Abstract
Chromatin immunoprecipitation followed by sequencing (ChIP-Seq) is a technique for genome-wide profiling of DNA-binding proteins, histone modifications, or nucleosomes. Enabled by the tremendous progress in next-generation sequencing technology, ChIP-Seq offers higher resolution, less noise, and greater coverage than its array-based predecessor ChIP-chip. With the decreasing cost of sequencing, ChIP-Seq has become an indispensable tool for studying gene regulation and epigenetic mechanisms. In this review, we describe the benefits as well as the challenges in harnessing this technique, with an emphasis on issues related to experimental design and data analysis. ChIP-Seq experiments generate large quantities of data, and effective computational analysis will be critical for uncovering biological mechanisms.
Introduction
Genome-wide mapping of protein-DNA interactions and epigenetic marks is essential for full understanding of transcriptional regulation. A precise map of binding sites for transcription factors, core transcriptional machinery and other DNA-binding proteins is vital for deciphering gene regulatory networks that underlie various biological processes [1]. The combination of nucleosome positioning and dynamic modification of DNA and histones plays a key role in gene regulation [2–4] and guides development and differentiation [5]. Chromatin states can influence transcription directly by altering the packaging of DNA to allow or prevent access to DNA-binding proteins; or they can modify the nucleosome surface to enhance or impede recruitment of effector protein complexes. Recent advances suggest that this interplay between chromatin and transcription is dynamic and more complex than previously appreciated [6], and there has been a growing recognition that systematic profiling of the epigenomes in multiple cell types and stages may be needed for understanding developmental processes and disease states [7].
The main tool for investigating these mechanisms is chromatin immunoprecipitation (ChIP), a technique that enriches DNA fragments to which a specific protein or a certain class of nucleosomes is bound [8]. With the introduction of microarrays, fragments obtained from ChIP could be identified by hybridization to a microarray (ChIP-chip), thus enabling a genome-scale view of DNA-protein interactions [9, 10]. On high-density tiling arrays, oligonucleotide probes can now be placed across an entire genome or across selected regions of a genome - for instance, promoter regions, specific chromosomes, or gene families - at a preferred resolution.
With the rapid technological developments in next-generation sequencing (NGS), the arsenal of genomic assays available to the biologist has been transformed [11–13]. With the ability to sequence tens or hundreds of millions of short DNA fragments in a single run, an increasingly large set of experiments, which could only be imagined a few years ago, is becoming possible. NGS has already been applied in a number of areas including whole-genome sequencing [14, 15], mRNA-sequencing for gene expression profiling[16–18], characterization of structural variation [19], profiling of DNase I hypersensitive sites [20], detection of fusion genes from mRNA transcripts [21], and discovery of new classes of small RNAs [22]. If the ‘third-generation’ sequencing technologies that are under development deliver as promised, they will enable another epoch of genome-scale investigations [23].
Chromatin immunoprecipitation followed by sequencing (ChIP-Seq) has been one of the early applications of NGS, with the first publications in 2007 [24–27]. In ChIP-Seq, the DNA fragments of interest are sequenced directly instead of being hybridized on an array. With single base-pair resolution, fewer artifacts, greater coverage, and a larger dynamic range, ChIP-Seq offers significantly improved data compared to ChIP-chip. Although the short reads (~35 bp) generated on NGS platforms pose serious difficulties for certain applications, for example de novo genome assembly, they are acceptable for ChIP-Seq. The more precise mapping of protein binding sites provided by ChIP-Seq allows for a more accurate list of targets for transcription factors and enhancers as well as better identification of sequence motifs [24, 28]. Enhanced spatial resolution is particularly important for profiling post-translational modifications of chromatin and histone variants, as well as nucleosome positioning, and ChIP-Seq has enabled tremendous progress in these areas already (see BOX 1).
BOX 1. The contribution of ChIP-Seqto mapping epigenomes.
The enhanced spatial resolution afforded by next-generation sequencing improves the characterization of binding sites for transcription factors and other DNA-binding proteins, including identification of sequence motifs. The increased precision is especially important for profiling nucleosome-level features and it now allows one to systematically catalogue the patterns of histone modifications, histone variants, and nucleosome positioning. Here, we briefly describe recent chromatin immunoprecipitation (ChIP) studies that have enabled progress in characterizing epigenomes.
Histone modification maps
The first comprehensive genome-wide maps using ChIP-Seq were created in 2007. Twenty histone methylation marks, as well as the histone variant H2A.Z, RNA Polymerase II, and the DNA-binding protein CTCF, were profiled in human T cells [25], with an average of ~8 million tags per sample using Solexa 1G. This was followed by a map of 18 histone acetylation marks in the same cell type [90]. These studies suggested novel functions for histone modification and the importance of combinatorial patterns of modifications. To examine the role of histone modifications in differentiation, embryonic stem (ES) cells have also been profiled. Several lysine trimethylation modifications were profiled in mouse ES cells and two types of differentiated cells in 2007 [27]. This study showed the role of bivalent domains [91] in lineage potential as well as marks for imprinting control. Prior to ChIP-Seq, genome-wide modification profiles were available for yeast using tiling arrays [92–94], but only selected regions had been profiled for mouse and human. See Ref [35] for further description of the techniques used.
Nucleosome maps
Using ChIP-chip, nucleosome depletion at active promoters in yeast was described in 2004 [95]. This was followed by a high-resolution study [96] in 2005 and a complete map of nucleosome positioning [97] in 2007. In C. elegans MNase digestion followed by sequencing was used in 2006 to map core nucleosomes [98]. ChIP-Seq with Roche 454 pyrosequencing was used to generate a map of the histone variant H2A.Z in yeast [99] in 2007, and in fly [100] in 2008. For human cells, epigenetically modified and bulk mono-nucleosome positions were profiled for T cells in 2007 and 2008 [25, 30], with >140 million Illumina/Solexa reads per experiment, (see Ref [2] for a review). These studies have revealed the role of nucleosomes in transcriptional regulation and hint at the principles that guide nucleosome positioning.
In this review, we will describe advantages as well as challenges in applying the ChIP-Seq technology. We will discuss various issues in experimental design, including sample quality, controls, depth of sequencing, and the number of replicates. Given the large quantities of data generated in ChIP-Seq, computational analysis, including identification of binding sites and subsequent analysis, poses a significant challenge for most laboratories. We will discuss the main issues in data processing and statistical analysis.
ChIP-Seq basics
In a ChIP experiment for DNA-binding proteins, DNA fragments associated with a protein are enriched (Figure 1). First, the DNA-binding protein is crosslinked to DNA in vivo by treating cells with formaldehyde and then the chromatin is sheared by sonication into small fragments, generally in the 200–600 bp range. Then an antibody specific to the protein of interest is used to immunoprecipitate the DNA-protein complex. Finally, the crosslinks are reversed and the released DNA is assayed to determine the sequences bound by the protein. In construction of a sequencing library, the immunoprecipitated DNA is subjected to size selection (typically in the ~150–300 bp range), although there appears to be a bias toward shorter fragments in sequencing.
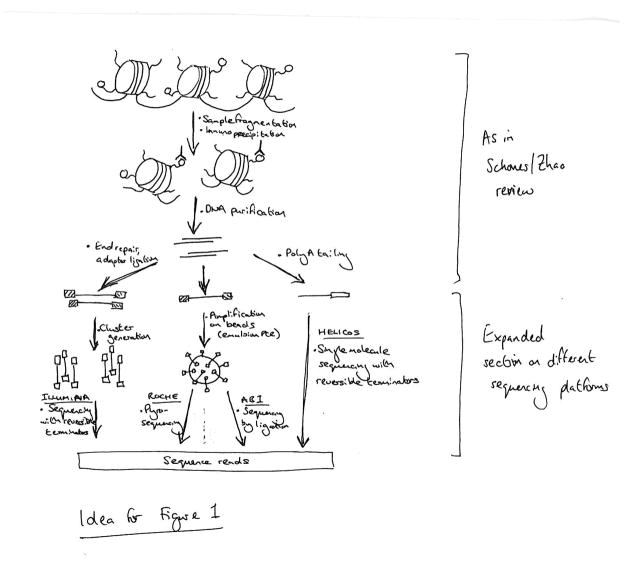
Figure 1. Overview of a ChIP-Seq experiment.
Specific DNA sites that interact with transcription factors or other chromatin-associated proteins as well as sites that correspond to modified chromatin can be profiled using chromatin immunoprecipitation (ChIP) followed by massively parallel sequencing. The ChIP process enriches crosslinked proteins or modified chromatin of interest using an antibody specific to the protein or the histone modification. Purified DNA can be sequenced on any of the next-generation platforms [12]. The basic concepts are similar on these platforms: common adaptors are ligated to the ChIP DNA, and clonally clustered amplicons are generated. The sequencing step involves enzyme-driven extension of all templates in parallel, alternating with detection of florescent labels incorporated with each extension by high-resolution imaging. On the Illumina/Solexa Genome Analyzer (bottom left), clusters of clonal sequences are generated by bridge PCR, and sequencing is performed by sequencing-by-synthesis. On the 454 and SOLiD platforms (bottom middle), clonal sequencing features are generated by emulsion PCR, with amplicons captured to the surface of μm-scale beads. Beads with amplicons are then recovered and immobilized to a planar substrate to be sequenced by pyrosequencing (454) or by DNA ligase-driven synthesis (SOLiD). On single-molecular sequencing platforms such as the HeliScope by Helicos (bottom right), fluorescent nucleotides incorporated into templates can be imaged at the level of single molecules, thus making clonal amplification unnecessary.
In a ChIP experiment to map nucleosome positions or histone modifications, micrococcal nuclease (MNase) digestion without crosslinking is most often used to fragment the chromatin. Although sonication has been used in this context [29], MNase treatment is generally preferred because it removes linker DNA more efficiently than sonication, allowing more precise mapping of each nucleosome [30]. On the other hand, MNase digestion is known to have a more pronounced sequence bias [31], as well as bias due to the solubility of chromatin [32]. There may also be changes in nucleosome position and histone modifications during the course of the experiment in the absence of crosslinking. ChIP with and without crosslinking is sometimes referred to as X-ChIP [33] and N-ChIP [34], respectively, with X denoting ‘crosslinking’ and N denoting ‘native.’
Nearly all ChIP-Seq data so far have been generated on the Illumina Genome Analyzer, although other platforms such as Applied Biosystems’ SOLiD and the Helicos platform are now available for ChIP-Seq (Figure 1). The Illumina and the SOLiD platforms currently generate 100–400 million reads in a single run, typically with 60–80% of reads that can be aligned uniquely to the genome.
Advantages and disadvantages
ChIP-Seq offers a number of advantages over ChIP-chip, as summarized in Table 1 (See also Ref [35]). Its base-pair resolution is perhaps the greatest improvement compared to ChIP-chip, as illustrated in Figure 2A. Although arrays could be tiled at high density, this requires a large number of probes and remains expensive for mammalian genomes [36]. Arrays also have fundamental limitations in resolution due to the uncertainties in the hybridization process. Second, ChIP-Seq does not suffer from noise due to the hybridization step in ChIP-chip. Nucleic acid hybridization is complex and is dependent on many factors including the GC-content, length, concentration, and secondary structure of both the target and probe sequences. Thus, cross-hybridization between imperfectly matched sequences frequently occurs and contributes to the noise. Third, the intensity signal measured on arrays may not be linear in its entire range and its dynamic range is limited below and above saturation points. In a recent study, distinct and biologically meaningful peaks seen in ChIP-Seq were obscured in the same experiment conducted with ChIP-chip [37]. Finally, the fourth significant advantage is that the genome coverage is not limited by the probe sequences fixed on the array. This is particularly important for analysis of repetitive regions of the genome, which are typically masked out on arrays. Studies involving heterochromatin or microsatellites, for instance, can be done much more effectively by ChIP-Seq. Sequence variations within repeat elements can be captured by sequencing and be used to map to the genome; unique sequences flanking repeats also are helpful in aligning the reads to the genome. For example, only 48% of the human genome is non-repetitive, but 80% is mappable with 30-bp reads and 89% is mappable with 70-bp reads [38].
Table 1.
Comparison of ChIP-chip and ChIP-Seq
| ChIP-chip | ChIP-Seq | |
|---|---|---|
| Resolution | Array-specific, generally 30–100bp | Single nucleotide |
| Coverage | Limited by sequences on the array; repetitive regions usually masked out | Limited only by alignability of reads to the genome; increases with read length; many repetitive regions can be covered |
| Cost | $400–$800 per array (1–6 million probes); multiple arrays may be needed for large genomes | $1000–$2000 per Illumina lane (6–15 million readsprior to alignment) |
| Source of platform noise | Cross-hybridization between probes and non-specific targets | Some GC-biasmay be present |
| Experimental design | Single- or double-channel, depending on platform | Single channel |
| Cost-effective cases | Large fraction enriched (broad binding), profiling of selected regions | Small fraction enriched (sharp binding), large genomes |
| Required amount of ChIP DNA | High (few μg) | Low (10–50 ng) |
| Dynamic range | Lower detection limit, saturation at high signal | Not limited |
| Amplification | More required | Less required; single molecule sequencing without amplification is available |
| Multiplexing | Not possible | Possible |
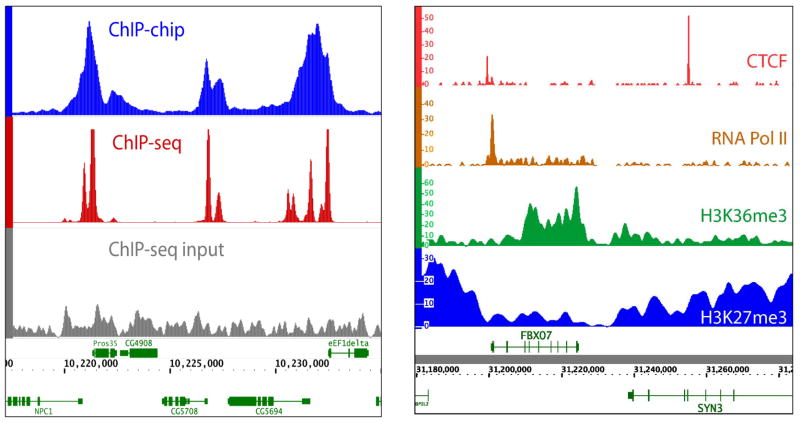
Figure 2. ChIP profiles.
A) An example of ChIP-Seq and ChIP-chip profiles. The figure shows a section of the binding profiles of the chromodomain protein Chromator measured by ChIP-chip (unlogged intensity ratio, blue) and ChIP-Seq (tag density, red) in the D. melanogaster S2 cell line. The tag density profile obtained by ChIP-Seq reveals specific positions of Chromator binding with higher spatial resolution and sensitivity. The ChIP-Seq input DNA (control experiment) tag density is shown (gray) for comparison. B) Examples of different types of ChIP-Seq tag density profiles. Profiles for different types of proteins and histone marks can have different types of features. For example: sharp binding sites, as shown for the insulator binding protein CTCF (red); a mixture of shapes, as shown for RNA Polymerase II (orange), which has a sharp peak followed by a broad region of enrichment; medium size broad peaks, as illustrated by H3K36me3 (green), which is associated with transcription elongation over the gene body; and large domains, as illustrated by H3K27me3 (blue), a repressive markindicative of Polycomb-mediated silencing. Data for part B are from human T-cells, from REF 20.
As with any technology, ChIP-Seq is not free from artifacts. Although sequencing errors have been reduced substantially, they are still present, especially toward the end of each read. This problem can be ameliorated by improvement in alignment algorithms (see below) and computational analysis. There is a bias toward high GC-rich content in fragment selection, both in library preparation and in amplification prior to sequencing [14, 39], although significant improvements have been made recently. When an insufficient number of reads are generated, there is loss of sensitivity or specificity (see the discussion below). There are also technical issues in performing the experiment, such as loading the correct amount of sample: too little sample will result in too few tags; too much sample will result in fluorescent labels too close to one another, causing lower data quality.
The main disadvantage for ChIP-Seq so far, however, has been cost and availability. Several groups have successfully developed and applied their own protocols for library construction, lowering this cost significantly. But the overall cost of ChIP-Seq, which includes machine depreciation and reagent cost, will have to be lowered furtherfor it to be comparable to ChIP-chipin every case. For high-resolution profiling of an entire large genome, ChIP-Seq is already less expensive than ChIP-chip; but depending on the genome size and the depth of sequencing needed, a ChIP-chip experiment on carefully selected regions using a customized microarray may yield as much biological understanding. The recent decrease in sequencing cost per base pair has not affected ChIP-seq as significantly as other applications, as it has come more from increased read lengths than the number of sequenced fragments. The gain in the fraction of reads that can be uniquely aligned to the genome decreases noticeably after ~25–35 bp and it is marginal beyond 70–100 nucleotides [40]. However, as the sequencing cost continues to decline and institutional support for sequencing platforms grows, ChIP-Seq will become the method of choice for most experiments in the near future.
Issues in experimental design
Antibody quality
The value of any ChIP data, including ChIP-Seq, depends critically on the quality of the antibody. A sensitive yet specific antibody will give a high level of enrichment compared to the background, making it easier to detect binding events. Many antibodies are commercially available, some noted as ChIP-grade, but their quality is highly variable, including lot-to-lot variation. Rigorous validation is a laborious process: for histone modifications, for instance, reactivity of the antibody with unmodified histones or non-histone proteins should be checked by Western blotting. Furthermore, cross-reactivity with similar histone modifications (for example, di- vs. tri-methylation at the same residue) should be checked by using two independent antibodies, combined with RNAi against enzymes predicted to deposit the modification or mass spectrometry of the precipitated peptides. As part of the model organism ENCODE project [41], the author has been involved in a large scale profiling of histone modifications for D. melanogaster. Our validation procedure with the steps described above has resulted in unsatisfactory findings for 20–35% of the commercially produced antibodies tested.
Sample quantity
One advantage of ChIP-Seq over ChIP-chip is the smaller amount of sample material needed. A typical ChIP experiment yields 10–100 ng of DNA, requiring on the order of 107 cells. Several ChIP protocols have been developed for a smaller number of cells, 104–105 for genome-wide profiling [42] or 102–103 for PCR quantification at specific loci [43–45], but so far they have been shown to work for abundant transcription factors or histone modifications (for example, RNA Polymerase II or histone H3 trimethylated at lysine 27, H3K27me3) pulled down by a high-quality antibody. For ChIP-chip, the ChIP sample is usually amplified to generate >2μg of DNA per array. In contrast, on the Illumina platform, 10–50 ng of ChIP DNA is recommended. With fewer rounds of amplification, the potential for artifacts due to PCR bias decreases for ChIP-Seq. The precise amount of ChIP DNA and the number of cells needed depend on the abundance of the chromatin-associated protein targets or histone modification, as well as the quality of the antibody. ChIP-Seq without amplification is possible on the Helicos Single-Molecule Sequencing platform [46] and other ‘third-generation’ platforms in development (see Figure 1).
Control experiment
The experimental steps in ChIP involve several potential sources of artifacts. Shearing of DNA, for example, does not result in uniform fragmentation of the genome: open chromatin regions tend to be fragmented more easily than closed regions, creating an uneven distribution of sequence tags across the genome. Also, repetitive sequences may appear enriched because the number of copies of the repeats is not accurately reflected in the calculation. Therefore, a peak in the ChIP-Seq profile must be compared to the same region in a matched control sample to determine its significance. There are three commonly used choices for this control: input DNA (that is, DNA prior to immunoprecipitation, IP); mock IP (treated the same as the IP but without any antibody); and non-specific IP (that is, using an antibody against a protein not known to be involved in DNA binding or chromatin modification, such as IgG). These controls test for different types of artifacts and there is no consensus on which is most appropriate. Input DNA has been used in nearly all ChIP-Seq studies so far; it corrects mainly for bias related to the variable solubility of different regions, shearing of the DNA, and amplification. One problem with mock IP is that very little material may be pulled down in the absence of an antibody and therefore results of multiple mock IPs may not be consistent. In one set of ChIP-chip experiments, mock IP was found to contribute little to the overall result when data are properly normalized [47]. For histone modifications, using the ratio between ChIP sample and bulk nucleosomes is also informative, as this ratio corresponds to the fraction of nucleosomes with the particular modification at that location, averaged over all the cells assayed.
One of the difficulties for a ChIP-Seq control experiment is the amount of sequencing necessary. For input DNA or bulk nucleosomes, many of the sequenced tags would be spread out evenly across the genome. To obtain accurate estimates along the genome, sufficient numbers of tags are needed at each point; otherwise, fold enrichment at the peaks will be have large errors due to sampling bias. Thus the total number of tags to be sequenced is potentially very large. Alternatively, it is possible to avoid sequencing of a control sample if one is only interested in differential binding patterns between conditions or time points and the variation in chromatin preparations is small.
Depth of sequencing
One critical difference between ChIP-chip and ChIP-Seq is that the number of tiling arrays used in a ChIP-chip experiment is the same regardless of the protein or modification of interest, whereas the number of fragments to be sequenced in ChIP-Seq is determined by the investigator. In published ChIP-Seq experiments, a single lane of the Illumina Genome Analyzer was the basic unit of sequencing. Initially, a single lane generated 4–6 million reads prior to alignment but now a lane generates 8–15 million or more. Given the cost of each experiment, many early data sets contained reads from a single lane regardless of what the specific experiment was. Intuitively, when a large number of binding sites are present in the genome for a DNA-binding protein or when a histone modification covers a large fraction of the genome, a correspondingly large number of tags are needed to cover each bound region at the same tag density. One reasonable criterion for determining sufficient sequencing depth would be that the results of a given analysis do not change when more reads are obtained. In terms of the number of binding sites, this criterion translates to the presence of a ‘saturation’ point after which no further binding sites are discovered with additional reads.
The issue of saturation points has been examined in a recent manuscript through simulation studies [48]. In three example data sets, a reference set of sites was generated based on the full set of sequencing reads in each case. Then, a wide range of different read counts was sampled (with multiple random selections for each sample size) from the complete data set, binding sites were determined for each sample with a threshold p-value, and the results for each sample size averaged. The fraction of the reference set recovered as a function of number of reads is shown in Figure 3A. If there were a saturation point, the number of sites found would increase up to a certain point and then plateau, signifying that the rate at which new sites are discovered has slowed down sufficiently to make any further increase in the number of reads inefficient. When the simulation was performed, however, the results indicated that more and more sites continued to be found at a steady pace with additional sequencing (the lower curve). In another study [38], human RNA polymerase II targets were shown to saturate quickly while for the transcription factor STAT1 the number targets continued to rise steadily. This suggests that, at least in some cases, there may not be a saturation point that can be used to determine the number of tags to be sequenced if peaks are found based on statistical significance.
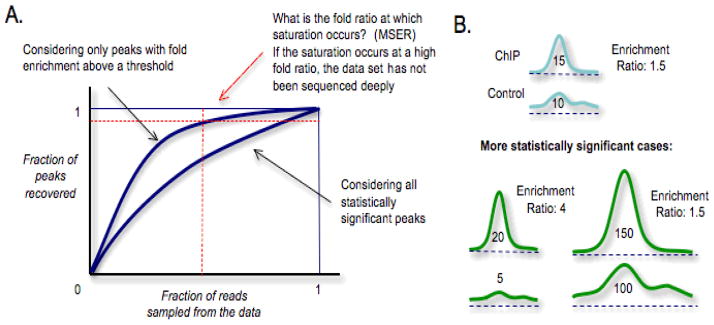
Figure 3. Depth of Sequencing.
(A) To determine whether enough tags have been sequenced, simulation can be carried out to characterize the fraction of the peaks that would be recovered if a smaller number of tags had been sequenced. In many cases, new statistically significant peaks are discovered at a steady rate with an increasing number of tags (solid curve), i.e., there is no saturation of binding sites. However, when a minimum threshold is imposed for the enrichment ratio between ChIP and input DNA peaks, the rate at which new peaks are discovered slows down (dashed curve). That is, saturation of detected binding sites can occur when sufficiently prominent binding positions are considered. For a given data set, multiple curves corresponding to different thresholds can be examined to identify the threshold at which the curve becomes sufficiently flat to meet the desired saturation criteria (upper right box defined by the red lines). We refer to such threshold as the Minimum Saturation Enrichment Ratio (MSER). MSER can serve as a measure for the depth of sequencing achieved in a data set: A high MSER, for example, indicates that the data set may be under-sampled, as only the more prominent peaks were saturated. See REF Kharchenko et al for details. (B) There are two ways in which a peak can be more statistically significant than another (lower panels compared to upper panels): higher enrichment ratio in ChIP compared to control for the same number of tags (shown under the curve in each case) (lower left) or the same enrichment ratio but a larger number of tag counts (lower right). As the latter case illustrates, there may not be saturation of binding sites when more sequencing leads to less prominent peaks becoming more statistically significant.
A saturation point does exist, however, if a fixed threshold is imposed on the fold enrichment between the peaks in the ChIP experiment and the control experiment. That is, when only prominent peaks (as defined by minimum fold enrichment) are considered, saturation is likely to occur. When all peaks are considered, even peaks with small enrichment can become statistically significant as more tags are accumulated, as illustrated in Figure 3B. This is similar to what happens in genome-wide association studies and other genomic investigations where a large sample size increases the statistical power and causes features of small effect sizes to attain statistical significance. The authors [48] proposed that each ChIP-Seq data set can be annotated with a Minimal Saturated Enrichment Ratio (MSER) at which saturation occurs to give a sense of the sequencing depth achieved. They also found that there is a linear relationship between the number of reads and MSER when properly scaled. This makes it possible to predict how many more reads are needed when a particular level of MSER is desired. While these concepts and tools should be tested on more data sets, they provide a framework for understanding depth-of-sequencing issues in ChIP-Seq experiments.
Multiplexing
For small genomes, including S. cerevisiae, C. elegans and D. melanogaster, the number of reads generated in a sequencing unit (for example, one of eight lanes on an Illumina Genome Analyzer) may be several times greater than the number of reads needed to provide sufficient coverage of the genome at a suitable depth for the ChIP-Seq experiment. As the number of reads per run continues to increase, the ability to sequence multiple samples at the same time (referred to as ‘multiplexing’) becomes important for cost-effectiveness. In theory, multiplexing of samples is not difficult, only requiring different barcode adaptors to be ligated to different samples during sample preparation. Even allowing for sequencing error, a few bases are sufficient to serve as unique identifiers for many samples. In practice, however, multiplexing has not been widely used so far on the Illumina platform, due to uneven coverage of the samples and other technical problems. Some recent protocols, however, show promise [49] and multiplexing is likely to be utilized frequently in the future.
Paired-ends sequencing
The ChIP fragments are generally sequenced at the 5′ ends, but they can also sequenced at both ends, as is frequently done for detection of structural variations in the genome [19]. Paired-ends sequencing can be used in conjunction with ChIP for additional specificity in mapping, especially to repetitive regions, and to map long-range chromatin interactions [50].
Number of replicates
Replicate experiments are needed for ensuring reproducibility of the data. For microarrays, platforms and protocols have improved substantially so that technical replicates of the same samples are generally not done anymore. While this is likely to be the case for ChIP-Seq [51], biological replicates are still strongly recommended to account for variation between samples and to verify the fidelity of experimental steps. Assuming that they are sequenced deeply, two concordant replicates would generally be sufficient, as a third replicate appears to add little value [38].
Challenges in Data Analysis
As NGS platforms and ChIP-Seq protocols mature, data generation is gradually becoming routine and the limiting factor in a study is shifting to computational analysis of the data and validation experiments. In this section, we discuss the key issues and concepts involved in data analysis. These will serve as a basis for a full range of ChIP-Seq analyses, which are too varied and complex to be discussed in this review. A flowchart of the steps involved in ChIP-Seq analysis is shown in Figure 4.
Figure 4. Overview of ChIP-Seq analysis.
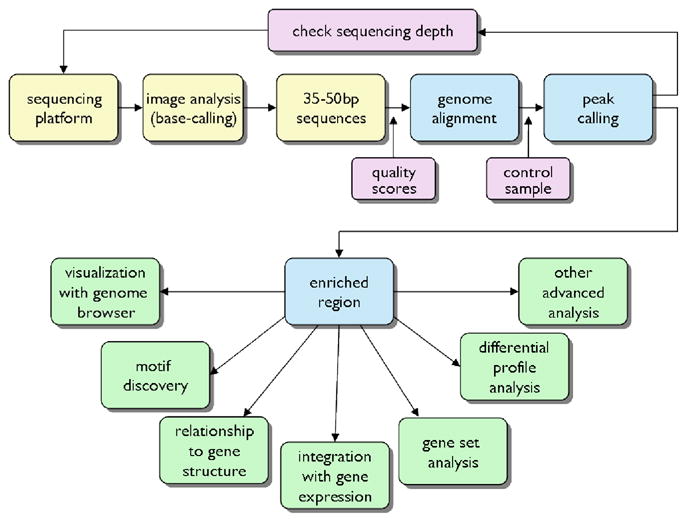
The raw data for ChIP-Seq analysis are images from the next generation sequencing platform (top left). A base-caller converts the image data to sequence tags, which are then aligned to the genome, on some platforms with the aid of quality scores that indicate the reliability of each base call. Peak calling using the ChIP and a control profile (usually input DNA) are used to generate a list of enriched regions ordered by false discovery rate as a statistical measure. Subsequently, the profiles of enriched regions are viewed with a browser and a variety of advanced analyses are performed.
Data management
NGS produces an unprecedented amount of data. Raw data and images are on the order of terabytes per machine run, making data storage a challenge even for facilities with considerable expertise in management of genomic data. Data can be stored at three levels: image data, sequence tags, and alignment data. Ideally, one would like to keep the raw data so that if a new base-caller is developed, one can re-process the raw data. Sequence tags can be used to map the data when an improved aligner is available or when a reference genome assembly is updated. Alignment data can be useful for generating summary statistics and can be used to generate SNP or copy number variation calls. There is no consensus in the community regarding which data type must be stored, but many feel that the image data are too expensive to maintain and that a reasonable approach at this point is to discard the raw data after a short period of time and keep only the sequence-level data.
In microarray-based studies, investigators are encouraged, and often required, to submit their data upon publication to a public database such as Gene Expression Omnibus [52]. For NGS data, data transfer and maintenance are more complicated due to the large file sizes. Depositing data via standard ftp or http protocols, for instance, is likely to fail when many gigabytes are to be uploaded. To meet this challenge, National Center for Biotechnology Information (NCBI) in the US, the European Bioinformatics Institute and the DNA Databank of Japan havedeveloped the Sequence Read Archive (SRA) [53, 54]. Meta-data describing all experimental details should be submitted at the same time for the data in the repositories to be useful to the community.
Genome Alignment
Image processing and base-calling are platform-specific and are mostly done using the software provided by the manufacturer, although some new base callers have been proposed recently [55, 56] for the Illumina platform. More important is the choice of strategy for [54]genome alignment, as all subsequent results are based on the aligned reads. Due to the large number of reads, the use of conventional alignment algorithms can take hundreds or thousands of processor hours. Thus, a new generation of aligners has been developed recently [57] and more are expected soon. Every aligner is a balance between accuracy, speed, memory, and flexibility, and no aligner can be best suited for all applications. Alignment for ChIP-Seq should allow for a small number of mismatches due to sequencing errors, single nucleotide polymorphism (SNPs) and indels, or the difference between the genome of interest and the reference genome. This is simpler than in RNA-seq, for example, where large gaps corresponding to introns must be considered. Currently, popular aligners include: Eland, an efficient and fast aligner for short reads that was developed by Illumina and is the default on that platform; MAQ [58], a widely-used aligner with a more exhaustive algorithm and excellent capabilities for detecting SNPs; and Bowtie [59], an extremely fast mapper based on an algorithm originally developed for file compression. These methods utilize the quality score that accompanies each base call to indicate its reliability. For the SOLiD dibase sequencing technology, in which two consecutive bases are read at a time, modified aligners have been developed [60, 61]. Many current analysis pipelines discard non-unique tags, but studies involving the repetitive regions of the genome [27, 62–64] require careful handling of these non-unique tags.
Identification of enriched regions
After sequenced reads are aligned to the genome, the next step is to identify regions that are enriched in the sample relative to the control with statistical significance. Several ‘peak callers’ that scan along the genome to identify the enriched regions are currently available [24, 26, 38, 48, 65–70]. In early algorithms, regions were scored firstly by the number of tags in a window of given size, and then assessed by a set of criteria on such factors as enrichment over the control and minimum tag density. Subsequent algorithms take advantage of the directionality of the reads [71]. As illustrated in Figure 5, the fragments are sequenced at the 5′ end, and the locations of mapped reads should form two distributions, one on the positive strand and the other on the negative strand, with a consistent distance between the peaks of the distributions. In these methods, a smoothed profile on each strand is first constructed [65, 72] and then the combined profile is calculated, either by shifting each distribution toward the center or by extending each mapped position into a ‘fragment’ with appropriate orientation and then summing the fragments. The latter approach should result in a more accurate profile with respect to the width of the binding, but it requires an estimate of the fragment size as well as the assumption that fragment size is uniform.
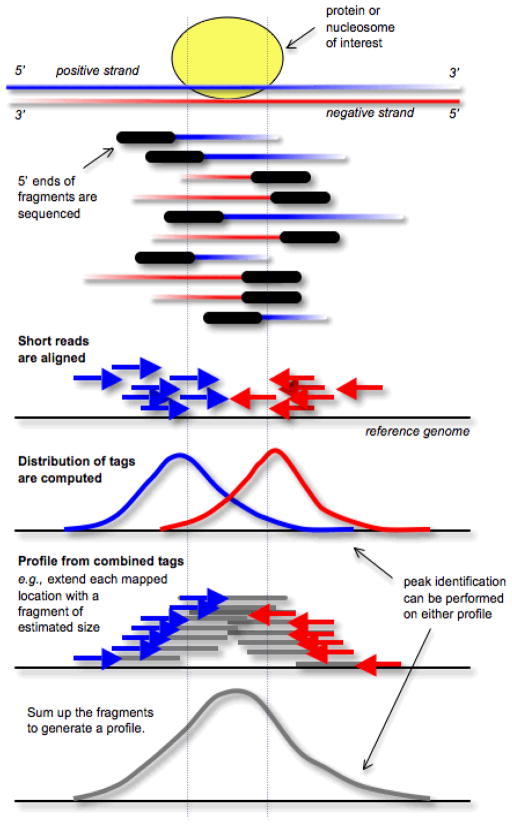
Figure 5. Strand-specific profile at enriched sites.
DNA fragments from a chromatin immunoprecipitation experiment are sequenced from the 5′ end. Thus, alignment of these tags to the genome results in two peaks, one on each strand, flanking the location where the protein or nucleosome of interest was bound. This strand-specific pattern can be used for optimal detection of enriched regions. To approximate the distribution of all fragments, each tag location can be extended by an estimated fragment size in the appropriate orientation and the number of fragments is counted
Given a combined profile, peaks can be scored in a number of ways. A simple fold ratio of the signal for the ChIP sample relative to that of the control sample around the peak (Figure 3B) provides important information, but it is not adequate. A fold ratio of 5 estimated from 50 and 10 tags (ChIP/control) has a different statistical significance from the same ratio estimated from 500 and 100 tags, for example. A Poisson model for the tag distribution is an effective approach that accounts for the ratio as well as the absolute tag numbers [27], especially with a correction for regional bias in tag density due to chromatin structure, copy number variation, or amplification bias [67]. A binomial distribution or other models can also be used [38]. In another approach, the peaks are scored before a combined profile is generated, by considering how well the tag distributions on the two strands resemble each other and whether the distance between the peaks is close to the expected number of base pairs [48]. Another important local correction, regardless of the peak detection method, is to adjust for sequence alignability. Depending on how the non-uniquely mapped reads are processed, regions of the genome containing repetitive elements will have a different expected tag count. By keeping track of how many times each k-mer along a segment appears in the rest of the genome, one can correct for the variation in mappability among segments [27, 38].
A major difficulty in identification of enriched regions is that there are three types: sharp, broad, and mixed (Figure 2B). Sharp peaks are generally found for protein-DNA binding or histone modifications at regulatory elements, whereas broad regions are often associated with histone modifications marking domains, for example, transcribed or repressed regions. Most current algorithms have been designed for sharp peaks, with coalescing of adjacent peaks post hoc for broad regions, but many techniques from ChIP-chip and DNA copy number analysis [73] will soon be modified for ChIP-seq as well as new ones developed [74, 75]. A powerful method would incorporate elements of both types of methods and apply a technique appropriate for the features found without knowing the type of enrichment a priori.
The performance of a peak caller can be tested by validating a large set of sites via qPCR or by computing the distribution of distances from each peak to a nearby known protein-binding sequence motif. While a careful comparison of the algorithms is still being carried out, it is clear that the best methods should at least take advantage of the strand-specific pattern expected at a binding location and adjust for local variation as measured by input DNA and, to a lesser extent, correct for sequence alignability. Statistical significance of enriched sites is generally measured by false discovery rate (FDR) [76, 77], which is the expected proportion of incorrectly identified sites among those found to be significant. Determining significance for a multitude of features in the data results in a ‘multiple hypothesis problem,’ in which features that appear to be significant arise simply due to the large number of features being considered. The q-value of a peak is the minimum FDR at which the peak is deemed significant and is analogous of the p-value in a single hypothesis test setting. As in analysis of other genomic data types, it is important to note that the accuracy of statistical significance computed in these algorithms depends on how realistic the underlying null distribution is. For ChIP-Seq, an FDR derived from a null distribution based on randomization of ChIP reads can be off by an order of magnitude [48], because tags in the same or neighboring positions are not completely independent even without true binding, as can be seen in the input control profile.
Downstream analysis
For protein-DNA binding, the most common follow-up analysis is discovery of binding sequence motifs [78]. The sequences of the top-scoring sites can be entered into motif-finding algorithm programs such as MEME [79], MDScan [80], Weeder [81] and TAMO [82], and potential motifs are returned along with their statistical significance. In some cases, a single motif clearly stands out with much higher statistical significance than the subsequent matches and is largely insensitive to the number of the sites used to search. In other cases, there is a series of motifs with gradual decrease in statistical significance, and further analysis on combinatorial occurrences of the motifs may be informative in identifying cooperative interactions among transcription factors or other more complex relationships among the motifs. The process of computing statistical significance is not straightforward and algorithms that are available use different null models and multiple-testing adjustment; thus it is important to validate functionally any motifs that are found. While ChIP-chip has been used successfully in numerous occasions for motif discovery, analysis of the distances between ChIP-Seq peaks and the nearby motifs clearly demonstrate that ChIP-Seq data are superior for this application [48, 65]. For some factors, most of the ChIP-Seq peaks are within 10–30bp of the known motif [48]. After a motif is found, searching for the sequence in the genome generally reveals that there are many more sites with the motif than those identified by ChIP-Seq. Why some occurrences of a motif are functional and others are not is at least partially related to presence or absence of nucleosomes or a specific histone modification; this can be explored with nucleosome profiles obtained by sequencing [29, 37].
Another basic analysis that can be performed using ChIP-Seq data is to annotate the location of the peaks on the genome in relation to known genomic features, such as the transcriptional start site (TSS), exon/intron boundaries, and the 3′ ends of genes. TSS of active genes, for instance, are known to be enriched with histone H3 trimethylated at lysine 4 (H3K4me3) and while enhancers are enriched with lysine 4 monomethylation (H3K4me1) [25, 83]. It is generally informative to view this type of data both in absolute and relative scale, for example, by rescaling all genes to have the same length so that the average profile over the gene body can be viewed. To find relationships between the profiles, correlation analysis can be performed, as well as more advanced clustering methods [84]. ChIP-chip and ChIP-Seq data from the same experiments are generally comparable but have subtle differences; thus, combining both platforms requires careful attention, especially to the amount of smoothing applied to profiles. Incorporating other data types into the analysis is alsonecessary for biological interpretation. Classifying the ChIP-Seq patterns by their relationship to expression data, for example, is an important first step. Expression levels correlated with the binding status of a transcriptional activator would indicate that the gene may be a target of the activator; a chromatin mark with enrichment at the promoter of genes with high expression can be inferred to be related to transcriptional activation. For a group of genes with a common feature - for example, binding of the same transcription factor or presence of the same modification - Gene Ontology analysis [85] can be performed to see whether a particular molecular function or biological process is over-represented in those genes [86]. More advanced analysis includes discovery of novel elements based on ChIP-Seq data. For example, the location of H3K4me3 and H3K36me3, which are known to be found at promoters and over transcribed regions, respectively, can be used to identify large non-coding RNAs [87]. Combined with SNP information, ChIP-Seq data can also be used to investigate allele-specific binding and modification [27].
Available software
Many of the algorithms for alignment and peak detection discussed earlier are accompanied by software. Some are available as a plug-in package for the statistical language R, a powerful system for data analysis that is popular among bioinformaticians [88], while others are based on standard compiled languages such as C/C++. Most programs generate a list of enriched sites as well as the binding profile to be viewed on a genome browser. One program with a menu-driven user interface is CisGenome [69], featuring a ChIP-chip and ChIP-Seq analysis pipeline with support for interactive analysis and visualization. More user-friendly software tools designed for biologists will be developed in the future, but it is unlikely that tools available in a single software package will meet all analysis needs. This is particularly the case when the experimental design is more complicated or advanced analysis involving integration of other data types is needed. Thus, as is in most genomics projects, it is imperative to have a bioinformatics expert as a member of the research team.
Conclusion and future directions
ChIP has become a principal tool for understanding transcriptional cascades and for deciphering information encoded in chromatin. With the remarkable progress in high-throughput sequencing platforms in the recent years, ChIP-Seq is poised to become the dominant approach in the coming years. The cost of sequencing and lack of easy access to platforms are still the limiting factors for most investigators, but the situation is expected to improve in the near future. ChIP-Seq already offers higher-resolution and cleaner data at lower cost than the array-based alternatives for genome-wide profiling of large genomes. Improved spatial resolution has already resulted in significant progress in several areas, most notably in genome-wide characterization of chromatin modifications at the nucleosome level and in accurate identification of DNA sequence elements involved in transcriptional regulation. Enhanced sequencing capabilities in the future will allow profiling of a large number of DNA-binding proteins as well as a more complete set of chromatin marks in a myriad of epigenomes across multiple tissues, cell types, conditions, and developmental stages. The human Encyclopedia of DNA Elements (ENCODE) [89], the model organism ENCODE [41], and the NIH Epigenome Roadmap Program are a first step in large-scale profiling, and lessons from these projects will spur more detailed characterizations in specific systems. To extract most information from ChIP-Seq data, integrative analysis with other data types will be essential. Integrated with RNA-Seq data, for example, ChIP-Seq data may result in elucidation of gene regulatory networks and characterization of the interplay between the transcriptome and the epigenome. Experimental challenges for the future include careful validation of antibodies, development of methods for working with a small number of cells, and single-cell level characterization. Even greater challenges for many laboratories are likely to be effective management and analysis of the immense amount of sequencing data. This will require development of user-friendly and robust software tools for data analysis and closer interaction between experimentalists and bioinformaticians.
Acknowledgments
I thank P. Kharchenko, M. Tolstorukov, A. Alekseyenko and other members of the Park and the Kuroda laboratories for their insights. I gratefully acknowledge support from the National Institutes of Health grants R01GM082798, U01HG004258 and RL1DE019021.
Footnotes
Weblinks
http://seqanswers.com A community forum for discussion of issues related to next-generation sequencing
References
- 1.Farnham PJ. Insights from genomic profiling of transcription factors. Nat Rev Genet. 2009 doi: 10.1038/nrg2636. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10(3):161–72. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9(1):15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 4.Li B, et al. The role of chromatin during transcription. Cell. 2007;128(4):707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Allis CD, et al., editors. Epigenetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2007. [Google Scholar]
- 6.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein BE, et al. The mammalian epigenome. Cell. 2007;128(4):669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 8.Solomon MJ, et al. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53(6):937–47. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 9.Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98(2):249–59. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 10.Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290(5500):2306–9. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 11.Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16(6):545–52. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–45. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 13.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 14.Hillier LW, et al. Whole-genome sequencing and variant discovery in C. elegans. Nat Methods. 2008;5(2):183–8. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 15.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456(7218):66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JB, et al. Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science. 2007;316(5830):1481–4. doi: 10.1126/science.1137325. [DOI] [PubMed] [Google Scholar]
- 17.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320(5881):1344–9. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453(7199):1239–43. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 19.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318(5849):420–6. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132(2):311–22. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maher CA, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458(7234):97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau NC, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313(5785):363–7. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 23.Branton D, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26(10):1146–53. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson DS, et al. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316(5830):1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 25.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4(8):651–7. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 27.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457(7231):854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robertson AG, et al. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 2008;18(12):1906–17. doi: 10.1101/gr.078519.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132(5):887–98. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tolstorukov MY, et al. Comparative analysis of H2A.Z nucleosome organization in the human and yeast genomes. Genome Res. 2009 doi: 10.1101/gr.084830.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henikoff S, et al. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19(3):460–9. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem Sci. 2000;25(3):99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- 34.O’Neill LP, Turner BM. Immunoprecipitation of native chromatin: NChIP. Methods. 2003;31(1):76–82. doi: 10.1016/s1046-2023(03)00090-2. [DOI] [PubMed] [Google Scholar]
- 35.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9(3):179–91. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim TH, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436(7052):876–80. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alekseyenko AA, et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134(4):599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rozowsky J, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol. 2009;27(1):66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quail MA, et al. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5(12):1005–10. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whiteford N, et al. An analysis of the feasibility of short read sequencing. Nucleic Acids Res. 2005;33(19):e171. doi: 10.1093/nar/gni170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Celniker SE, et al. Unlocking the secrets of the genome. Nature. 2009;459(7249):927–30. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Acevedo LG, et al. Genome-scale ChIP-chip analysis using 10,000 human cells. Biotechniques. 2007;43(6):791–7. doi: 10.2144/000112625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dahl JA, Collas P. MicroChIP--a rapid micro chromatin immunoprecipitation assay for small cell samples and biopsies. Nucleic Acids Res. 2008;36(3):e15. doi: 10.1093/nar/gkm1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu AR, et al. Automated microfluidic chromatin immunoprecipitation from 2,000 cells. Lab Chip. 2009;9(10):1365–70. doi: 10.1039/b819648f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Neill LP, et al. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat Genet. 2006;38(7):835–41. doi: 10.1038/ng1820. [DOI] [PubMed] [Google Scholar]
- 46.Harris TD, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320(5872):106–9. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 47.Peng S, et al. Normalization and experimental design for ChIP-chip data. BMC Bioinformatics. 2007;8:219. doi: 10.1186/1471-2105-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kharchenko PV, et al. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26(12):1351–9. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lefrancois P, et al. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics. 2009;10:37. doi: 10.1186/1471-2164-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fullwood MJ, et al. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009;19(4):521–32. doi: 10.1101/gr.074906.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marioni JC, et al. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18(9):1509–17. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barrett T, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37(Database issue):D885–90. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sayers EW, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37(Database issue):D5–15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cochrane G, et al. Petabyte-scale innovations at the European Nucleotide Archive. Nucleic Acids Res. 2009;37(Database issue):D19–25. doi: 10.1093/nar/gkn765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erlich Y, et al. Alta-Cyclic: a self-optimizing base caller for next-generation sequencing. Nat Methods. 2008;5(8):679–82. doi: 10.1038/nmeth.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rougemont J, et al. Probabilistic base calling of Solexa sequencing data. BMC Bioinformatics. 2008;9:431. doi: 10.1186/1471-2105-9-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trapnell C, Salzberg SL. How to map billions of short reads onto genomes. Nat Biotechnol. 2009;27(5):455–7. doi: 10.1038/nbt0509-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, et al. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18(11):1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ondov BD, et al. Efficient mapping of Applied Biosystems SOLiD sequence data to a reference genome for functional genomic applications. Bioinformatics. 2008;24(23):2776–7. doi: 10.1093/bioinformatics/btn512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rumble SM, et al. SHRiMP: accurate mapping of short color-space reads. PLoS Comput Biol. 2009;5(5):e1000386. doi: 10.1371/journal.pcbi.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bourque G, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18(11):1752–62. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pauler FM, et al. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res. 2009;19(2):221–33. doi: 10.1101/gr.080861.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng D. Asymmetric histone modifications between the original and derived loci of human segmental duplications. Genome Biol. 2008;9(7):R105. doi: 10.1186/gb-2008-9-7-r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Valouev A, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5(9):829–34. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fejes AP, et al. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24(15):1729–30. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jothi R, et al. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36(16):5221–31. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ji H, et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26(11):1293–300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nix DA, et al. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics. 2008;9:523. doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmid CD, Bucher P. ChIP-Seq data reveal nucleosome architecture of human promoters. Cell. 2007;131(5):831–2. doi: 10.1016/j.cell.2007.11.017. author reply 832–3. [DOI] [PubMed] [Google Scholar]
- 72.Boyle AP, et al. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24(21):2537–8. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lai WR, et al. Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics. 2005;21(19):3763–70. doi: 10.1093/bioinformatics/bti611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu H, et al. An HMM approach to genome-wide identification of differential histone modification sites from ChIP-seq data. Bioinformatics. 2008;24(20):2344–9. doi: 10.1093/bioinformatics/btn402. [DOI] [PubMed] [Google Scholar]
- 75.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25(15):1952–8. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 77.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tompa M, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol. 2005;23(1):137–44. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 79.Bailey TL, et al. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34(Web Server issue):W369–73. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu XS, et al. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol. 2002;20(8):835–9. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- 81.Pavesi G, et al. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32(Web Server issue):W199–203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gordon DB, et al. TAMO: a flexible, object-oriented framework for analyzing transcriptional regulation using DNA-sequence motifs. Bioinformatics. 2005;21(14):3164–5. doi: 10.1093/bioinformatics/bti481. [DOI] [PubMed] [Google Scholar]
- 83.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 84.Hon G, et al. ChromaSig: a probabilistic approach to finding common chromatin signatures in the human genome. PLoS Comput Biol. 2008;4(10):e1000201. doi: 10.1371/journal.pcbi.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Orford K, et al. Differential H3K4 methylation identifies developmentally poised hematopoietic genes. Dev Cell. 2008;14(5):798–809. doi: 10.1016/j.devcel.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120(2):169–81. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 92.Kurdistani SK, et al. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117(6):721–33. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 93.Liu CL, et al. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3(10):e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pokholok DK, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122(4):517–27. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 95.Lee CK, et al. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36(8):900–5. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 96.Yuan GC, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309(5734):626–30. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 97.Lee W, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39(10):1235–44. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 98.Johnson SM, et al. Flexibility and constraint in the nucleosome core landscape of Caenorhabditis elegans chromatin. Genome Res. 2006;16(12):1505–16. doi: 10.1101/gr.5560806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Albert I, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446(7135):572–6. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 100.Mavrich TN, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453(7193):358–62. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]