1 | (TCA)9 | 239 | TTCTGTTCACACATCATTTCCC | 59.8 | TGTGGCTGTAATTTGACTGGAG | 60.2 |
| PPEST 17 | FG116712.1 | (TGC)9 | 208 | CTGTTCAGCAAAACGAGACG | 59.6 | TCCCAAGTACAAAGACGGAACT | 60.0 |
| PPEST 18 | EY216123.1 | (CT)9 | 276 | ACTTGCATACTCTTTCGCACAA | 59.9 | AAATTCATGGAAAACCTCCCTC | 60.5 |
| PPEST 19 | EY210796.1 | (AC)9 | 211 | ACCCATCACCGTCTCTGC | 59.6 | TTTCCCTTGAACAACAACCAC | 59.9 |
| PPEST 20 | EY210288.1 | (TC)9 | 293 | ACAGAAGAAACCATCCATTTGC | 60.4 | CCATATTCCCGATTGAGAGAGA | 60.4 |
| PPEST 21 | EX474074.1 | (AG)9 | 244 | GGATTAGTGTGGCTCAAGATGG | 60.9 | GCAGGAAAATAGCAAAAGGGAT | 60.8 |
| PPEST 22 | ES347170.1 | (GT)9 | 229 | ATGGGGTATTAAGGGAGAATGC | 60.4 | GGGACGTGTGTGAGTGAGATAG | 59.6 |
| PPEST 23 | FG120710.1 | (GA)8 | 223 | CTGCATTTCTAATTTCGCGG | 60.7 | GGAGGAAGTGGACAGTGAAAAC | 60.0 |
| PPEST 24 | FG118096.1 | (TC)8 | 129 | AGGCACATTTTGGTTGTCTTCT | 60.0 | ATTTAGGGAGTCAATAGCGCAG | 59.8 |
| PPEST 25 | EY218678.1 | (TTTA)8 | 132 | CCATTCACTTCAAATCCATCCT | 60.2 | AACTGGGTGGTTGGTTGTTTT | 60.5 |
| PPEST 26 | EY216313.1 | (AC)8 | 217 | GATTCCCCAGGCAAAATAAA | 58.9 | TAATCAATATGGTGGGTTCCG | 59.5 |
| PPEST 27 | EY213284.1 | (TAAA)8 | 150 | TTGCTAAAGACAAGCGCAACT | 60.2 | CCATTCACTTCAAATCCATCCT | 60.2 |
| PPEST 28 | EY209870.1 | (CAA)8 | 245 | GATCAAGGCGGTTAATTTCAAG | 60.0 | ACAATCCAGAAGGACGATGC | 60.1 |
| PPEST 29 | FK814636.1 | (GGT)6 | 228 | TTTGTGGAGTTCGATGACTACG | 60.2 | GGACACATTCCTGTTCCAATTC | 60.6 |
| PPEST 30 | EY218675.1 | (CT)9 | 373 | TTCGCTCTTTCTCTCTCTCTCC | 59.5 | ATTCTGTACGTTACCTGCCCTG | 60.4 |
| PPEST 31 | EY215872.1 | (TA)11 | 400 | AAACGTGCATTCTCTGCCTAAT | 60.2 | CTCGATATTTATTTCCCCGCT | 59.5 |
| PPEST 32 | EY210648.1 | (TTTA)6 | 123 | ATTCACTTCAAACCCCTCCTTT | 60.2 | AACTGGGTGGTTGGTTGTTTT | 60.5 |
| PPEST 33 | FG115100.1 | (GAA)15 | 251 | ATACTCCCTCAGAACTAGCCCC | 60.0 | TTCGTCTTCTTCTTCTTCCTCC | 59.1 |
| PPEST 34 | EY215687.1 | (AAAG)5 | 137 | CACCTACAGAGATGCTGGATTG | 59.8 | GGGCTAAAATGTGTCTTGACTTG | 60.0 |
| PPEST 35 | EY214242.1 | (GT)15 | 245 | TAGTCACAACACACGAACCACA | 60.1 | TTAACCGTGAGAGTACCAGCAA | 59.8 |
| PPEST 36 | EY203279.1 | (TTTA)6 | 123 | ATTCACTTCAAACCCCTCCTTT | 60.2 | AACTGGGTGGTTGGTTGTTTT | 60.5 |
| PPEST 37 | EX474189.1 | (AT)7 | 208 | ACCGTGCAACCATTTTAAGTTC | 60.3 | AGTTATTCTTCTTCTTACTGCGCC | 59.5 |
| PPEST 38 | FK811878.1 | (AG)5 | 256 | TCCAGATACTCAAGTTCCAGCC | 60.6 | TATAGCGTTCAGATCCACCAGA | 59.7 |
| PPEST 39 | FG107375.1 | (CT)6 | 292 | CCCCAAAGAGAGTACACCAAAG | 60.0 | ATCAGCCAGTGTCGTATGAATG | 60.0 |
| PPEST 40 | FG114532.1 | (AG)5 | 322 | TCCCAAGGCTATTAAGTCTGGT | 59.2 | GGCTATCGTGCAATTTTCTTCT | 59.8 |
Discussion
Molecular markers are central for investigating genetic variability and for understanding genome dynamics. In the case of sand flies, the development of molecular markers, however, has remained slow. Microsatellites or SSRs have proven to be useful markers in population genetic studies of sand flies [16]. The presence of SSRs in coding regions suggests their importance as functional markers. While the development of microsatellite markers for sand flies from genomic libraries has been relatively costly, labour intensive and time consuming [14-16,18], the mining of microsatellite markers from EST data overcomes these disadvantages.
The ESTs used in the present study were normalized. Hence, redundancy in the EST database was minimized and a wealth of unique cDNA sequences (unigenes) for marker development was found. Examining the distribution of SSR motifs can assist in gaining insights into genome composition and genetic makeup [24,25]. Although, SSR motifs with more than five repeats were considered here, shorter SSRs were identified. The maximum length achieved was 16 repeats; this is consistent with studies that revealed shorter SSRs in Drosophila [26,27].
Dimeric repeat motifs were more abundant than trimeric repeats. However, this observation was expected, as the frequency and distribution of SSRs depend on several factors, such as size of dataset, and tools and criteria used for SSR discovery. Tetra- and penta-repeat motifs were considerably less represented.
The most abundant SSRs were of dinucleotide type (Figures 1 and 2), in which homopurine-homopyrimidine stretches, such as AG and CT, have the highest frequency. Dinucleotide repeats are typically more frequent in noncoding regions [28-30]; however, they occur occasionally in coding regions as well [31]. Some dinucleotides, such as (AG)n/(CT)n, are not selectively neutral and may have functional roles. These repetitive sequences occur in the 5'-UTR and are likely to be involved in gene regulation [32-34]. The (GC)n repeats were absent from P. papatasi, even though they are numerically abundant SSR loci in most eukaryotes [35,36]. However, dinucleotide SSR motifs in P. papatasi ESTs are relatively biased toward an excess of (AX)n repeats and a low GC base content, the broader implications of this observation are unclear.
The high frequency of dinucleotide motifs (AT, AG, and CT) could be explained by their abundance in several codons with different nucleotide arrangements. This observation is in agreement with previous reports [32,37]. EST-derived SSRs; AG/CT repeats have been studied widely in eukaryotes, particularly in plants, and found to be highly abundant and highly polymorphic [38,39]. For P. papatasi, the number of published SSR markers is very limited compared with other major insect vectors, including species of Anopheles and Aedes [14]. In the present study, an in-depth analysis of microsatellites, in terms of density, resulted in the development of a new set of 40 SSR markers (Table 2). Thus, we have shown that the mining of ESTs is an effective strategy to identify functional microsatellites, with perfect repeats, in P. papatasi.
The prevalence of trinucleotide SSRs in P. papatasi ESTs was expected, since they do not interrupt triplet codons, whereas other repetitive stretches, such as mono-, di-, or tetra-nucleotides lead to frame-shift mutations, which would result in severe adverse effects in coding regions. However, the abundance of trinucleotide SSRs in coding regions of various organisms was much higher than in non-coding regions of the genome [37,40-45]. In contrast, the present study showed that trinucleotides were the second most abundant SSRs in P. papatasi ESTs (31.1%) compared with dinucleotides (67%). This observation could be explained by the SSR mining tool used here and its preset criteria, such as identification of a minimum number of repeats, which could have led to the identification of more repeats. This approach could have led to the identification of more dinucleotides and fewer tri- and tetra-nucleotides, with this bias contributing to the over-representation of dinucleotides compared with tri- and tetra-nucleotides. Another possible explanation is that P. papatasi EST data do not contain many trinucleotide SSRs compared with dinucleotides.
Conclusions
This is the first large-scale survey of 1,499 unique EST-SSRs of P. papatasi. Despite the number of EST sequences surveyed, SSR loci do not appear to be particularly dense or frequent in P. papatasi (3.5%). SSR repeats characterized are mainly of dinucleotide type and heterogeneously distributed across all potential base compositions, with a small number of GC-rich repeat motifs. The DNA replication machinery likely contributes to the elevated abundance of dinucleotide AT-, and AG- rich repeat motifs and to lesser extent trinucleotide motifs, suggesting that future screens of P. papatasi and other sand fly molecular markers may benefit by focusing on SSR motifs. The utility of the microsatellite markers characterized in this study should be evaluated in the near future. More microsatellite markers should be characterized for P. papatasi and other key sand flies of major importance as vectors of Leishmania.
Methods
Retrieval of EST sequences
P. papatasi EST sequences used were directly retrieved from NCBI database http://www.ncbi.nlm.nih.gov/projects/dbEST/ on May 10, 2011. A total of 42,784 P. papatasi ESTs were listed and annotated. These ESTs were derived from three cDNA libraries constructed from uninfected sugar fed, uninfected blood fed, and L. major infected blood fed P. papatasi sand flies. All the sequences were saved in FASTA-formatted text files that were used for further analysis.
Characterization of SSRs
PolyA and polyT tracts were removed, leaving no (T)10 or (A)10 in any 10 bp window at either end of the sequences. The dataset was divided into small files, each containing 100 FASTA formatted sequences. SSR-containing sequences were identified using SSRIT web based SSR identification tool [46] available at http://www.gramene.org/db/markers/ssrtool. Any sequence was considered as an SSR where a repeat motif of one to six nucleotides in length was repeated at least five times for dinucleotide, trinucleotide, tetranucleotide and pentanucleotide SSRs. Redundant sequences were filtered by BLAST analysis, using each individual sequence as a query against the total set of selected sequences. Homologous sequences were aligned using MEGA 5 and scanned manually in the sequence editor window [47]. The criteria for redundancy were: (i) where a cluster contained two or more identical sequences, the longest was retained; (ii) sequences which were composed entirely of SSR motif, lacking any flanking sequence, were discarded since their uniqueness could not be established and in any event, primer design was not possible.
Sequence analysis
Total number of characters, sequence composition frequency and A+T and G+C contents were carried out by CLC Genomics Workbench program, v.3.7 (CLC bio, Denmark). The EST sequences were screened for the presence of perfect SSRs, and repeat motifs ≥ 5, these sequences were selected, annotated and filed for primer design.
Primer design
The non-redundant EST-SSRs were used for primer design to flanking sequences using PRIMER3 [48]. PRIMER3 was calibrated to the following parameters: (i) Primer length from 18-27 bases, the optimal annealing temperature (Tm) from 55 to 60°C, the target amplicon size 100-300 bp, and GC content between 30 and 70% (50% as the optimum). All other parameters were set to default values. The output from PRIMER3 was further analyzed in order to lessen the chance of encompassing tandem repeats in primer sequences and self- and pair-complementarity.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
OMH designed the study, conducted data analysis and drafted the manuscript. AHA participated in data analysis and drafting the manuscript. Both authors approved the final version of the manuscript.
Contributor Information
Omar Hamarsheh, Email: ohamarsheh@science.alquds.edu.
Ahmad Amro, Email: ahmadymm@hotmail.com.
Acknowledgements
This work was partly financed by COMSTECH. Ref: RAB & GH 10-11/16 in collaboration with Dr. Meryem Lemrani from Institut Pasteur du Maroc, and by Al-Quds University.
References
- Sawalha SS, Shtayeh MS, Khanfar HM, Warburg A, Abdeen ZA. Phlebotomine sand flies (Diptera: Psychodidae) of the Palestinian West Bank: potential vectors of leishmaniasis. J Med Entomol. 2003;40:321–328. doi: 10.1603/0022-2585-40.3.321. [DOI] [PubMed] [Google Scholar]
- Killick-Kendrick R. Phlebotomine vectors of the leishmaniases: a review. Med Vet Entomol. 1990;4:1–24. doi: 10.1111/j.1365-2915.1990.tb00255.x. [DOI] [PubMed] [Google Scholar]
- Schlötterer C. Evolutionary dynamics of microsatellite DNA. Chromosoma. 2000;109:365–371. doi: 10.1007/s004120000089. [DOI] [PubMed] [Google Scholar]
- Tautz D, Schlötterer. Simple sequences. Curr Opin Genet Dev. 1994;4:832–837. doi: 10.1016/0959-437X(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Lovin DD, Washington KO, deBruyn B, Hemme RR, Mori A, Epstein SR, Harker BW, Streit TG, Severson DW. Genome-based polymorphic microsatellite development and validation in the mosquito Aedes aegypti and application to population genetics in Haiti. BMC Genomics. 2009;10:590. doi: 10.1186/1471-2164-10-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongnoparut P, Yaicharoen S, Sirichotpakorn N, Rattanarithikul R, Lanzaro GC, Linthicum KJ. Microsatellite polymorphism in Anopheles maculatus, a malaria vector in Thailand. Am J Trop Med Hyg. 1996;55:589–594. doi: 10.4269/ajtmh.1996.55.589. [DOI] [PubMed] [Google Scholar]
- Fonseca DM, Atkinson CT, Fleischer RC. Microsatellite primers for Culex pipiens quinquefasciatus, the vector of avian malaria in Hawaii. Mol Ecol. 1998;7:1617–1619. [PubMed] [Google Scholar]
- Schonian G, El Fari M, Lewin S, Schweynoch C, Presber W. Molecular epidemiology and population genetics in Leishmania. Med Microbiol Immunol. 2001;190:61–63. doi: 10.1007/s004300100081. [DOI] [PubMed] [Google Scholar]
- Njiokou F, Cuny G, Asonganyi T. Trypanosoma brucei s.l.: Microsatellite markers revealed high level of multiple genotypes in the mid-guts of wild tsetse flies of the Fontem sleeping sickness focus of Cameroon. Exp Parasitol. 2011;128:272–278. doi: 10.1016/j.exppara.2011.02.023. [DOI] [PubMed] [Google Scholar]
- Donnelly MJ, Cuamba N, Charlwood JD, Collins FH, Townson H. Population structure in the malaria vector, Anopheles arabiensis patton, in East Africa. Heredity. 1999;83(Pt 4):408–417. doi: 10.1038/sj.hdy.6885930. [DOI] [PubMed] [Google Scholar]
- Amro A, Schonian G, Al-Sharabati MB, Azmi K, Nasereddin A, Abdeen Z, Schnur LF, Baneth G, Jaffe CL, Kuhls K. Population genetics of Leishmania infantum in Israel and the Palestinian Authority through microsatellite analysis. Microbes Infect. 2009;11:484–492. doi: 10.1016/j.micinf.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Seridi N, Amro A, Kuhls K, Belkaid M, Zidane C, Al-Jawabreh A, Schonian G. Genetic polymorphism of Algerian Leishmania infantum strains revealed by multilocus microsatellite analysis. Microbes Infect. 2008;10:1309–1315. doi: 10.1016/j.micinf.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Chargui N, Amro A, Haouas N, Schonian G, Babba H, Schmidt S, Ravel C, Lefebvre M, Bastien P, Chaker E. et al. Population structure of Tunisian Leishmania infantum and evidence for the existence of hybrids and gene flow between genetically different populations. Int J Parasitol. 2009;39:801–811. doi: 10.1016/j.ijpara.2008.11.016. [DOI] [PubMed] [Google Scholar]
- Hamarsheh O, Presber W, Abdeen Z, Sawalha S, Al-Lahem A, Schoenian G. Isolation and characterization of microsatellite loci in the sand fly Phlebotomus papatasi (Diptera: Psychodidae) Mol Ecol Notes. 2006;6:826–828. doi: 10.1111/j.1471-8286.2006.01359.x. [DOI] [Google Scholar]
- Aransay AM, Malarky G, Ready PD. Isolation (with enrichment) and characterization of trinucleotide microsatellites from Phlebotomus perniciosus, a vector of Leishmania infantum. Mol Ecol Notes. 2001;1:176–178. [Google Scholar]
- Hamarsheh O, Presber W, Al-Jawabreh A, Abdeen Z, Amro A, Schonian G. Molecular markers for Phlebotomus papatasi (Diptera: Psychodidae) and their usefulness for population genetic analysis. Trans R Soc Trop Med Hyg. 2009;103:1085–1086. doi: 10.1016/j.trstmh.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Hamarsheh O, Presber W, Yaghoobi-Ershadi MR, Amro A, Al-Jawabreh A, Sawalha S, Al-Lahem A, Das ML, Guernaoui S, Seridi N. et al. Population structure and geographical subdivision of the Leishmania major vector Phlebotomus papatasi as revealed by microsatellite variation. Med Vet Entomol. 2009;23:69–77. doi: 10.1111/j.1365-2915.2008.00784.x. [DOI] [PubMed] [Google Scholar]
- Hamarsheh O. Distribution of Leishmania major zymodemes in relation to populations of Phlebotomus papatasi sand flies. Parasit Vectors. 2011;4:9. doi: 10.1186/1756-3305-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpelainen H, Kostamo K, Virtanen V. Microsatellite marker identification using genome screening and restriction-ligation. Biotechniques. 2007;42:479–486. doi: 10.2144/000112415. [DOI] [PubMed] [Google Scholar]
- Merkel A, Gemmell N. Detecting microsatellites in genome data: variance in definitions and bioinformatic approaches cause systematic bias. Evol Bioinform. 2008;4:1–6. doi: 10.4137/ebo.s420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Korol A, Fahima T, Beiles A, Nevo E. Microsatellites: genomic distribution, putative functions, and mutational mechanisms: a review. Mol Ecol. 2002;11:2453–2465. doi: 10.1046/j.1365-294X.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- Ellis JR, Burke JM. EST-SSRs as a resource for population genetic analyses. Heredity. 2007;99:125–132. doi: 10.1038/sj.hdy.6801001. [DOI] [PubMed] [Google Scholar]
- Behura SK. Molecular marker systems in insects: current trends and future avenues. Mol Ecol. 2006;15:3087–3113. doi: 10.1111/j.1365-294X.2006.03014.x. [DOI] [PubMed] [Google Scholar]
- Ju Z, Wells MC, Martinez A, Hazlewood L, Walter RB. An in silico mining for simple sequence repeats from expressed sequence tags of zebrafish, medaka, Fundulus, and Xiphophorus. In Silico Biol. 2005;5:439–463. [PubMed] [Google Scholar]
- Serapion J, Kucuktas H, Feng J, Liu Z. Bioinformatic mining of type I microsatellites from expressed sequence tags of channel catfish (Ictalurus punctatus) Mar Biotechnol (NY) 2004;6:364–377. doi: 10.1007/s10126-003-0039-z. [DOI] [PubMed] [Google Scholar]
- Schug MD, Regulski EE, Pearce A, Smith SG. Isolation and characterization of dinucleotide repeat microsatellites in Drosophila ananassae. Genet Res. 2004;83:19–29. doi: 10.1017/S0016672303006542. [DOI] [PubMed] [Google Scholar]
- Pascual M, Chapuis MP, Mestres F, Balanya J, Huey RB, Gilchrist GW, Serra L, Estoup A. Introduction history of Drosophila subobscura in the New World: a microsatellite-based survey using ABC methods. Mol Ecol. 2007;16:3069–3083. doi: 10.1111/j.1365-294X.2007.03336.x. [DOI] [PubMed] [Google Scholar]
- Gao H, Cai S, Yan B, Chen B, Yu F. Discrepancy variation of dinucleotide microsatellite repeats in eukaryotic genomes. Biol Res. 2009;42:365–375. [PubMed] [Google Scholar]
- Calabrese P, Durrett R. Dinucleotide repeats in the Drosophila and human genomes have complex, length-dependent mutation processes. Mol Biol Evol. 2003;20:715–725. doi: 10.1093/molbev/msg084. [DOI] [PubMed] [Google Scholar]
- McNeil JA, Smith KP, Hall LL, Lawrence JB. Word frequency analysis reveals enrichment of dinucleotide repeats on the human × chromosome and [GATA]n in the × escape region. Genome Res. 2006;16:477–484. doi: 10.1101/gr.4627606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VK, Kumar N, Brahmachari SK, Ramachandran S. Abundance of dinucleotide repeats and gene expression are inversely correlated: a role for gene function in addition to intron length. Physiol Genomics. 2007;31:96–103. doi: 10.1152/physiolgenomics.00183.2006. [DOI] [PubMed] [Google Scholar]
- Li B, Xia QY, Lu C, Zhou ZY. Analysis of microsatellites derived from bee Ests. Yi Chuan Xue Bao. 2004;31:1089–1094. [PubMed] [Google Scholar]
- Grasela JJ, McIntosh AH. Application of inter-simple sequence repeats to insect cell lines: identification at the clonal and tissue-specific level. In Vitro Cell Dev Biol Anim. 2003;39:353–363. doi: 10.1290/1543-706X(2003)039<0353:AOISRT>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Han YJ, de Lanerolle P. Naturally extended CT. AG repeats increase H-DNA structures and promoter activity in the smooth muscle myosin light chain kinase gene. Mol Cell Biol. 2008;28:863–872. doi: 10.1128/MCB.00960-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Mishra RK, Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astolfi P, Bellizzi D, Sgaramella V. Frequency and coverage of trinucleotide repeats in eukaryotes. Gene. 2003;317:117–125. doi: 10.1016/s0378-1119(03)00659-0. [DOI] [PubMed] [Google Scholar]
- Li B, Xia Q, Lu C, Zhou Z, Xiang Z. Analysis on frequency and density of microsatellites in coding sequences of several eukaryotic genomes. Geno Prot Bioinfo. 2004;2:24–31. doi: 10.1016/S1672-0229(04)02004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M, Hanafey M, Powell W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet. 2002;30:194–200. doi: 10.1038/ng822. [DOI] [PubMed] [Google Scholar]
- Yi G, Lee JM, Lee S, Choi D, Kim BD. Exploitation of pepper EST-SSRs and an SSR-based linkage map. Theor Appl Genet. 2006;114:113–130. doi: 10.1007/s00122-006-0415-y. [DOI] [PubMed] [Google Scholar]
- Jung S, Abbott A, Jesudurai C, Tomkins J, Main D. Frequency, type, distribution and annotation of simple sequence repeats in Rosaceae ESTs. Funct Integr Genomics. 2005;5:136–143. doi: 10.1007/s10142-005-0139-0. [DOI] [PubMed] [Google Scholar]
- Kaushik N, Malaspina A, de Belleroche J. Characterization of trinucleotide- and tandem repeat-containing transcripts obtained from human spinal cord cDNA library by high-density filter hybridization. DNA Cell Biol. 2000;19:265–273. doi: 10.1089/10445490050021177. [DOI] [PubMed] [Google Scholar]
- Ashley CT Jr, Warren ST. Trinucleotide repeat expansion and human disease. Annu Rev Genet. 1995;29:703–728. doi: 10.1146/annurev.ge.29.120195.003415. [DOI] [PubMed] [Google Scholar]
- Clark RM, Bhaskar SS, Miyahara M, Dalgliesh GL, Bidichandani SI. Expansion of GAA trinucleotide repeats in mammals. Genomics. 2006;87:57–67. doi: 10.1016/j.ygeno.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Schlötterer C, Harr B. Drosophila virilis has long and highly polymorphic microsatellites. Mol Biol Evol. 2000;17:1641–1646. doi: 10.1093/oxfordjournals.molbev.a026263. [DOI] [PubMed] [Google Scholar]
- Harr B, Schlötterer C. Long microsatellite alleles in Drosophila melanogaster have a downward mutation bias and short persistence times, which cause their genome-wide underrepresentation. Genetics. 2000;155:1213–1220. doi: 10.1093/genetics/155.3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001;11:1441–1452. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011. in press . [DOI] [PMC free article] [PubMed]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]