Abstract
Impaired flexibility in the use of substrates for energy production in the heart is implicated in cardiomyopathy. We investigated the effect of maternal protein restriction during pregnancy in rats on the transcription of key genes in cardiac lipid and carbohydrate metabolism in the offspring. Rats were fed protein-sufficient or protein-restricted (PR) diets during pregnancy. Triacylglycerol concentration in adult (day 105) heart was altered by maternal protein intake contingent on post-weaning fat intake and sex. mRNA Expression of peroxisomal proliferator-activated receptor (PPAR)-α and carnitine palmitoyltransferase-1 was increased by the maternal PR diet in adult, but not neonatal, offspring. PPARα promoter methylation was lower in adult and neonatal heart from PR offspring. These findings suggest that prenatal nutrition alters the future transcriptional regulation of cardiac energy metabolism in the offspring through changes in epigenetic regulation of specific genes. However, changes in gene functional changes may not be apparent in early life.
Keywords: Heart, protein-restriction, PPAR, epigenetic, cardiomyopathy
Introduction
The mammalian heart uses several substrates, including fatty acids, glucose, lactate and ketone bodies, to meet its constant demand for energy to support contractile function. Such flexibility in substrate choice facilitates adequate energy production despite changing availability of substrates in blood. In the fasting state fatty acids are the preferred substrate for energy production in the adult heart. Conversely, in the fed state, when the concentration of non-esterified fatty acids in blood is low, glucose is the main energy substrate. Type 2 diabetes mellitus is associated with accumulation of triacylglycerol (TAG) in the heart and remodelling of the myocardium, left ventricular hypertrophy, myocardial fibrosis and progressive left ventricular systolic dysfunction; this is termed diabetic cardiomyopathy.1,2 These structural changes may reflect increased availability of non-esterified fatty acids in blood due to impaired regulation of fatty acid secretion by adipose tissue and increased demands for fatty acids for energy production to offset impaired glucose utilisation due to insulin resistance.3-5 The resulting increase in fatty acid β-oxidation may induce oxidative damage to myocardial cell membranes leading to remodelling and apoptosis.6,7
Cardiac energy production from fatty acids is regulated by specific transcription factors including PPARα, PPARγ8-10 and sterol regulatory element binding protein (SREBP)-1C.11 Dysregulation of PPARα has been implicated in the pathogenesis of diabetic cardiomyopathy.12 Activation of PPARα increases fatty acid β-oxidation by binding to specific response elements in the promoter regions of carnitine:palmitoyl transferase (CPT)-1 and acyl-CoA oxidase (AOX), which encode the enzymes that catalyse their respective rate-limiting reactions in mitochondria and peroxisomes, and the fatty acid transporter CD36. PPARα also increases transcription of pyruvate dehydrogenase kinase 4 which, in turn, decreases activity of the glycolytic pathway. The evidence for a role for PPARα in diabetic cardiomyopathy is largely based on the findings of animal studies. PPARα null mice were protected from increased cardiac fatty acid utilisation following induction of diabetes by streptozotocin.13 Cardiac-specific over-expression of PPARα was associated with increased fatty acid β-oxidation and cardiac remodelling that was exacerbated by feeding a high fat diet which caused accumulation of TAG within the myocardium13, and left-ventricular dysfunction and premature death.6 SREBP-1C and PPARγ together promote insulin-mediated lipogenesis and TAG biosynthesis.11 SREBP-1C and PPARγ expression is increased in the ventricles of patients with the metabolic syndrome14 which is consistent with increased deposition of TAG.
Environmental constraint before birth is associated with increased risk of the metabolic syndrome in later life.15 Rodent models of maternal under-nutrition during pregnancy induce in the offspring metabolic changes which resemble the metabolic syndrome in humans.16 Feeding pregnant rats a protein-restricted (PR) diet induces in the offspring increased hepatic fatty acid β-oxidation which is marked by raised plasma β-hydroxybutyrate concentration.17 This was associated with increased PPARα, AOX and CPT-1 expression in the liver associated with induced hypomethylation of the PPARα promoter.18 Feeding a PR diet to pregnant rats also induced impaired cardiac function and recovery from ischaemia in the offspring.19,20 However, the effect of maternal under-nutrition during pregnancy on the transcription of genes involved in cardiac energy homeostasis has not been reported.
We investigated in rats the effect of maternal protein restriction during pregnancy and fat intake after weaning on the mRNA expression of critical genes in cardiac fatty acid and carbohydrate metabolism, and on the methylation status of the PPARα promoter in the offspring. We also determined the effect of maternal and post-weaning (PW) diets on TAG concentration in adult heart.
Materials and methods
Animal procedures
The study was carried out in accordance with the Home Office Animals (Scientific Procedures) Act (1986). The hearts used in this study were collected from animals which have been described previously17. Briefly, virgin female Wistar rats (approximately 220g) (n = 6 per dietary group) were mated and fed either protein-sufficient (PS) (180 g protein/kg feed) or PR (90 g protein/kg feed) diet from conception until delivery (Special Diets Services, Witham, Essex). The composition of these diets has been described.17 After spontaneous delivery on approximately post-conceptional day 21, litters were reduced to eight pups, equal males and females, within 24 hours of birth, and the hearts from excess animals were frozen in liquid nitrogen and stored at −80°C (Neonatal group). Offspring were weaned at postnatal day 28 on to either a diet containing lard and soybean oil (9:1, w/w) to provide 40 g fat/kg feed (LF) or a diet containing 100 g fat/kg feed (HF) on postnatal day 28.17 Food intake and body weights of these rats have been reported previously.17 On postnatal day 105, food was withdrawn at about 08.00 hours, but water was provided ad libitum. Offspring were killed by asphyxiation with CO2 6 hours later and hearts were removed immediately, frozen in liquid nitrogen and stored at −80°C.
Measurement of triacylglycerol composition
Portions of right and left ventricle from adult hearts (approximately 100mg) were pulverised under liquid nitrogen. Internal standard (triheptadecanoin, 70μg) was added, and total lipids were extracted and TAG was purified by solid phase extraction using 100mg aminopropylsilica cartridges.21 Fatty acid methyl esters (FAMEs) were synthesised by incubation with methanol containing 2% (v/v) sulphuric acid at 50°C for 2 hours.21 FAMEs were resolved using a 6890 gas chromatograph (Agilent, Cheshire, UK) equipped with a 30m × 0.25 μm × 0.25mm BPX-70 fused silica capillary column (SGE, Milton Keynes, UK) and flame ionization detection. The concentrations of individual fatty acids were determined by comparing the peak area to the peak area of the internal standard, corrected for the mass of extracted tissue. Total TAG concentration was calculated from the sum of the areas of the identified peaks.
Analysis of gene mRNA expression by real time RTPCR
Measurement of the levels of specific mRNA transcripts was carried out essentially as described22 using the primers detailed previously,18,22 with the exception of DGAT2 (forward ATCTTCTCTGTCACCTGGCT and reverse ACCTTTCTTGGGCGTGTTCC) and SREBP-1C (Qiagen QuantiTect primer assay QT00432684). Briefly, total RNA was isolated from left and right ventricles which had been pulverised under liquid nitrogen using Tri Reagent (Sigma) according to the manufacturer’s instructions. cDNA was prepared and amplified using real-time RT-PCR.22 Samples were analyzed in duplicate and the expression of the individual transcripts was normalised to ATP synthase subunit beta (ATP5B) and Calnexin (CANX) which did not differ in transcript level between groups.
Measurement of DNA methylation of putative 5′-regulatory region of PPARa
PPARα promoter methylation was measured using PCR primers described previously.22 Genomic DNA was isolated from left and right ventricles as described.22 Purified DNA was incubated with the methylation-sensitive restriction endonucleases AciI and HpaII according to the manufacturer’s instructions (New England Biolabs).22 The resulting DNA was amplified in duplicate using real-time PCR as described previously.22 A region of the PPARγ2 promoter that does not contain AciI or HpaII cleavage sites was used as an internal control.22
Statistical analysis
Data are presented as mean ± 1 SD. Statistical analysis was carried out using SPSS (SPSS Inc., Chicago, Illinois, USA). Comparisons between groups of adult offspring were by a General Linear Model with sex, maternal diet and PW diet as fixed factors using Bonferroni’s post hoc correction. Comparison between adult and neonatal hearts was by Students unpaired t-test. Analysis using the Kolmogorov-Smirnov test showed the results of measurements of mRNA expression and DNA methylation were not normally distributed, and so data were log transformed prior to statistical analysis. The relationship between the methylation status of the PPARα promoter and its expression was assessed by linear regression. While it would have been preferable to analyse litters rather than individual offspring, this was prevented by the cost and labour-intensive nature of the molecular biology analyses.
Results
Triacylglycerol concentration
There were significant interactive effects of maternal diet (P=0.004), and interactive effects of sex*maternal diet (P=0.006) and sex*post-weaning diet (P=0.015) on TAG concentration in adult heart. In males, feeding the HF diet increased total TAG concentration in the heart of the offspring of dams fed the PS diet, but did not alter TAG concentration in offspring of PR dams compared to PS LF offspring (Table 1). Total TAG concentration did not differ between PS LF and PR LF offspring. In female offspring, there was no effect in PS offspring of feeding the HF diet on total TAG concentration in heart compared to PS LF offspring. Total TAG concentration was higher in PR LF offspring than PS LF offspring, and was significantly greater in PR HF offspring compared to all other groups of female offspring (Table 1).
Table 1.
Triacylglycerol concentration in adult heart, and mRNA expression of genes involved in carbohydrate and fatty acid metabolism
| mRNA expression (log normalised ct) |
||||||||
|---|---|---|---|---|---|---|---|---|
| PS LF | PS HF | PR LF | PR HF | PS LF | PS HF | PR LF | PR HF | |
|
|
||||||||
| Male | Female | |||||||
|
|
||||||||
| Adult | ||||||||
| TAG concentration (μg/g) | 2.4±1.1a | 4.2±1.6b | 2.8±1.2a | 2.3±0.7a | 1.6±0.4a | 2.4±1.1a | 3.1±1.6b | 4.2±2.7c |
| AOX | 2.0±0.4 | 1.7±0.2 | 1.6±0.6 | 2.2±0.5 | 2.2±0.5 | 1.9±0.3 | 1.9±0.3 | 1.9±0.4 |
| CPT-1 | 0.4±0.2a | 0.4±0.2a | 0.8±0.2b | 0.8±0.2b | 0.5±0.2a | 0.4±0.2a | 0.7±0.2b | 0.8±0.2b |
| LPL | 1.5±0.2 | 1.9±0.7 | 1.6±0.4 | 1.7±0.2 | 1.8±0.4 | 1.5±0.2 | 1.7±0.6 | 1.7±0.4 |
| DGAT2 | 3.1±0.3 | 3.2±0.5 | 2.5±0.7 | 2.9±0.2 | 2.9±0.6 | 3.2±0.4 | 3.0±0.8 | 2.8±0.8 |
| PPARγ | 2.9±0.8 | 2.8±0.5 | 2.9±1.1 | 2.5±0.8 | 2.7±0.7 | 2.6±0.2 | 2.6±0.7 | 2.1±0.3 |
| SREBP-1C | 0.7±0.2 | 0.6±0.2 | 0.7±0.4 | 0.5±0.2 | 0.7±0.4 | 0.5±0.2 | 0.7±0.4 | 0.7±0.2 |
| Neonate | ||||||||
| AOX | 1.4±0.1* | 1.5±0.3 | 1.5±0.1* | 1.4±0.5 | ||||
| CPT-1 | 0.8±0.2* | 0.9±0.2 | 0.9±0.2* | 1.0±0.1* | ||||
| LPL | 1.3±0.7 | 1.3±0.5 | 1.8±0.6 | 1.3±0.5 | ||||
| DGAT2 | 1.8±0.4* | 1.4±0.2 | 1.2±0.1 | 1.6±0.1 | ||||
| PPARγ | 0.9±0.6* | 0.6±0.4* | 0.3±0.5* | 0.5±0.4* | ||||
| SREBP-1C | 0.3±0.2* | 0.3±0.2* | 0.2±0.1* | 0.2±0.1* | ||||
Values are mean ± 1SD, n = 7 - 15. Overall effects of maternal and post-weaning diet, and sex on gene expression and on PPARα methylation in adult and neonatal hearts were tested by a General Linear Model and the results are given in the text. Different superscripts indicate values which differ significantly (P <0.05) for each gene among adult or neonatal hearts.
Indicates statistically significant differences (P < 0.001) by Student’s unpaired t-test between hearts from adults fed the LF diet after weaning and neonatal heart from the same maternal dietary group.
mRNA expression in adult and neonatal heart
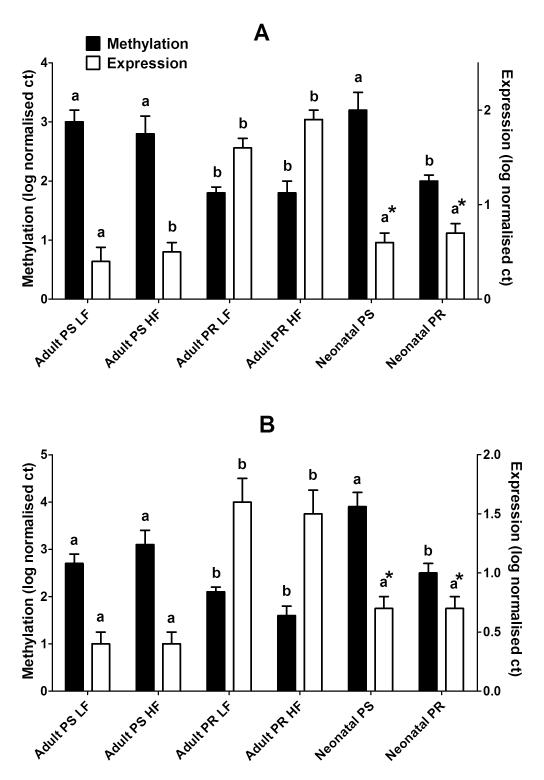
There was a significant effect of maternal diet (P < 0.0001), but not of sex or PW diet, on PPARα expression in adult heart. PPARα expression was increased in adult male and female offspring of PR dams compared to PS offspring, irrespective of PW diet (Figure 1). There was no effect of maternal or PW diet, or offspring sex on AOX expression (Table 1). CPT-1 expression was increased in both adult male and female offspring of PR dams, irrespective of post-weaning diet, compared to PS offspring. There was no effect of maternal or PW diets, or offspring sex on the mRNA expression of LPL, PPARγ, SREBP-1C or DGAT2 in adult heart (Table 1).
Fig. 1.
PPARα mRNA expression (open bars) and promoter methylation (solid bars) in (a) male and (b) female adult and neonatal heart. Values are mean ± 1SD, n = 7 – 15 samples per group. LF, low fat post-weaning diet; HF; PS, maternal protein-sufficient diet; PR, maternal protein-restricted diet. Values which are significantly different (P < 0.05) within an age group for each outcome are indicated by different letters. *Indicates statistically significant differences (P < 0.001) by Student’s unpaired t-test between hearts from adults fed the LF diet after weaning and neonatal heart within the same maternal dietary group.
There were no statistically significant differences in the expression of any of the genes measured between neonates from different maternal dietary groups or between sexes (Table 1, Figure 1A). There was a significant effect of age (P=0.014) on PPARα expression. PPARα expression was similar in all male and female neonates to adult PS offspring, but lower than adult PR offspring (Figure 1A). There was a significant effect of age on AOX expression (P<0.0001), and a significant effect of age (P<0.0001) and of age*maternal diet (P<0.0001) on CPT-1 expression. AOX expression was lower and CPT-1 mRNA level of was higher in hearts from neonatal compared to adult PS offspring irrespective of sex (Table 1). There was no significant difference between male and females in AOX expression in hearts from neonatal or adult offspring (Table 1). There was a significant effect of age on DGAT2, SREBP-1C and PPARγ (all P<0.0001) such that the expression of these genes was lower in neonatal compared to adult heart irrespective of sex or maternal diet (Table 1). LPL expression did not differ significantly between neonatal and adult hearts.
PPARα promoter methylation in adult and neonatal heart
Analysis of PPARα methylation in adult and neonatal hearts is summarised in Figure 1B. There was a significant effect of maternal diet (P < 0.0001), but not age, sex or post-weaning diet, on PPARα methylation. The methylation status of the PPARα promoter was significantly lower in both neonatal and adult hearts from offspring of PR dams compared to offspring of PS dams. There were no significant differences in the level of PPARα methylation between hearts from adult and neonatal offspring from the same maternal dietary group. When analysed across dietary groups, the methylation status of the PPARα promoter was associated negatively (r = −0.34, P = 0.01) with its expression in adult offspring, but these was no significant association with the mRNA level in neonatal heart.
Discussion
Poor nutrition before birth induced dysregulation of lipid metabolism in the heart leading to increased TAG concentration. However, the nature of this effect was contingent on the sex of the offspring and fat intake after weaning. We have shown previously in these adult rats that in dyslipidaemia and impaired glucose homeostasis induced by maternal dietary protein restriction were similar for male and female offspring.17 The present findings suggest that the changes in total TAG concentration in the heart were not simply dependent upon supply of fatty acids from blood and so imply that the protein content of the maternal diet specifically modifies the metabolic partitioning of fatty acids in the heart of the offspring.
There were no differences between maternal dietary groups in the mRNA expression of any of the genes measured in neonatal hearts. The expression of all of the genes measured, except LPL, differed between adult and neonatal hearts which is consistent with maturation of specific regulatory pathways in lipid and carbohydrate metabolism during postnatal development.23 Hearts from the adult offspring of dams fed the PR diet showed a specific increase in PPARα mRNA expression irrespective of fat intake after weaning. This was accompanied by increased CPT-1 expression with no change in AOX which suggests increased mitochondrial, but not peroxisomal, fatty acid β-oxidation. In the fasting state, fatty acids are the preferred substrate for energy production in the heart. Thus the offspring of PR dams may have greater capacity for energy production from fatty acids. PPARα also increases pyruvate dehydrogenase kinase 4 expression which decreases capacity to generate energy by glycolysis.9 Thus one implication of these findings is that the hearts from PR offspring are at risk of increased oxidative damage due to higher capacity for fatty acid β-oxidation, but may also have reduced flexibility to change substrate use in different nutritional states. Such metabolic changes are consistent with impaired recovery form ischaemia-reperfusion injury19 and the protective effects of antioxidants.20 There was no effect of maternal or post-weaning diet on the mRNA expression of DGAT2, SREBP-1C or PPARγ which suggests that accumulation of TAG within the heart in this model does not appear to involve these genes.
The presence of changes in the mRNA expression of PPARα and CPT-1 associated with differences maternal protein intake in adult, but not neonatal heart, suggests two possible mechanisms. First, the mechanism which underlies altered cardiac gene expression was induced before birth but did not exert an effect until later in life. Alternatively, the effects maternal protein restriction persisted beyond the period of feeding the restricted diet and induced altered gene regulation at some later point. To investigate which of these processes was likely to occur in the hearts of these offspring we measured the methylation status of the PPARα promoter. PPARα promoter methylation was reduced in both adult and neonatal hearts from the offspring of PR dams. This suggests that altered regulation of PPARα was induced before birth, but the effects on its mRNA expression were only detected later, possibly as a result of developmentally programmed induction of PPARα expression in response to increased fatty acid availability after birth.24
Together these findings show for the first time that poor maternal nutrition during pregnancy induces in the offspring altered epigenetic regulation of a key transcription factor that controls energy homeostasis which may have important consequences for future cardiac function. One implication in the context of human disease is that risk of developing cardiomyopathy as a complication of diabetes mellitus may be contingent on the quality of nutrition before birth. Furthermore, appropriate epigenetic probes may provide a means of detecting such vulnerability before the onset of disease.
Acknowledgements
This study was funded by a grant from the British Heart Foundation (PG/06/098/21347) which also provided salary support for MAH.
Footnotes
Statement of Interest None.
References
- 1.Factor SM, Minase T, Sonnenblick EH. Clinical and morphological features of human hypertensive-diabetic cardiomyopathy. Am Heart J. 1980;99:446–458. doi: 10.1016/0002-8703(80)90379-8. [DOI] [PubMed] [Google Scholar]
- 2.Bell DS. Heart failure: the frequent, forgotten, and often fatal complication of diabetes. Diabetes Care. 2003;26:2433–2441. doi: 10.2337/diacare.26.8.2433. [DOI] [PubMed] [Google Scholar]
- 3.Denton RM, Randle PJ. Concentrations of glycerides and phospholipids in rat heart and gastrocnemius muscles. Effects of alloxan-diabetes and perfusion. Biochem J. 1967;104:416–422. doi: 10.1042/bj1040416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murthy VK, Shipp JC. Accumulation of myocardial triglycerides ketotic diabetes; evidence for increased biosynthesis. Diabetes. 1977;26:222–229. doi: 10.2337/diab.26.3.222. [DOI] [PubMed] [Google Scholar]
- 5.Rizza RA, Crass MF, III, Shipp JC. Effect of insulin treatment in vivo on heart glycerides and glycogen of alloxan-diabetic rats. Metabolism. 1971;20:539–543. doi: 10.1016/0026-0495(71)90002-3. [DOI] [PubMed] [Google Scholar]
- 6.Chiu HC, Kovacs A, Ford DA, et al. A novel mouse model of lipotoxic cardiomyopathy. J Clin Invest. 2001;107:813–822. doi: 10.1172/JCI10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finck BN, Lehman JJ, Leone TC, et al. The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus. J Clin Invest. 2002;109:121–130. doi: 10.1172/JCI14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watanabe K, Fujii H, Takahashi T, et al. Constitutive regulation of cardiac fatty acid metabolism through peroxisome proliferator-activated receptor alpha associated with age-dependent cardiac toxicity. J Biol Chem. 2000;275:22293–22299. doi: 10.1074/jbc.M000248200. [DOI] [PubMed] [Google Scholar]
- 9.Bishop-Bailey D. Peroxisome proliferator-activated receptors in the cardiovascular system. Br J Pharmacol. 2000;129:823–834. doi: 10.1038/sj.bjp.0703149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finck BN, Kelly DP. Peroxisome proliferator-activated receptor alpha (PPARalpha) signaling in the gene regulatory control of energy metabolism in the normal and diseased heart. J Mol Cell Cardiol. 2002;34:1249–1257. doi: 10.1006/jmcc.2002.2061. [DOI] [PubMed] [Google Scholar]
- 11.Shimano H. SREBPs: physiology and pathophysiology of the SREBP family. FEBS J. 2009;276:616–621. doi: 10.1111/j.1742-4658.2008.06806.x. [DOI] [PubMed] [Google Scholar]
- 12.Boudina S, Abel ED. Diabetic cardiomyopathy revisited. Circulation. 2007;115:3213–3223. doi: 10.1161/CIRCULATIONAHA.106.679597. [DOI] [PubMed] [Google Scholar]
- 13.Finck BN, Han X, Courtois M, et al. A critical role for PPARalpha-mediated lipotoxicity in the pathogenesis of diabetic cardiomyopathy: modulation by dietary fat content. Proc Natl Acad Sci U S A. 2003;100:1226–1231. doi: 10.1073/pnas.0336724100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marfella R, Di FC, Portoghese M, et al. Myocardial lipid accumulation in patients with pressure-overloaded heart and metabolic syndrome. J Lipid Res. 2009 doi: 10.1194/jlr.P900032-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Armitage JA, Khan IY, Taylor PD, Nathanielsz PW, Poston L. Developmental programming of the metabolic syndrome by maternal nutritional imbalance: how strong is the evidence from experimental models in mammals? J Physiol. 2004;561:355–377. doi: 10.1113/jphysiol.2004.072009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burdge GC, Lillycrop KA, Jackson AA, Gluckman PD, Hanson MA. The nature of the growth pattern and of the metabolic response to fasting in the rat are dependent upon the dietary protein and folic acid intakes of their pregnant dams and post-weaning fat consumption. Br J Nutr. 2008;99:540–549. doi: 10.1017/S0007114507815819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135:1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- 19.Elmes MJ, Gardner DS, Langley-Evans SC. Fetal exposure to a maternal low-protein diet is associated with altered left ventricular pressure response to ischaemia-reperfusion injury. Br J Nutr. 2007;98:93–100. doi: 10.1017/S000711450769182X. [DOI] [PubMed] [Google Scholar]
- 20.Elmes MJ, McMullen S, Gardner DS, Langley-Evans SC. Prenatal diet determines susceptibility to cardiac ischaemia-reperfusion injury following treatment with diethylmaleic acid and N-acetylcysteine. Life Sci. 2008;82:149–155. doi: 10.1016/j.lfs.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 21.Burdge GC, Wright P, Jones AE, Wootton SA. A method for separation of phosphatidylcholine, triacylglycerol, non-esterified fatty acids and cholesterol esters from plasma by solid-phase extraction. Br J Nutr. 2000;84:781–787. [PubMed] [Google Scholar]
- 22.Burdge GC, Lillycrop KA, Phillips ES, et al. Folic Acid Supplementation during the Juvenile-Pubertal Period in Rats Modifies the Phenotype and Epigenotype Induced by Prenatal Nutrition. J Nutr. 2009;139:1054–1060. doi: 10.3945/jn.109.104653. [DOI] [PubMed] [Google Scholar]
- 23.Rajabi M, Kassiotis C, Razeghi P, Taegtmeyer H. Return to the fetal gene program protects the stressed heart: a strong hypothesis. Heart Failure Reviews. 2007;12:331–343. doi: 10.1007/s10741-007-9034-1. [DOI] [PubMed] [Google Scholar]
- 24.Panadero M, Herrera E, Bocos C. Different sensitivity of PPAR[alpha] gene expression to nutritional changes in liver of suckling and adult rats. Life Sciences. 2005;76:1061–1072. doi: 10.1016/j.lfs.2004.10.018. [DOI] [PubMed] [Google Scholar]