SUMMARY
RNase R, an Escherichia coli exoribonuclease important for degradation of structured RNAs, increases 3- to 10-fold under certain stress conditions due to an increased half-life for this usually unstable protein. Components of the trans-translation machinery, tmRNA and its associated protein, SmpB, are essential for RNase R instability. However, it is not understood why exponential phase RNase R is unstable or how it becomes stabilized in stationary phase. We show here that these phenomena are regulated by acetylation catalyzed by YfiQ protein. One residue, Lys544, is acetylated in exponential phase RNase R, but not in the stationary phase protein, resulting in tighter binding of tmRNA-SmpB to the C-terminal region of exponential phase RNase R, and subsequent proteolytic degradation. Removal of the positive charge at Lys544 or a negative charge in the C-terminal region likely disrupts their interaction facilitating tmRNA-SmpB binding. These findings indicate that acetylation can regulate the stability of a bacterial protein.
INTRODUCTION
Ribonucleases (RNases) play important roles in all aspects of RNA metabolism including maturation of RNA precursors, turnover of mRNA, and degradation of stable RNAs under stress conditions or as a consequence of RNA quality control (reviewed in Deutscher, 2003, 2006; Andrade et al., 2009; Carpousis et al., 2009; Deutscher, 2009). However, despite their importance, relatively little is known about whether RNase levels might change in response to physiological conditions or how such changes might be regulated. One RNase known to be regulated is Escherichia coli RNase R, whose levels increase 3- to 10-fold under a variety of stress conditions, such as cold shock and stationary phase (Cairrão et al., 2003; Chen and Deutscher, 2005; Andrade et al., 2006). RNase R is a ubiquitous, processive exoribonuclease (Zuo and Deutscher, 2001; Cheng and Deutscher, 2002) that together with polynucleotide phosphorylase participates in the degradation of structured RNAs (Cheng et al., 1998; Cheng and Deutscher, 2005; Vincent and Deutscher, 2006, 2009a, 2009b). Hence, its elevation may be important for a cell’s response to stress conditions (Chen and Deutscher, 2005; Andrade et al., 2006). Based on these considerations, understanding the mechanisms by which RNase R is regulated is of considerable interest.
In earlier work, we showed that in contrast to most E. coli proteins, RNase R is extremely unstable in the exponential phase of growth with a half-life of ~10 min due to action of Lon protease (Chen and Deutscher, 2010). In contrast, it becomes stabilized upon entry into stationary phase, and it is this stabilization that results in RNase R elevation in stationary phase compared to exponential growth (Chen and Deutscher, 2005, 2010). We also showed (Liang and Deutscher, 2010) that the instability of RNase R in exponential phase cells is due to the binding of two components of the trans-translation system, tmRNA and its associated protein SmpB (Karzai et al., 1999; Karzai and Sauer, 2001; Withey and Friedman, 2003), to the C-terminal region of the RNase. In the absence of either tmRNA or SmpB, the half-life of RNase R increases from ~10 min to >60 min (Liang and Deutscher, 2010); likewise, removal of the C-terminal region of RNase R also stabilizes the protein (Liang and Deutscher, 2010). In both situations, the level of RNase R in exponential phase cells increases to close to that in stationary phase (Liang and Deutscher, 2010). However, it is still not clear what is responsible for the difference in stability between exponential and stationary phase RNase R.
Here, we demonstrate that the difference is due to post-translational modification of exponential phase RNase R by acetylation of Lys544. Acetylation leads to tighter binding of tmRNA-SmpB to the exponential phase protein compared to its counterpart in stationary phase. We also show that mutation of Lys544 to another positively-charged amino acid, arginine, which is not acetylated, stabilizes exponential phase RNase R. In contrast, converting Lys544 to the uncharged amino acid, alanine, maintains the protein’s instability in exponential phase even though in this case residue 544 is not acetylated. Most interestingly, the alanine mutation now renders the stationary phase protein unstable. Likewise, mutation of two acidic amino acid residues in the C-terminal region, Glu764 and Asp766, also leads to instability of stationary phase RNase R. Additionally, we find that the product of the yfiQ gene is responsible for acetylation of Lys544. These findings provide a clear example in which acetylation regulates the half-life of a bacterial protein. A model is presented to explain how acetylation can lead to instability of RNase R.
RESULTS
RNase R displays a dramatic difference in stability between exponential phase and stationary phase; the half-life of the exponential phase protein is ~10 min, whereas the protein in stationary phase is completely stable (Chen and Deutscher, 2010; Liang and Deutscher, 2010). The mechanisms responsible for these differences have been unclear, and are explored in the studies described below.
Levels of tmRNA and SmpB Increase in Stationary Phase Cells
The instability of RNase R in exponential phase is dependent on the binding of both tmRNA and its associated protein, SmpB (Liang and Deutscher, 2010). One possibility to explain the stabilization of RNase R in stationary phase is that the levels of either or both of these factors are lowered resulting in decreased binding. To examine this possibility, we directly compared the amount of tmRNA and SmpB in exponential and stationary phase cells. Interestingly, both tmRNA and SmpB actually increased in stationary phase to levels almost 4-fold higher than those present in exponential phase (Figure S1). Consequently, it is unlikely that stabilization of RNase R in stationary phase is due to effects on tmRNA and SmpB. These findings suggest that a change in RNase R itself might be responsible for the difference in stability of the two forms of the enzyme.
Purification of RNase R from Exponential and Stationary Phase Cells
In order to examine RNase R in more detail, we extensively purified the protein from cells in exponential and stationary phase taking advantage of a His-tag placed at the N-terminus of the protein. Chromosomally-encoded RNase R, expressed from its own promoter, was used to avoid any effects that might ensue due to overexpression from a plasmid-encoded gene. Each form of the protein was purified to apparent homogeneity for use in further studies (Figure S2). Both proteins retained nuclease activity against poly(A) (data not shown).
tmRNA and SmpB Bind More Tightly to Exponential Phase RNase R
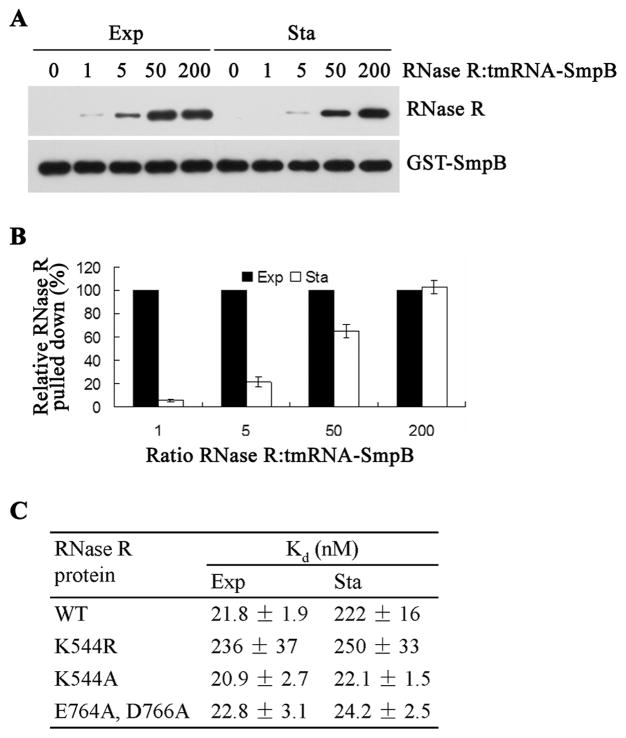
Binding of tmRNA-SmpB to the C-terminal region of RNase R is responsible for the latter’s instability in exponential phase cells (Liang and Deutscher, 2010). To determine whether altered binding might account for the stability of RNase R in stationary phase, we measured directly the binding of each form of RNase R to tmRNA-SmpB in a pull-down experiment (see “Experimental Procedures”). To do this, increasing amounts of purified exponential or stationary phase RNase R were added to a constant amount of tmRNA-GST-tagged SmpB, and the amount of RNase R bound was determined by immunoblotting after elution. The results are presented in Figure 1A and are quantified in Figure 1B. The data are presented as the relative amount of each RNase R bound at each ratio of RNase R to tmRNA-SmpB complex, which varied from 1 to 200. The data show that binding is weaker with RNase R isolated from stationary phase cells compared to RNase R from exponential phase cells. For example, at a 1: 1 ratio of RNase R to tmRNA-SmpB, less than 10% as much stationary phase RNase R is pulled down compared to the exponential phase enzyme, which has been set at 100% at each RNase R amount tested. As more stationary phase RNase R is added, its relative percent bound increases such that at the highest amount examined its binding equals that of the exponential phase protein. Using the data from the two lowest ratios of RNase R:tmRNA-SmpB, we calculated apparent Kd values for binding (Figure 1C). These data indicate that binding between tmRNA-SmpB and stationary phase RNase R is ~10-fold weaker than binding to the exponential phase protein, and they help to explain why RNase R is stabilized in stationary phase. Moreover, these findings suggest that there is a structural difference between the two forms of RNase R that would explain their different affinities for the tmRNA-SmpB complex.
Figure 1.
Binding of exponential phase (Exp) and stationary phase (Sta) RNase R to tmRNA-SmpB complex. (A) Increasing amounts of purified RNase R were mixed with a constant amount of GST-SmpB and tmRNA for pull-down assays as described in “Experimental Prodedures”. RNase R and GST-SmpB in the eluant from each pull-down were resolved on 8% SDS-PAGE and detected by purified RNase R antibody and anti-GST mAb, respectively. (B) Quantification of 3 independent experiments carried out as shown in Panel (A). The amount of exponential phase RNase R pulled-down was set at 100% for each amount of RNase R added, and the corresponding value for the stationary phase protein is shown. Error bars indicate SEM. (C) Binding of exponential or stationary phase RNase R to tmRNA-SmpB complex. The Kd values for the wild type or the mutant RNase R proteins were determined from data as in panel (A). The Kd values shown represent the mean of three independent experiments for WT and two for the mutant proteins. Kd values were calculated using only the two lowest ratios (1:1 and 5:1) of RNase R:tmRNA-SmpB from the concentrations of bound and free RNase R and tmRNA-SmpB.
Exponential Phase RNase R is Acetylated
One possible structural difference between the exponential and stationary phase RNase R proteins is some type of post-translational modification. We initially focused our attention on acetylation because one recent proteomic analysis included RNase R among 91 acetylated proteins in E. coli (Zhang et al., 2009). Therefore, using an antibody directed against N-acetyl-lysine, we examined whether RNase R from either phase of growth is acetylated. The data in Figure 2A indicate that RNase R from exponential phase contains N-acetyl-lysine, whereas that from stationary phase lacks the modification.
Figure 2.
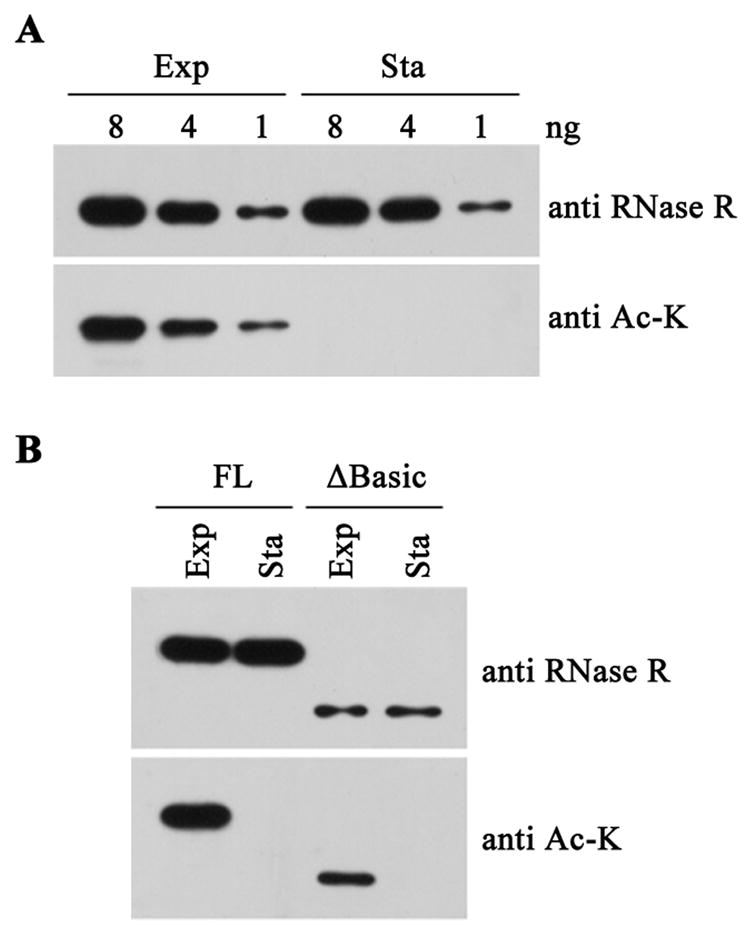
Structural analysis of exponential phase (Exp) and stationary phase (Sta) RNase R. (A) Immunological analysis for acetyl-lysine. Varying amounts of purified exponential and stationary phase RNase R were resolved on 8% SDS-PAGE and probed with RNase R antibody and anti acetylated-lysine monoclonal antibody (Ac-K). (B) Analysis of acetylation in C-terminal-truncated RNase R. Truncated RNase R (ΔBasic) was immunoprecipitated from extracts of exponential phase and stationary phase cells using purified RNase R antibody and then detected with RNase R antibody and anti acetylated-lysine monoclonal antibody, respectively, after separation on 8% SDS-PAGE. Purified full-length (FL) exponential and stationary phase RNase R were loaded as controls. Note that different amounts of RNase R were present in the FL purified protein lanes and the ΔBasic extract lanes. Shown are representative gels from experiments carried out twice.
To confirm and extend this observation, the purified exponential and stationary phase proteins were analyzed by mass spectrometry following digestion with chymotrypsin as described in “Experimental Procedures”. A single altered peptide (residues 541–548) was observed in the exponential phase RNase R whose mass increase (42 Da) corresponded to the presence of one acetyl group (Figure S3 and Table S1). In contrast, the identical peptide in the stationary phase protein was unmodified (Figure S3 and Table S1). No unmodified peptide was observed in the exponential phase preparation. Moreover, no other modification was observed in either protein upon analysis of peptides covering ~90% of the residues in RNase R (Table S1). Based on these data, we propose that acetylation of Lys544 is important for the tighter binding of the tmRNA-SmpB complex to exponential phase RNase R, and thus for the instability of the exponential phase protein.
Truncated RNase R is Acetylated
To determine whether acetylation is independent of binding of the tmRNA-SmpB complex, we examined RNase R lacking its C-terminal 83 residues (ΔBasic) to ascertain if it is acetylated in vivo. Since earlier work had shown that the C-terminal region is required for binding of tmRNA and SmpB (Liang and Deutscher, 2010), acetylation of the truncated protein would eliminate any role for tmRNA-SmpB in the acetylation process. The data in Figure 2B indicate that truncated RNase R is acetylated in exponential phase cells, and as with the full-length protein, it is unmodified in stationary phase cells. These data show that neither the C-terminal region nor binding of tmRNA-SmpB is required for acetylation of RNase R.
Mutation of Lys544 Affects RNase R Stability
To further test the proposal that acetylation affects RNase R stability, Lys544 was mutated to either Arg or Ala. In the K544R mutant, the positive charge on Lys would be maintained, whereas in the K544A mutant, the positive charge would be eliminated just as it is when the Lys residue is acetylated. For each mutant RNase R, we determined its level in exponential and stationary phase cells as well as its half-life. Moreover, we purified each of the mutant proteins from exponential and stationary phase cells and measured its binding to tmRNA-SmpB. We also determined their catalytic activity using poly(A) as substrate and found no reduction in specific activity compared to their wild type counterparts (data not shown).
The data presented in Figure 3A show that substituting Arg for Lys544 dramatically increases the amount of RNase R in exponential phase cells to a level essentially identical to that in stationary phase cells. This increase is due to stabilization of the mutant protein in exponential phase cells (Figure 3B) such that its half-life increases from ~10 min to >120 min, comparable to that of the wild type (Chen and Deutscher, 2010) and mutant proteins in stationary phase (Figure 3B). In both the K544R mutant protein and in the wild type protein in stationary phase there is a positive charge at position 544, and in each case, this is associated with a stable RNase R. Binding measurements indicate that the K544R mutant protein binds tmRNA-SmpB weakly (Figure 1C), providing an explanation for why it is stabilized in exponential phase cells.
Figure 3.
Amount and stability of RNase R mutant proteins. (A) Amount of wild type (WT) and K544R mutant RNase R protein in exponential phase (Exp) and stationary phase (Sta) cells. (B) Half-life of wild type and K544R mutant RNase R in exponential and stationary phase cells. (C) Amount of wild type and K544A mutant RNase R protein in exponential phase and stationary phase cells. (D) Half-life of K544A mutant RNase R in exponential and stationary phase cells. (E) Amount of wild type and E764A, D766A mutant RNase R protein in exponential phase and stationary phase cells. (F) Half-life of E764A, D766A mutant RNase R in exponential and stationary phase cells. The indicated cells were grown in YT medium, treated with chloramphenicol and assayed for RNase R protein by immunoblotting as described in “Experimental Procedures”. For panels (A) through (F), 5 μg of total protein were added to each lane. (G) Acetylation analysis of RNase R mutant proteins. Equal amounts of purified exponential phase and stationary phase wild type, K544R, K544A and E764A, D766A mutant proteins (1 ng) were resolved on 8% SDS-PAGE and then probed with RNase R antibody and anti acetylated-lysine monoclonal antibody (Ac-K), respectively. Each gel shown in panels (A) through (G) is a representative experiment carried out at least twice.
In contrast, the K544A mutant protein behaves very differently. In this protein the positive charge at position 544 is no longer present, and the protein is present in exponential phase at the same decreased level as the wild type protein (Figure 3C), which also is uncharged at position 544 due to acetylation. Most interestingly, the K544A mutant protein also becomes destabilized in stationary phase such that its half-life decreases from >240 min for the wild type protein (Chen and Deutscher, 2010) to ~28 min (Figure 3D). This shorter half-life results in a reduced amount of mutant protein in stationary phase compared to the wild type protein (Figure 3C). Moreover, binding experiments show that the mutant stationary phase protein binds as tightly to tmRNA-SmpB as its exponential phase counterpart (Figure 1C), explaining its instability in stationary phase cells. These data suggest that the charge of the residue at position 544 plays an important role in the stability of RNase R.
Mutation of Acidic Residues in the C-terminal Region Affects RNase R Stability
tmRNA-SmpB binds to the C-terminal region of RNase R, and as shown, this binding is enhanced in forms of RNase R in which the positive charge on Lys544 is eliminated by acetylation or mutation (Figure 1C). One possibility to explain these findings is that Lys544 normally interacts with certain acidic residues in the C-terminal region making the latter less accessible for binding of tmRNA-SmpB. If this idea is correct, then mutation of these negatively-charged residues might disrupt the interaction with Lys544 leading to a more exposed C-terminal region and destabilizing RNase R even in stationary phase cells.
To test this prediction and to identify residues in the C-terminal tail of RNase R that might interact with Lys544, we first carried out a bioinformatics analysis of this region. The C-terminal region of E. coli RNase R is over 80 residues long and is overall highly positively charged. Based on sequence conservation among C-terminal tails of RNase R homologs (Figure S4), and homology modeling of this region (described in more detail in Supplemental Data), we identified two residues, Glu764 and Asp766, as potential candidates for interaction with Lys544 (Figure S5). These residues were simultaneously mutated to Ala, and the properties of the resulting protein examined (Figure 3E and 3F). As can be seen, the mutant protein continues to be acetylated in exponential phase cells (Figure 3G), in contrast to the K544R and K544A mutant proteins, which cannot be acetylated (Figure 3G). Most importantly, the mutant protein accumulates to a lower level in stationary phase cells (1.5-fold increase) than does wild type RNase R (4-fold increase) (Figure 3E). This is due to its shorter half-life in stationary phase cells (~30 min) (Figure 3F) compared to wild type RNase R (>120 min) (Figure 3B), resulting from tighter binding of tmRNA-SmpB to the mutant protein (Figure 1C). At present, we do not know whether both Glu764 and Asp766, or only one of these residues, are important for the altered properties of RNase R. Nevertheless, these findings are consistent with an interaction between Lys544 and the C-terminal region that can be disrupted either by removal of the positive charge on Lys544 or of a negative charge in the C-terminal region.
Acetylation of Lys544 is Dependent on the Product of the yfiQ Gene
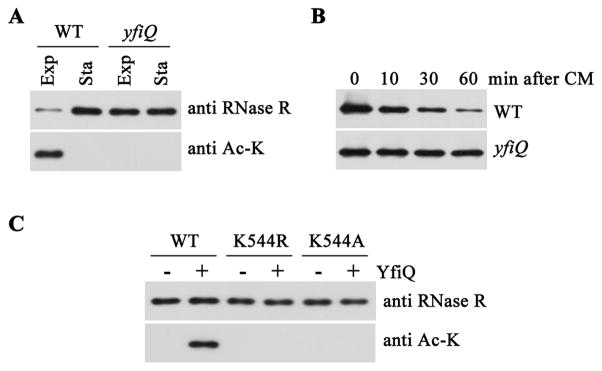
To examine the relation between acetylation of Lys544 and RNase R stability in more detail, it was of interest to identify and eliminate the enzyme responsible for acetylation of the RNase. Although E. coli contains multiple genes that may encode the putative acetyltranferase (Escalante-Semerena, 2010), we initially focused on the yfiQ gene. YfiQ is highly similar (92% identity) to the Pat protein of Salmonella which was recently shown to participate in acetylation of multiple proteins in that organism (Wang et al., 2010). Accordingly, we determined the level of RNase R in a strain in which the yfiQ gene is interrupted with a kanamycin cassette. Removal of YfiQ eliminates acetylation of exponential phase RNase R (Figure 4A, bottom panel), indicating that YfiQ is the enzyme which acetylates Lys544. Concomitantly, the amount of RNase R in exponential phase cells is elevated to that present in stationary phase cells (Figure 4A, top panel) because RNase R is now stabilized (Figure 4B). Moreover, purified YfiQ acetylates wild type RNase R in vitro, whereas it displays no activity against either the K544R or the K544A mutant RNase R protein (Figure 4C). These data demonstrate that YfiQ is responsible for acetylation of Lys544 in RNase R, and they show that acetylation directly regulates the level of RNase R in exponential phase cells. Based on these results, we propose that yfiQ be renamed pla (protein lysine acetyltransferase) since the gene symbol, pat, is alreadly used in E. coli.
Figure 4.
Acetylation of RNase R by YfiQ. (A) Amount (top panel) and acetylation (bottom panel) of RNase R in wild type and yfiQ mutant strains. Exponential (Exp) and stationary (Sta) phase cells were lysed and subjected to immunoprecipitation with RNase R antibody. Precipitates were then analyzed by immunoblotting using RNase R antibodies or anti acetylated-lysine monoclonal antibody (Ac-K). (B) Half-life of RNase R in wild type and yfiQ mutant strains. Cells were grown in YT medium to exponential phase, and the half-life of RNase R was determined as in Figure 3. Five μg of total protein was added to each lane in panels (A) and (B). (C) YfiQ directly acetylates RNase R in vitro. Purified stationary phase WT, K544R or K544A mutant RNase R proteins (50 ng) were incubated with or without 50 ng of purified YfiQ. Products were then analyzed by immunoblotting using RNase R antibodies or anti acetylated-lysine monoclonal antibody. Each gel shown is a representative experiment carried out twice.
DISCUSSION
Although RNases are major participants in essentially all aspects of RNA metabolism, relatively little is known about how these enzymes might be regulated. The information presented here indicates that an RNase can be regulated by post-translational modification. Moreover, the modification does not affect the RNase’s catalytic activity, as is often the case, but rather, the protein’s stability. As such, these findings greatly expand our understanding of RNase regulation and open up new possibilities for investigation in the RNase field.
The data presented here also provide a likely explanation for the known difference in stability between RNase R in exponential phase and stationary phase cells (Chen and Deutscher, 2010). Thus, we have shown that exponential phase RNase R is acetylated at a single lysine residue at position 544, whereas the stationary phase enzyme lacks the modification, and we suggest that this acetylation is responsible for regulating the instability of the exponential phase protein. We propose a simple model (Figure S6) to explain how acetylation regulates RNase R stability. In this model, Lys544 interacts with acidic residues within the C-terminal region of the protein, and this interaction maintains the C-terminal region in a position that is poorly accessible to tmRNA-SmpB whose binding is essential for RNase R instability (Liang and Deutscher, 2010). Addition of the acetyl group would remove the positive charge on lysine, breaking this interaction, thereby exposing the C-terminal region, and enabling tmRNA-SmpB to bind tightly, which is necessary for proteolysis by Lon to occur (Chen and Deutscher, 2010; Liang and Deutscher, 2010). Although the X-ray structure of E. coli RNase R is not yet available, homology modeling to its ortholog, RNase II, suggests that Lys544 would be on the surface of the protein, near the entrance to the catalytic channel (Frazão et al., 2006; Zuo et al., 2006). The C-terminal region of RNase R, which is at least 80 residues in length (Liang and Deutscher, 2010), would have no difficulty in reaching and interacting with Lys544 (see structural model in Figure S5).
The proposed model is supported by multiple observations: 1) RNase R in stationary phase cells is not acetylated, and therefore, position 544 is positively charged. As a consequence, the C-terminal region would not be accessible, and as we have shown, tmRNA-SmpB binds weakly, resulting in a stable RNase R; 2) The K544R mutant protein is not acetylated. Thus, position 544 retains a positive charge, and this exponential phase enzyme is now stable; 3) The K544A mutant protein also is not acetylated, but in this case the protein has lost the positive charge at position 544, and would not be able to interact with the C-terminal region. As a result, the exponential phase protein remains unstable, but most importantly, the stationary phase mutant protein also becomes unstable; 4) The E764A, D766A mutant protein, in which the C-terminal region would not be able to interact with Lys544, also is unstable in both exponential and stationary phase cells; and 5) Removal of YfiQ, which eliminates acetylation of Lys544, stabilizes exponential phase RNase R, and shows directly that acetylation regulates RNase R stability. Taken together, these observations lend support to the model and would explain the difference in stability between RNase R in exponential phase and stationary phase cells. Further structural analysis of exponential and stationary phase RNase R will be necessary to prove the validity of the proposed model.
These studies also provide a clear example in which acetylation affects the stability of a protein in bacteria. Acetylation has been shown to affect many proteins and processes in eukaryotic systems (reviewed in Kouzarides, 2000; Blander and Guarente, 2004; Cohen and Yao, 2004; Yang, 2004; Bordone and Guarente, 2005), but to date, only a few instances of a specific effect on a bacterial protein have been uncovered (Escalante-Semerena, 2010; Hu et al., 2010; Liarzi et al, 2010; Thao et al., 2010). The discovery here that acetylation of one specific lysine residue in RNase R increases binding of the tmRNA-SmpB complex resulting in protein instability provides a clear example of a direct effect of acetylation on a regulatory process in bacteria. With the recent discovery that a large number of E. coli proteins are acetylated in vivo (Yu et al., 2008; Zhang et al., 2009; Wang et al., 2010), it is likely that the findings reported here are just the beginning of what will be a widespread phenomenon in bacteria.
EXPERIMENTAL PROCEDURES
Detailed information about experimental procedures not provided here can be found in Supplemental Information.
Bacterial Strains and Growth Conditions
All strains used were derivatives of E. coli K12 strain MG1655(Seq)rph+ and were previously described (Liang and Deutscher, 2010). Cells were grown at 37°C in liquid culture in YT medium. Antibiotics, when present, were at the following concentrations: kanamycin, 50 μg/ml; ampillicin, 100 μg/ml; chloramphenicol, 34 μg/ml. Exponential phase cells were collected at an A550 of ~0.3, and cells grown overnight were used as stationary phase samples.
Purification of RNase R
His-tagged RNase R was purified on a Ni-NTA column followed by chromatography on MonoS. Details are presented in Supplemental Information.
Pull-down Assay
GST-SmpB (0.2 nM) was mixed with an equal amount of tmRNA and incubated at room temperature for 15 min in buffer A [140 mM NaCl, 8 mM NaH2PO4, 2 mM K2HPO4, 10 mM KCl, 0.1% NP-40, and 1 mM phenylmethanesulfonyl fluoride (PMSF)]. Different amounts of exponential phase or stationary phase RNase R were then added and incubated with gentle rocking for 60 min at 4°C. Following addition of 50 μl of GST resin, the mixture was incubated for an additional 30 min at 4°C. The GST resin was recovered by centrifugation and washed five times with 300 μl of buffer A. Under these wash conditions, no SmpB is removed from the GST resin. Bound proteins were then eluted with 50 μl of buffer B (50 mM Tris-HCl, pH 8.0, 10 mM reduced glutathione). Eluted proteins were separated by SDS-PAGE and probed with RNase R antibody and anti-GST mAb.
Mass Spectrometry Analysis and Protein Sequence Database Search
Chymotrypsin digestion of purified exponential phase and stationary phase RNase R and LC/MS/MS analysis were performed at the W. M. Keck Mass Spectrometry and Proteomics Laboratory, Yale University. Tandem mass spectra data were searched against the NCBI-nr database using the MASCOT search engine (Matrix Science, London, U.K.), which allowed acetylation sites to be identified. Detailed procedures are described in Supplemental Information.
Immunoprecipitation of RNase R
RNase R was incubated with purified RNase R antibody and the complex was adsorbed with Protein A agarose beads. Eluted RNase R was determined by Western blot analysis. Details are provided in Supplemental Information.
Measurement of RNase R Half-life
Cells were grown under normal conditions to an A550 of ~0.3 or overnight. A portion of cells was collected for the zero time point and chloramphenicol was added to the remaining culture at 200 μg/ml. Cells were collected at the indicated times, lysed by sonication (Chen and Deutscher, 2005), and assayed by immunoblotting to determine the amount of RNase R remaining.
RNase R Activity Assay
The activity of RNase R was determined by release of acid-soluble material from [3H]poly(A) (Chen and Deutscher, 2005). Details are provided in Supplemental Information.
YfiQ-mediated in vitro Acetylation
The in vitro reaction was performed as described previously (Wang et al., 2010) with 50 ng purified stationary phase WT, K544R or K544A RNase R proteins in the absence or presence of 50 ng purified YfiQ. Reaction mixtures were incubated at 37 °C for l h, and then analyzed by immunoblotting using RNase R antibody or acetylated-lysine antibody.
Supplementary Material
HIGHLIGHTS.
RNase R is acetylated in growing cells, but not in stationary phase cells
Acetylation is catalyzed by YfiQ protein
Acetylation leads to tighter binding of tmRNA-SmpB complex and instability of RNase R
Acetylation regulates protein stability in bacteria
Acknowledgments
This work was supported by Grant GM16317 from the National Institute of Health. We thank Dr. Kenneth Rudd for bacterial strains and Dr. Richard Myers for assistance with the recombineering protocol. We also thank members of the laboratory for critical comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrade JM, Cairrão F, Arraiano CM. RNase R affects gene expression in stationary phase: Regulation of ompA. Mol Microbiol. 2006;60:219–228. doi: 10.1111/j.1365-2958.2006.05092.x. [DOI] [PubMed] [Google Scholar]
- Andrade JM, Pobre V, Silva IJ, Domingues S, Arraiano CM. The role of 3′-5′ exoribonucleases in RNA degradation. Prog Mol Biol Transl Sci. 2009;85:187–229. doi: 10.1016/S0079-6603(08)00805-2. [DOI] [PubMed] [Google Scholar]
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- Cairrão F, Cruz A, Mori H, Arraiano CM. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol Microbiol. 2003;50:1349–1360. doi: 10.1046/j.1365-2958.2003.03766.x. [DOI] [PubMed] [Google Scholar]
- Carpousis AJ, Luisi BF, McDowall KJ. Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog Mol Biol Transl Sci. 2009;85:91–135. doi: 10.1016/S0079-6603(08)00803-9. [DOI] [PubMed] [Google Scholar]
- Chen C, Deutscher MP. Elevation of RNase R in response to multiple stress conditions. J Biol Chem. 2005;280:34393–34396. doi: 10.1074/jbc.C500333200. [DOI] [PubMed] [Google Scholar]
- Chen C, Deutscher MP. RNase R is a highly unstable protein regulated by growth phase and stress. RNA. 2010;16:667–672. doi: 10.1261/rna.1981010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- Cheng ZF, Deutscher MP. An important role for RNase R in mRNA decay. Mol Cell. 2005;17:313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- Cheng ZF, Zuo Y, Li Z, Rudd KE, Deutscher MP. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J Biol Chem. 1998;273:14077–14080. doi: 10.1074/jbc.273.23.14077. [DOI] [PubMed] [Google Scholar]
- Cohen T, Yao TP. AcK-knowledge reversible acetylation. Sci STKE. 2004;245:42. doi: 10.1126/stke.2452004pe42. [DOI] [PubMed] [Google Scholar]
- Deutscher MP. Degradation of Stable RNA in Bacteria. J Biol Chem. 2003;278:45041–45044. doi: 10.1074/jbc.R300031200. [DOI] [PubMed] [Google Scholar]
- Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. Maturation and degradation of ribosomal RNA in bacteria. Prog Mol Biol Transl Sci. 2009;85:369–91. doi: 10.1016/S0079-6603(08)00809-X. [DOI] [PubMed] [Google Scholar]
- Escalante-Semerena JC. Nε-lysine acetylation control conserved in all three life domains. Microbe. 2010;5:340–344. doi: 10.1128/microbe.5.340.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazão C, McVey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, Carrondo MA. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–114. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- Hu LI, Lima BP, Wolfe AJ. Bacterial protein acetylation: the dawning of a new age. Mol Microbiol. 2010;77:15–21. doi: 10.1111/j.1365-2958.2010.07204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai AW, Susskind MM, Sauer RT. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Acetylation: A regulatory modification to rival phosphorylation. EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, Li C, Liu F, Jiang H, Li S, Sun J, Wu X, Li C. The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res. 2009;19:307–316. doi: 10.1038/cr.2008.317. [DOI] [PubMed] [Google Scholar]
- Liang W, Deutscher MP. A novel mechanism for ribonuclease regulation: transfer-messenger RNA (tmRNA) and its associated protein SmpB regulate the stability of RNase R. J Biol Chem. 2010;17:29054–29058. doi: 10.1074/jbc.C110.168641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liarzi O, Barak R, Bronner V, Dines M, Sagi Y, Shainskaya A, Eisenbach M. Acetylation represses the binding of CheY to its tatget proteins. Mol Microbiol. 2010;76:932–943. doi: 10.1111/j.1365-2958.2010.07148.x. [DOI] [PubMed] [Google Scholar]
- Thao S, Chen CS, Zhu H, Escalante-Semerena JC. Nε Lysine acetylation of a bacterial transcription factor inhibits its DNA-Binding activity. PLoS ONE. 2010;5:1–9. doi: 10.1371/journal.pone.0015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- Vincent HA, Deutscher MP. The roles of individual domains of RNase R in substrate binding and exoribonuclease activity. The nuclease domain is sufficient for digestion of structured RNA. J Biol Chem. 2009a;284:486–494. doi: 10.1074/jbc.M806468200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent HA, Deutscher MP. Insights into how RNase R degrades structured RNA: analysis of the nuclease domain. J Mol Biol. 2009b;387:570–583. doi: 10.1016/j.jmb.2009.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning ZB, Zeng R, Xiong Y, Guan KL, Zhao S, Zhao GP. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withey JH, Friedman DI. A salvage pathway for protein structures: tmRNA and trans-translation. Annu Rev Microbiol. 2003;57:101–123. doi: 10.1146/annurev.micro.57.030502.090945. [DOI] [PubMed] [Google Scholar]
- Yang XJ. The diverse superfamily of lysine acetyltranferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BJ, Kim JA, Moon JH, Ryu SE, Pan JG. The diversity of lysine-acetylated proteins in Escherichia coli. J Microbiol Biotechnol. 2008;18:1529–1536. [PubMed] [Google Scholar]
- Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics. 2009;8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Deutscher MP. Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Vincent HA, Zhang J, Wang Y, Deutscher MP, Malhotra A. Structural basis for processivity and single-strand specificity of RNase II. Mol Cell. 2006;24:149–156. doi: 10.1016/j.molcel.2006.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.