Abstract
A complex of ExbB, ExbD, and TonB couples cytoplasmic membrane (CM) proton motive force (pmf) to the active transport of large, scarce, or important nutrients across the outer membrane (OM). TonB interacts with OM transporters to enable ligand transport. Several mechanical models and a shuttle model explain how TonB might work. In the mechanical models, TonB remains attached to the CM during energy transduction, while in the shuttle model the TonB N terminus leaves the CM to deliver conformationally stored potential energy to OM transporters. Previous studies suggested that TonB did not shuttle based on the activity of a GFP–TonB fusion that was anchored in the CM by the GFP moiety. When we recreated the GFP–TonB fusion to extend those studies, in our hands it was proteolytically unstable, giving rise to potentially shuttleable degradation products. Recently, we discovered that a fusion of the Vibrio cholerae ToxR cytoplasmic domain to the N terminus of TonB was proteolytically stable. ToxR–TonB was able to be completely converted into a proteinase K-resistant conformation in response to loss of pmf in spheroplasts and exhibited an ability to form a pmf-dependent formaldehyde crosslink to ExbD, both indicators of its location in the CM. Most importantly, ToxR–TonB had the same relative specific activity as wild-type TonB. Taken together, these results provide conclusive evidence that TonB does not shuttle during energy transduction. We had previously concluded that TonB shuttles based on the use of an Oregon Green® 488 maleimide probe to assess periplasmic accessibility of N-terminal TonB. Here we show that the probe was permeant to the CM, thus permitting the labeling of the TonB N-terminus. These former results are reinterpreted in the context that TonB does not shuttle, and suggest the existence of a signal transduction pathway from OM to cytoplasm.
Keywords: TonB, energy transduction, membrane proteins, OGM membrane permeability
Introduction
The TonB system energizes the transport of large, scarce, and important nutrients such as vitamin B12 and siderophores complexed with iron across the outer membrane (OM) of Escherichia coli. A complex of the cytoplasmic membrane (CM) proteins ExbB and ExbD harnesses energy derived from the proton motive force (pmf) and transmits this energy to TonB. TonB then physically contacts a ligand-loaded TonB-gated transporter (TGT) in the OM, thus enabling release of the ligand into the periplasmic space. The per-cell copy number ratio of the TonB:ExbB:ExbD proteins is 1:7:2, although the precise ratio in an energy-transducing complex is unknown (Higgs et al., 2002). In E. coli, one set of tonB–exbB–exbD genes supports transport for multiple TGTs. But in certain other species, logistics are more complex; for example, Xanthomonas campestris has eight tonB genes and genes for 48 different transporters (Schauer et al., 2008).
Escherichia coli TonB has a topology consistent with that of an inter-membrane energy transducer: a short cytoplasmic region (residues 1–11), an N-terminal transmembrane domain (TMD, residues 12–32), a periplasmic linker region from 33–149, and a C terminus (residues 150–239) capable of interacting with TGTs. The TMD is essential for localization and maintaining interactions with integral CM proteins ExbB/D (Karlsson et al., 1993; Larsen et al., 1999; Ollis et al., 2009). The periplasmic linker domain is predicted to be largely disordered and contains a non-essential proline-rich domain from 70–102 (Larsen et al., 1993, 2007; Peacock et al., 2005). The C terminus is essential for interaction with TGTs, but none of the 90 residues in the C terminus are individually essential for function (Postle et al., 2010). The molecular mechanism by which TonB transduces energy from the CM to TGTs and facilitates subsequent ligand transport is largely unknown.
The question of how TonB participates in energy transduction has been fertile ground for researchers’ imaginations since the idea of energy transduction from the CM pmf to the OM first emerged (Hancock and Braun, 1976). In 1980, the Bradbeer laboratory suggested the possibility that TonB might synthesize a mobile messenger that would diffuse from CM to OM (Reynolds et al., 1980). Wookey (1982) proposed that TonB might be a periplasmic permease. Kadner (1990) proposed the idea that TonB might shuttle in a review article. Subsequent evidence for the shuttle model came from sucrose density gradient fractionation experiments where our lab showed TonB to localize in a CM:OM ratio of 60:40%, and exclusively at the OM in conditions in which ExbB/D and TolQ/R were absent (Letain and Postle, 1997). These initial findings were supported by in vivo demonstrations that a cys substitution in the extreme TonB N terminus, L3C, could be labeled with the probe Oregon Green® 488 maleimide (OGM; Larsen et al., 2003). As a control, truncated TonB L3C, consisting of the N-terminal 168 amino acids, was unable to interact with TGTs and could not be labeled although it formed a normal complex with ExbB. The degree of labeling for TonB L3C also increased substantially in the absence of the CM proteins ExbB/D and TolQ/R. Taken together, the results suggested that the extreme N terminus of TonB was labeled in the periplasm after it had departed from the CM. We suggested that energy transduction involved the physical cycling between the CM and OM compartments, a process called “shuttling.”
Alternative models have arisen since that time, including a propeller model, a pulling model, and a PBP-assisted model (reviewed in Krewulak and Vogel, 2011). What the later models have in common is that the TonB TMD never leaves the CM while accomplishing its functions. None of the models for TonB-dependent energy transduction has been widely embraced or ruled out, because of a lack of conclusive supporting evidence.
Here we show that in our hands a GFP–TonB fusion was too proteolytically unstable to allow meaningful interpretation of the data. In contrast, a fusion of the cytoplasmic domain of Vibrio cholerae ToxR (aa 1–181) to the N terminus of TonB was proteolytically stable, localized to the CM and had a relative specific activity that was essentially the same as wild-type TonB. These data indicated that TonB does not shuttle. We also discovered that our previous interpretation that TonB shuttled had been compromised by the fact, shown in these studies, that OGM could leak across the CM and label a cys substitution in a cytoplasmically confined TonB derivative. In light of these findings, our previous data are reinterpreted here.
Materials and Methods
Media, culture conditions
Saturated overnight cultures grown in Luria-Bertani (LB) broth were subcultured 1:100 into M9 minimal medium or MOPS medium with chloramphenicol added to 34 μg/ml as necessary to maintain plasmids (Shedlovsky and Brenner, 1963; Miller, 1972; Neidhardt et al., 1974). For M9 minimal medium, M9 salts were supplemented with 0.5% glycerol, 0.2% casamino acids (wt/vol), 40 μg/ml tryptophan, 0.4 μg/ml thiamine, 1 mM MgSO4, 0.5 mM CaCl2, and 1.85 μM FeCl3 (Postle, 2007). When necessary, l-arabinose or sodium propionate was added to induce expression of the TonB derivatives to normal chromosomal levels. For all assays, cells were grown to mid-exponential phase of 0.4–0.5 A550 units as measured on a Spectronic 20 spectrophotometer using a 1.5 cm pathway. A sample of each culture was precipitated with an equal volume of ice cold 20% w/v trichloro acetic acid (TCA) and protein levels were visualized on immunoblots of SDS-polyacrylamide gels.
Strains and construction of plasmids
Escherichia coli K-12 strains W3110 [F− IN(rrnD–rrnE)1; Hill and Harnish, 1981)], KP1344 (W3110 ΔtonB::blaM; Larsen et al., 1999), KP1440 (W3110 ΔtonB::blaM exbB::Tn10, tolQam; Larsen et al., 2003), KP1035 (W3110 entA::mini-kan), KP1229 (W3110 Δ ana–tonB–trpC; Ahmer et al., 1995), and KP1231 [W3110 Δ(ana–tonB–trpC) entA::mini-kan] were the principal strains used in this study. KP1231 was constructed by sequential P1vir transduction of the entA::mini-kan cassette from MK3, a generous gift from Mark McIntosh, into W3110 to generate KP1035 and finally from KP1035 into KP1229 to yield KP1231 (Ahmer et al., 1995). Plasmids used in this study are shown in Table 1. Attempts were made to replicate construction of a GFP–TonB fusion protein previously described (Kaserer et al., 2008), where sggfp is connected to tonB by a BamHI site. The plasmid encoding the exact GFP variant (sggfp) used in that study was no longer available (discontinued by the manufacturer), so a gfpmut2 variant was used (encoded on pDR107B, a generous gift from P. de Boer). sggfp and gfpmut2 both encode derivatives of Aequorea victoria green fluorescent protein. Each has three substitutions to increase or enhance GFP fluorescence, but the substitutions are different – for sggfp, F64L, S65C, and I167T (Kaserer et al., 2008) and for gfpmut2, S65A, V68L, and S72A (Cormack et al., 1996) gfpmut2–tonB was constructed in three steps. First, tonB including the native RBS, from pKP325, was cloned into pPro33 using a 5′ KpnI site and 3′ SphI site, creating pKP1141 (primer sequences available upon request). Digestion of pPro33 with these restriction enzymes excised the unique BamHI site in the pPro33 MCS, since a unique BamHI site was needed in the final construct, as described above. In the second step, a BamHI site was added immediately before the tonB start codon in pKP1141, creating pKP1159. Forward and reverse primers were designed with the desired insertion flanked on both sides by 12–15 homologous bases. DpnI digestion was used to remove the template plasmid. Clones were screened for the presence of the BamHI site. In the third step, gfpmut2 from pDR107B was amplified, from the start codon through the final codon for Lys238 (no stop codon) and introducing flanking BamHI sites. This was cloned into the BamHI site of pKP1159, creating pKP1160 (gfpmut2–tonB). Clones were screened for insert orientation with with NcoI. With the exception of the GFP modifications, this construct was the same as pGT from Kaserer et al. (2008).
Table 1.
Plasmids used in this study.
| Plasmid name | Relevant genotype/ phenotype | Reference |
|---|---|---|
| pDR107B | pET21B derivative encoding GFP mut 2-T7tag-his fusion protein; Ampr | Raskin and De Boer (1997) |
| pBAD33 | PBAD promoter; AraC; Camr | Guzman et al. (1995) |
| pPro33 | pBAD33 2-methyl-citrate inducible; Camr | Lee and Keasling (2005) |
| pASAToxRTonB1 | pHK toxR(1–181) – tonB(1–239) Camr | Sauter et al. (2003) |
| pKP325 | TonB (araBAD regulated); AraC Camr | Larsen et al. (1999) |
| pKP477 | pKP325 ΔTonB | Ghosh and Postle (2004) |
| pKP478 | pKP325 TonB L3C C18G | Larsen et al. (2003) |
| pKP544 | pKP325 TonB L3C ΔTMD | This study |
| pKP1141 | pPro33 tonB | This study |
| pKP1159 | pPro33 tonB with novel BamHI site | This study |
| pKP1160 | pPro33 gfpmut2–tonB | This study |
| pKP1529 | pBAD33 ToxR–TonB | This study |
Plasmid pKP1529 was generated by cloning PCR-amplified ToxR–TonB (including the ToxR Shine–Dalgarno sequences) from pASAToxRTonB1 (Sauter et al., 2003) into the unique XbaI site in pBAD33 (Guzman et al., 1995). As a result, ToxR–TonB expression was now regulated by the PBAD promoter. All other plasmids are derivatives of pKP325 (Larsen et al., 2007). Plasmid pKP544 was constructed by extra-long PCR, resulting in the removal of TonB residues 13–35. Plasmid DNA sequences were verified at the Penn State Genomics Core Facility – University Park, PA, USA.
In vivo formaldehyde crosslinking
Formaldehyde crosslinking assays were performed essentially as described previously (Postle, 2007; Ollis et al., 2009). Mid-exponential phase cells were resuspended in sodium phosphate buffer pH 6.8. A portion of each culture served as a control for TonB expression levels. To examine crosslinking of ToxR–TonB in the absence of pmf, carbonylcyanide-m-chlorophenylhydrazone (CCCP) was added to a final concentration of 50 μM. As a control for solvent addition, dimethyl sulfoxide (DMSO) was added to each sample that was not treated with CCCP. Samples were incubated at 37°C for 5 min. and treated with 16% paraformaldehyde (final concentration 1%) for 15 min at room temperature. After centrifugation for 5 min at room temperature, crosslinked samples were solubilized in sample buffer (Laemmli, 1970) at 60°C for 5 min., and uncrosslinked samples were boiled at 95°C for 7 min. Complexes were visualized on immunoblots with the anti-TonB monoclonal (4F1) and anti-ExbD polyclonal antibodies (Larsen et al., 1996; Higgs et al., 2002).
[55Fe]ferrichrome transport
Mid-exponential phase cells were harvested, suspended in buffer and initial rates of [55Fe]ferrichrome transport were determined in triplicate as described (Postle, 2007). A sample of each culture was TCA precipitated immediately prior to assay to determine TonB expression levels. Proteins were visualized on immunoblots of SDS-polyacrylamide gels with the anti-TonB monoclonal antibody 4F1. This experiment was performed independently three times and the same results obtained.
Proteinase K accessibility
Strains expressing TonB derivatives were converted to either intact or lysed spheroplasts and were treated with or without proteinase K and with or without carbonylcyanide-m-chlorophenylhydrazone (CCCP) as described previously (Larsen et al., 1999). Proteinase K-resistant forms were visualized on immunoblots with the monoclonal antibody 4F1.
Electron microscopy preparations
Plasmid-bearing mid-exponential phase KP1344 cells were pelleted and fixed with glutaraldehyde and paraformaldehyde at concentrations of 2% w/v, each according to standard protocols from the Franceschi Microscopy and Imaging Center, Washington State University, Pullman, WA, USA. Samples were dehydrated in an ethanol series and then embedded in LR-White resin. Cured samples were thin sectioned and adhered to nickel grids. Immunolabeling was performed with 1:100 4F1, followed by 1:100 goat anti-mouse antibody conjugated with 20 nm gold particles. Grids were stained with uranyl acetate and lead citrate and visualized with a JEOL transmission electron microscope (JEOL USA Inc.) with images captured with a MegaViewIII Digital camera at the Franceschi Microscopy and Imaging Center.
Oregon Green® maleimide labeling
In vivo labeling was performed as described previously (Larsen et al., 2003). Briefly, 75 ml cells were grown in LB broth with 34 μg/ml chloramphenicol to an A550 of 0.50. Buffer A (50 mM KPO4, 100 mM K2SO4, pH 7.0) was added to 25 ml of cells and OGM in DMSO was added to a final concentration of 172 μM. Samples were incubated for 1 h on ice with shaking and were quenched with 2-mercapoethanol. After washing three times with Buffer A, samples were suspended in Tris–LSB, boiled for 10 min and frozen at −30°C.
In vitro labeling was performed in a manner similar to the in vivo labeling protocol but differed in that RIPA buffer (1% Triton X-100, 1% sodium deoxycholate, 1% SDS in 150 mM NaCl 50 mM Tris-acetate pH 7.6) was added to harvested cells, the cells were passed twice through a French Pressure cell, and lysate was collected. Samples were incubated with OGM and frozen as above except that no LSB–Tris was added prior to freezing. The results did not change for either in vivo or in vitro labeling regardless of whether or not 2-mercaptoethanol was added to quench the reaction.
Immunoprecipitation
In vivo and in vitro samples were thawed, and TonB was immunoprecipitated with a cocktail of 4F1 and 4H4 monoclonal antibody as described previously (Larsen et al., 2003). Samples for GroEL analysis were prepared the same way as the TonB samples but immunoprecipitated with anti-GroEL antibody (Stressgen Bioreagents, Victoria, BC, Canada). Samples were divided, resolved on SDS-polyacrylamide gels and immunoblots were subsequently developed with either anti-TonB or anti-GroEL antibodies and anti-OGM (Pierce) antibodies.
Results
GFP–TonB can give rise to “shuttleable” degradation products
A recent study fused GFP from A. victoria to the N terminus of TonB and asked whether or not this GFP–TonB construct was active (Kaserer et al., 2008). The fusion of the TonB cytoplasmic N terminus to a large cytoplasmically localized protein should prevent potential shuttling by keeping TonB retained in the CM. The GFP–TonB construct supported TonB activity and the authors concluded that TonB did not shuttle. To extend this work, we reconstructed a GFP–TonB fusion with the same linker sequence between GFP and TonB as was used in that study and used TCA to precipitate bacteria immediately upon harvest to prevent the degradation of GFP–TonB (and presumably GFP–TonB fragments) that could occur in gel sample buffer at 95°C (Skare and Postle, 1991). This allowed for accurate determination of the steady-state levels of all TonB-specific peptides. In our hands, expression of GFP–TonB at chromosomal levels resulted in the detection of several prominent degradation products (Figure 1). The results did not change regardless of whether M9 or MOPS medium was used, nor if GFP–TonB was expressed in the tonB entA strain KP1231 (data not shown). The degradation products that were the size of full-length TonB or slightly larger might support TonB activity by shuttling. Because of the challenges and problems with proteolysis and because the mechanism of TonB energy transduction is still considered an open question (Krewulak and Vogel, 2011), we wanted to identify a system that could answer the question of shuttling more definitely.
Figure 1.
GFP–TonB exhibits prominent degradation in vivo. Steady-state levels were determined as described in Section “Materials and Methods” with the following strains: cTonB (W3110 expressing chromosomally encoded TonB); ΔTonB (KP1344); pPro33-TonB (KP1344/pKP1159); and pPro33 GFP–TonB (KP1344/pKP1160). Sodium propionate (*) was added to induce expression of plasmid-encoded TonB or GFP–TonB as follows: 0, 1, 5, 10 mM. TonB expression levels were visualized on immunoblots of 11% SDS-polyacrylamide gels with the TonB monoclonal antibody 4F1. Mass standards (kDa) are shown on the left. Full-length TonB migrates at 36 kDa, and the position of full-length GFP–TonB is indicated by the arrow. The most likely proteins to serve as the source of activity if TonB shuttles are indicated by the bracket.
ToxR (1–181) – TonB is stable and fully active
During the course of studies on TonB dimerization, we identified a fusion of full-length TonB to the cytoplasmic domain (aa 1–181) of V. cholerae ToxR (Miller et al., 1987), that was extremely stable as expressed from pASAToxRTonB1 and analyzed by immunoblot (data not shown). This plasmid was a kind gift of V. Braun from earlier studies on TonB dimerization where the expression levels and activities of the ToxR–TonB constructs were not reported (Sauter et al., 2003). Because ToxR–TonB was overexpressed from pASAToxRTonB1 (data not shown), we cloned toxR–tonB, including its putative Shine–Dalgarno sequences into pBAD33 so that its expression could be regulated by the PBAD promoter (Guzman et al., 1995).
Like wild-type TonB, virtually all of the ToxR–TonB could be converted into a proteinase K-resistant conformation by collapse of the pmf due to CCCP addition (Figure 2). Because ExbB and ExbD are required for this proteinase K-resistant conformation (Larsen et al., 1999), it suggested that ToxR–TonB was located in the CM. The pmf is also required for TonB and ExbD to formaldehyde crosslink in vivo through their periplasmic domains (Ollis et al., 2009). Likewise, ToxR–TonB was also able to form this functionally significant crosslink with ExbD (Figure 3), and this complex was undetected when pmf was collapsed with CCCP. Thus, ToxR–TonB could be energized in the same way as wild-type TonB.
Figure 2.
ToxR–TonB is conformationally responsive to pmf. Strains were converted to spheroplasts and treated with or without proteinase K and CCCP as described in Section “Materials and Methods” with the following strains: cTonB (W3110 expressing chromosomally encoded TonB) and pToxR–TonB [KP1344 (ΔtonB)/pKP1529]. l-Arabinose was added to induce expression of ToxR–TonB to chromosomal levels (0.00046%). Samples were resolved on 13% SDS-polyacrylamide gels, and immunoblotting was performed with the α-TonB monoclonal antibody 4F1. Mass standards are shown on the left. Abbreviations: WC, whole cells; Sph, spheroplasts; PKRF, proteinase K-resistant fragment.
Figure 3.
ToxR–TonB makes the pmf-dependent crosslink with ExbD. Crosslinking was performed as described in Section “Materials and Methods” with the following strains: cTonB (W3110 expressing chromosomally encoded TonB and ExbD); ΔTonB (KP1344); pTonB (KP1344/pKP325); pToxR–TonB (KP1344/pKP1529); and pToxR–TonB CCCP (KP1344/pKP1529 treated with 50 μM CCCP). l-Arabinose was added to induce expression of plasmid-encoded TonB as follows: 0.00075% for pTonB and 0.00046% for pToxR–TonB. Complexes were detected on immunoblots of 13% SDS-polyacrylamide gels using ExbD polyclonal antibodies (upper two panels). Uncrosslinked monomeric TonB and ToxR-TonB levels were visualized with the monoclonal anti-TonB antibody 4F1 (bottom panel). The molecular mass standards (kDa) are shown on the left.
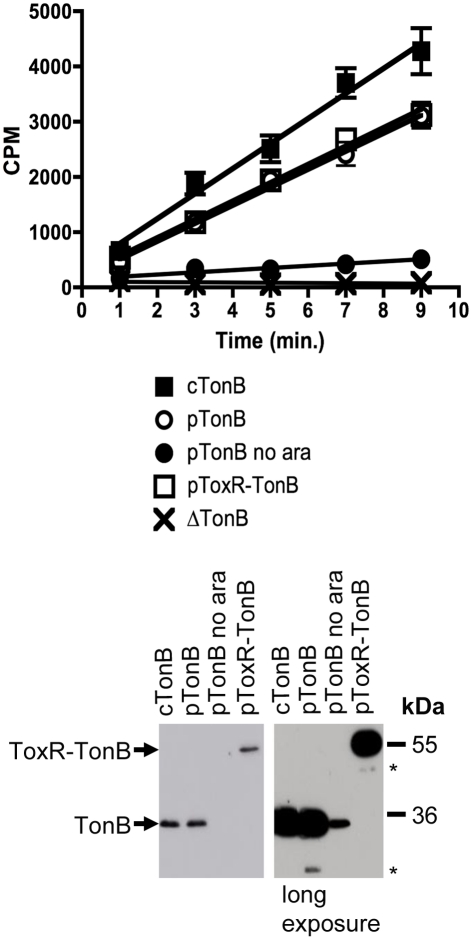
Finally, [55Fe]ferrichrome transport assays revealed that ToxR–TonB, when expressed to chromosomal levels, supported ∼100% of the wild-type plasmid-encoded TonB ferrichrome transport rate (Figure 4). ToxR–TonB supported 84 and 92% of wild-type activity in additional triplicate trials (data not shown). The key observation was that this activity could only have arisen from full-length ToxR–TonB. The amount of TonB in the uninduced sample (pTonB no ara) far exceeded the amount of the only detectable degradation product that derived from ToxR–TonB and supported only 12% activity (Figure 4). Thus, stable, full length ToxR–TonB had the same relative specific activity as chromosomally encoded wild-type TonB.
Figure 4.
ToxR–TonB supports as much activity as plasmid-encoded wild-type TonB. Ferrichrome transport assays were performed as described in Section “Materials and Methods” with the following strains: cTonB (W3110 expressing chromosomally encoded TonB); ΔTonB (KP1344); pTonB (KP1344/pKP325); pTonB no ara (KP1344/pKP325 uninduced); and pToxR–TonB (KP1344/pKP1529). l-Arabinose was added to induce expression of plasmid-encoded TonB as follows: 0.00075% for pTonB and 0.00045% for pToxR–TonB. Initial ferrichrome transport rates are as follows, expressed as counts per minute (cpm)/0.05 A550 ml cells/minute: cTonB 453 ± 31; pTonB 326 ± 13 (standard to 100%); pTonB no ara 39 ± 7 (12%); pToxR–TonB 333 ± 15 (102%); ΔTonB −4 ± 4 (0%). TonB expression levels were visualized on immunoblots of 11% SDS-polyacrylamide gels with anti-TonB monoclonal antibody 4F1. A longer exposure is shown to provide a comparison between uninduced pKP325 and the lack of full-length degradation product from pKP1529. Mass standards are shown on the right. Degradation products are marked with an asterisk (*).
Membrane permeability of OGM explains previous results
The strongest prior evidence for in vivo shuttling was the demonstration that a cys substitution predicted to be cytoplasmically localized could be labeled by OGM in full-length TonB but not in a truncated TonB derivative missing much of its C terminus and incapable of interaction with the OM (Larsen et al., 2003). OGM has been considered a “generally membrane impermeant” probe, and other groups have used this reagent to discriminate between free cysteines in the periplasm or cytoplasm of Gram-negative bacteria (Ye et al., 2001; Ye and Maloney, 2002; Yang et al., 2005; Liu et al., 2007; Nanatani et al., 2007, 2009). Given the contrast in findings between the ToxR fusions and the OGM labeling, we tested the possibility that OGM leakage across the CM could account for our previous findings (Larsen et al., 2003).
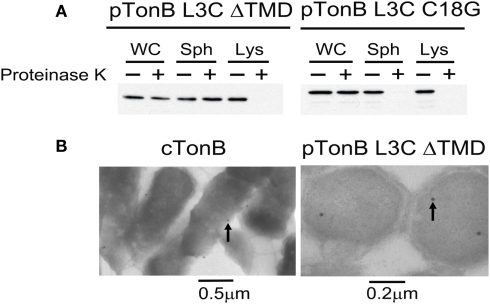
To assess the potential for OGM to leak across the CM, we constructed a TonB variant lacking its transmembrane domain but retaining the L3C substitution of the original OGM labeling study. TonB L3C ΔTMD expressed from pKP544 should be cytoplasmically localized (Karlsson et al., 1993). The cytoplasmic localization of the TonB L3C ΔTMD construct was tested by two methods: proteinase K accessibility assays in intact spheroplasts and transmission electron microscopy immunolocalization. Whereas wild-type TonB (TonB L3C C18G) is sensitive to proteinase K in intact spheroplasts, the TonB L3C ΔTMD was sequestered from proteinase K (Figure 5A). When spheroplasts were lysed, both TonB L3C C18G and TonB L3C ΔTMD were sensitive to proteinase K, indicating that TonB L3C ΔTMD was cytoplasmically localized (Figure 5A). The same results were also obtained in KP1344 (data not shown). Consistent with that result, electron microscopy revealed that the TonB L3C ΔTMD was confined to the cytoplasm whereas gold particles detecting wild-type TonB localized to the envelope (Figure 5B).
Figure 5.
TonB L3C ΔTMD is cytoplasmically localized. (A) TonB L3C ΔTMD is resistant to proteinase K in intact spheroplasts but sensitive to proteinase K in lysed spheroplasts. Strains pTonB L3C ΔTMD (KP1440 (ΔtonB, exbB::Tn10, tolQam)/pKP544) and pTonB L3C C18G (KP1440/pKP478) were converted to either intact or lysed spheroplasts and treated with or without proteinase K as described in Section “Materials and Methods.” Proteins were resolved on 11% SDS-polyacrylamide gels and immunoblotted with anti-TonB 4F1 antibodies. Abbreviations: WC, whole cells; Sph, spheroplast; Lys, lysed spheroplast. (B) Electron micrographs of strains cTonB (W3110 expressing chromosomally encoded TonB) or pTonB L3C ΔTMD (KP1344/pKP544). Cells were fixed and labeled with immunogold particles as described in Section “Materials and Methods.” Examples of labeled TonB derivatives are indicated with arrows. Size bars are included at the bottom.
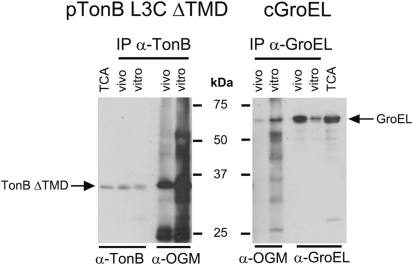
Oregon Green® 488 maleimide was able to label the cytoplasmically localized TonB L3C ΔTMD both in vitro and in vivo. This showed that in our previous experiments, OGM was indeed capable of crossing the CM and TonB L3C did not need to shuttle to become labeled by it (Figure 6). In addition, OGM labeled the cytoplasmic chaperone GroEL in vitro and in vivo, confirming its ability to leak across the CM (Figure 6).
Figure 6.
Oregon Green maleimide labels cytoplasmic proteins. OGM labeling and subsequent immunoprecipitation were performed as described in Section “Materials and Methods” with the following strains: cGroEL (W3110 expressing chromosomally encoded GroEL) and pTonB L3C ΔTMD (KP1344/pKP544). Samples of cells were also lysed by passage through a French pressure cell and labeled in vitro; these served as a standard for 100% labeling. l-Arabinose was added to induce expression of TonB L3C ΔTMD (0.002%). Proteins were also harvested by treatment with TCA to serve as a total protein control. Samples were divided and resolved on 11% SDS-polyacrylamide gels and immunoblotted with anti-TonB or anti-GroEL antibodies for half of the samples and with anti-OGM antibodies for the other half. Positions of TonB L3C ΔTMD and GroEL are indicated. Molecular mass standards are shown in the center. Abbreviation: IP, immunoprecipitation.
Discussion
The fate of the TonB N terminus during energy transduction has been speculated about and investigated for many years. Today, the primary models that explain its role include several mechanical models where TonB remains continuously associated with the CM and the shuttle model, where TonB cyclically dissociates from the CM and shuttles to the OM. The mechanical models suggest direct use of the pmf in energizing TGTs with TonB as a bridge between the two. The shuttle model suggests that TonB somehow stores potential energy from the pmf prior to shuttling to the OM. The TonB TMD was proposed to store the potential energy through response of residues S16 and H20 to the pmf (Larsen and Postle, 2001). Because these two models have different consequences and make different predictions, it is important to determine which is correct.
Since the emergence of the shuttle model, we have learned that the TonB TMD is highly tolerant to mutation. In particular, the only sidechain that cannot be replaced by ala and retain activity is H20, yet H20 can be replaced by asn and retain full activity, suggesting that the TonB TMD is not on a proton translocation pathway (Larsen et al., 2007; Swayne and Postle, 2011). These results cast doubt on the shuttle hypothesis by eliminating the TMD as the region of TonB that needed to be delivered to the OM. Here we demonstrate that, in contrast to GFP–TonB, a fusion of the cytoplasmic domain of ToxR (1–181) to TonB produced virtually no degradation products when expressed at levels characteristic of native TonB. ToxR–TonB appeared to be anchored in the CM at all times because it could be fully converted to a proteinase K-resistant form in de-energized cells. ToxR–TonB could also form a pmf-dependent crosslink with ExbD and was fully active in ferrichrome transport assays. This eliminated the possibility that somehow the entire ∼180 residue folded cytoplasmic domain of ToxR could be pulled out of the CM to shuttle. Even if the ToxR domain could be pulled out of the CM, it would not be possible for it to recross back into the cytoplasm and allow the TonB portion to assume its rightful relationship with ExbB and ExbD such that it had 100% activity. Because ToxR–TonB had an equivalent relative specific activity to TonB, we conclude that TonB does not shuttle. These results confirm and expand the conclusions of Kaserer et al. (2008).
The permeability of OGM to the CM explained the apparent contradiction between the ToxR–TonB results, and the previous ones suggesting that TonB shuttles in vivo (Larsen et al., 2003). The data from our 2003 study can be reinterpreted in the light of this new information. The labeling seen for full-length TonB L3C can be explained by the observation that OGM diffuses into the cytoplasm. A more interesting explanation may pertain to the observation that TonBam168 L3C did not become labeled with OGM, even though it still retained the ability to respond conformationally to pmf in spheroplasts and formaldehyde crosslink to ExbB, indicating that the truncation did not radically interfere with assembly with ExbB/D. The reinterpretation of the data raises the question of whether there is signal transduction occurring from the OM to the CM ExbB/D complex through TonB. It could be that when full-length TonB is present, the cytoplasmic domains of ExbB allow access of OGM to the extreme N terminus of TonB. In this model, when the C terminus of TonB is deleted and TonB does not associate with the OM, then the cytoplasmic domains of ExbB are conformationally rearranged such that the TonB L3C residue is sequestered. Consistent with that idea, TonB L3C was not labeled by OGM in the inside-out vesicles arising from French-pressed cells that not only lacked pmf but also any connection to the OM (data not shown).
The observation that TonB remains attached to the CM during energy transduction has ramifications for the meaning of the solved TonB–FhuA and TonB–BtuB crystal structures (Pawelek et al., 2006; Shultis et al., 2006). The conformation of the TonB C terminal domain solved in complex with the TGTs accounts only for ∼25 Å, but arises from ∼1/3 of the length of TonB (residues 153–233, with 239 being the terminal residue; Figure 7). While the proline-rich domain can span up to 100 Å of periplasmic space, it is non-essential (Evans et al., 1986; Larsen et al., 1993; Seliger et al., 2001). Based on the assembled structure of the periplasmic-spanning AcrAB/TolC complex, the distance between the CM and OM appears to be approximately 180 Å (Symmons et al., 2009). This suggests that the C terminus of TonB must be more disordered than is represented by the solved structures as it spans the periplasm and identifies a ligand-loaded transporter. Consistent with that, the TonB primary amino acid sequence predicts a highly disordered protein, and the C terminus is conformationally dynamic in vivo (Larsen et al., 1999, 2007; Postle et al., 2010). If TonB must be disordered in order to span the periplasm, then it is likely that TonB C terminus participates in an energy transduction cycle of association and dissociation from the TGTs rather than being constantly associated with the OM. In support of this idea, the number of OM transporters in a given cell exceeds the number of available TonB molecules (Kadner and Heller, 1995; Higgs et al., 2002; Postle et al., 2010).
Figure 7.
TonB cannot reach OM transporters with the conformation depicted by crystal structures of its C terminus. TonB residues 33–69 and 103–149, predicted to be disordered regions (Larsen et al., 2007), are depicted as yellow rectangles. The span of the periplasmic space was estimated based on crystal structure reconstructions of the AcrA/B/TolC complex (Symmons et al., 2009). Abbreviations: CM, cytoplasmic membrane; OM, outer membrane.
In a detailed analysis of four TGT structures, Chimento et al. (2005) noted that the β-strands of the cork domain that fills the TGT barrel are mostly parallel to the periplasmic space. They propose that only a modest perpendicular force supplied by TonB would be necessary to unfold the cork (Chimento et al., 2005). Now that we know that TonB does not shuttle, and that it may contact the TGT in a disordered conformation, it suggests that the mere folding of TonB upon contact might be sufficient to allow transport.
These new data also raised the question of what the 30–40% of TonB that fractionates with the OM represents. It could possibly represent the fraction of TonB that is strongly associated with the OM transporters at any one point in the energy transduction cycle. Alternatively it could represent TonB that has temporarily dissociated from the ExbB/D complex while its C terminus is associated with the OM. In either case, it must mean that TonB is being artifactually pulled out of the CM because ExbB and ExbD remain associated with the CM (Letain and Postle, 1997).
A remaining mystery is why, in the absence of ExbB/D and TolQ/R, nearly all of the TonB is found associated with the OM. It could be that the conformation of the TonB C terminus is such that its default interaction is at the OM. Alternatively it may be that the action of ExbB/D is primarily required to assist the TonB C terminus in dissociating from an OM transporter after an energy transduction event. The demonstration that TonB does not shuttle eliminates debate concerning how energy would be used to physically remove TonB from a CM complex of ExbB/D, and also how the N terminus would somehow target TonB back to an ExbB/D complex at the CM (Larsen et al., 2003; Postle and Kadner, 2003).
Conflict of Interest Statement
The authors are aware of no personal or monetary issues which arose during preparation of this manuscript that could represent a conflict of interest.
Acknowledgments
We thank Volkmar Braun and Silke Patzer for pASAToxRTonB1. We thank Jay Keasling for pPro33, Piet de Boer for pDR107B, and Mark McIntosh for strain MK3. Penelope Higgs created strain KP1231. This work was supported by grant GM42146 to K.P. from the National Institute of General Medical Sciences.
References
- Ahmer B. M. M., Thomas M. G., Larsen R. A., Postle K. (1995). Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. J. Bacteriol. 177, 4742–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimento D. P., Kadner R. J., Wiener M. C. (2005). Comparative structural analysis of TonB-dependent outer membrane transporters: implications for the transport cycle. Proteins 59, 240–251 10.1002/prot.20416 [DOI] [PubMed] [Google Scholar]
- Cormack B. P., Valdivia R. H., Falkow S. (1996). FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38 10.1016/0378-1119(95)00685-0 [DOI] [PubMed] [Google Scholar]
- Evans J. S., Levine B. A., Trayer I. P., Dorman C. J., Higgins C. F. (1986). Sequence-imposed structural constraints in the TonB protein of E. coli. FEBS Lett. 208, 211–216 10.1016/0014-5793(86)81020-1 [DOI] [PubMed] [Google Scholar]
- Ghosh J., Postle K. (2004). Evidence for dynamic clustering of carboxy-terminal aromatic amino acids in TonB-dependent energy transduction. Mol. Microbiol. 51, 203–213 10.1046/j.1365-2958.2003.03816.x [DOI] [PubMed] [Google Scholar]
- Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose P-BAD promoter. J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E. W., Braun V. (1976). Nature of the energy requirement for the irreversible adsorption of bacteriophages T1 and Φ80 to Escherichia coli. J. Bacteriol. 125, 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs P. I., Larsen R. A., Postle K. (2002). Quantitation of known components of the Escherichia coli TonB-dependent energy transduction system: TonB, ExbB, ExbD, and FepA. Mol. Microbiol. 44, 271–281 10.1046/j.1365-2958.2002.02880.x [DOI] [PubMed] [Google Scholar]
- Hill C. W., Harnish B. W. (1981). Inversions between ribosomal RNA genes of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 78, 7069–7072 10.1073/pnas.78.1.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadner R. J. (1990). Vitamin B12 transport in Escherichia coli: energy coupling between membranes. Mol. Microbiol. 4, 2027–2033 10.1111/j.1365-2958.1990.tb00562.x [DOI] [PubMed] [Google Scholar]
- Kadner R. J., Heller K. J. (1995). Mutual inhibition of cobalamin and siderophore uptake systems suggests their competition for TonB function. J. Bacteriol. 177, 4829–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson M., Hannavy K., Higgins C. F. (1993). A sequence-specific function for the N-terminal signal-like sequence of the TonB protein. Mol. Microbiol. 8, 379–388 10.1111/j.1365-2958.1993.tb01582.x [DOI] [PubMed] [Google Scholar]
- Kaserer W. A., Jiang X., Xiao Q., Scott D. C., Bauler M., Copeland D., Newton S. M., Klebba P. E. (2008). Insight from TonB hybrid proteins into the mechanism of iron transport through the outer membrane. J. Bacteriol. 190, 4001–4016 10.1128/JB.00135-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krewulak K. D., Vogel H. J. (2011). TonB or not TonB: is that the question? Biochem. Cell Biol. 89, 87–97 10.1139/O10-141 [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Larsen R. A., Deckert G. E., Kastead K. A., Devanathan S., Keller K. L., Postle K. (2007). His20 provides the sole functionally significant side chain in the essential TonB transmembrane domain. J. Bacteriol. 189, 2825–2833 10.1128/JB.01925-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen R. A., Myers P. S., Skare J. T., Seachord C. L., Darveau R. P., Postle K. (1996). Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J. Bacteriol. 178, 1363–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen R. A., Postle K. (2001). Conserved residues Ser(16) and His(20) and their relative positioning are essential for TonB activity, cross-linking of TonB with ExbB, and the ability of TonB to respond to proton motive force. J. Biol. Chem. 276, 8111–8117 10.1074/jbc.M007479200 [DOI] [PubMed] [Google Scholar]
- Larsen R. A., Thomas M. G., Postle K. (1999). Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 31, 1809–1824 10.1046/j.1365-2958.1999.01317.x [DOI] [PubMed] [Google Scholar]
- Larsen R. A., Wood G. E., Postle K. (1993). The conserved proline-rich motif is not essential for energy transduction by Escherichia coli TonB protein. Mol. Microbiol. 10, 943–953 10.1111/j.1365-2958.1993.tb00966.x [DOI] [PubMed] [Google Scholar]
- Larsen R. A., Letain T. E., Postle K. (2003). In vivo evidence of TonB shuttling between the cytoplasmic and outer membrane in Escherichia coli. Mol. Microbiol. 49, 211–218 10.1046/j.1365-2958.2003.03579.x [DOI] [PubMed] [Google Scholar]
- Lee S. K., Keasling J. D. (2005). A propionate-inducible expression system for enteric bacteria. Appl. Environ. Microbiol. 71, 6856–6862 10.1128/AEM.71.12.7888-7896.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letain T. E., Postle K. (1997). TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Gram-negative bacteria. Mol. Microbiol. 24, 271–283 10.1046/j.1365-2958.1997.3331703.x [DOI] [PubMed] [Google Scholar]
- Liu F., Culham D. E., Vernikovska Y. I., Keates R. A., Boggs J. M., Wood J. M. (2007). Structure and function of transmembrane segment XII in osmosensor and osmoprotectant transporter ProP of Escherichia coli. Biochemistry 46, 5647–5655 10.1021/bi700201w [DOI] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Miller V. L., Taylor R. K., Mekalanos J. J. (1987). Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell 48, 271–279 10.1016/0092-8674(87)90430-2 [DOI] [PubMed] [Google Scholar]
- Nanatani K., Fujiki T., Kanou K., Takeda-Shitaka M., Umeyama H., Ye L., Wang X., Nakajima T., Uchida T., Maloney P. C., Abe K. (2007). Topology of AspT, the aspartate: alanine antiporter of Tetragenococcus halophilus, determined by site-directed fluorescence labeling. J. Bacteriol. 189, 7089–7097 10.1128/JB.00088-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanatani K., Maloney P. C., Abe K. (2009). Structural and functional importance of transmembrane domain 3 (TM3) in the aspartate: alanine antiporter AspT: topology and function of the residues of TM3 and oligomerization of AspT. J. Bacteriol. 191, 2122–2132 10.1128/JB.00830-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Smith D. F. (1974). Culture medium for enterobacteria. J. Bacteriol. 119, 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis A. A., Manning M., Held K. G., Postle K. (2009). Cytoplasmic membrane protonmotive force energizes periplasmic interactions between ExbD and TonB. Mol. Microbiol. 73, 466–481 10.1111/j.1365-2958.2009.06785.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelek P. D., Croteau N., Ng-Thow-Hing C., Khursigara C. M., Moiseeva N., Allaire M., Coulton J. W. (2006). Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science 312, 1399–1402 10.1126/science.1128057 [DOI] [PubMed] [Google Scholar]
- Peacock S. R., Weljie A. M., Peter Howard S., Price F. D., Vogel H. J. (2005). The solution structure of the C-terminal domain of TonB and interaction studies with TonB box peptides. J. Mol. Biol. 345, 1185–1197 10.1016/j.jmb.2004.11.026 [DOI] [PubMed] [Google Scholar]
- Postle K. (2007). TonB system, in vivo assays and characterization. Meth. Enzymol. 422, 245–269 [DOI] [PubMed] [Google Scholar]
- Postle K., Kadner R. J. (2003). Touch and go: tying TonB to transport. Mol. Microbiol. 49, 869–882 10.1046/j.1365-2958.2003.03629.x [DOI] [PubMed] [Google Scholar]
- Postle K., Kastead K. A., Gresock M. G., Ghosh J., Swayne C. D. (2010). The TonB dimeric crystal structures do not exist in vivo. MBio 1. 10.1128/mBio.00307-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D. M., De Boer P. A. (1997). The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell 91, 685–694 10.1016/S0092-8674(00)80455-9 [DOI] [PubMed] [Google Scholar]
- Reynolds P. R., Mottur G. P., Bradbeer C. (1980). Transport of vitamin B12 in Escherichia coli. Some observations on the roles of the gene products of btuC and tonB. J. Biol. Chem. 255, 4313–4319 [PubMed] [Google Scholar]
- Sauter A., Howard S. P., Braun V. (2003). In vivo evidence for TonB dimerization. J. Bacteriol. 185, 5747–5754 10.1128/JB.185.19.5747-5754.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer K., Rodionov D. A., De Reuse H. (2008). New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem. Sci. 33, 330–338 10.1016/j.tibs.2008.04.012 [DOI] [PubMed] [Google Scholar]
- Seliger S. S., Mey A. R., Valle A. M., Payne S. M. (2001). The two TonB systems of Vibrio cholerae: redundant and specific functions. Mol. Microbiol. 39, 801–812 10.1046/j.1365-2958.2001.02273.x [DOI] [PubMed] [Google Scholar]
- Shedlovsky A., Brenner S. (1963). A chemical basis for the host-induced modification of T-even bacteriophages. Proc. Natl. Acad. Sci. U.S.A. 50, 300–305 10.1073/pnas.50.2.300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultis D. D., Purdy M. D., Banchs C. N., Wiener M. C. (2006). Outer membrane active transport: structure of the BtuB: TonB complex. Science 312, 1396–1399 10.1126/science.1127694 [DOI] [PubMed] [Google Scholar]
- Skare J. T., Postle K. (1991). Evidence for a TonB-dependent energy transduction complex in Escherichia coli. Mol. Microbiol. 5, 2883–2890 10.1111/j.1365-2958.1991.tb01848.x [DOI] [PubMed] [Google Scholar]
- Swayne C., Postle K. (2011). Taking the Escherichia coli TonB transmembrane domain “offline”? Non-protonatable Asn substitutes fully for TonB His20. J. Bacteriol. 193, 3693–3701 10.1128/JB.05219-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons M. F., Bokma E., Koronakis E., Hughes C., Koronakis V. (2009). The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc. Natl. Acad. Sci. U.S.A. 106, 7173–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wookey P. (1982). The tonB gene of E. coli: Energy-coupling or molecular processing of permeases? FEBS Lett. 139, 145–154 10.1016/0014-5793(82)80838-7 [DOI] [PubMed] [Google Scholar]
- Yang Q., Wang X., Ye L., Mentrikoski M., Mohammadi E., Kim Y. M., Maloney P. C. (2005). Experimental tests of a homology model for OxlT, the oxalate transporter of Oxalobacter formigenes. Proc. Natl. Acad. Sci. U.S.A. 102, 8513–8518 10.1073/pnas.0406904102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L., Jia Z., Jung T., Maloney P. C. (2001). Topology of OxlT, the oxalate transporter of Oxalobacter formigenes, determined by site-directed fluorescence labeling. J. Bacteriol. 183, 2490–2496 10.1128/JB.183.8.2490-2496.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L., Maloney P. C. (2002). Structure/function relationships in OxlT, the oxalate/formate antiporter of Oxalobacter formigenes: assignment of transmembrane helix 2 to the translocation pathway. J. Biol. Chem. 277, 20372–20378 10.1074/jbc.M111140200 [DOI] [PubMed] [Google Scholar]