Abstract
Microcalcifications are an early mammographic sign of breast cancer and a target for stereotactic breast needle biopsy. We present here a Raman spectroscopic tool for detecting microcalcifications in breast tissue based on their chemical composition. We collected ex vivo Raman spectra from 159 tissue sites in fresh stereotactic breast needle biopsies from 33 patients, including 54 normal sites, 75 lesions with microcalcifications and 30 lesions without microcalcifications. Application of our Raman technique resulted in a positive predictive value of 97% for detecting microcalcifications. This study shows that Raman spectroscopy has the potential to detect microcalcifications during stereotactic breast core biopsies and provide real-time feedback to radiologists, thus reducing non-diagnostic and false negative biopsies.
OCIS Codes: (170.1610), Clinical applications; (170.6935), Tissue characterization; (300.6450) Spectroscopy, Raman
1. Introduction
Breast cancer is the second leading cause of cancer death in women, with one in eight women likely to develop breast cancer in her lifetime. In the United States alone 254,650 new cases of breast cancer occurred in 2010 [1]. The exact cause of breast cancer is still under debate although breast cancer surgery and treatment have been extensively studied. However, the most effective approach for preventing morbidity and mortality is early detection. X-ray mammography is currently the only accepted routine screening method for early detection of breast cancer [2]. Microcalcifications geographically target the most clinically significant abnormality within the breast and are considered an early mammographic sign of breast cancer [3]. They are currently graded radiographically as typically benign, of intermediate concern or of higher probability of malignancy, based on their size, shape, number, roughness and degree of clustering [4]. Microcalcifications can also be divided into two types based on their chemical composition and histopathologic appearance in breast biopsy tissue: type I microcalcifications composed of calcium oxalate (CAO) crystals; and type II microcalcifications composed of calcium phosphate concretions, mainly calcium hydroxyapatite (CHA). Microcalcification type also correlates with disease state [5]. Type I microcalcifications are seen almost exclusively in benign duct cysts and are rarely found in foci of breast cancer, while type II microcalcifications are most often seen in proliferative lesions, including invasive and in situ cancer. While microcalcifications can be detected by x-ray mammography, this procedure cannot distinguish type I and II microcalcifications based on their chemical composition. Thus, a tissue biopsy must be performed to determine whether the microcalcifications are associated with cancer.
Currently, most patients undergo vacuum-assisted stereotactic core needle biopsy for microcalcifications. Despite stereotactic guidance, recent studies have documented the failure to retrieve microcalcifications in up to 15% of these biopsies [6]. Failure to retrieve the microcalcifications results in non-diagnostic or false negative biopsies, requiring the patient to undergo repeat biopsy, often as a surgical procedure. Therefore, there is a clinical need for a tool that can detect microcalcifications in the tissue to be biopsied and provide real-time feedback to the radiologist during stereotactic biopsies as to whether the microcalcifications seen on mammography will be retrieved.
As a non-destructive, chemical-specific technique, Raman spectroscopy is particularly amenable to in vivo studies and an ideal choice for detecting microcalcifications during breast core needle biopsies. In this process, incident photons transfer energy to and from the molecular vibrational and/or rotational modes of chemical species in the tissue, giving rise to shifts in the energy of the emitted photons. Since the energy level is unique for each such vibrational/rotational mode of the molecule, these Raman shifts provide fingerprint details on the chemical composition of the tissue.
Raman spectroscopy has been successfully applied to the study of calcifications in various organ systems, including kidney and other urinary tract stones [7], atherosclerotic plaque [8–10] and calcified heart valves [11,12]. There are several groups currently studying breast microcalcifications using Raman spectroscopy [13–16]. Previously we have explored Raman spectroscopy [17–22] and other minimally invasive spectroscopic optical techniques for the diagnosis of breast cancer [23]. Our group was the first to demonstrate the potential of Raman spectroscopy to detect and distinguish type I and II microcalcifications and to differentiate type II calcifications associated with benign and malignant breast lesions [13], in Raman microscopy studies of formalin-fixed, paraffin-embedded breast biopsies. Baker et al in 2010 confirmed these findings in studies of breast microcalcifications using FTIR spectroscopy [16].
In this work, we present for the first time ex vivo studies for real-time detection of microcalcifications in core needle breast biopsy specimens using Raman spectroscopy. Raman spectra were collected from freshly excised tissue from 33 patients undergoing stereotactic core needle breast biopsy procedures and fit to our previously developed model of breast chemical and morphological components to determine quantitatively the contribution of each component to the measured Raman spectrum. We then developed algorithms to detect the presence of microcalcifications in the breast tissue based on model fit coefficients (FC) for total calcium (CAH plus CAO), collagen and fat. In addition to detecting microcalcifications, this technique could also be used to distinguish type I from type II microcalcifications. This technique has the potential to detect and classify microcalcifications, even smaller than those detected by radiology, in real-time, and thus to guide core needle biopsy procedures with the goal of reducing non-diagnostic and false negative biopsies.
2. Instrument
The portable, compact clinical Raman spectroscopy system (instrument and optical fiber probes) used in these studies was developed at the Laser Biomedical Research Center at the Massachusetts Institute of Technology (MIT). The instrument delivers excitation light from an 830 nm InGaAs diode laser (Process Instruments, Salt Lake City, UT) to the tissue via an optical fiber probe, which is in the form of a flexible catheter with a total length of about 4m and diameter of approximately 2 mm, previously described in detail [24]. The probe consists of a single excitation fiber surrounded by nine collection fibers, each with a diameter of 200 µm. The excitation fiber terminates in a short-pass filter centered at 830 nm which transmits the laser excitation light while blocking the fiber background (i.e. the Raman and fluorescence signal generated by the excitation fiber). The fiber is gently brought into contact with the tissue in a light tight black box while collecting data. The Raman signal is collected using the nine collection fibers which terminate in a long pass filter to suppress the elastically scattered light emanating from the tissue. This filter transmits the tissue Raman scattered light while effectively rejecting any undesirable fiber background that may have been caused by the high intensity elastic scattering component. The distal end of the probe has a sapphire ball lens which focuses the excitation light onto the tissue and efficiently collects the scattered light from the tissue into the collection fibers. The linear array of collection fibers at the proximal end of the fiber is coupled to a spectrograph for dispersion onto a back illuminated, deep depletion, thermoelectrically-cooled CCD detector (PIXIS 256, 26x26μm pixels, 1024x256 array, Princeton Instruments). Laser power was 98-105 mW. The Raman spectra were acquired by vertical binning prior to averaging 10 successive frames, each with an acquisition time of 0.25 s, for a total collection time of 2.5 s. The data is analyzed in real-time, with the fitting algorithm taking another 1.8 s to generate the FC and render a diagnosis using a FC-based decision algorithm. The whole process is automated and takes less than 5 s per measurement point.
3. Data analysis
The instrument was calibrated daily prior to data collection. For wavenumber calibration, spectra from 4-acetamidophenol (Tylenol) with known Raman peak positions are collected. To correct the Raman spectra for the system wavelength response, light is collected from a wavelength-calibrated tungsten halogen lamp diffusely scattered from a reflectance standard, barium sulfate (BaSO4). To get rid of the probe background, spectra are collected from a roughened aluminum surface. Different weightings of the aluminum spectra are subtracted, choosing the optimal one depending on the tissue optical properties. To remove background fluorescence, the data is fitted with a sixth order polynomial and subtracted from the raw data. The software stores these calibration spectra, which allows real-time data analysis and display of the calibrated Raman spectra and model fits and, thus, assessment of the quality of the Raman spectroscopic data acquired. This allows Raman data to be re-collected from the same spot if, for example, the CCD is saturated as it was on initial spectral acquisition for 6 tissue sites. In these instances, the data was collected again by averaging 20 frames, each with an acquisition time of 0.125 sec, maintaining a total collection time of 2.5 sec.
Once the Raman spectra are acquired, they are fit with a breast model we developed [18], in which the Raman tissue spectrum is considered a linear combination of its constituent basis spectra, Pi, and their concentrations, Ci, at wavenumber ω (Eq. (1)):
| (1) |
The model includes basis spectra for epithelial cell nuclei, epithelial cell cytoplasm, fat cells, cholesterol-like extracellular deposits, beta-carotene, stromal collagen fibers, type I microcalcifications (calcium oxalate) and type II microcalcifications (largely calcium hydroxyapatite).
Ordinary least squares fitting is used to find the contribution of each basis spectrum to the observed spectrum by minimizing the sum of squared residuals, where the residual represents the difference between the acquired spectrum and the fitted value provided by our model. The main advantage of this process is that it yields FC that correlate with the presence or absence of spectral contributions from the basis spectra, providing information about the morphological and chemical composition of the tissue. The goodness of the fit is qualitatively estimated from visual inspection of the residual after fit and quantitatively from the standard deviation of the residual. Specifically, a Raman spectrum is eliminated from analysis if the standard deviation of the residual is greater than 0.2 (as this results in a poor model fit) or if it is determined to be an outlier using a Student's t-test assessment of the Mahalanobis distance(s) as detailed in the Results section.
4. Patient population and tissue preparation
Breast tissue was obtained from 33 patients (all female; ages 38-79) undergoing vacuum-assisted stereotactic core needle breast biopsy procedures in the Breast Health Center at University Hospitals-Case Medical Center. All studies were approved by the Case Cancer Institutional Review Board and the Massachusetts Institute of Technology Committee On the Use of Humans as Experimental Subjects. Informed consent was obtained from all subjects prior to their biopsy procedures.
All data were collected from freshly excised biopsy specimens in a room just outside the biopsy suite within 30 minutes of excision. The tissue cores collected during the needle biopsy procedure were placed inside a Petri dish moistened with normal saline and were typically of approximate dimension 1.7mm × 2.0 cm, due to the geometry of the needle. A tissue core of interest was then selected by identifying a focus of microcalcifications within the core on the specimen radiograph. The selected core was then placed on an Aluminum block inside a light-tight black box for data collection. Spectra were collected from several tissue sites of interest on each core (typically a focus of microcalcifications, a lesion (grossly abnormal tissue) without microcalcifications and normal tissue) identified again by comparison with the specimen radiograph. Spectra were also collected from different cores in each biopsy, so the number of spectra varied from patient to patient. Real time data analysis was used to help confirm data collection from tissue sites with microcalcifications. After spectral acquisition, the sites interrogated spectroscopically on the tissue cores were marked with multicolored colloidal inks to uniquely identify each tissue site. The tissue was then fixed in 10% neutral buffered formalin, routinely processed, paraffin embedded, cut into tissue sections and stained with hematoxylin and eosin (H&E). The tissue cores were oriented at embedding and the cut tissue sections mounted on the glass slides so that the colored ink spots marking the sites interrogated spectroscopically were visible on microscopic examination, for comparison of histopathology and spectroscopy results. A specimen radiograph of a biopsy tissue core with microcalcifications and corresponding histopathology photomicrograph illustrating the overlying colored ink spot are shown in Fig. 1 . The H&E stained tissue sections are next examined by an experienced breast pathologist. Both the radiographic appearance based on the specimen radiograph and the histopathology diagnosis were considered as gold standards for comparison to the spectroscopic results. Breast lesions were classified as having microcalcifications in two ways. First, lesions were classified based on the radiographic appearance alone, if microcalcifications were seen at that tissue site on the specimen radiograph, as these are the lesions targeted by the radiologist for biopsy. And second, based on the radiographic appearance plus histopathology, if microcalcifications were seen at that tissue site either on the specimen radiograph or on the H&E stained tissue sections. This was done because microcalcifications were seen on the H&E stained tissue sections at 13 sites that were not seen on the specimen radiograph [25].
Fig. 1.
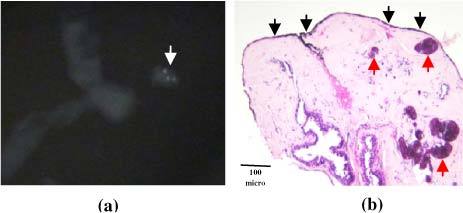
(a) Radiography and (b) histopathology of breast biopsy tissue site with type II microcalcifications. The microcalcifications are seen as small white densities (white arrow) in the specimen radiography in (a), and as blue concretions (red arrows) in the photomicrograph in (b) (H&E; 4X), which shows black ink (black arrows) marking the site interrogated spectroscopically.
5. Results
Raman spectra were collected from 228 breast biopsy tissue sites. The signal-to-noise rates (SNR) of the Raman spectra varied based on the histopathology category, with diseased tissue exhibiting a lower SNR than normal tissue due to higher fluorescence. Never-the-less, the model fits were reasonably good for both normal and diseased tissues. Figures 2a and 2b show typical Raman spectra of normal breast tissue and a breast lesion (fibrocystic change) with type II microcalcifications, with model fits and residuals. A prominent band at 960 cm−1 due to hydroxyapatite (arrow) is present in the Raman spectrum of the lesion with microcalcifications.
Fig. 2.
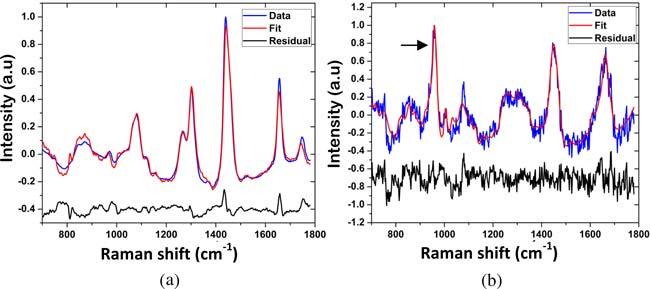
Typical Raman spectra of (a) normal breast tissue and (b) breast lesion (fibrocystic change) with type II microcalcifications, with model fits and residuals. A prominent band at 960 cm-1 due to hydroxyapatite (arrow) is present in the Raman spectrum of the lesion with microcalcifications.
The Raman spectrum of only 1 tissue site was excluded from data analysis due to poor SNR that resulted in a poor model fit. Raman spectra of 9 tissue sites were excluded as spectral outliers using a Student’s t-test employing a Mahalanobis distance function (95% probability; p<0.05) [26,27]. Raman spectra of 38 tissue sites were excluded because a histopathology diagnosis could not be rendered as the corresponding ink spots were not seen on the H&E stained tissue sections due to improper paraffin embedding of the biopsy. Three additional tissue sites were eliminated from data analysis due to contamination of the tissue core with an unidentified birefringent foreign material seen on the H&E stained tissue sections. The radiographic appearance was uncertain for 18 tissue sites and these were also excluded. Therefore, the final data set consisted of Raman spectra from 159 tissue sites. The 159 tissue sites were classified as follows based on the radiographic appearance: 54 normal, 75 lesions with microcalcifications, and 30 lesions without microcalcifications. Based on radiography plus histopathology, the tissue sites were classified as follows: 51 normal, 88 lesions with microcalcifications, and 20 lesions without microcalcifications. Table 1 details the histopathology diagnoses of the breast lesions with and without microcalcifications, which included fibrocystic change, fibroadenoma, ductal carcinoma in situ and invasive ductal carcinoma.
Table 1. Histopathology Diagnoses of Breast Tissue Sites classified by Radiography plus Histopathology.
| Lesion Classification | Normal | Fibro- cystic Change | Fibro- Adenoma | Ductal Carcinoma in Situ | Invasive Ductal Carcinoma | Other* | Total |
|---|---|---|---|---|---|---|---|
| Normal | 51 | 0 | 0 | 0 | 0 | 0 | 51 |
| Lesions with microcalcifications | 4 | 49 | 17 | 13 | 1 | 4 | 88 |
| Lesions without microcalcifications | 0 | 18 | 0 | 1 | 1 | 0 | 20 |
*Other, fat necrosis or healing biopsy site.
Seventy-four of the 88 lesions classified as lesions with microcalcifications on radiography plus histopathology had microcalcifications confirmed on histopathology, 14 did not. Seventy had type II and only 4 had type I microcalcifications. Some lesions with microcalcifications contained only a single microcalcification; others contained greater than 25 microcalcifications. Individual microcalcifications ranged from 10 to 1030 µm in diameter (mean 218 µm). The microcalcifications were 0-2150 µm deep in the tissue cores (distant from the probe-tissue interface), which ranged from 1075 to 2850 µm in maximum thickness.
5.1. Detection of breast lesions with microcalcifications
Raman spectra collected from all 159 tissue sites were analyzed for the FC of CAH, CAO and total calcium (CAH + CAO), as well as the other Raman active model constituents present in the breast tissue. We visualized the spread in the FCs for all of the model constituents using box plots. Boxplots of the FC of total calcium for breast lesions classified by radiographic appearance alone and by radiography plus histopathology are shown in Fig. 3 . The box plots are a graphical representation of the data, in which the lower limit and upper limit of the box represent the first and third quartiles, and the center line representing the second quartile (median). The extreme values are defined by values which exceed or are below the median by more than 1.5 times the interquartile range. Data points outside of this range are defined as outliers and indicated by crosses. From the box plot shown in Figs. 3a and 3b it is clearly evident that normal tissue, lesions with microcalcifications and lesions without microcalcifications can be easily distinguished based on total calcium content.
Fig. 3.
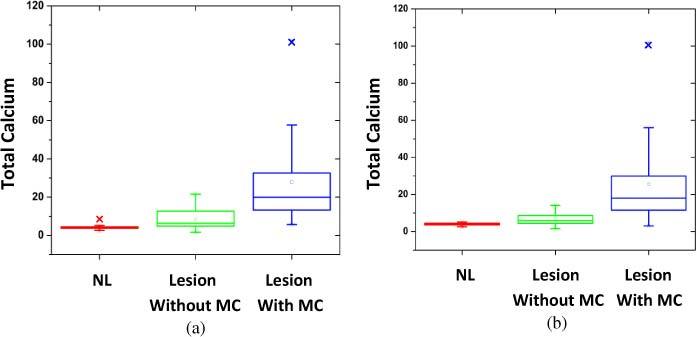
Box plots of the distribution of FC for total calcium for breast tissue sites classified by (a) radiographic appearance alone and (b) radiography plus histopathology (MC, microcalcifications).
Based on the results of the boxplot analysis, we devised an empirical Raman spectral algorithm to detect breast lesions with microcalcifications based on the FC of total calcium. The performance of the algorithm varied with different threshold FC values of total calcium. We selected the threshold FC for total calcium of 11 in order to maximize the positive predictive value (PPV), which is the most clinically significant measure of algorithm performance for the intended clinical application (Figs. 4a and 4b). With this decision threshold value, the algorithm yielded a PPV of 89% for the detection of lesions with microcalcifications, with a negative predictive value (NPV) of 86%, sensitivity (SE) of 84%, specificity (SP) of 86% and overall accuracy of 87%, when the tissue sites are classified by radiographic appearance alone. Performance of the algorithm improves significantly to a PPV of 97%, with an NPV of 78%, SE of 77%, SP of 97% and overall accuracy of 89%, when the tissue sites are classified by radiography plus histopathology. Using this decision threshold, we were able to detect microcalcifications as small as 10 µm and as deep as 2150 µm from the probe-tissue interface.
Fig. 4.
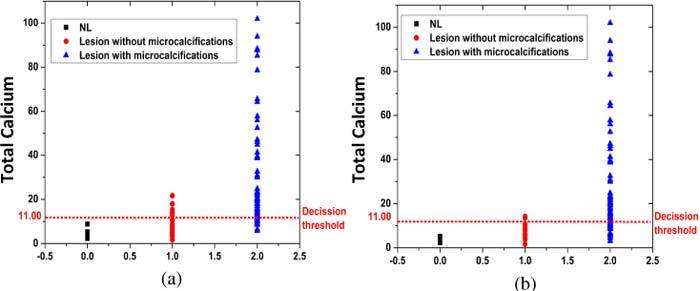
Empirical Raman algorithm using decision threshold of FC total calcium = 11 for detection of breast lesions with microcalcifications, classified by (a) radiographic appearance alone and (b) radiography plus histopathology.
We next devised a Raman spectral algorithm to detect breast lesions with microcalcifications using logistic regression, a discriminate analysis technique, which we used to correlate the FC with the diagnostic categories for all combinations of the 12 components in the model (plus total calcium), and determine the probability thresholds that correctly classified the most tissue sites [20]. A likelihood ratio test was used to determine the fit coefficients most significant for diagnosis, which are: CAH, CAO, fat and collagen. The optimal algorithm (Fig. 5 ) was based on three parameters, total calcium, collagen, and fat, and defined by the following plane equation (Eq. (2)):
Fig. 5.
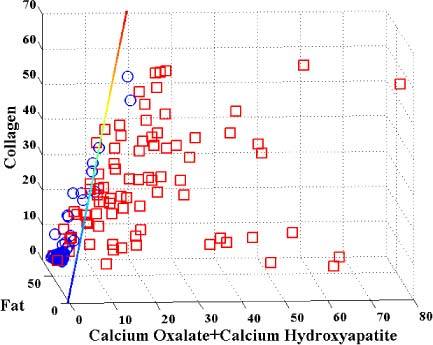
Logistic regression Raman algorithm distinguishing lesions with microcalcifications (red squares) from normals + lesions without microcalcifications (blue circles), classified by radiography plus histopathology.
| (2) |
where FCCAO, FCCAH, FCFat and FCCollagen are the model fit coefficients for CAO, CAH, collagen and fat, resp. This logistic regression algorithm yielded a PPV of 96%, NPV of 85%, SE of 86%, SP of 96% and overall accuracy of 91%, when the tissue sites are classified by radiography plus histopathology. (These numbers were obtained by employing the logistic regression algorithm on the training data set itself, i.e. auto-prediction.) The PPV and SP of this algorithm (96% and 96%, resp.) are comparable to those of the empirical algorithm (97% and 97%), but there is significant improvement in NPV and SE (85% and 86%, resp.) compared to those of the empirical algorithm (78% and 77%, resp.).
Subsequently, the logistic regression Raman algorithm was also validated using a traditional leave-one-out cross validation technique [20]. In this technique, the data from a particular tissue site is eliminated, and logistic regression is used to form a decision line that classifies the remaining tissue sites optimizing agreement with the histopathology diagnoses. The resulting decision threshold is then used to classify the excluded site. This process is successively applied to each of the sites. Leave-one-out cross validation yielded a PPV of 95%, NPV of 85%, SE of 86%, SP of 94% and overall accuracy of 90%. Only one additional tissue site was misclassified (as compared to the aforementioned autoprediction case), shifting from a true negative to a false positive. Since cross-validation is widely accepted to be a good indicator of the quality of modeling (in contrast to autoprediction which may force an unrealistically optimistic answer on a system), the cross-validation results obtained here provide a high level of confidence in the viability of this approach and its outlook for true prospective application. A comparison of the performance of the empirical and logistic regression algorithms and leave-one-out cross validation is shown in Table 2 .
Table 2. Comparison of Performance of Raman Algorithms for Detecting Breast Lesions with Microcalcifications, with Tissue Sites Classified using Radiography plus Histopathology.
| Raman Algorithm | PPV (%) | NPV (%) | SE (%) | SP (%) | Overall Accuracy (%) |
|---|---|---|---|---|---|
| Empirical | 97 | 78 | 77 | 97 | 89 |
| Logistic Regression | 96 | 85 | 86 | 96 | 91 |
| Cross Validation | 95 | 85 | 86 | 94 | 90 |
5.2. Distinction of type I and type II microcalcifications
As a chemically sensitive technique, Raman spectroscopy also has the potential to distinguish type I and type II microcalcifications based on the presence or absence of vibrational intensity at specific wavenumbers. In this study, microcalcifications in 4 of 74 lesions with microcalcifications were type I. The spectra of these breast lesions were characterized by the presence of Raman bands at 912 cm−1 and 1477 cm-1 (Fig. 6a ), characteristic of calcium
Fig. 6.
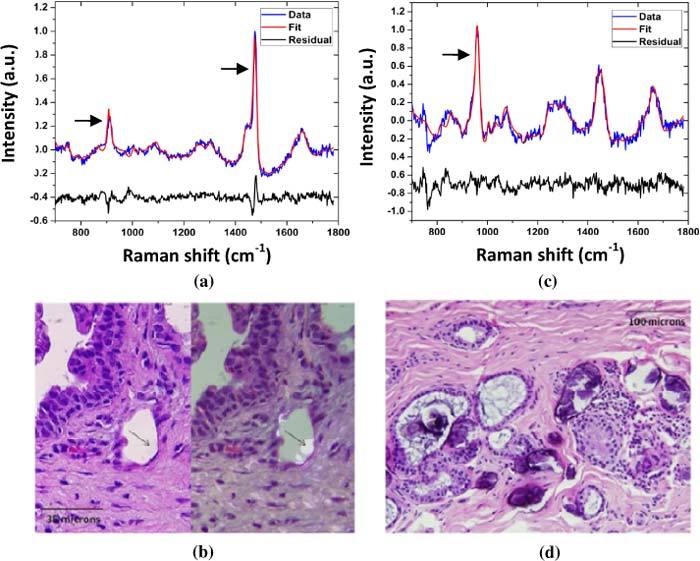
Typical Raman spectra and histopathology of breast lesions (fibrocystic change) with type I and II microcalcifications. The Raman spectrum of the breast lesion with type I microcalcifications in (a) shows prominent bands at 912 cm-1 and 1477 cm-1 (arrows) characteristic of calcium oxalate; the calcium oxalate crystals comprising the type I microcalcifications (b) do not bind H&E (left panel) and appear as colorless crystals (arrows) that are birefringent when viewed under polarized light (right panel). In contrast, the Raman spectrum of the breast lesion with type II microcalcifications in (c) shows a prominent band at 960 cm_1 (arrow) characteristic of calcium hydroxyapatite; the calcium hydroxyapatite rich type II microcalcifications appear as basophilic concretions on the H&E stain (d) and are non-birefringent.
oxalate (Fig. 6b). In contrast, Raman spectra of the 70 lesions with type II microcalcifications on histopathology were dominated by a Raman band at 960 cm_1 (Fig. 6c), characteristic of calcium hydroxyapatite (Fig. 6d). These results show that Raman spectroscopy has the potential to not only detect microcalcifications, but also distinguish type I and type II microcalcifications. Because very few type I microcalcifications are present in the data set, a spectral algorithm to distinguish type I and II microcalcifications was not developed.
6. Discussion
The results of this study demonstrate that Raman spectroscopy has the potential to both detect microcalcifications, and potentially distinguish type I and type II microcalcifications, in tissue cores obtained during stereotactic breast needle biopsies. We initially classified the tissue sites based on the radiographic appearance, as the lesions targeted by the radiologist for biopsy are identified radiographically. Using a Raman algorithm with a total calcium FC decision threshold = 11, all normal tissue sites were correctly identified as negative for microcalcifications. When the tissue sites were classified based on the radiographic appearance alone, there were only 8 false positives, that is breast lesions without radiographic microcalcifications that were diagnosed by the Raman algorithm as having microcalcifications. Interestingly, 4 of these breast lesions had microcalcifications on histopathology, suggesting that the Raman diagnosis was in fact correct for these lesions.
Because of this finding, we also classified the tissue sites using radiography plus histopathology, so that lesions with histologic microcalcifications not seen by radiography would also be classified as lesions with microcalcifications. In this case, the number of false positives decreased to 2 for the empirical algorithm and 3 for the logistic regression algorithm. The 4 tissue sites with microcalcifications seen only on histopathology were correctly diagnosed by both Raman algorithms as having microcalcifications. The average size of the microcalcifications seen on histopathology for these 4 lesions was 65 µm (average depth (distance from the probe-tissue interface) 575 µm), which is smaller than can be detected on mammography (detection limit 100 µm) [25], suggesting that at the selected decision threshold our Raman technique is more sensitive to the presence of microcalcifications than mammography. The most likely explanation for the remaining false positives is histopathology sampling error [28], although additional H&E stained tissue sections were examined in an effort to find microcalcifications in these tissue sites.
This Raman technology is being developed to guide the radiologist during stereotactic core needle biopsy procedures to retrieve the target tissue with microcalcifications for pathology diagnosis. PPV is the most important measure of algorithm performance for this clinical application, because false positive results represent a greater risk to the patient than false negative results [29]. If the results of the Raman algorithm indicate that the tissue to be biopsied harbors a microcalcification when it does not, the radiologist may retrieve only that one tissue core and thus miss the target microcalcification. In this case, the biopsy may be non-diagnostic or false negative and the patient may have to undergo a second stereotactic or surgical biopsy. On the other hand, if the results of the Raman algorithm indicate that the tissue to be biopsied does not harbor a microcalcification when it does, the radiologist may retrieve additional tissue cores that were not needed. While undesirable, this does not represent a significant risk to the patient.
Our empirical Raman algorithm was based on the FC of total calcium as it must detect both CAO-rich type I and CAH-rich type II microcalcifications. It detected breast lesions with microcalcifications with a PPV of 89% when the tissue sites were classified by radiographic appearance and 97% when the tissue sites were classified by radiography plus histopathology, reflecting its sensitivity to microcalcifications seen on histopathology that were not seen on radiography. The PPV was comparable (96%) for the logistic regression algorithm based on the FC of total calcium, fat and collagen, again when the tissue sites were classified by radiography plus histopathology. In addition, there was significant improvement in NPV and SE (85% and 86%, resp.) compared to those of the empirical algorithm (78% and 77%, resp.). Therefore, the logistic regression algorithm appears more robust. This is not surprising since the algorithm employs not only the FC for CAH and CAO found in the microcalcifications themselves (as total calcium), but also the FC for fat and collagen. Using the logistic regression algorithm, in order for a tissue site to be classified as a lesion with microcalcifications, it must have not only increased total calcium (FC CAH + FC CAO) but also an increased FC for collagen and a decreased FC of fat. This reflects the increase in collagen-rich stroma at the expense of fat cells seen in the breast lesions associated with microcalcifications, including fibrocystic change, fibroadenoma, ductal carcinoma in situ and invasive ductal carcinoma. Interestingly, the FC for fat and collagen are the key diagnostic parameters in our previously devised Raman algorithm to distinguish breast cancer from benign breast lesions such as fibrocystic change and fibroadenoma in the absence of microcalcifications [20,22]. Leave-one-out cross validation yielded only one additional misclassification, indicating that the logistic regression framework is not overfitting the acquired data set. This in turn suggests that the logistic regression algorithm can be applied successfully for prospective predictions in larger scale clinical studies, as it is more likely to be generalizable for larger data sets than the empirical algorithm.
One surprising result was the detection of microcalcifications up to 2150 µm deep in the breast biopsy tissue cores (distant from the probe-tissue interface). Previous studies have suggested a penetration depth of only ~1mm (1000 µm) for Raman spectroscopy using our optical fiber probe [17]. There are several possible explanations for the apparent deeper tissue penetration depth. One is that the target microcalcifications are highly active Raman scatterers and thus may be detectable with fewer effective photons at greater depths than less Raman active breast constituents. Second, our optical fiber probe makes use of separate excitation and collection fibers. Thus there is a small (800 µm) source-detector separation, which may also increase penetration depth. Tissue simulating phantom studies by Stone and associates have shown that microcalcifications can be detected to a depth of 8.7 mm depth using spatially offset Raman spectroscopy (SORS), in which Raman signals are collected from a region of interest that is spatially offset from the point of illumination on the sample surface (by 2cm in their studies) [15]. Therefore it is not unreasonable to suggest that we can detect microcalcifications at depths greater than 1mm.
7. Conclusion
We present here a Raman spectroscopic tool for detection of microcalcifications in breast tissue obtained during stereotactic breast core biopsies, based on their chemical composition. Application of our optimal, logistic regression-derived Raman diagnostic algorithm resulted in a PPV of 96% for detecting microcalcifications. This study shows that Raman spectroscopy has the potential to detect microcalcifications at stereotactic breast core biopsy and provide real-time feedback to radiologists, reducing non-diagnostic and false negative biopsies.
Acknowledgments
This research was supported by the National Institute of Health National Center for Research Resources (P41-RR02594) and the National Cancer Institute (R01-CA140288). The authors would like to acknowledge Abigail Haka for her invaluable advice on data analysis.
References and links
- 1.Breast Cancer Statistics , http://www.breastcancer.org/symptoms/understand_bc/statistics.jsp\
- 2.Rim A., Chellman-Jeffers M., Fanning A., “Trends in breast cancer screening and diagnosis,” Cleve. Clin. J. Med. 75(Suppl 1), S2–S9 (2008). 10.3949/ccjm.75.Suppl_1.S2 [DOI] [PubMed] [Google Scholar]
- 3.Johnson J. M., Dalton R. R., Wester S. M., Landercasper J., Lambert P. J., “Histological correlation of microcalcifications in breast biopsy specimens,” Arch. Surg. 134(7), 712–716 (1999). 10.1001/archsurg.134.7.712 [DOI] [PubMed] [Google Scholar]
- 4.Burnside E. S., Ochsner J. E., Fowler K. J., Fine J. P., Salkowski L. R., Rubin D. L., Sisney G. A., “Use of microcalcification descriptors in BI-RADS 4th edition to stratify risk of malignancy,” Radiology 242(2), 388–395 (2007). 10.1148/radiol.2422052130 [DOI] [PubMed] [Google Scholar]
- 5.Radi M. J., “Calcium oxalate crystals in breast biopsies. An overlooked form of microcalcification associated with benign breast disease,” Arch. Pathol. Lab. Med. 113(12), 1367–1369 (1989). [PubMed] [Google Scholar]
- 6.Jackman R. J., Rodriguez-Soto J., “Breast microcalcifications: retrieval failure at prone stereotactic core and vacuum breast biopsy--frequency, causes, and outcome,” Radiology 239(1), 61–70 (2006). 10.1148/radiol.2383041953 [DOI] [PubMed] [Google Scholar]
- 7.Pestaner J. P., Mullick F. G., Johnson F. B., Centeno J. A., “Calcium oxalate crystals in human pathology. Molecular analysis with the laser Raman microprobe,” Arch. Pathol. Lab. Med. 120(6), 537–540 (1996). [PubMed] [Google Scholar]
- 8.Motz J. T., Fitzmaurice M., Miller A., Gandhi S. J., Haka A. S., Galindo L. H., Dasari R. R., Kramer J. R., Feld M. S., “In vivo Raman spectral pathology of human atherosclerosis and vulnerable plaque,” J. Biomed. Opt. 11(2), 021003 (2006). 10.1117/1.2190967 [DOI] [PubMed] [Google Scholar]
- 9.Šćepanović O. R., Fitzmaurice M., Gardecki J. A., Angheloiu G. O., Awasthi S., Motz J. T., Kramer J. R., Dasari R. R., Feld M. S., “Detection of morphological markers of vulnerable atherosclerotic plaque using multimodal spectroscopy,” J. Biomed. Opt. 11(2), 021007 (2006). 10.1117/1.2187943 [DOI] [PubMed] [Google Scholar]
- 10.Rocha R., Silveira L., Jr, Villaverde A. B., Pasqualucci C. A., Costa M. S., Brugnera A., Jr, Pacheco M. T., “Use of near-infrared Raman spectroscopy for identification of atherosclerotic plaques in the carotid artery,” Photomed. Laser Surg. 25(6), 482–486 (2007). 10.1089/pho.2007.2111 [DOI] [PubMed] [Google Scholar]
- 11.Otero E. U., Sathaiah S., Silviera L., Jr, Pomerantzeff P. M. A., Pasqualucci C. A. G., “Raman spectroscopy for diagnosis of calcification in human heart valves,” Spectroscopy 18(1), 75–84 (2004). [Google Scholar]
- 12.Rocha R., Villaverde A. B., Pasqualucci C. A., Silveira L., Jr, Brugnera A., Jr, Costa M. S., Pacheco M. T., “Identification of calcifications in cardiac valves by near infrared Raman spectroscopy,” Photomed. Laser Surg. 25(4), 287–290 (2007). 10.1089/pho.2007.2100 [DOI] [PubMed] [Google Scholar]
- 13.Haka A. S., Shafer-Peltier K. E., Fitzmaurice M., Crowe J., Dasari R. R., Feld M. S., “Identifying microcalcifications in benign and malignant breast lesions by probing differences in their chemical composition using Raman spectroscopy,” Cancer Res. 62(18), 5375–5380 (2002). [PubMed] [Google Scholar]
- 14.Matousek P., Stone N., “Prospects for the diagnosis of breast cancer by noninvasive probing of calcifications using transmission Raman spectroscopy,” J. Biomed. Opt. 12(2), 024008 (2007). 10.1117/1.2718934 [DOI] [PubMed] [Google Scholar]
- 15.Stone N., Baker R., Rogers K., Parker A. W., Matousek P., “Subsurface probing of calcifications with spatially offset Raman spectroscopy (SORS): future possibilities for the diagnosis of breast cancer,” Analyst (Lond.) 132(9), 899–905 (2007). 10.1039/b705029a [DOI] [PubMed] [Google Scholar]
- 16.Baker R., Rogers K. D., Shepherd N., Stone N., “New relationships between breast microcalcifications and cancer,” Br. J. Cancer 103(7), 1034–1039 (2010). 10.1038/sj.bjc.6605873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manoharan R., Shafer K., Perelman L., Wu J., Chen K., Deinum G., Fitzmaurice M., Myles J., Crowe J., Dasari R. R., Feld M. S., “Raman spectroscopy and fluorescence photon migration for breast cancer diagnosis and imaging,” Photochem. Photobiol. 67(1), 15–22 (1998). 10.1111/j.1751-1097.1998.tb05160.x [DOI] [PubMed] [Google Scholar]
- 18.Shafer-Peltier K. E., Haka A. S., Fitzmaurice M., Crowe J., Myles J., Dasari R. R., Feld M. S., “Raman microspectroscopic model of human breast tissue: implications for breast cancer diagnosis in vivo,” J. Raman Spectrosc. 33(7), 552–563 (2002). 10.1002/jrs.877 [DOI] [Google Scholar]
- 19.Shafer-Peltier K. E., Haka A. S., Motz J. T., Fitzmaurice M., Dasari R. R., Feld M. S., “Model-based biological Raman spectral imaging,” J. Cell. Biochem. Suppl. 87(S39), 125–137 (2002). 10.1002/jcb.10418 [DOI] [PubMed] [Google Scholar]
- 20.Haka A. S., Shafer-Peltier K. E., Fitzmaurice M., Crowe J., Dasari R. R., Feld M. S., “Diagnosing breast cancer by using Raman spectroscopy,” Proc. Natl. Acad. Sci. U.S.A. 102(35), 12371–12376 (2005). 10.1073/pnas.0501390102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haka A. S., Volynskaya Z., Gardecki J. A., Nazemi J., Lyons J., Hicks D., Fitzmaurice M., Dasari R. R., Crowe J. P., Feld M. S., “In vivo margin assessment during partial mastectomy breast surgery using raman spectroscopy,” Cancer Res. 66(6), 3317–3322 (2006). 10.1158/0008-5472.CAN-05-2815 [DOI] [PubMed] [Google Scholar]
- 22.Haka A. S., Volynskaya Z., Gardecki J. A., Nazemi J., Shenk R., Wang N., Dasari R. R., Fitzmaurice M., Feld M. S., “Diagnosing breast cancer using Raman spectroscopy: prospective analysis,” J. Biomed. Opt. 14(5), 054023 (2009). 10.1117/1.3247154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volynskaya Z., Haka A. S., Bechtel K. L., Fitzmaurice M., Shenk R., Wang N., Nazemi J., Dasari R. R., Feld M. S., “Diagnosing breast cancer using diffuse reflectance spectroscopy and intrinsic fluorescence spectroscopy,” J. Biomed. Opt. 13(2), 024012 (2008). 10.1117/1.2909672 [DOI] [PubMed] [Google Scholar]
- 24.Motz J. T., Hunter M., Galindo L. H., Gardecki J. A., Kramer J. R., Dasari R. R., Feld M. S., “Optical fiber probe for biomedical Raman spectroscopy,” Appl. Opt. 43(3), 542–554 (2004). 10.1364/AO.43.000542 [DOI] [PubMed] [Google Scholar]
- 25.Dahlstrom J. E., Sutton S., Jain S., “Histologic-radiologic correlation of mammographically detected microcalcification in stereotactic core biopsies,” Am. J. Surg. Pathol. 22(2), 256–259 (1998). 10.1097/00000478-199802000-00016 [DOI] [PubMed] [Google Scholar]
- 26.Qi D., Berger A. J., “Chemical concentration measurement in blood serum and urine samples using liquid-core optical fiber Raman spectroscopy,” Appl. Opt. 46(10), 1726–1734 (2007). 10.1364/AO.46.001726 [DOI] [PubMed] [Google Scholar]
- 27.Dingari N. C., Barman I., Kang J. W., Kong C. R., Dasari R. R., Feld M. S., “Wavelength selection-based nonlinear calibration for transcutaneous blood glucose sensing using Raman spectroscopy,” J. Biomed. Opt. 16(8), 087009 (2011). 10.1117/1.3611006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grimes M. M., Karageorge L. S., Hogge J. P., “Does exhaustive search for microcalcifications improve diagnostic yield in stereotactic core needle breast biopsies?” Mod. Pathol. 14(4), 350–353 (2001). 10.1038/modpathol.3880314 [DOI] [PubMed] [Google Scholar]
- 29.Fitzmaurice M., “Principles and pitfalls of diagnostic test development: implications for spectroscopic tissue diagnosis,” J. Biomed. Opt. 5(2), 119–130 (2000). 10.1117/1.429978 [DOI] [PubMed] [Google Scholar]


