Abstract
Potassium ions are required for gastric acid secretion. Several potassium channels have been implicated in providing K+ at the apical membrane of parietal cells. In examining the mRNA expression levels between gastric mucosa and liver tissue, KCNJ15 stood out as the most highly specific K+ channel in the gastric mucosa. Western blot analysis confirmed that KCNJ15 is abundant in the stomach. Immunofluorescence staining of isolated gastric glands indicated that KCNJ15 was expressed in parietal cells and chief cells, but not in mucous neck cells. In resting parietal cells, KCNJ15 was mainly found in puncta throughout the cytoplasm but was distinct from H+-K+-ATPase. Upon stimulation, KCNJ15 and H+-K+-ATPase become colocalized on the apical membranes, as suggested by immunofluorescence staining. Western blot analysis of the resting and the stimulated membrane fractions confirmed this observation. From nonsecreting preparations, KCNJ15-containing vesicles sedimented after a 4-h centrifugation at 100,000 g, but not after a 30-min spin, which did sediment most of the H+-K+-ATPase-containing tubulovesicles. Most of the KCNJ15 containing small vesicle population was depleted upon stimulation of parietal cells, as indicated by the fact that the KCNJ15 signal was shifted to a large membrane fraction that sedimented at 4,000 g. Our results demonstrate that, in nonsecreting parietal cells, KCNJ15 is stored in vesicles distinct from the H+-K+-ATPase-enriched tubulovesicles. Furthermore, upon stimulation, KCNJ15 and H+-K+-ATPase both translocate to the apical membrane for active acid secretion. Thus KCNJ15 can be added to the family of apical K+ channels in gastric parietal cells.
Keywords: potassium channel, apical membrane, membrane trafficking
potassium ions are needed in the generation of acidity by the stomach (9). For every proton pumped into the gastric lumen by the H+-K+-ATPase (H-K-ATPase), K+ is equivalently pumped into the cytosol of gastric parietal cells. To provide a continuous supply of K+ for the proper function of the proton pump it is necessary to have some leak or channel mechanism for the flow of K+ from cytoplasm to the luminal side of the proton pump. These two apical membrane transporters, the K+ channel and the H+/K+ exchange pump, together with an apical Cl− channel, account for the continuous recycling of K+ across the apical membrane and the net secretory flow of HCl acid (8, 29).
Many potassium channels or transporters have been implicated in gastric acid secretion. KCNQ1 was the first K+ channel suggested for apical K+ recycling in human and rat parietal cells (13). Soon, KCNJ10 (Kir4.1) was suggested for a similar role in rat parietal cells (12). Fujita et al. (12) also detected the mRNAs of KCNJ15 (Kir4.2) and Kir7.1 in parietal cells but failed to detect the proteins of these two channels. In addition, Cuppoletti and colleagues (5, 19) found that the mRNA of Kir2.1 (KCNJ2) is more abundant than that of Kir4.1 and Kir7.1 in rabbit gastric mucosa. They demonstrated that Kir2.1 is colocalized with H-K-ATPase and ClC-2 in parietal cells and this potassium channel can be activated by PKA and low pH. It is also noteworthy that gastric glands isolated from ROMK knockout mice (ROMK is also an inwardly rectifying K+ channel) exhibit no H-K-ATPase activity (26).
The number of K+ transporters expressed in parietal cells continued to grow, when the K+-Cl− cotransporter (KCC) 4 (KCC4, SLC12A7) was reported to be an apical protein, in both resting and stimulated cells (11). Interestingly, a KCC inhibitor suppressed H-K-ATPase activity in an in vitro assay.
It is known that certain tissues [such as rat pituitary (31) and mouse cardiac cells (20)] express a large diversity of potassium channels (19). To test the hypothesis that multiple potassium channels may be involved in the apical recycling of potassium ions, we examined microarray data sets generated from mouse stomach and liver tissue. Surprisingly, microarray analysis indicated that KCNJ15 is the most highly specific K channel in the stomach. This K+ channel translocated from cytoplasm to apical membrane upon stimulation, supporting a role in the production of gastric acid.
MATERIALS AND METHODS
Animals.
The procedures and treatments for handling animals were reviewed and approved by the Institutional Animal Care and Use Committee of the State University of New York at Buffalo. C57BL/6 mice were anesthetized with pentobarbital before whole body perfusions were performed, with the catheter inserted into the apex of the left ventricle and the right atrium snipped to vent the perfusate (PBS with 1 mM CaCl2 and 1 mM of MgCl2, bubbled with pure oxygen). About 20 ml of perfusate was used for each mouse to achieve a complete whole-body perfusion. Mouse tissues were then stored in RNAlater (Qiagen) for the preparation of RNA or snap frozen for protein analyses.
Gastric glands (3) and gastric parietal cells (4) were prepared from New Zealand White rabbits following established methods.
Microarray data analysis.
The microarray data sets for mouse stomach are taken from a recently published LASP1 knockout study (33). The dataset used here were generated from the gastric mucosa of wild-type C57BL/6 mice. The Gene Expression Omnibus (GEO, website: http://www.ncbi.nlm.nih.gov/geo/index) accession numbers for these stomach samples are GSM370907, GSM370908, and GSM370909. Similar datasets generated on the same microarray platform for livers of C57BL/6 mice were downloaded from GEO for comparison. The GEO accession numbers for these liver samples are GSM388169, GSM388170, and GSM388171. The gene expression levels in all these datasets are median normalized.
qRT-PCR.
Primer pairs were manually designed for kcnj15, kcnq1, polrA2, and β-actin, with the latter two housekeeping genes serving as internal references. For all primer pairs, the forward and reverse primers were designed to hybridize to sequences in different exons and to produce an amplicon of ∼100 bp. Single correct PCR products were confirmed by melting-curve analysis, 3% agarose gel electrophoresis, and direct sequencing of the PCR products. All PCR reactions had efficiencies around 1.9, as determined experimentally with four-times serial-diluted samples. The primer pairs used for quantitative real-time PCR (qRT-PCR) analysis are 1) kcnj15, 5′-agcagagccccatggtagccaggtgggagaa-3′ (sense), 5′-gccttgagtccaaccccgttggtatgct-3′ (antisense); 2) kcnq1, 5′-gccgagtctacaacttcctcgag-3′ (sense), 5′-catactgctcaatagtggacaggacac-3′ (antisense); 3) actb, 5′-gatctggcaccacaccttctaca-3′ (sense), 5′-gcatacagggacagcacagcctggat-3′ (antisense); and 4) polr2a, 5′-ggctgaggagtttcggctgagtggagag-3′ (sense); 5′-ggaaagtgttcagggtcatctgagtgg-3′ (antisense).
Tissue slices of ∼5 mm thick were stored in RNAlater before total RNA was extracted with RNeasy (Qiagen, Valencia, CA). Complementary DNA was synthesized from 0.8 μg of RNA in a volume of 20 μl, by use of the iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA). A three-step quantitative real-time PCR was performed (95°C, 30 s; 55°C, 30 s; 72°C, 10 s; 50 cycles) on an iCycler iQ real-time detection system (Bio-Rad Laboratories), using SYBR Green (iQ SYBR Green Supermix; Bio-Rad Laboratories) for real-time monitoring of the PCR.
Threshold cycles (Ct) for each sample were determined by Bio-Rad iQ5 optical system software (Bio-Rad Laboratories). The concentration of mRNA ([mRNA]) is represented by the following equation: [mRNA] = M/ECt, where constant M is an arbitrary threshold, E is the efficiency of PCR, and Ct is the threshold cycle. The relative mRNA concentration of each target gene was calculated as the mRNA concentration of target gene normalized against that of a housekeeping gene, as represented by the equation [mRNA]target/[mRNA]housekeeping = EhousekeepingCt.housekeeping/EtargetCt.target.
Plasmids.
Mouse total RNA was isolated from mouse gastric mucosa by use of a RNeasy mini kit (Qiagen). With this total RNA as template, KCNJ15 cDNA was synthesized using iScript cDNA synthesis kit (Bio-Rad Laboratories). The following primer pair was used to amplify KCNJ15 fragment carrying EcoRI and SalI restriction sites at each end of the fragment: forward primer, 5′-CCGCGAATTCAATGGTAGCCAGGTGGGAGAAGGG-3′; and reverse primer, 5′-CTAGAGTCGACTCAGACATTGCTCTGCTGTAGCAGGAGGC-3′. The PCR product was digested with EcoRI and SalI and inserted into similarly digested pCMV2-FLAG vector (Sigma-Aldrich) to produce pCMV2-FLAG-KCNJ15.
To generate a plasmid expressing KCNJ15 with a cyan fluorescent protein (CFP)-tagged KCNJ15, the following primer pair carrying XhoI and BamHI sites was used: forward primer, 5′-ACACCTCGAGCCACCATGGTAGCCAGGTGGGAGAAGGG-3′; reverse primer, 5′-CGACCGGTGGATCCACGACATTGCTCTGCTGTAGCAGGAGG-3′. KCNJ15 fragment was PCR-amplified with these primer pair, digested with XhoI and BamHI, and inserted into similarly digested pDC311/Ez-C (36), producing pDC311-KCNJ15-CFP. The open reading frames of these plasmids were verified by sequencing.
Deglycosylation analysis.
Gastric mucosa or gastric glands were resuspended in resuspension buffer containing 50 mM Na2HPO4, pH 7.5, 10 mM EDTA, 0.5% SDS, and 2% 2-mercaptoethanol. After boiling for 5 min, the lysate was diluted with resuspension buffer and supplemented with 2-mercaptoethanol and Triton X-100 to obtain final concentrations of 0.1% SDS, 0.5% Triton X-100, and 1% 2-mercaptoethanol. To half of the lysate, PNGase F (NEB, 1 μl/100 μg total protein) was added and incubated for 2 h at 37°C; the other half of the lysate was incubated similarly without adding PNGase F.
Western blot analysis.
Western blots were performed as described previously (34). The primary antibodies used in this study are mouse monoclonal anti-H-K-ATPase β antibody (1:5,000, Affinity Bioreagents), rabbit anti-KCNJ15 (1:2,000, Sigma-Aldrich, catalog no. HPA016702), and mouse monoclonal anti-β-actin (1:3,000, clone C4, MP Biomedicals, Solon, OH). Horseradish peroxidase-conjugated goat-anti-mouse IgG (1:10,000) or goat-anti-rabbit IgG (1:10,000) were from Jackson ImmunoResearch Laboratories (West Grove, PA).
Immunoprecipitation.
Gastric mucosa was isolated from rabbit stomach and homogenized in phosphate-buffered saline, supplemented with 1 mM PMSF, 1 mM EDTA, and 1% Triton X-100. The lysate was cleared by centrifugation at 10,000 g for 10 min and 4°C before immunoprecipitation with rabbit anti-KCNJ15 following our published method (36).
Immunofluorescence and confocal microscopy.
Gastric glands were fixed with 3.7% formaldehyde (room temperature/15 min) and then attached to glass slides coated with poly-l-lysine. The glands were then perforated with 0.5% Triton X-100 in PBS, before probing with primary antibodies and fluorescent secondary antibodies. The glands were then covered with a coverglass with a drop of the mounting media (VectorShield, Burlingame, CA). The primary antibodies used in this study are mouse monoclonal anti-H-K-ATPase β antibody (1:50, Affinity Bioreagents), goat anti-pepsin C (I-19) antibody (1:50, Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-KCNJ15 antibody (1:20, Sigma-Aldrich) and mouse anti-Na+-K+-ATPase antibody (1:50, Santa Cruz Biotechnology). Although raised with a human KCNJ15 peptide, this KCNJ15 antibody is deemed suitable for immunofluorescence staining of rabbit tissues for three reasons: 1) the antibody only recognized a single band from rabbit gastric glands in Western blots (Fig. 1B, lane 9); 2) this antibody could immunoprecipitate KCNJ15 (unpublished data), indicating capability of recognizing native KCNJ15; 3) the sequence of the peptide used to raise this antibody is conserved among the KCNJ15 peptides of human, cow, and mouse and therefore likely conserved in rabbit KCNJ15 as well. In addition, normal rabbit serum did not generate any image similar to KCNJ15 staining. Alexa 568-phalloidin (1 unit/100 μl), Alexa 568, and Alexa 488-conjugated secondary antibodies (1:1,000 for both) were products of Invitrogen (Eugene, OR). Alexa 488-conjugated lectin GS II (1:200) was from Invitrogen.
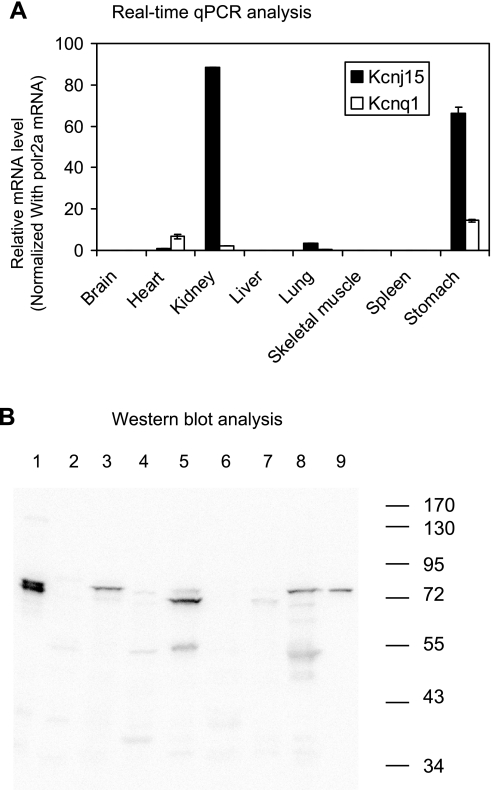
Fig. 1.
Tissue distribution of kcnj15 and kcnq1. A: real-time quantitative real-time PCR (qPCR) analyses of expression of kcnj15 and kcnq1 in mouse tissues. The plotted relative mRNA levels were calculated with polr2a mRNA levels serving as internal references. Error bars indicate standard deviations. High-level expression of kcnj15 was observed in kidney and stomach. Similar results were obtained with β-actin serving as the normalizing housekeeping gene. B: Western blot analysis of the expression of kcnj15 in mouse tissues (lanes 1-8) and rabbit gastric glands (lane 9). The same amount of total protein (∼20 μg) was loaded for each sample. 1, brain; 2, heart; 3, kidney; 4, liver; 5, lung; 6, skeletal muscle; 7, spleen; 8, stomach.
Gastric parietal cells grown on Matrigel-coated coverglasses were treated with cimetidine (100 μM), histamine (100 μM)/IBMX (3-isobutyl-1-methyl-xanthine, 50 μM), or histamine (100 μM)/IBMX (50 μM)/SCH28080 (50 μM) for 30 min at 37°C. The cells were then immediately fixed with 3.7% formaldehyde for 15 min at room temperature, followed by a similar procedure of perforation and antibody reactions. The coverslips were then mounted onto glass slides with a drop of the mounting media.
Cryosections of fasting mouse stomach were prepared and immunostained as described previously (35).
Images were collected with a Zeiss LSM 510 meta confocal microscope at one airy unit pinhole with a Plan-Neofluar 40×/1.3 oil differential interference contrast objective.
Indirect measurement of acid secretion by isolated gastric glands with [14C]aminopyrine.
The aminopyrine (AP) assay measures the accumulation of aminopyrine in acidic spaces caused by the proton-pumping enzyme H-K-ATPase. A detailed description of this assay was published previously (2). When cimetidine, histamine, and IBMX were used, final concentrations of these drugs were 100, 100, and 50 μM, respectively.
Membrane fractionation by differential centrifugation.
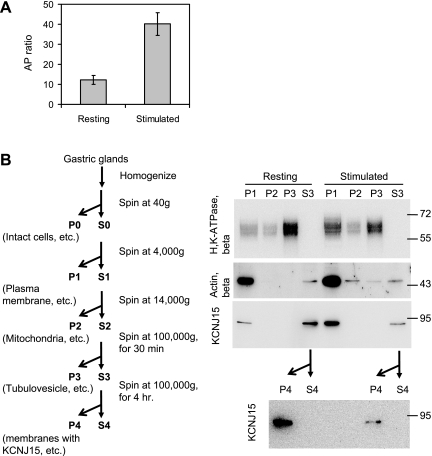
Membrane fractionation was performed as previously described (27) with minor modifications. Purified gastric glands were used as the starting materials in this study. Gastric glands treated with cimetidine or histamine/IBMX for 30 min were homogenized in a buffer containing 125 mM mannitol, 40 mM sucrose, 1 mM EDTA, and 5 mM PIPES-Tris, pH 6.7. The nuclei and unbroken cells were removed by centrifugation at 40 g and 4°C for 5 min. The resulting supernatants were sequentially centrifuged at 4,000 g and 4°C for 10 min, 14,000 g and 4°C for 10 min, and 100,000 g and 4°C for 30 min to obtain membrane fractions P1, P2, and P3. The final supernatant fractions are S3. To pellet KCNJ15, S3 fractions were further centrifuged at 100,000 g and 4°C for 4 h, producing pellets (P4) and supernatants (S4).
RESULTS
KCNJ15 mRNA is highly expressed in the stomach.
To identify stomach-specific potassium channels, we compared the microarray datasets generated from mouse gastric mucosa to that generated from mouse livers. The high quality of these datasets was exemplified by the facts that 1) mRNA levels of the majority of the 45,000 genes are similar between gastric mucosa and liver as represented by the housekeeping genes listed in Table 1; 2) high expression levels of stomach-specific genes were detected in the gastric mucosa compared with the liver tissues (Table 1, stomach genes); and 3) high expression levels of liver specific genes were detected in the liver tissues compared with the gastric mucosa (Table 1, liver genes).
Table 1.
Microarray analysis of gene expression in mouse stomachs and livers
| GenBank Accession No. | Gene Description | Mouse Stomach (n = 3) | Mouse Liver (n = 3) | Stomach/Liver | P Value | |
|---|---|---|---|---|---|---|
| Potassium channels | NM_001039057.1 | Potassium inwardly rectifying channel, subfamily J, member 15 (Kcnj15) | 32.62 ± 4.31 | 0.97 ± 0.04 | 33.57 | 0.002 |
| NM_008430.1 | Potassium channel, subfamily K, member 1 (Kcnk1) | 21.81 ± 3.40 | 1.05 ± 0.01 | 20.69 | 0.004 | |
| NM_134110.1 | Potassium voltage-gated channel, Isk-related subfamily, gene 2 (Kcne2) | 17.86 ± 4.07 | 1.06 ± 0.04 | 16.81 | 0.015 | |
| NM_010604.3 | Potassium inwardly rectifying channel, subfamily J, member 16 (Kcnj16) | 10.87 ± 0.47 | 1.20 ± 0.03 | 9.09 | 0.000 | |
| NM_011390.2 | Solute carrier family 12, member 7 (Slc12a7, KCC4) | 8.15 ± 1.16 | 3.30 ± 0.23 | 2.47 | 0.015 | |
| NM_019659.2 | Potassium inwardly rectifying channel, subfamily J, member 1 (Kcnj1) | 0.82 ± 0.03 | 0.80 ± 0.02 | 1.03 | 0.545 | |
| NM_008434.2 | Potassium voltage-gated channel, subfamily Q, member 1 (Kcnq1) | 0.98 ± 0.07 | 0.97 ± 0.02 | 1.00 | 0.957 | |
| NM_020269.3 | Potassium inwardly rectifying channel, subfamily J, member 10 (Kcnj10) | 0.99 ± 0.01 | 1.05 ± 0.05 | 0.94 | 0.259 | |
| NM_008425.2 | Potassium inwardly rectifying channel, subfamily J, member 2 (Kcnj2) | 1.06 ± 0.10 | 1.20 ± 0.05 | 0.89 | 0.299 | |
| Housekeeping genes | NM_019468.1 | Glucose-6-phosphate dehydrogenase 2 (G6pd2) | 0.85 ± 0.03 | 0.88 ± 0.06 | 0.96 | 0.669 |
| NM_138953.2 | Elongation factor RNA polymerase II 2 (Ell2) | 2.53 ± 0.09 | 2.25 ± 0.20 | 1.13 | 0.263 | |
| NM_177342.3 | TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor (Taf5) | 1.23 ± 0.07 | 1.14 ± 0.05 | 1.08 | 0.377 | |
| NM_019647.5 | Ribosomal protein L21 (Rpl21) | 1.52 ± 0.26 | 1.27 ± 0.11 | 1.20 | 0.428 | |
| NM_011297.2 | Ribosomal protein S24 (Rps24) | 319.49 ± 7.38 | 274.73 ± 3.48 | 1.16 | 0.005 | |
| Liver genes | NM_007409.2 | alcohol dehydrogenase 1 (class I) (Adh1) | 13.06 ± 2.91 | 439.14 ± 2.42 | 0.03 | 0.000 |
| NM_011996.2 | Alcohol dehydrogenase 4 (class II), pi polypeptide (Adh4). | 0.79 ± 0.04 | 85.77 ± 2.70 | 0.01 | 0.000 | |
| NM_021282.2 | Cytochrome P-450, family 2, subfamily e, polypeptide 1 (Cyp2e1) | 0.97 ± 0.04 | 748.98 ± 41.13 | 0.00 | 0.000 | |
| Stomach genes | NM_009724.2 | ATPase, H+/K+ exchanging, beta polypeptide (Atp4b) | 488.02 ± 23.11 | 0.87 ± 0.05 | 558.64 | 0.000 |
| NM_018731.2 | ATPase, H+/K+ exchanging, gastric, alpha polypeptide (Atp4a) | 485.91 ± 23.81 | 1.15 ± 0.01 | 422.64 | 0.000 | |
| NM_025973.3 | Progastricsin (pepsinogen C) (Pgc) | 547.01 ± 34.29 | 1.02 ± 0.07 | 538.65 | 0.000 |
Gene expression levels (sample mean ± SE) shown were median normalized. The Gene Expression Omnibus (GEO, website: http://www.ncbi.nlm.nih.gov/geo/index) accession numbers are GSM370907, GSM370908, GSM370909 for stomach samples and GSM388169, GSM388170, GSM388171 for liver samples. Stomach/Liver values are fold difference of gene expression levels (sample mean) between mouse stomachs and mouse livers. P values were determined by 2-tailed Student t-test.
We then examined the gene expression levels of all potassium channels and transporters with these microarray datasets. Out of the ∼150 genes related to potassium transportation, the relative expression of KCNJ15 in the stomachs vs. liver stood out among all other potassium transporters and channels. The KCNJ15 mRNA level in the stomach is 33.6-fold of that in the liver (Table 1), followed by kcnk1, the stomach mRNA level of which is 20.7-fold of that in the liver. The relative expression level of KCNJ15 was much greater than all other potassium channels implicated in the apical recycling of potassium in parietal cells (Table 1): kcne2 (MIRP1), kcnj16 (kir5.1), slc12a7 (KCC4), kcnj1 (kir1.1, ROMK), kcnq1 (LTQ1), kcnj10 (kir4.1), and kcnj2 (kir2.1). High-level gene expression of KCNJ15 was also observed when the stomach microarray datasets were compared with the lymphocyte datasets generated on the same platform (data not shown). Therefore, efforts were then focused on the possible role of KCNJ15 in the parietal cells. We first sought to confirm the KCNJ15 expression level in the stomach by qRT-PCR.
Well-characterized primer pairs for kcnj16 and kcnq1 were used for qRT-PCR analysis, with housekeeping gene polrA2 as internal reference. The qRT-PCR results (Fig. 1A) indicated that KCNJ15 was most highly expressed in the kidney, followed by the stomach, and then the lung. All other tissues (including the liver) examined in this study exhibited little expression of kcnj15 mRNA, consistent with the microarray datasets. The mRNA level for kcnq1 in the stomach is apparently much lower than that of kcnj15. However, different from the microarray datasets, the qRT-PCR results suggested that the kcnq1 level in the stomach is much higher than that in the liver. The reason for this discrepancy is a subject of speculation. For example, the particular kcnq1 probe on the microarray chip might be unspecific. Higher expression of kcnj15 mRNA compared with kcnq1 mRNA was also observed when β-actin mRNA level was used as internal references (data not shown).
Expression of KCNJ15 protein in the stomach.
The above results suggested the possibility that KCNJ15 is expressed in the stomach. Western blot analyses were then performed to examine the KCNJ15 protein expression in the stomach and several other tissues (Fig. 1B). Protein bands of expected ∼80 kDa (32) were observed in the kidney and the stomach. Little expression was observed in the heart, the liver, the skeletal muscle, and the spleen. Unexpectedly, the brain showed high levels of KCNJ15 signal despite that the kcnj15 mRNA level was low, suggesting a slow turnover of this protein in the brain. It is of interest that the major KCNJ15 band in the lung is slightly smaller than that in the stomach and kidney.
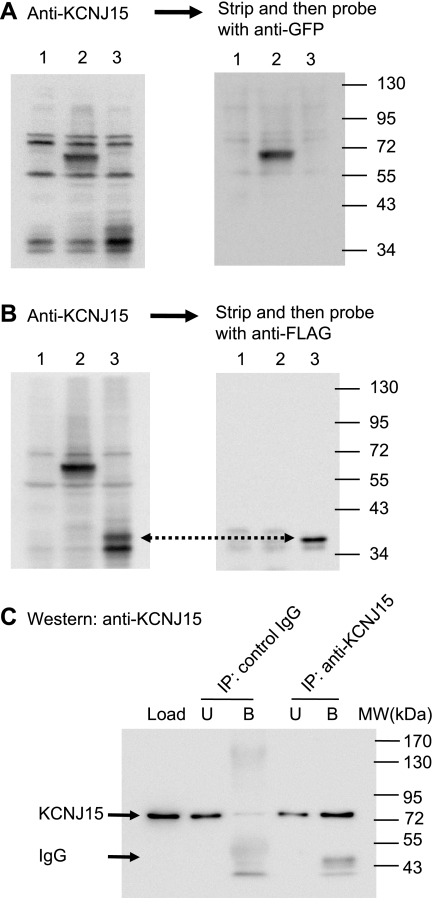
Although KCNJ15 appeared at the expected ∼80 kDa (32), the theoretical molecular mass for this protein is ∼40 kDa. To be sure of the specificity of the KCNJ15 antibody, Western blots were performed with two recombinant KCNJ15 constructs: one with a CFP tag at the COOH-terminus of KCNJ15, and the other with a FLAG tag ahead of the NH2-terminal signal peptide of KCNJ15. HEK-293 cells expressing KCNJ15CFP exhibited a KCNJ15 antibody specific band at ∼66 kDa (Fig. 2A), as expected from the peptide sizes of KCNJ15 (40 kDa) and CFP (26 kDa). Importantly, this 66-kDa band was the major signal detected by the green fluorescent protein (GFP) antibody (Fig. 2A), which recognizes GFP and its derivatives including CFP. The FLAG-tagged KCNJ15 construct exhibited signals at two slightly different sizes, which were specifically detected by KCNJ15 antibody, compared with the nontransfected cells (Fig. 2B). The larger product was detected by a FLAG antibody (Fig. 2B), indicating that it was the unprocessed KCNJ15. The smaller product was not recognized by the FLAG antibody, indicating mature product with the FLAG tag and the signal peptide removed.
Fig. 2.
Characterization of anti-KCNJ15 antibody. HEK-293 cells were transfected to express KCNJ15 with a COOH-terminal cyan fluorescent protein (CFP) tag (lane 2 in all blots) or an NH2-terminus FLAG tag (lane 3). Cell lysates were analyzed by Western blot together with nontransfected 293 cell lysates (lane 1). A: blot was probed with anti-KCNJ15, stripped, and then probed with anti-green fluorescent protein (GFP). B: blot was probed with anti-KCNJ15, stripped, and then probed with anti-FLAG. Note that the majority of exogenous FLAG-KCNJ15 was processed to remove signal peptide and the FLAG tag altogether, therefore not recognized by anti-FLAG. Incomplete removal of signal peptide is one of the reasons for multiple KCNJ15 bands. C: lysate made from rabbit gastric mucosa was immunoprecipitated (IP) with KCNJ15 and a control rabbit IgG, respectively. The lysate (Load), bound (B), and unbound (U) fractions were analyzed by Western blots with anti-KCNJ15. KCNJ15 was precipitated by the anti-KCNJ15 (signals at ∼80 kDa). Signals at ∼50 kDa are IgG molecules. MW, molecular mass.
To further characterize both the KCNJ15 protein and the KCNJ15 antibody, immunoprecipitation was performed with rabbit gastric lysate by use of KCNJ15 antibody. A similar procedure was performed with a control rabbit IgG. The lysate (Load), bound, and unbound fractions from the immunoprecipitations were analyzed by Western blot as shown in Fig. 2C. KCNJ15 band of 80 kDa was precipitated with the KCNJ15 antibody, but not with the control IgG, suggesting that this antibody is able to recognize native KCNJ15.
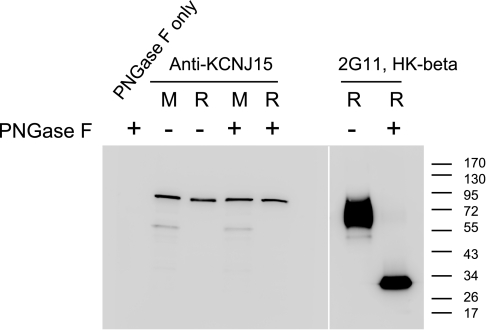
These Western blots demonstrated the specificity of the KCNJ15 antibody. But it remained a puzzle as to why the endogenous KCNJ15 appeared at 80 kDa. One explanation could be glycosylation: two theoretical N-glycosylation sites in KCNJ15 sequence have been cited (21, 22), yet no experimental evidence was given. To provide experimental evidence for N-glycosylation, gastric gland lysates were treated with PNGase F and analyzed by Western blot. According to data presented in Fig. 3, H-K-ATPase β showed a clear shift from the broad 60- to 80-kDa band to ∼32 kDa after PNGase F treatment, whereas no change was detected for KCNJ15, indicating that this channel protein is not significantly N-glycosylated.
Fig. 3.
Deglycosylation analysis of KCNJ15. Lysates prepared from mouse gastric mucosa (M) and rabbit gastric glands (R) were digested by PNGase F to remove N-glycosylations on proteins. PNGase F-digested samples (+), mock-digested samples (−), and a sample of only PNGase F were analyzed by Western blot with anti-KCNJ15 antibody (left). To ascertain the efficiency of the PNGase F digestion, the rabbit samples were probed with anti-H-K-β antibody (right).
Cellular and subcellular distribution of KCNJ15 in the gastric glands.
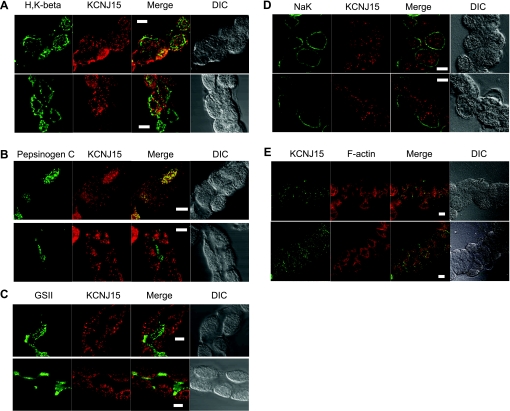
Gastric glands are composed mainly of three types of cells: parietal cells, chief cells, and mucous neck cells. Immunofluorescence staining was performed with resting glands to identify the cell type(s) expressing KCNJ15 and location within the cells. When glands were costained with KCNJ15 antibody and an antibody exclusive for parietal cells (H-K-ATPase β subunit), it was clear that positive signals for both antibodies existed in the same cells, indicating the expression of KCNJ15 in parietal cells (Fig. 4A). KCNJ15 staining appeared as variable sized puncta throughout the parietal cell. It was also noticed that some H-K-ATPase-positive cells exhibited lower KCNJ15 signals. The punctate KCNJ15 staining was distinct from the H-K-ATPase staining, recalling the early observation that there is little potassium channel activity on the H-K-ATPase-rich tubulovesicles (29).
Fig. 4.
Expression of KCNJ15 in rabbit gastric glands. Rabbit gastric glands were fixed with 3.7% formaldehyde, permeabilized with 0.5% Triton, and then probed for KCNJ15 (red) and a marker for either parietal cells (A: H-K-ATPase, green), or chief cells (B: pepsinogen C), or mucous neck cells (C: glycoproteins specific for GSII lectin, green). DIC, differential interference contrast. To examine the basolateral expression, KCNJ15 (red) and Na+-K+-ATPase (green) were costained (D). For better visualization of KCNJ15 localization, dual staining of KCNJ15 (green) and F-actin (red) were also performed (E). Images were collected with a Zeiss LSM510 confocal microscope. Bar = 10 μm.
Glands were also costained with an antibody for pepsinogen C as well as the KCNJ15 antibody (Fig. 4B), to test whether KCNJ15 is expressed in the pepsinogen-secreting chief cells. Results indicated that some chief cells expressed KCNJ15 whereas others did not. It appeared that ∼50% chief cells were KCNJ15 positive whereas ∼80% parietal cells were expressing KCNJ15. No KCNJ15 expression was detected in the mucous neck cells (Fig. 4C) identified by positive GSII staining (7, 35).
To further characterize the subcellular localization of the channel protein, dual staining of KCNJ15 and Na+-K+ATPase was performed. With Na+-K+-ATPase as a marker for basolateral membrane of the gastric epithelial cells (25), it is clear that most of the KCNJ15 signal is intracellular or associated with the “intracellular” canalicular apical membrane structures (Fig. 4D). Only a very small amount of KCNJ15 signal was found close to Na-K-ATPase at the basolateral membrane. The lack of KCNJ15 association with basolateral membrane is further supported by the histamine/SCH28080-treated parietal cells (Fig. 6C), where the basolateral membrane is clearly separated from apical membrane and KCNJ15 signal was not detected on basolateral membrane.
Fig. 6.
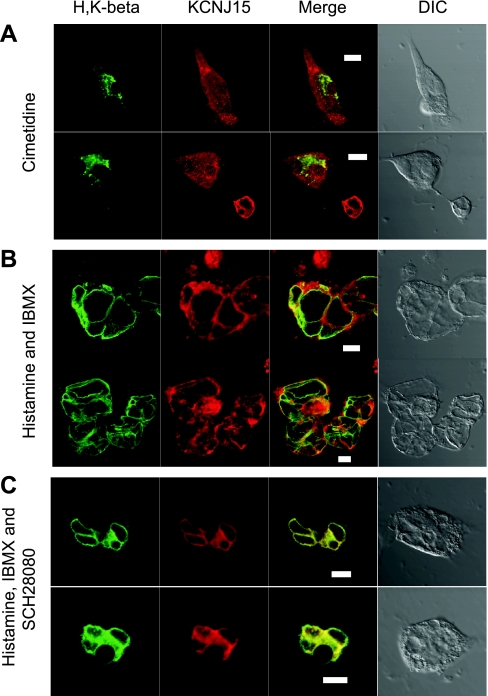
Stimulation-associated translocation of KCNJ15 in rabbit parietal cells. Primary cultures of rabbit parietal cells were treated differently to achieve 3 different physiological states before immunofluorescence staining with anti-KCNJ15 (red) and anti-H-K-ATPase (green) antibodies. A: cimetidine. B: histamine and IBMX. C: histamine, IBMX, and SCH28080. Images were collected with a Zeiss LSM510 confocal microscope. Bar = 10 μm.
For further insight about subcellular localization of KCNJ15, dual staining of KCNJ15 and F-actin was performed (Fig. 4E). In gastric glands, F-actin signal is dense at the canalicular apical membrane of parietal cells, in addition to relatively weaker signal at other locations of plasma membrane. KCNJ15 was found in these parietal cells, but exhibiting a distinct pattern compared with F-actin staining.
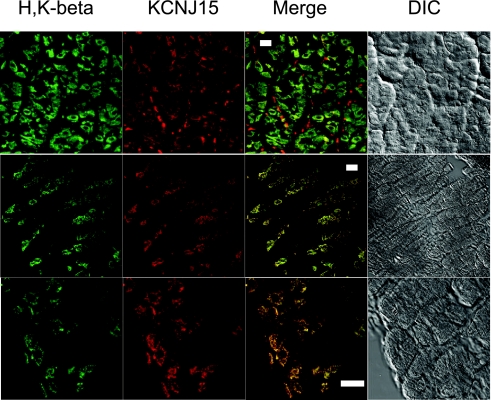
To examine the KCNJ15 in intact tissue, cryosections of mouse gastric mucosa were dual stained for KCNJ15 and H-K-ATPase (Fig. 5). KCNJ15 was detected in all H-K-ATPase-positive parietal cells. Similar to the observations made with rabbit glands, KCNJ15 was also found in other cells. In mouse gastric mucosa, the KCNJ15 seemed to be better colocalized with H-K-ATPase compared with rabbit glands. This may be explained by the differences in sample preparation or differences in species.
Fig. 5.
Expression of KCNJ15 in mouse gastric mucosa. Stomachs from whole-body-perfused mice were fixed with 3.7% formaldehyde and cryosectioned. The cryosections were dual stained with anti-KCNJ15 (red) and anti-H-K-ATPase antibodies (green). Images were collected with a Zeiss LSM510 confocal microscope. Bar = 10 μm.
Stimulation-associated translocation of KCNJ15 in the parietal cells.
The fact that membrane protein KCNJ15 existed in parietal cells, and most of the KCNJ15 not on the basolateral membrane, suggested some role for this channel, other than basolateral K+ regulation, in the physiology of parietal cells. Experiments were then performed with cultured parietal cells to ascertain whether KCNJ15 channels undergo stimulation-dependent translocation, similar to the recycling pathway well known for the H-K-ATPases (10). Upon culture, the parietal cells take on a modified morphology convenient for the studies of membrane trafficking: the invaginated apical canalicular membrane becomes completely sequestered to form intracellular vacuoles so that the apical membrane and basolateral membranes remain distinctly separated (4).
In some cases parietal cells were treated with cimetidine to maintain the resting state. In typical resting cells, H-K-ATPase exhibited staining throughout the cytosol because it is mainly present on the small cytoplasmic tubulovesicles. In these cells, KCNJ15 staining also appeared cytosolic although it was distinct from the H-K-ATPase staining (Fig. 6A). In cells treated with histamine and IBMX to achieve a maximum stimulation of acid secretion, HCl and water are pumped into the apical vacuoles. Without a mechanism to vent the acid, the vacuoles continue expanding until they occupy the majority of space in the cell. Under this condition, both H-K-ATPase and KCNJ15 appeared on the apical membrane (Fig. 6B). To exclude the possibility that the KCNJ15 staining pattern in stimulated parietal cells was a result of KCNJ15 being close to the apical membrane instead of being incorporated into it, cells were stimulated with histamine/IBMX in the presence of a proton pump inhibitor SCH28080. This treatment allows the trafficking and assembly of the machineries required for acid secretion without the actual production of acid since the pump is effectively inhibited by SCH28080 (1). Under this condition, as demonstrated in Fig. 6C, there was a clear translocation of H-K-ATPase from cytoplasmic tubulovesicles onto the apical vacuolar membrane. Since acid secretion was inhibited by SCH28080, the expansion of the apical vacuoles was rather limited, which allowed a clear observation of the translocation and apparent colocalization of both H-K-ATPase and KCNJ15.
Fractionation of the different membranes by differential centrifugation was performed as an alternative test for the stimulation-associated translocation of KCNJ15. Isolated gastric glands were either treated with cimetidine or histamine/IBMX to achieve the two distinct physiological states: resting or stimulated. The secretory status of the glands was measured by the [14C]AP-accumulation assay (Fig. 7A). The glands were then homogenized and membranes of different sizes were fractionated by differential centrifugation. Plasma membranes including the canalicular apical membrane sediment at a lower centrifugal force whereas the smaller tubulovesicle membranes sediment at higher speed. Similar portions of each pellet and the final supernatant from both resting and stimulated preparations were analyzed by Western blots probed for signals of H-K-ATPase, KCNJ15, and β-actin (Fig. 7B). A clear redistribution of H-K-ATPase accompanied stimulation of the gastric glands. In the resting preparations, the majority of the H-K-ATPase signal was found in the smaller and/or less dense P3 microsomal membrane fraction, whereas the stimulated preparations had a large increase in H-K-ATPase in the larger and/or more dense P1 membrane fraction.
Fig. 7.
Stimulation-triggered redistribution of KCNJ15 from small vesicles to larger membrane fractions. Rabbit gastric glands were treated with either cimetidine or histamine/IBMX to achieve 2 different physiological states: resting and stimulated. Active acid secretion of the stimulated glands were monitored by [14C]aminopyrine (AP)-accumulation assay (A). AP ratios for resting and stimulated glands were plotted with error bars representing the standard errors of the means. The ascertained resting and stimulated gastric glands were then homogenized and subjected to differential centrifugation according to the scheme illustrated (B). The same proportion of each fraction was loaded for Western blot analysis with antibodies against H-K-ATPase β subunit, β-actin, and KCNJ15.
The KCNJ15 blot was a surprise. In both resting and stimulated preparations the KCNJ15 signal was detected in the larger membrane P1 and in the supernatant S3, but not in P2 or P3. The KCNJ15 signal was strong in resting S3, but weak in the stimulated S3, with a concomitant increase of KCNJ15 signal in the stimulated P1 fraction. Given the fact that KCNJ15 has a transmembrane domain in its peptide sequence, we considered the possibility that this protein may exist in a light vesicular membrane in the S3 fraction. Therefore, the S3 fractions from both resting and stimulated preparations were centrifuged for an extended 4 h at 100,000 g. The resulting pellets, designated P4, and supernatants (S4) were analyzed by Western blot for KCNJ15. Most of the KCNJ15 signal was found in the P4 pellet, confirming that the KCNJ15 vesicles are distinct from the H-K-ATPase enriched tubulovesicles. These fractionation results and the immunofluorescence staining results suggested a stimulation associated translocation of KCNJ15 from a light vesicular membrane onto the apical membrane of parietal cells.
DISCUSSION
Discovery of KCNJ15 in the gastric parietal cells.
Sachs's group (17) pioneered the microarray study on gastric parietal cells. In comparing the gene expressions between purified parietal cells and gastric mucosa, the KCNQ1 mRNA level in pure parietal cells was found to be 7.66-fold higher than in total gastric mucosa, whereas other K+ channels showed little or no increase in comparative mRNA. One drawback of this approach is that any protein highly expressed in cells other than parietal cells might be missed even if they are also highly expressed in parietal cells. This is exactly the case for KCNJ15, which is expressed in both parietal cells and chief cells (Fig. 4B) and may be expressed in surface mucous cells (see Ref. 12). Moreover, from the data of Lambrecht et al. (17), the overall KCNJ15 signal seemed to be higher in parietal cells than that in other gastric cells, with KCNJ15 mRNA in pure parietal cells being 1.84-fold over that in total gastric mucosa.
We took a different approach when analyzing the microarray data that cover the whole mouse genome. The gene expression levels of the 45,000 transcripts were compared between gastric mucosa and a different tissue. KCNJ15 clearly stood out among other K+ channels in this comparison with a 33-fold higher expression level in the gastric mucosa. Confirmed by qRT-PCR analysis, this observation suggested specific expression of KCNJ15 in gastric mucosa. Immunofluorescence staining of stomach cryosections, isolated gastric glands, and parietal cells suggested that KCNJ15 was expressed in both parietal cells and chief cells, but not mucous neck cells.
It is noteworthy that KCNJ15 has been implicated in acid secretion in pulmonary epithelial cells, which use a nongastric H-K-ATPase to generate a low pH and KCNJ15 to recycle K+ at the apical membrane (16, 30). In these acid-secreting cells, the KCNJ15 channels are resistant to low pH, with mild inhibition seen after a long-time treatment.
A new member in the family of parietal cell apical K+ channels.
In this study, evidence is presented to support a role for KCNJ15 in stimulated gastric acid secretion. In resting parietal cells, KCNJ15 is mainly located in the cytoplasm as indicated by immunofluorescence staining. But this cytoplasmic staining is clearly different from that of H-K-ATPase, according to both immunofluorescence staining results and the cellular fractionation results: whereas H-K-ATPase is mainly found in the P3 fraction, KCNJ15 was not detected in P3. Instead, most of the KCNJ15 remained in the “classical” S3 supernatant. Only after an extended centrifugation did the KCNJ15 sediment in newly designated P4 fraction. The separation of two types of small vesicles with distinct sedimentation properties may allow interesting studies to elucidate other differences between these two types of vesicles. The KCNJ15 signal found in the resting P1 fraction may come from other gastric cells, or even entrapment of KCNJ15 in larger membrane fragments. Nevertheless, upon stimulation, there is a clear translocation of KCNJ15 from a cytosolic compartment to the fraction rich in apical membranes. This stimulation-associated translocation strongly suggests a role for KCNJ15 although more direct evidence is needed.
A brief review of K+ channels in parietal cells.
Several other K+ channels have been implicated in the apical K+ recycling. Although the initial study on KCNQ1 (13) suggested its colocalization with H-K-ATPase (namely, every parietal cell expressed KCNQ1 on apical membrane and tubulovesicles), close examination of immunostained tissue sections suggests that many parietal cells have little or no expression of KCNQ1 (14). This is in concert with the observation that treatment of gastric glands and isolated mouse stomach with supermaximal doses (1,000-fold higher than IC50) of a highly specific inhibitor, HMR1556, of KCNQ1 channels still allowed stimulated acid secretion (15). Kaufhold et al. (15) also reported that most of the KCNQ1 in parietal cells did not translocate to the apical membrane upon stimulation. Therefore, the observation that KCNQ1-knockout mice exhibited complete inhibition of stimulated acid secretion suggested that KCNQ1 might play some additional role in parietal cell biology and development besides apical K+ recycling.
Seidler's group (15) observed colocalization of Kir4.1 (KCNJ10) and H-K-ATPase on tubulovesicles and the translocation of this channel to apical membrane upon stimulation. However, Fujita et al. (12) reported that Kir4.1 is always localized to the apical membrane. The latter observation is in concert with the fact that K+ channel activity is absent from the tubulovesicles by electrophysiological measurements (18, 28). A possible role for Kir4.1 is also supported by its resistance to low pH (12).
Different from the observation of Fujita et al. (12) with rat, Cuppoletti, Malinowski, and colleagues (5, 19) have reported abundant Kir2.1 (KCNJ2) mRNA and protein in rabbit gastric parietal cells. They also found that Kir2.1 channel activity (open time) can be stimulated by low pH and cAMP, strongly suggesting that Kir2.1 could be involved in stimulated acid secretion.
More recently, the KCl cotransporter KCC4 in parietal cells was found to be associated with a large size membrane fraction called stimulation-associated membranes, but not with a microsomal membrane fraction referred to as tubulovesicles (11). Moreover, both H+ uptake and 36Cl− uptake by the stimulation-associated membrane vesicles were severely inhibited by the KCC inhibitor DIOA, whereas DIOA had virtually no effect on uptake in tubulovesicles. These authors suggested that KCC4 was a resident apical KCl pathway that was supplemented in vivo by additional K+ and Cl− channels recruited to the greatly expanded apical surface after stimulation.
Several groups have hypothesized that multiple K+ channels may cooperate for the task of apical K+ recycling (11, 14, 15). Our observation that both KCNJ15 and KCNQ1 mRNA are highly expressed in the stomach is consistent with this hypothesis. To explain the observation that both KCNQ1 and Kir4.1 (KCNJ10) exist at the same cellular location, Seidler and colleagues (15) suggested that KCNQ1 is important when the parietal cell is depolarized, whereas KCNJ10 is more active when the cell is hyperpolarized, but it is really the channel activation or recruitment that is responsible for the polarization state of the cell. Another explanation for multiple K+ pathways in apical K+ recycling is that a cotransporter such as KCC4 might be needed for basal acid secretion whereas conductive K+ channels might be responsible (recruited) for stimulated acid secretion (11). In fact, it may be naive to expect that a single K+ channel could account for all the requisite properties to accomplish HCl secretion. The channel has to operate at enormous variation of pH and membrane voltage; it has to have characteristics of conductance activation and/or voltage dependence; it is likely to be a subject of membrane recruitment; and there must be flexibility of activation to account for variations in secretory output. There are wide differences among species for observed relative rates of basal vs. stimulated acid secretion (6), and it would be of interest to correlate variations in the apparent levels of different K+ transporters. The discovery of Kir2.1 and KCNJ15 at the apical membrane does not conflict with these possibilities. Yet the reason for so many K+ channels directed toward one general function could be more complicated. For example, considering the inconsistent expression levels of KCNQ1 and KCNJ15 in parietal cells, and the developmentally regulated expression of KCNJ15 in various epithelia (24), it could be hypothesized that the expression levels of certain K+ channels are associated with the development of parietal cells both in early ontogeny and with continued development and differentiation of adult gastric glands.
The data presented here support the participation of another K+ transport candidate, KCNJ15, in membrane recruitment associated with enhanced apical K+ recycling and HCl secretion. At the present time it is not possible to establish which, if not several, of the various candidate transporters participate in the apical K+ conductance that is necessary for K+ recycling and resultant HCl secretion.
GRANTS
This work was supported by a start-up fund from SUNY Buffalo (L. Zhu) and NIH grants DK010141-42 (J. G. Forte) and DK31900 (C. S. Chew).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGMENTS
The authors thank Dr. Wade Sigurdson (Confocal and Flow Cytometry Facility, University at Buffalo) for helping with confocal microscopy and Dr. Lisa Martin and Dr. Sasha Black (University at Buffalo Laboratory Animal Facilities) for helping with proper animal protocols.
Contact information for J. G. Forte: 241 LSA, Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720 (e-mail: jforte@berkeley.edu).
REFERENCES
- 1. Agnew BJ, Duman JG, Watson CL, Coling DE, Forte JG. Cytological transformations associated with parietal cell stimulation: critical steps in the activation cascade. J Cell Sci 112: 2639–2646, 1999 [DOI] [PubMed] [Google Scholar]
- 2. Berglindh T. Effects of common inhibitors of gastric acid secretion on secretagogue-induced respiration and aminopyrine accumulation in isolated gastric glands. Biochim Biophys Acta 464: 217–233, 1977 [DOI] [PubMed] [Google Scholar]
- 3. Chew CS, Hersey SJ, Sachs G, Berglindh T. Histamine responsiveness of isolated gastric glands. Am J Physiol Gastrointest Liver Physiol 238: G312–G320, 1980 [DOI] [PubMed] [Google Scholar]
- 4. Chew CS, Ljungstrom M, Smolka A, Brown MR. Primary culture of secretagogue-responsive parietal cells from rabbit gastric mucosa. Am J Physiol Gastrointest Liver Physiol 256: G254–G263, 1989 [DOI] [PubMed] [Google Scholar]
- 5. Cuppoletti J, Tewari KP, Sherry AM, Malinowska DH. Kir2.1 K+ channels of the gastric parietal cell. In: Mechanisms and Consequences of Proton Transport, edited by Urushidani T, Forte JG, Sachs G. Boston, MA: Kluwer Academic, 2002, p. 255–264 [Google Scholar]
- 6. DeBas HT. Peripheral regulation of gastric acid secretion. In: Physiology of the Gastrointestinal Tract, edited by L. J.New York: Raven, 1987, p. 931–945 [Google Scholar]
- 7. Falk P, Roth KA, Gordon JI. Lectins are sensitive tools for defining the differentiation programs of mouse gut epithelial cell lineages. Am J Physiol Gastrointest Liver Physiol 266: G987–G1003, 1994 [DOI] [PubMed] [Google Scholar]
- 8. Forte JG. K+ channels in the secretory membrane of the parietal cell. Focus on “Gastric parietal cell secretory membrane contains PKA- and acid-activated Kir21 K+ channels.” Am J Physiol Cell Physiol 286: C478–C479, 2004 [DOI] [PubMed] [Google Scholar]
- 9. Forte JG, Forte GM, Saltman P. K+-stimulated phosphatase of microsomes from gastric mucosa. J Cell Physiol 69: 293–304, 1967 [DOI] [PubMed] [Google Scholar]
- 10. Forte JG, Zhu L. Apical recycling of the gastric parietal cell H-K-ATPase. Annu Rev Physiol 72: 273–296, 2010 [DOI] [PubMed] [Google Scholar]
- 11. Fujii T, Takahashi Y, Ikari A, Morii M, Tabuchi Y, Tsukada K, Takeguchi N, Sakai H. Functional association between K+-Cl− cotransporter-4 and H+-K+-ATPase in the apical canalicular membrane of gastric parietal cells. J Biol Chem 284: 619–629, 2009 [DOI] [PubMed] [Google Scholar]
- 12. Fujita A, Horio Y, Higashi K, Mouri T, Hata F, Takeguchi N, Kurachi Y. Specific localization of an inwardly rectifying K+ channel, Kir4.1, at the apical membrane of rat gastric parietal cells; its possible involvement in K+ recycling for the H+-K+-pump. J Physiol 540: 85–92, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grahammer F, Herling AW, Lang HJ, Schmitt-Graff A, Wittekindt OH, Nitschke R, Bleich M, Barhanin J, Warth R. The cardiac K+ channel KCNQ1 is essential for gastric acid secretion. Gastroenterology 120: 1363–1371, 2001 [DOI] [PubMed] [Google Scholar]
- 14. Heitzmann D, Warth R. No potassium, no acid: K+ channels and gastric acid secretion. Physiology (Bethesda) 22: 335–341, 2007 [DOI] [PubMed] [Google Scholar]
- 15. Kaufhold MA, Krabbenhoft A, Song P, Engelhardt R, Riederer B, Fahrmann M, Klocker N, Beil W, Manns M, Hagen SJ, Seidler U. Localization, trafficking, and significance for acid secretion of parietal cell Kir4.1 and KCNQ1 K+ channels. Gastroenterology 134: 1058–1069, 2008 [DOI] [PubMed] [Google Scholar]
- 16. Krouse ME, Talbott JF, Lee MM, Joo NS, Wine JJ. Acid and base secretion in the Calu-3 model of human serous cells. Am J Physiol Lung Cell Mol Physiol 287: L1274–L1283, 2004 [DOI] [PubMed] [Google Scholar]
- 17. Lambrecht NW, Yakubov I, Scott D, Sachs G. Identification of the K efflux channel coupled to the gastric H-K-ATPase during acid secretion. Physiol Genomics 21: 81–91, 2005 [DOI] [PubMed] [Google Scholar]
- 18. Lee HC, Breitbart H, Berman M, Forte JG. Potassium-stimulated ATPase activity and hydrogen transport in gastric microsomal vesicles. Biochim Biophys Acta 553: 107–131, 1979 [DOI] [PubMed] [Google Scholar]
- 19. Malinowska DH, Sherry AM, Tewari KP, Cuppoletti J. Gastric parietal cell secretory membrane contains PKA- and acid-activated Kir2.1 K+ channels. Am J Physiol Cell Physiol 286: C495–C506, 2004 [DOI] [PubMed] [Google Scholar]
- 20. Nerbonne JM, Nichols CG, Schwarz TL, Escande D. Genetic manipulation of cardiac K+ channel function in mice: what have we learned, and where do we go from here? Circ Res 89: 944–956, 2001 [DOI] [PubMed] [Google Scholar]
- 21. Pearson WL, Dourado M, Schreiber M, Salkoff L, Nichols CG. Expression of a functional Kir4 family inward rectifier K+ channel from a gene cloned from mouse liver. J Physiol 514: 639–653, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shuck ME, Piser TM, Bock JH, Slightom JL, Lee KS, Bienkowski MJ. Cloning and characterization of two K+ inward rectifier (Kir) 1.1 potassium channel homologs from human kidney (Kir12 and Kir13). J Biol Chem 272: 586–593, 1997 [DOI] [PubMed] [Google Scholar]
- 23. Song P, Groos S, Riederer B, Feng Z, Krabbenhoft A, Smolka A, Seidler U. KCNQ1 is the luminal K+ recycling channel during stimulation of gastric acid secretion. J Physiol 587: 3955–3965, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thiery E, Gosset P, Damotte D, Delezoide AL, de Saint-Sauveur N, Vayssettes C, Creau N. Developmentally regulated expression of the murine ortholog of the potassium channel KIR4.2 (KCNJ15). Mech Dev 95: 313–316, 2000 [DOI] [PubMed] [Google Scholar]
- 25. Vagin O, Turdikulova S, Tokhtaeva E. Polarized membrane distribution of potassium-dependent ion pumps in epithelial cells: different roles of the N-glycans of their beta subunits. Cell Biochem Biophys 47: 376–391, 2007 [DOI] [PubMed] [Google Scholar]
- 26. Vucic E, Radebold K, MacGregor G, Shull GE, Spicer Z, Wang T, Hebert S, Geibel JP. ROMK is the apical potassium regulatory channel in the parietal cell (Abstract). Gastroenterology 122: A244, 2002. [Google Scholar]
- 27. Wolosin JM, Forte JG. Changes in the membrane environment of the (K+ + H+)-ATPase following stimulation of the gastric oxyntic cell. J Biol Chem 256: 3149–3152, 1981 [PubMed] [Google Scholar]
- 28. Wolosin JM, Forte JG. Functional differences between K+-ATPase rich membranes isolated from resting or stimulated rabbit fundic mucosa. FEBS Lett 125: 208–212, 1981 [DOI] [PubMed] [Google Scholar]
- 29. Wolosin JM, Forte JG. Stimulation of oxyntic cell triggers K+ and Cl− conductances in apical H+-K+-ATPase membrane. Am J Physiol Cell Physiol 246: C537–C545, 1984 [DOI] [PubMed] [Google Scholar]
- 30. Wu JV, Krouse ME, Rustagi A, Joo NS, Wine JJ. An inwardly rectifying potassium channel in apical membrane of Calu-3 cells. J Biol Chem 279: 46558–46565, 2004 [DOI] [PubMed] [Google Scholar]
- 31. Wulfsen I, Hauber HP, Schiemann D, Bauer CK, Schwarz JR. Expression of mRNA for voltage-dependent and inward-rectifying K channels in GH3/B6 cells and rat pituitary. J Neuroendocrinol 12: 263–272, 2000 [DOI] [PubMed] [Google Scholar]
- 32. Yang D, Zhang X, Hughes BA. Expression of inwardly rectifying potassium channel subunits in native human retinal pigment epithelium. Exp Eye Res 87: 176–183, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang H, Chen X, Bollag WB, Bollag RJ, Sheehan DJ, Chew CS. Lasp1 gene disruption is linked to enhanced cell migration and tumor formation. Physiol Genomics 38: 372–385, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu L, Crothers J, Jr, Zhou R, Forte JG. A possible mechanism for ezrin to establish epithelial cell polarity. Am J Physiol Cell Physiol 299: C431–C443, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhu L, Hatakeyama J, Zhang B, Makdisi J, Ender C, Forte JG. Novel insights of the gastric gland organization revealed by chief cell specific expression of moesin. Am J Physiol Gastrointest Liver Physiol 296: G185–G195, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu L, Liu Y, Forte JG. Ezrin oligomers are the membrane-bound dormant form in gastric parietal cells. Am J Physiol Cell Physiol 288: C1242–C1254, 2005 [DOI] [PubMed] [Google Scholar]