Abstract
The impact of digestive diseases, which include disorders affecting the oropharynx and alimentary canal, ranges from the inconvenience of a transient diarrhoea to dreaded conditions such as pancreatic cancer, which are usually fatal. Currently, the major limitation for the diagnosis of such diseases is sampling error because, even in the cases of rigorous adherence to biopsy protocols, only a tiny fraction of the surface of the involved gastrointestinal tract is sampled. Optical coherence tomography (OCT), which is an interferometric imaging technique for the minimally invasive measurement of biological samples, could decrease sampling error, increase yield, and even eliminate the need for tissue sampling provided that an automated, quick and reproducible tissue classification system is developed. Segmentation and quantification of ophthalmologic pathologies using OCT traditionally rely on the extraction of thickness and size measures from the OCT images, but layers are often not observed in nonopthalmic OCT imaging. Distinct mathematical methods, namely Principal Component Analysis (PCA) and textural analyses including both spatial textural analysis derived from the two-dimensional discrete Fourier transform (DFT) and statistical texture analysis obtained independently from center-symmetric autocorrelation (CSAC) and spatial grey-level dependency matrices (SGLDM), have been previously reported to overcome this problem. We propose an alternative approach consisting of a region segmentation according to the intensity variation along the vertical axis and a pure statistical technique for feature quantification, i.e. morphological analysis. Qualitative and quantitative comparisons with traditional approaches are accomplished in the discrimination of freshly-excised specimens of gastrointestinal tissues to exhibit the feasibility of the proposed method for computer-aided diagnosis (CAD) in the clinical setting.
OCIS codes: (110.4500) Optical coherence tomography, (170.3880) Medical and biological imaging, (110.2960) Image analysis
1. Introduction
Optical coherence tomography (OCT) is an optical technique based on low-coherence interferometry that provides noninvasive, subsurface, high-resolution imaging of biological microstructure [1–3]. It was first described by Huang et al. [1] and is becoming an established technique through posterior developments such as those in the spectral-domain (SD) [4] including the use of swept-sources (SS) [5] that have considerably improved its sensitivity. These have also helped to increase the speed of the analysis [6]. Traditionally, opthalmology has been the main application of OCT, but new OCT applications that include noninvasive “optical biopsy” in areas such as cardiology, pulmonology, skin imaging, orthopedics and gynecology are emerging. The generation of OCT images of the gastrointestinal (GI) tract is another area where it is predicted to be used in the following years since it could decrease sampling error, increase yield, and ultimately it could even eliminate the need for tissue sampling [3]. This would solve the limitation of current clinical management for patients with gastrointestinal diseases, such as Barrett’s esophagus (BE) and its associated adenocarcinoma, in terms of the reduced tissue fraction sampled during standard endoscopic examinations. Previous works have reported fiber-based OCT imaging systems that can be readily integrated with standard endoscopes for minimally invasive diagnosis [7, 8] of Barrett’s oesophagus [9] and oesophageal dysplasia [10] and even detection of colon dysplasia [11] has been established. However, the need for tissue sampling would only be eliminated if an automated tissue classification is developed that is quicker than visually assessing images and likely to be more reproducible.
Computer analysis of ophthalmologic pathologies using OCT, such as segmentation and quantification, traditionally relies on the extraction of thickness and size measures from the OCT images. However, for imaging in the gastrointestinal tract, such defined layers are usually not observed. In this regard, texture analysis of OCT images has shown promising results [3, 12–15]. These approaches are based on the assumption that the loss of structure associated with normal histological organization presumably resulting from the altered tissue architecture of the dysplastic tissue can be quantified as texture features, such as smoothness, coarseness and homogeneity, etc. in OCT images [15]. Therefore, these features could be employed for tissue classification at a later stage. Among these alternatives, feature extraction derived from the two-dimensional discrete Fourier transform (DFT) has proved to be a feasible option [13], since DFT features can detect texture periodicity and orientation. A region of interest (ROI) is selected from the OCT images and the complex two-dimensional Fourier transform of the region is obtained. The latter is divided into four concentric square rings based on frequency (with the outermost ring representing the highest spatial frequency content) and then the magnitudes of the spatial frequencies in each ring are integrated and normalized to the total signal magnitude, such that each feature value represents the percentage of signal within a certain range of spatial frequencies. Images that contain large relatively homogenous areas, such as the epithelial region in normal oesophagus, would have high values for DFT features associated with lower spatial rings while images with lots of smaller inhomogeneous areas as the crypt-like glandular structures in Barrett’s oesophagus, would have higher values in the DFT features that correspond to higher spatial frequency rings [13, 14].
More sophisticated approaches propose the employment of other texture analysis procedures, specifically the spatial gray-level dependence matrices (SGLDM) [12,14] and the center-symmetric auto-correlation method (CSAC) [3]. A SGLDM is a spatial histogram of an image that quantifies the distribution of grey-scale values and allows the computation of the statistical textural features for the selected region including energy, entropy, correlation, local homogeneity and inertia (also called contrast). On the other hand, CSAC texture features relate to local intensity variations and can capture local structure variation because this method quantifies the relationships between each pixel and its neighboring pixels [3]. The covariance of local centre-symmetric patterns is measured employing two local center-symmetric auto-correlations, linear and rank-order (SAC and SRAC), together with a related covariance measure (SCOV) and variance ratio (SVR), within-pair variance (WVAR) and between-pair variance (BVAR) [15]. Unlike SGLDM features these are rotation-invariant measures [16]. In the last years, several publications have dealt with refinements of these techniques through the simultaneous computation of different types of texture features and the further processing of the combined features using principal components analysis (PCA) [17] to reduce the variable dimensions and increase the discriminative power. Then the scores of the principal components, i.e. the projections of the original variables to the principal component space, are used as variables for linear discriminant analysis (LDA) [14]. More recently a PCA- and LDA-based prediction algorithm constructed directly from the depth intensity profiles in the OCT images has been proposed [18] as well as the study of the skewness of the intensity distribution in distinct regions in the axial direction using physiological information known a priori [19]. Within the same framework, the performance of diverse multivariate analysis for the automated classification of dysplasia in Barrett’s oesophagus has been compared yielding an accuracy of 84% by Qi et al. [15].
One of the key issues is that healthcare processes and decision making will be favored by near real time computer-aided diagnosis (CAD) and, consequently, texture analysis procedures that simultaneously perform feature quantification and data compression (each tomogram is in the end represented by a short series of numbers depending on the particular textural approach employed) are preferable to a straightforward PCA analysis of the depth intensity profiles because the latter has a much higher computational load. However, both the distinct textural analysis techniques previously proposed and also the first attempt in a statistical study of the intensity distribution are subject to the selection of an appropriate ROI for the quantification of the image features or the disposal of physiological information for region segmentation. A two-step methodology is reported to overcome this problem. First, an automated region segmentation of every OCT image according to the intensity variation along the vertical direction is proposed. In the second step, a morphological analysis of the segmented OCT images is employed for quantifying the features that could serve for tissue classification. This way of extracting features from the original images has been successfully employed for feature extraction for breast tissue density classification in mammographic CAD systems [20] or automated segmentation based upon remitted scatter spectra from pathologically distinct tumor regions [21]. To the authors’ knowledge, morphological analysis, however, has not previously been performed on OCT images. The proposed methodology will be qualitatively and quantitatively compared with textural analyses to show that it surpasses their capabilities in gastrointestinal tissue discrimination.
2. Materials and methods
2.1. OCT instrumentation
Surgical specimens were imaged employing a commercial SS-OCT system (OCS1300SS, Thorlabs Incorporated, Newton, New Jersey), which incorporates a high-speed frequency swept external cavity laser (1325 nm central wavelength) having a 3 dB spectral bandwidth (> 100 nm) and an average output power of 10 mW. The frequency clock for the laser is provided by a built-in Mach-Zehnder Interferometer (MZI, Thorlabs INT-MZI-1300) and the main output of the laser is coupled into a fiber-based Michelson interferometer and split into the reference and sample arm using a 50/50 coupler (Thorlabs FC1310-70-50-APC).
In the reference arm of the interferometer, the light is reflected back into the fiber by a stationary mirror. In the sample arm, it is fiber coupled into the microscope head, collimated and then directed by the XY galvo scanning mirrors towards the sample. The axial scans (A-scans) are performed at 16 kHz, which is the sweeping frequency of the laser. The transverse scan (B-scan) is controlled by the galvo scanning mirrors and determines the frame rate of the OCT imaging. The sample is placed on a stage, providing XY and rotational translation. A pair of XY galvo mirrors sequentially scans the probe beam across the sample surface area, and the 3D volume data set under this area is acquired (C-scan).
This OCT system produces high-resolution cross-sectional images of the gastrointestinal tissues with axial and transverse resolution of 9 and 15 μm, respectively. The interference signal is detected using a high-impedance gain balance photodetector having a provision for noise correction. The fast Fourier transform (FFT) is used to convert the time to frequency of the interference signal. However, raw OCT interference fringe signals in the time domain are recorded using the software package within the SS-OCT system. Time to frequency domain conversion to obtain the depth-dependent reflectivity profile for the OCT image is subsequently performed with Matlab 7.9.0.529 (R2009b) that is also utilized off-line for image enhancement and further processing.
2.2. Gastrointestinal tissue surgical specimens
Data was collected at St Mary’s Hospital, Paddington, London from February to September 2010. Patients undergoing elective gastrointestinal surgery, who where able to provide written informed consent, were included in the study. Suitable patients were identified after liasing with the surgical teams and written informed consent was obtained, and those under the age of 18 were excluded from the study. The purpose of the research and its implications were outlined during a pre-operative consultation. The suitability of both the patients and resected tissue was assessed in conjunction with the surgeons and the histopathologists, to ensure that external handling and OCT imaging did not adversely affect tissue quality for histopathological investigation.
Specimens from gastrointestinal surgery are generally quite large, as they consist of large parts of - or even whole - organs, such as the oesophagus, stomach, large bowel or rectum. In advanced stages, where the tumour may have spread to neighbouring organs, they, or parts thereof, may also be included, such as the tip of the pancreas or even the whole spleen. Normally, these specimens are placed in formalin as soon as they are excised, along with any other tissues, such as lymph nodes, unless the surgeon wishes to open the specimen, for example to visually assess the completeness of excision.
For the purposes of this study, specimens were collected from theaters in warm normal saline (0.9% sodium chloride) to maintain hydration. Formalin was not used until after imaging, as it is a fixative, causing cross-linking of proteins and effectively changing the structural properties of tissues and, consequently, their optical properties too. The specimens were collected as soon as they were excised and immediately taken to the histology lab. There, they were gently rinsed exposing the mucosa and any lesions. In order to stabilise them for imaging and mark sites of interest, specimens were pinned onto corkboards. Large pins were used to secure the specimen and smaller ones to mark areas from which OCT imaging was carried out, apart from tumour sites, where the thickness of the wall and tumour would not allow the pins to reach the cork-board. The specimens were then carried to the OCT lab in a closed tray. Since they are much thicker than the penetration depth of the OCT beam, they do not require any special mounting and were imaged directly on the corkboard. After imaging was complete (within 30 minutes of resection), tissues were fixed with 10% formalin and returned to pathology within one hour of excision, for routine histological processing, which included paraffin embedding, sectioning and Hematoxylin/Eosin (H/E) staining.
For each site that was imaged, a letter was appended to a unique patient code, starting from “A” for the first site imaged and carrying on in alphabetical order. This allowed posterior correlation with the histological and automated classification results, as well as any intended patient per patient analysis. A total of 35 sites from 11 patients that were 3 × 3 mm in size were imaged. Nine of them corresponded to tumour sites (belonging to 7 different patients), while the remaining imaged sites included stomach (20 sites from 9 patients), and oesophagus (6 sites from 6 patients). The sample population is, therefore, unevenly distributed across patients, meaning that not only the number of imaged sites per diagnostic category varied, but also the tissue types imaged per patient. A 3D volume data set (C-scan) was obtained per imaged site. The OCT software always generated data to a fixed depth of 3 mm, regardless of the on-screen depth set by the user, which was for viewing purposes only. Consequently, the other two dimensions were fixed to 3 mm length to obtain a cube-shaped C-scan. The lateral resolution was set to 512 pixels which implies that each C-scan consists of 512 transverse OCT images (B-scans). As the axial resolution of the system was also fixed at 512 pixels, the resulting OCT images contains 512 axial scans (A-scans) each.
2.3. Image enhancement
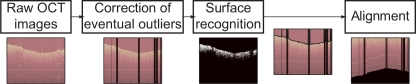
Raw OCT images suffer from eventual outliers (e.g. reflection artifacts) that need to be corrected or removed. Additionally, intensity information above the surface also has to be discarded. Reflections caused by the beam angle provoke brighter pixels at certain depth positions such as the top of the image as well as on the surface, and these were employed for detecting and removing the intensity depth profiles at these pixel localizations. A ribbon of “air” at the top of the image was considered and the mean intensity of this ribbon was calculated for every A-scan. Only those A-scans whose mean intensity in the ribbon was higher than the 75th quartile plus 1.5× the interquartile range of the mean intensities along the whole B-scan were discarded as reflection artifacts but the rest of the axial scans that comprise the OCT image were maintained. Regarding surface detection, there is no established method for unsupervised surface recognition [18]. The highest intensity within one A-scan is not necessarily correlated with the sample surface, and neither is the largest change in intensity if the first derivative is taken. Consequently, a variety of methods for surface recognition in OCT have been previously reported, such as erosion/dilation techniques based on a binary threshold image [3], shapelet-based boundary recognition [22] or rotation kernel transformations [23]. A binary thresholding approach as reported in [18] was selected because of its accuracy and simplicity. Again a ribbon of “air” above the surface was considered and its mean plus 0.75× its standard deviation was used as the intensity threshold. This threshold cuts off low intensity signals from deeper areas, therefore providing noise-free images. The 95th quartiles of these noise-free images were determined and employed for surface detection. Finally, every OCT image was wrapped, i.e. aligned with respect to the orientation of the extracted surface. Figure 1 summarizes the whole image preprocessing and shows the obtained enhanced image in a sample tomogram.
Fig. 1.
Flow diagram of OCT image preprocessing stages and visualization of the full process on a sample image.
2.4. Morphological analysis in optical coherence tomography images
Texture features such as smoothness, coarseness, homogeneity, etc. in OCT images have been previously employed for quantifying a loss of structure associated with normal histological organization since this structure loss is a hallmark of dysplastic mucosa [15]. A morphological analysis of OCT images is proposed in this work as an alternative for the quantification of image features that could equally serve for the classification of gastrointestinal tissues. This morphological analysis consists in a statistical study of the intensity distribution of the tomograms. A previous stage to the computation of intensity statistics is a region segmentation of every B-scan according to the intensity variation along the vertical direction. The so-called k-means method [24], which is a nonhierchical clustering procedure, is utilized in this regard. Although different versions have been reported as implementations, the k-means algorithm basically involves three steps. An initial set of k clusters are first determined. Then each observation is moved to the cluster whose centroid/mean is closest in distance and finally the cluster centroids/means are recalculated and the second step is repeated until no observation is reassigned to a new cluster. The segmentation performance of this algorithm, as any other partitioning clustering procedure, depends on the initial seed centroids [20]. Additionally, random initialization of cluster centroids could result in the creation of empty clusters at the first iteration, which consequently implies that different OCT images would be segmented into different number of regions in the vertical direction. To avoid these issues, a study of the intensity distribution of all the pixels within the B-scan was performed and k percentiles (where k is the number of regions) evenly distributed across the intensity range were determined and employed as the seed centroids. Corresponding percentiles are equally distributed across the intensity values depending on the number of regions, since the tissue discrimination capability will be precisely evaluated as a function of the number regions in this preliminary step. The employment of such a segmentation strategy avoids the necessity of previously known information about the tissue structure as in [19] and, therefore, makes the approach more extensible for tissue classification in urology or gynecology for example, where OCT is expected to gain popularity. To study the intensity distribution of the segmented regions, the first four statistical moments of the intensity values per region are computed as follows:
| (1) |
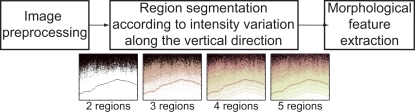
where Ii is the intensity value of a pixel within the region and N the number of pixels of the region. Along with the first four statistical moments of the intensity distribution, the relative area of the region with respect to the total area of the B-scan is employed as the fifth feature of the region. Accordingly, the total number of extracted features from the OCT images for tissue classification is five times the number of segmented regions. Figure 2 depicts a block diagram of the proposed two-step methodology for the quantification of morphological features of OCT images for tissue classification. The same tomogram is employed as a sample image to illustrate the region segmentation procedure. Because of computational performance, the optimum number of regions would be the smallest one that provides an assumable classification error.
Fig. 2.
Schematic of the proposed two-step methodology for feature quantification of OCT images.
The ability of the proposed morphological approach to extract the features that serve for tissue classification is qualitatively and quantitatively compared with textural approaches previously reported in this regard. The qualitative comparison is accomplished in terms of the clustering degree of the classes observed in the scatter plots. However, class separabilities measured in terms of the quotient between the traces of the between- and within-class scatter matrices are also indicated in the plots:
| (2) |
where “tr” denotes the trace of a matrix, i.e. the sum of the diagonal elements, Qb is the between-class scatter matrix, and Qw is the within-class scatter matrix, estimated as follows:
| (3) |
where xik (k = 1,...,ni) are the elements from the ith class, vi is the mean of the vectors in the ith class, v the mean of the centers and C the number of classes which possess a priori class probability Pi (i = 1,...,C) and cardinality ni (i = 1,...,C).
Separability measures based on scatter matrices are normally preferable to probabilistic distances because they do not require an estimation of the probability density function of the classes or that those probability density functions are known a priori. The average distance between the elements of the classes can be expressed as the sum of the traces of their between-and within-class scatter matrices [25]. However, the quotient between the traces is employed instead of their sum because it reflects more the intuitive notion of maximizing the trace of the between-class matrix while simultaneously minimizing the trace of the within-class matrix [26]. The quantitative comparison is based on a classification criterion, i.e. the objective function is the classification accuracy attained with a pattern recognition algorithm, as described in Section 2.5.
2.5. Classification and validation
A KNN classifier [27] is employed for ready discrimination between gastrointestinal tissues employing the morphological features of the segmented OCT images. For comparison, textural features are also interpreted for tissue characterization. In both cases, an unclassified tomogram (herein referred to as the query point) represented by a vector in a d-dimensional space, where d depends on the number of segmented regions if morphological features are employed or on the number of concatenated textural features, is assigned to the majority diagnosis of its K-nearest vectors found in the feature space. This approach can naturally deal with multiclass data while some of the more advanced classifiers, such as support vector machines (SVM) [28], require the bridging of results from a combinatorial set of such classifiers to simulate multiclass parameters [20]. Accuracy of the KNN classifier mostly depends on the metric used to compute distances between the query point and all training pixels in the feature space. Extracted features varied greatly and, therefore, they were normalized to prevent some features from being more strongly weighted than others. All features were statistically normalized to zero mean and unit variance employing a combined mean feature vector (μ) and a combined standard deviation vector (σ) from the training data set. The employment of the Euclidean distance,
| (4) |
where x and y are the two compared tomograms and M is the identity matrix, is the most common and simplest approach for measuring the separation between the query point and the training data when no prior knowledge about the probability density function of a particular class (tissue type) is available. Since it assumes that all features that define a tomogram are equally important and independent from others [29], it will not be the ideal metric when diagnostic categories are, for example, elongated in some directions. This difficulty has been overcome alternatively employing the Mahalanobis distance which is given in Eq. 4, where M is the covariance matrix for the extracted features. If all the features were uncorrelated and they had the same variance, the computation of the Mahalanobis distance would be equivalent to the Euclidean metric.
Apart from the distance criterion, the behaviour of the classifier depends on the number of nearest neighbors (K). Classification accuracy should be expected to increase with K because this reduces the influence of training data points assigned to a wrong diagnostic category. However, the error percentage is also influenced by the spreading of the extracted features within classes [21].
In order to accurately estimate the performance of the feature extraction methodology in comparison with textural approaches a threefold cross-validation technique or procedure [30, 31] was applied. B-scans across the whole data set were randomly divided into three nonoverlapping sets, with roughly equal size. Two of these sets were employed as a training set (used to populate feature space), and the other was employed as a validation set (query points) to compute the accuracy, sensitivity, specificity, negative predictive value (NPV) and positive predictive value (PPV) per diagnostic category from all others. This procedure is repeated three times, each time with different training and validation sets. Finally, the estimated performance of the classifier was calculated by averaging the three resulting errors. This approach eliminates the dependency of classification results on the training or test sets. Additionally, a leave-one-patient-out procedure is accomplished to show that the proposed disease markers, i.e. the morphological features, are greater than interpatient variation and the approach is feasible for gastrointestinal tissue classification in the clinical setting. Because of the moderate sample size, the validation set consists of all the C-scans from one patient while C-scans from other patients populate the feature space. Therefore, B-scans pertaining to the same C-scan were split across training and validation sets in the cross-validation while all B-scans comprising all C-scans from one patient were kept together and classified using all C-scans from other patients in the leave-one-patient-out procedure. As in the aforementioned cross-validation analysis, the leave-one-patient-procedure is, however, repeated as many times as the number of patients and reported measures were calculated by averaging the resulting sensitivity and specificity values per tissue type. In this second validation process, B-scans are not equally distributed in either the training or testing sets. Classification measures are, therefore, influenced by the heterogeneity of the sample population, but it is more realistic in a clinical scenario than the cross-validation employed in the comparison of image parameter extraction approaches.
3. Results
3.1. Qualitative comparison of feature extraction strategies
Morphological analysis of OCT images is initially compared with the multiple image feature quantification approaches previously reported in a pilot study using a restricted patient population, i.e. all B-scans belonging to the same patient across the distinct diagnostic categories. This comparison is accomplished in a semi-qualitative manner, i.e. their capabilities to minimize the within-class variance and to maximize the between-class variance. The aim of this is to estimate the ability of the proposed methodology to deal with intrapatient variability (tissue belonging to the same patient but different diagnostic types) and it first requires the determination of the number of regions for OCT image segmentation. As depicted in Figure 3 higher clustering degrees within the same tissue type and greater separation among types are obtained when the tomograms are divided in 3 axial regions. This number will later be confirmed in terms of the tissue discrimination accuracy obtained with the KNN classifier and the computational expensiveness of the methodology. For visualization purposes only, the extracted morphological features are further processed with PCA to allow the presentation of the discriminative power of the features but in fewer dimensions. Figure 4 finally depicts a qualitative comparison in terms of the clustering degree between the proposed methodology (3 segmented regions) and previous textural alternatives and indicates that the morphological approach surpasses the traditional ones when dealing with the intrapatient variability, i.e. data from the same type are better grouped together and the distance between different classes is also higher.
Fig. 3.
Grouped scatter plots and interclass separabilities (J) for the distinct gastrointestinal tissues depending on the number of segmented regions (only B-scans comprising all C-scans from a sample patient are included).
Fig. 4.
Grouped scatter plots and interclass separabilities J for the distinct gastrointestinal tissues depending on the approach employed for image feature quantification (The map is populated with all data points from a sample patient).
The technique employed in the quantification of features also needs to deal with interpatient variability because these features need to serve for tissue classification in the clinical setting. As shown in Fig. 4 (a and c), individual spatial frequency texture analysis, i.e. based on the DFT, and CSAC-based statistical texture analysis exhibit limited performance when dealing with the intrapatient variability and, consequently, different types are combined before their employment in computer-aided diagnosis of GI diseases as described in the Introduction. The behaviour of SGLDM-based statistical texture analysis was significantly better (J = 9.58) and it approaches to the interclass separability attained with morphological features (J = 15.07), which maximize the ratio between the within-class and between-class variances in this pilot study using a restricted patient population. Therefore, it is expected that they will have a better performance when data from different patients are considered. The validity of this intuitive assumption is demonstrated in a clinical investigation with a larger population of patients, i.e. the whole data set described above, and the results of this investigation are depicted in Figure 5. A tendency to group is still observed in the scatter plot, indicating the ability of the approach to reasonably differentiate diagnostic categories. Although the overlap among these categories is also noticeable, and, as a consequence, the interclass separability drops significantly, the quantitative comparison in the following section will demonstrate that the further processing of the morphological features with the KNN classifier provides reliable tissue categorization in clinical settings.
Fig. 5.
3D feature space assembled with the PCA-processed morphological features (3 segmented regions) from all B-scans that comprise the whole data set and its corresponding interclass separability (J).
3.2. Quantitative comparison of feature extraction strategies
The accuracy of the proposed morphological approach for the quantification of OCT image features fundamentally depends on the number of segmented regions in the axial direction. If KNN classification is applied to the extracted features to understand the relationship between the image parameters, and to classify new data, there is an additional tunable variable that influences tissue discrimination capability, i.e. the number of neighbors (K to differentiate it from the number of regions for axial segmentation (k)). The best choice of the number of neighbors depends upon the data; generally, larger values of neighbors reduce the effect of noise on the classification, but make boundaries between classes less distinct [15]. As shown in the grouped scatter plot in Fig. 5, different gastrointestinal tissues exhibit a tendency to group but their degree of overlap is also relevant. Therefore, a multivariate analysis that establishes sharp boundaries among classes is sought and, accordingly, one nearest neighbor is employed. Once the number of neighbors is fixed, a direct comparison of the number of segmented regions can be accomplished.
Figure 6 graphically compares the attained specificity and sensitivity values for varying number of segmented regions. As described previously, these measures are based on the ability to discriminate a given tissue type from all other categories evaluated and were averaged for all possible permutations of training and test sets in the cross validation procedure. Classification accuracy first increases when OCT images are vertically segmented into two regions since these predictions are closer to the representation of the perfect classifier (sensitivity:specificity of 100%:100%). Differences in tissue discrimination capability are hardly noticeable for a number of segmented regions varying between 2 and 5, and then accuracy decreases again for a higher number of regions. In terms of the computational load, a reduced number of regions is preferable, because this indicates that accurate classification is performed with a smaller number of image parameters. Consequently, only two regions will be further considered for the comparison with textural approaches.
Fig. 6.
Specificity and sensitivity values per tissue type as a function of the number of segmented regions.
An identical approach is followed for comparing morphological analysis with previously reported textural approaches and this comparison is shown in Figure 7. SGLDM features offer the best discrimination capability of gastrointestinal tissues among textural approaches but all of them are greatly surpassed by the proposed two-step methodology. Table 1 presents the complete results of this quantitative comparison measured as the obtained sensitivity, specificity, PPV, NPV and accuracy for discriminating a tissue type from all others.
Fig. 7.
Comparison of morphological and textural prediction in terms of their sensitivity and specificity values in the discrimination of each tissue type.
Table 1.
Summary of the quantitative comparison between morphological and textural approaches for feature quantification of OCT images in the classification of gastrointestinal tissues.
| Feature extraction approach | Sensitivity | Specificity | PPV | NPV | Accuracy |
|---|---|---|---|---|---|
| Tumor | |||||
| Morphological (2 segmented regions) | 99.97% | 99.85% | 99.57% | 99.99% | 99.88% |
| DFT features | 58.02% | 46.81% | 27.30% | 76.40% | 49.69% |
| CSAC features | 94.20% | 70.08% | 52.06% | 97.25% | 76.27% |
| SGLDM features | 89.58% | 88.95% | 73.64% | 96.12% | 89.11% |
| DFT + CSAC features | 96.17% | 50.95% | 40.35% | 97.55% | 62.53% |
| DFT + SGLDM features | 91.14% | 69.34% | 50.58% | 95.78% | 74.91% |
| Stomach | |||||
| Morphological (2 segmented regions) | 100% | 99.74% | 99.87% | 100% | 99.91% |
| DFT features | 55.64% | 55.77% | 72.14% | 37.92% | 55.67% |
| CSAC features | 88.17% | 84.20% | 92.11% | 77.98% | 86.86% |
| SGLDM features | 93.53% | 87.92% | 94.09% | 86.85% | 91.70% |
| DFT + CSAC features | 78.89% | 83.75% | 90.90% | 65.83% | 80.48% |
| DFT + SGLDM features | 83.16% | 85.60% | 92.24% | 71.18% | 83.96% |
| Oesophagus | |||||
| Morphological (2 segmented regions) | 100% | 99.97% | 99.64% | 100% | 99.97% |
| DFT features | 80.80% | 33.36% | 8.47% | 95.80% | 36.72% |
| CSAC features | 99.63% | 77.85% | 25.66% | 99.96% | 79.39% |
| SGLDM features | 100% | 90.23% | 43.82% | 100% | 90.92% |
| DFT + CSAC features | 99.89% | 53.00% | 13.95% | 99.98% | 56.32% |
| DFT + SGLDM features | 100% | 61.14% | 16.42% | 100% | 63.90% |
Table 2 finally summarizes the classification efficacy, in terms of the sensitivity:specificity values, when performing leave-one-patient-out cross-validation. This means that training and validation datasets are not randomly generated anymore, as in the cross-validation procedure described above, but all C-scans from each patient are intentionally left out as the validation set. Then classification measures from the different patients are averaged. This is the relevant situation in a clinical setting, because data from a patient is classified using a feature space populated by data from other patients, and whose C-scans where acquired at different time points. The latter means that variations due to system artifacts are also taken into account. Still, the enhancement in tissue discrimination capability provided by the proposed two-methodology is noticeable and consequently it is more suited to clinical investigations than previous approaches.
Table 2.
Summary of the efficacy of the KNN classifier (K = 1) to understand the relationship between the image features, and to predict new data. Reported measures are the sensitivity:specificity values in the discrimination of a given gastrointestinal tissue type from all other evaluated types.
| Feature extraction approach | Tumor | Stomach | Oesophagus |
|---|---|---|---|
| Morphological (2 segmented regions) | 86.9%:73.0% | 93.4%:75.0% | 100%:72.0% |
| DFT features | 56.1%:45.0% | 54.3%:56.4% | 76.3%:31.9% |
| CSAC features | 67.4%:45% | 75.6%:62.6% | 88.5%:68.3% |
| SGLDM features | 55.7%:81.8% | 82.8%:72.5% | 98.6%:76.2% |
| DFT + CSAC features | 79.3%:39.8% | 66.8%:68.3% | 99.7%:49.6% |
| DFT + SGLDM features | 67.9%:62.0% | 72.7%:76.2% | 100%:35.0% |
4. Discussion and conclusions
In this contribution, we propose a novel two-step methodology for feature extraction from OCT images that solves the limitations of previously reported strategies in this regard and demonstrates that gastrointestinal tissue types may be ascertained from the extracted image parameters. OCT images are first segmented in the axial direction in an automated manner according to intensity. Therefore, there is no need for previous physiological information for region segmentation or any uncertainty regarding the selection of the ROI for the quantification of the image features as in previous textural approaches found in the literature. Then, the parameters that serve for tissue classification are the relative area of each segmented region with respect to the total area of the B-scan along with the first four statistical moments (mean, standard deviation, skewness and kurtosis) of the intensity distribution within the region. The validity of this morphological analysis for feature quantification has been previously demonstrated for breast tissue density classification in mammographic CAD systems and scattering image analysis, but, according to the authors’ knowledge, it has not been previously performed on OCT images.
The feasibility of texture analysis of optical coherence tomography images for tissue classification relies on the quantification of the loss of structure associated with normal histological organization. Meanwhile, the proposed method is based on an evaluation of the intensity distribution in different regions along the vertical axis, which allows a statistical comparison of the extent of signal in different tissue types. Recent studies have proposed to employ the attenuation coefficient (μt), which describes the decay of detected light intensity with depth [32,33]. In malignant tissue displaying larger and irregularly shaped nuclei compared with normal tissue, light scattering is expected to be larger resulting in changes in μt. Statistically significant differences between normal renal tissue and renal cell carcinoma were reported in [33], but the differences encountered between benign bladder tissue and bladder urothelial carcinoma [32] were not significant. Several environmental factors (orientation of the biopsy, cauterization effects, etc.) that might account for this lack of difference in μt among the different pathological types are mentioned. In this study, we assessed signal attenuation using a more sophisticated approach that is independent of environmental artifacts.
This ability to discriminate different tissue types is achieved without requiring the a priori information required for previous techniques. In fact, classification efficacy even clearly surpasses that obtained with textural analysis both in cross-validation comparisons, which have been accomplished to eliminate the dependence on biased sample populations, as well as leave-one-patient-out procedures. Sensitivity and specificity values higher than 99.97%:99.85% are achieved in the discrimination of all tissue types in the validation set from all other categories evaluated in the cross validation procedure when a KNN classifier (one nearest neighbor) is employed in the further processing of the extracted features. Correlation coefficients among extracted features as high as 0.98 were experienced and, consequently, the Mahalanobis distance metric was employed instead of the Euclidean distance to account for correlations among extracted features. A significant improvement is expected in the KNN classifier’s performance using the Mahalanobis metric instead of the Euclidean for distance calculations. However, a full comparison of the classifier’s performance using both distance criteria, such as the one accomplished in [34], would be required to conclude the real necessity of the Mahalanobis distance. Textural features, specifically SGLDM features, provide the best sensitivity and specificity values of 100% and 90.23%, respectively, for oesophagus identification when the same multivariate algorithm is used for classify new data. But oesophagus is precisely the easiest one to identify because it is brighter in comparison to other considered tissue types. B-scans belonging to the same C-scan are split between training/test sets in this cross validation procedure to attain an even distribution of tissue categories among these sets. In this way, the dependence of classification on training and test sets is removed but it could incur over-training of the classifier. However, if the nearly perfect sensitivity and specificity values attained were only due to this fact and not to the suitability of the extracted features to be employed as tissue markers, near-perfect classification would also have been attained with textural features because the cross-validation is also performed across the same training and validation sets. The performance of textural approaches is however much worse, which demonstrates the significant improvement attained using morphological features as compared to textural.
This better performance when using morphological features can also be appreciated in the results presented in Table 2 from the ‘clinical’ investigation. This has been accomplished through a leave-one-patient-out procedure and, consequently, is more relevant for establishing the validity of the proposal for the clinical environment. A sensitivity and specificity of 86.9% and 73.0% is obtained, which again greatly enhances that attained with textural approaches. The clinical reliability of the approach could be limited by the employment of the KNN classifier as the multivariate analysis to understand the relationship between the image parameters, and to predict new data. More sophisticated classification algorithms for the further processing of the extracted parameters, such as artificial neural networks (ANN) or classification trees, could aid to enhance achieved sensitivity and specificity values. However, KNN is sufficient to demonstrate the enhanced behaviour of morphological analysis with respect to textural strategies in extracting the OCT image parameters for tissue classification, as demonstrated both in the qualitative and quantitative comparisons accomplished in this study. This enhanced behaviour is presumably due to a greater sensitivity of the extent of light penetration into tissues to the morphological changes occurring in malignant tissue than the loss of structure quantified by texture features. This sensibility is strengthened by the region segmentation performed in the first stage of the proposed method. While texture features only quantify the loss of structure in a previously determined ROI, the variation of signal strength with depth is quantified independently in all the segmented regions allowing more detailed signal attenuation information to be retained. The benefit of this might be barely noticeable in the sensitivity and specificity values attained in the cross-validation procedure as a consequence of the overfitting of the algorithm, but it is particularly relevant, however, for the ’clinical’ investigation, i.e. the leave-one-patient-out study. If no segmentation of the OCT images is accomplished specificity:sensitivity values of 69.74%:42.6% (tumor), 77.02%:59.97% (stomach) and 100%:36% (oesophagus) are obtained, which are much worse than if the OCT images are segmented into two regions, as indicated in Table 2. This confirms the necessity of segmenting the tomograms in the vertical direction to enhance the effect of smaller light penetration in tumours, or the brightness of oesophagus on the extracted features beyond the inter-patient variability.
Ongoing studies are further investigating the clinical validity of the methodology. This includes the accomplishment of a blind study with a much larger population of patients. Apart from that, the discrimination capability has only been assessed for gastrointestinal tissues. Accordingly, future research lines are also intended to demonstrate the suitability of the proposed feature extraction approach from OCT images for tissue classification in other medical applications, specifically urology.
Acknowledgments
Funding is gratefully acknowledged from the ERC grant StG 242991. This work was also partly funded by the National Physical Laboratory, Teddington, Middlesex, UK.
References and links
- 1.Huang D., Swanson E.A., Lin C.P., Schuman J.S., Stinson W.G., Chang W., Hee H.R., Flotte F., Gregory K., Puliafito C.A., Fujimoto J.G., “Optical coherence tomography,” Science 254, 1178–1181 (1991). 10.1126/science.1957169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Izatt J.A., Kulkarni M.D., Wang H.W., Kobayashi K., Sivak M.V., Jr., “Optical coherence tomography and microscopy in gastrointestinal tissues,” IEEE J. Sel. Top. Quantum Electron. 2, 1017–1028 (1996). 10.1109/2944.577331 [DOI] [Google Scholar]
- 3.Qi X., Sivak M.V., Jr., Isenberg G., Willis J.E., Rollins A.M., “Computer-aided diagnosis of dysplasia in Barrett’s esophagus using endoscopic optical coherence tomography,” J. Biomed. Opt. 11, 044010 (2006). 10.1117/1.2337314 [DOI] [PubMed] [Google Scholar]
- 4.Leitgeb R., Hitzenberger C., Fercher A., “Performance of Fourier domain vs. time domain optical coherence tomography,” Opt. Express 11, 889–894 (2003). 10.1364/OE.11.000889 [DOI] [PubMed] [Google Scholar]
- 5.Choma M., Sarunic M., Yang C., Izatt J.A., “Sensitivity advantage of swept source and Fourier domain optical coherence tomography,” Opt. Express 11, 2183–2189 (2003). 10.1364/OE.11.002183 [DOI] [PubMed] [Google Scholar]
- 6.Yun S., Tearney G., Bouma B., Park B., de Boer J., “High-speed spectral-domain optical coherence tomography at 1.3 μm wavelength,” Opt. Express 11, 3598–3604 (2003). 10.1364/OE.11.003598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tearney G.J., Brezinski M.E., Bouma B.E., Boppart S.A., Pitvis C., Southern J.F., Fujimoto J.G., “In vivo endoscopic optical biopsy with optical coherence tomography (OCT): a review,” Science 276, 2037–2039 (1997). 10.1126/science.276.5321.2037 [DOI] [PubMed] [Google Scholar]
- 8.Sergeev A.M., Gelikonov V.M., Gelikonov G.V., Feldchtein F.I., Kuranov R.V., Gladkova N.D., Shakhova N.M., Suopova L.B., Shakhov A.V., Kuznetzova I.A., Denisenko A.N., Pochinko V.V., Chumakov Y.P., Streltzova O.S., “In vivo endoscopic OCT imaging of precancer and cancer states of human mucosa,” Opt. Express I, 432–440 (1997). 10.1364/OE.1.000432 [DOI] [PubMed] [Google Scholar]
- 9.Poneros J.M., Brand S., Bouma B.E., Tearney G.J., Compton C.C., Nishiosa N.S., “Diagnosis of specialized intestinal metaplastia by optical coherence tomography,” Gastroenterology 120, 7–12 (2001). 10.1053/gast.2001.20911 [DOI] [PubMed] [Google Scholar]
- 10.Isenberg G., Sivak M.V., Jr., Chak A., Wong R.C.K., Willis J.E., Wolf B., Rowland D.Y., Das A., Rollins A., “Accuracy of endoscopic optical coherence tomography in the detection of dysplasia in Barrett’s esophagus: a prospective, doubled-blinded study,” Gastrointest. Endosc. 62, 825–831 (2005). 10.1016/j.gie.2005.07.048 [DOI] [PubMed] [Google Scholar]
- 11.Pfau P.R., Sivak M.V., Jr., Chak A., Kinnard M., Wong R.C., Isenberg G.A., Izatt J.A., Rollins A., Westphal V., “Criteria for diagnosis of dysplasia by endoscopic optical coherence tomography,” Gastrointest. Endosc. 58, 196–202 (2003). 10.1067/mge.2003.344 [DOI] [PubMed] [Google Scholar]
- 12.Gossage K.W., Rodriguez J.J., Barton J.K., “Texture analysis of optical coherence tomography images: feasibility for tissue classification,” J. Biomed. Opt. 8, 570–575 (2003). 10.1117/1.1577575 [DOI] [PubMed] [Google Scholar]
- 13.Gossage K.W., Smith C.M., Kanter E.M., Hariri L.P., Stone A.L., Rodriguez J.J., Williams S.K., Barton J.K., “Texture analysis of speckle in optical coherence tomography images of tissue phantoms,” Phys. Med. Biol. 51, 1563–1575 (2006). 10.1088/0031-9155/51/6/014 [DOI] [PubMed] [Google Scholar]
- 14.Chen Y., Aguirre A.D., Hsiung P.L., Huang S.W., Mashimo H., Schmitt J.M., Fujimoto J.G., “Effects of axial resolution improvement on optical coherence tomography (OCT) imaging of gastrointestinal tissues,” Opt. Express 16, 2469–2485 (2008). 10.1364/OE.16.002469 [DOI] [PubMed] [Google Scholar]
- 15.Qi X., Pan Y., Sivak S.V., Jr., Willis J.E., Isenberg G., Rollins A.M., “Image analysis for classification of dysplasia in Barrett’s esophagus using endoscopic optical coherence tomography,” Biomedical Optics Express 1, 825–847 (2010). 10.1364/BOE.1.000825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harwood D., Ojala T., Pietikainen M., Kelman S., Davis L., “Texture classification by center-symmetric auto-correlation using Kullback discrimination of distributions,” Pattern Recognit. Lett. 16, 1–10 (1995). 10.1016/0167-8655(94)00061-7 [DOI] [Google Scholar]
- 17.Jolliffe I.T., Principal Component Analysis, 2nd ed. (Springer, 2002). [Google Scholar]
- 18.Bazant-Hegemark F., Stone N., “Near real-time classifiation of optical coherence tomography data using principal component analysis fed linear discriminant analysis,” J. Biomed. Opt. 13, 034002 (2008). 10.1117/1.2931079 [DOI] [PubMed] [Google Scholar]
- 19.Barui A., Banerjee P., Patra R., Das R.K., Dhara S., Dutta P.K., Chatterjee J.,“Swept-source optical coherence tomography of lower limb wound healing with histopathological correlation,” J. Biomed. Opt. 16, 0260101–0260108 (2011). 10.1117/1.3535593 [DOI] [PubMed] [Google Scholar]
- 20.Olivier A., Freixenet J., Marti R., Pont J., Pere E., Denton E.R.E., Zwiggelaar R., “A novel breast tisue density classification methodology,” IEEE T. Inf. Technol. Biomed. 12, 55–64 (2008). 10.1109/TITB.2007.903514 [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Allende P.B., Krishnaswamy V., Hoopes P.J., Samkoe K.S., Conde O.M., Pogue B.W., “Automated identification of tumor microscopic morphology based on macroscopically measured scatter signatures,” J. Biomed. Opt. 14, 034034 (2009). 10.1117/1.3155512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogowska J., Hancewicz T., Kaplan P., “Optical coherence tomography of skin for measurement of epidermal thickness by shapelet-based image analysis,” Opt. Express 12, 5760–5769 (2004). 10.1364/OPEX.12.005760 [DOI] [PubMed] [Google Scholar]
- 23.Rogowska J., Bryant C.M., Breinski M.E., “Cartilage thicknes measurements from optical coherence tomography,” J. Opt. Soc. Am. A 20, 357–367 (2003). 10.1364/JOSAA.20.000357 [DOI] [PubMed] [Google Scholar]
- 24.Ghanadesikan R., Methods for Statistical Data Analysis of Multivariate Observation (Wiley, New York, 1997). 10.1002/9781118032671 [DOI] [Google Scholar]
- 25.Devijver P.A., Kittler J., Pattern Recognition: A Statistical Approach (Prentice Hall, London, 1982). [Google Scholar]
- 26.Tikk D., Wong K.W., “A feature ranking technique based on interclass separability for fuzzy modeling,” in Proceedings of IEEE International Computational Cybernetics ICCC 2007 (Academic, Gammarth, Tunisia, 2007), pp. 251–256 10.1109/ICCCYB.2007.4402044 [DOI] [Google Scholar]
- 27.Fukunaga K., Introduction to Statistical Pattern Recognition, 2nd ed. (Academic Press, New York, 1990). [Google Scholar]
- 28.Vapnik V., The nature of Statistical Learning Theory (Springer, New York, 1995). [Google Scholar]
- 29.Burns D. A., Ciurczak E.W., Handbook of Near-Infrared Analysis, 3rd ed. (CRC Press, 2008), Chap. 15. [Google Scholar]
- 30.Zhu H., Rohwer R., “No free lunch for cross-validation,” Neural Comput. 8, 1421–1426 (1996). 10.1162/neco.1996.8.7.1421 [DOI] [Google Scholar]
- 31.Goutte C., “Note on free lunches and cross validation,” Neural Comput. 9, 1211–1215 (1997). 10.1162/neco.1997.9.6.1245 [DOI] [Google Scholar]
- 32.Cauberg E.C., de Bruin D.M., Faber D.J., de Reijke T.M., Visser M., de la Rosette J.J., van Leeuwen T.G., “Quantitative measurement of attenuation coefficients of bladder biopsies using optical coherence tomography for grading urothelial carcinoma of the bladder,” J. Biomed. Opt. 15, 066013 (2010). 10.1117/1.3512206 [DOI] [PubMed] [Google Scholar]
- 33.Barwari K., de Bruin D.M., Cauberg E.C., Faber D.J., van Leeuwen T.G., Wijkstra H., de la Rosette J.J., Laguna M.P., “Advanced diagnostics in renal mass using optical coherence tomography: A preliminary report,” J. Endourol. 25, 311–315 (2011). 10.1089/end.2010.0408 [DOI] [PubMed] [Google Scholar]
- 34.Laughney A.M., Krishnaswamy V., Garcia-Allende P.B., Conde O.M., Wells W.A., Paulse K.D., Pogue B.W., “Automated classification of breast pathology using local measures of broadband reflectance,” J. Biomed. Opt. 15, 066019 (2010). 10.1117/1.3516594 [DOI] [PMC free article] [PubMed] [Google Scholar]