Abstract
AMP-activated protein kinase (AMPK) and peroxisome proliferator-activated receptor-α (PPAR-α) are critical regulators of short-term and long-term fatty acid oxidation, respectively. We examined whether the activities of these molecules were coordinately regulated. H4IIEC3 cells were transfected with PPAR-α and PPAR-γ expression plasmids and a peroxisome-proliferator-response element (PPRE) luciferase reporter plasmid. The cells were treated with PPAR agonists (WY-14,643 and rosiglitazone), AMPK activators 5-aminoimidazole-4-carboxamide riboside (AICAR) and metformin, and the AMPK inhibitor compound C. Both AICAR and metformin decreased basal and WY-14,643-stimulated PPAR-α activity; compound C increased agonist-stimulated reporter activity and partially reversed the effect of the AMPK activators. Similar effects on PPAR-γ were seen, with both AICAR and metformin inhibiting PPRE reporter activity. Compound C increased basal PPAR-γ activity and rosiglitazone-stimulated activity. In contrast, retinoic acid receptor-α (RAR-α), another nuclear receptor that dimerizes with retinoid X receptor (RXR), was largely unaffected by the AMPK activators. Compound C modestly increased AM580 (an RAR agonist)-stimulated activity. The AMPK activators did not affect PPAR-α binding to DNA, and there was no consistent correlation between effects of the AMPK activators and inhibitor on PPAR and the nuclear localization of AMPK-α subunits. Expression of either a constitutively active or dominant negative AMPK-α inhibited basal and WY-14,643-stimulated PPAR-α activity and basal and rosiglitazone-stimulated PPAR-γ activity. We concluded that the AMPK activators AICAR and metformin inhibited transcriptional activities of PPAR-α and PPAR-γ, whereas inhibition of AMPK with compound C activated both PPARs. The effects of AMPK do not appear to be mediated through effects on RXR or on PPAR/RXR binding to DNA. These effects are independent of kinase activity and instead appear to rely on the activated conformation of AMPK. AMPK inhibition of PPAR-α and -γ may allow for short-term processes to increase energy generation before the cells devote resources to increasing their capacity for fatty acid oxidation.
Keywords: nuclear receptors, fatty acid oxidation, compound C, AMP-activated protein kinase, peroxisome proliferator-activated receptor
amp-activated protein kinase (AMPK) and peroxisome proliferator-activated receptor-α (PPAR-α) are critical regulators of fatty acid oxidation. AMPK phosphorylates and inhibits acetyl CoA carboxylase and activates malonyl-CoA decarboxylase. As a result, levels of malonyl-CoA fall, relieving the inhibition of carnitine-palmitoyl transferase-1 (CPT-1), the rate-limiting step in fatty acid oxidation (9, 30). PPAR-α is a member of the nuclear hormone receptor family that dimerizes with retinoid X receptor (RXR) and binds to promoters at peroxisome-proliferator-responsive elements (PPRE). Binding of ligands such as fibrates or fatty acids leads to the activation of transcription of target genes such as fatty acid binding protein, CPT-1, acyl-CoA oxidase, and medium chain acyl-CoA dehydrogenase, which increases the capacity for fatty acid oxidation (11).
AMPK is a heterotrimeric kinase that contains two regulatory subunits, β and γ, and one of the isoforms of the α-catalytic subunit, α1 or α2. AMP was considered the classical activator of AMPK, binding the γ-subunit and inducing a conformational change, which causes movement of the autoinhibition domain of the α-regulatory subunit away from the kinase domain toward the γ-subunit. This conformational change allows phosphorylation at the Thr172 residue of the α-subunit or reduces the ability of phosphatases to remove the phosphate (19). Phosphorylation of this residue is now known to be mediated by numerous kinases and is essential for enzyme activity. Thus it can be used as a marker of AMPK activity. AMPK is activated by hypoxia, ischemia, hyperosmolality, reactive oxygen species, hypoglycemia, muscle contraction, and stimulation of signaling pathways (e.g., adiponectin, IL-6, and transforming growth factor-β) (5, 7, 13, 18, 24, 28, 29, 32, 34, 35, 39, 43). Regardless of the stimulus, once activated, AMPK turns on ATP-producing processes, such as fatty acid oxidation and glycolysis, and turns off ATP-consuming processes such as fatty acid and protein synthesis (20, 38).
From the perspective of metabolic regulation, AMPK and PPAR-α activation sometimes need to be coordinated. For instance, during fasting, as glucose levels fall and fatty acid levels rise, AMPK activation increases mitochondrial fatty acid uptake and PPAR-α activation increases the maximal fatty acid-oxidizing capacity in the liver. The phosphorylation cascades that regulate AMPK activity could also modify PPAR-α activity because activity of PPAR-α is affected by phosphorylation (4). There is evidence that p38, ERK, protein kinases A and C, and possibly AMPK can phosphorylate PPAR-α (the latter contention relies mainly on the effect of pharmacological interventions) (see below and Ref. 4). In most cases, activation of these kinases results in increased PPAR-α activity. However, p38 activation has been shown to induce activation of PPAR-α in some cells while inhibiting it in others, and phosphorylation of PPAR-α by glycogen synthase kinase leads to its degradation (1, 2, 4).
There is some evidence for coregulation of AMPK and PPAR-α. Activation of AMPK in myocytes with either 5-aminoimidazole-4-carboxamide riboside (AICAR) (an AMP analog and well-established AMPK activator) or adiponectin increased PPAR-α activity (26, 42). In kidney cells and hepatocytes, Bronner et al. (3) demonstrated that transfected AMPK-α subunits (including several kinase-deficient mutants) bound to and activated PPAR-α. This occurred independently of AMPK activity, was not associated with increased AMP levels, and instead was stimulated by increased Mg ATP levels. On the other hand, treatment with AICAR decreased PPAR-α activity, which was associated with translocation of the AMPK-α2 isoform out of the nucleus. The inhibitory effect of AICAR was observed with PPAR-α constructs in which the putative AMPK consensus phosphorylation site had been mutated. In a separate review of the role of AMPK in transcriptional regulation, Leff cited unpublished data showing similar inhibitory effects of AICAR on PPAR-α and -γ (27). Thus there are conflicting reports regarding the interaction of AMPK and PPAR-α, and it is uncertain whether the effects are attributable to kinase activity or to protein-protein interactions
Both AMPK and PPAR-α have been shown to play an important role in the pathogenesis of alcoholic liver disease. Chronic ethanol consumption leads to impaired fatty acid oxidation in the liver and hepatic steatosis. Our laboratory and those of others have demonstrated that ethanol impairs AMPK activity and decreases PPAR-α activity in mice (12, 15, 43). Administration of AMPK activators, such as AICAR, adiponectin, thiazolidinediones, and hepatocyte growth factor, or the PPAR-α activator WY-14,643 (WY) can ameliorate ethanol-induced fatty liver (12, 25, 36, 37, 41). Given the congruent effects of AMPK and PPAR-α on fatty acid oxidation, we hypothesized that the effects of ethanol on PPAR-α may be mediated by inhibition of AMPK. We thus sought to define the relationship between AMPK activation and PPAR-α activity.
MATERIALS AND METHODS
Chemicals and treatments.
Most chemicals were purchased from Sigma Aldrich Chemical (St. Louis, MO) or Calbiochem-Novabiochem (San Diego, CA). Compound C [6-(4-(2-piperidin-1-yl-etoxy)-phenyl)-3-pyridin-4-yl-pyrazolo(1,5-a) pyrimidine] an inhibitor of AMPK (24), was from Calbiochem. Trypsin and tissue culture media were purchased from GIBCO Invitrogen (Carlsbad, CA). Fetal bovine serum charcoal stripped of lipids was purchased from Hyclone Laboratories (Logan, UT). Nitrocellulose was from Schleicher and Schuell (Keene, NH). H4IIEC3 hepatoma cells were from the American Type Culture Collection. PPRE3-tk-luciferase [containing 3 copies of PPRE from the acyl-CoA oxidase gene ligated to a Herpes Simplex thymidine kinase promoter upstream of the firefly luciferase gene (23)], and the expression plasmid for murine PPAR-α and -γ were the kind gift of Dr. Ronald Evans (Salk Institute) (22). CYP26A1–3′ RARE-pGL4.23, a retinoic acid receptor (RAR)-responsive reporter plasmid cloned upstream of the luciferase gene, was kindly provided by Dr. Ralf Kittler (University of Chicago) (21). AMPK-α1312, a constitutively active form of AMPK lacking the autoinhibition domain, and α1-DN AMPK (D157A mutation), a dominant negative form of AMPK that associates with the β- and γ-subunits but is unable to bind ATP at the catalytic site, were kindly provided by Dr. David Carling (Hammersmith Hospital, London, United Kingdom) and by Dr. Jin-Zhong Zhang (Case Western Reserve University, Cleveland, Ohio), respectively (8, 40). Antibodies for Western blotting were from Santa Cruz Biotechnology (Santa Cruz, CA) and Cell Signaling Technology (Danvers, MA). All chemicals were dissolved in DMSO or double-distilled H2O, and treatments were added directly to cell culture medium.
Transfection of tissue-culture cells.
All cells were grown in MEM supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 250 ng/ml amphotericin B. H4IIEC3 cells were grown to 95% confluence by the day of transfection. For studies on PPAR-α and PPAR-γ the cells were transfected with 0.5 μg of reporter plasmid (PPRE3-tk-luc), 0.5 μg of receptor expression plasmid, and 0.1 μg of pRL-tk-luc (as an internal control for transfection efficiency) per well by Lipofectamine 2000 from GIBCO Invitrogen. For studies on RAR activity, cells were transfected with 0.5 μg of RAR reporter plasmid and 0.2 μg of pRL-tk-luc per well. For studies with the dominant negative AMPK-α1 (α1-DN AMPK) and the constitutively active AMPK-α1 (AMPK-α1312), 0.5 μg of the plasmids were included in the PPAR-α, PPRE reporter plasmid, and pRL-tk-luc transfection. We have previously demonstrated the effects of α1-DN AMPK and AMPK-α1312 on AMPK activity assays (43). AMPK-α1312 expression led to a twofold increase in AMPK activity, whereas α1-DN AMPK decreased AMPK activity ∼30–40% (43). Twenty-four hours later, the cells were incubated with fresh DMEM with 5% charcoal-stripped FBS. They were treated with 1 mM metformin, 40 μM compound C, 500 μM AICAR, 1 μM WY-14,643 [4-chloro-6-(2,3-xylidino)-2-pyrimidinylthio acetic acid], 30 μM rosiglitazone, 100 μM oleic acid, and 50 nM AM580 by addition directly to the medium for an additional 24 h. The cells were then washed twice with PBS and lysed in 100 μl passive lysis buffer. Cell extract (20 μl) was incubated with luciferase assay reagents from the Stop & Glo kit from Promega (Madison, WI). The number of relative light units was determined with a 2-s delay and a 10-s reading for each step with the TD 20/20 Luminometer (TurnerDesigns, Sunnyvale, CA).
Immunoblot analysis.
Aliquots of cell extracts prepared in RIPA buffer (containing 1% deoxycholate, 0.1% SDS, 1% NP-40, 1 mM iodoacetamide, 100 mM Na3VO4, 1 mM PMSF, and 1 mM aprotinin) were fractionated in an SDS-PAGE gel and electroblotted to nitrocellulose membranes. The proteins were detected by incubating the membranes with antibodies (overnight at 4°C). Antibodies for AMPK-α, AMPK-α2, phospho-AMPK-α (Thr172), RAR-α, and the secondary antibody (goat anti-rabbit) were from Cell Signaling Technology. PPAR-α and PPAR-γ antibodies were from Santa Cruz Biotechnology. Detection of the protein bands was performed using the ECL Western Blotting Detection System Kit (Amersham Biosciences, Piscataway, NJ).
Biotinylated DNA binding assay.
PPRE-containing oligonucleotides (from Invitrogen) consisted of the following sequence: 5′- GAACTAGGTCAAAGGTCATCCCCT-3 ′. The nonbiotinylated 3′ oligonucleotide was incubated with either the biotinylated 5′ oligonucleotide or the nonbiotinylated 5′ oligonucleotide (used as a competitive inhibitor in the binding assays) for 5 min in Roche Buffer M at 95°C to anneal the strands. The double-stranded DNA was then mixed with 100 μg of total cell lysate in the following buffer: 25 mM HEPES, 80 mM NaCl, 0.5 mM DTT, 0.5% NP-40, 0.1 mM EDTA (pH 7.5) and 10% glycerol filtered through a 0.45 μm membrane. The competitive binding assays contained nonbiotinylated double-stranded oligonucleotides at a concentration five times that of the biotinylated oligonucleotides. Samples were rotated overnight at 4°C. The next day 40 μl of streptavidin-agarose beads (Millipore, Billerica, MA) were added to each sample, after which they were rotated at 4°C for another 2 h. Samples were then spun down, the supernatant was discarded, and, after being washed twice, the beads were incubated with SDS-sample buffer for 5 min at 95°C before being loaded onto an SDS-PAGE gel.
Nuclear extraction.
Cells were grown to 95% confluency in 60-mm plates in MEM supplemented with 10% FBS and antibiotics as described above. Sixteen hours before treatment, cells were switched to MEM with antibiotics. Twenty-four hours after treatment, cells were harvested and nuclear extracts were prepared according to the Active Motif (Carlsbad, CA) nuclear extraction kit protocol. After being boiled with SDS-sample buffer, 15 μg of protein of each sample was loaded on SDS-PAGE gels and subjected to Western blotting.
Statistical analysis.
Luciferase assays were carried out using extracts from duplicate culture plates, and the results were averaged to represent a single data point for each transfection. Activity was expressed as a ratio of the firefly luciferase activity to the Renilla luciferase activity. Analysis of results demonstrated no effect of compound C on the transcriptional activity of the internal control Renilla luciferase activity. Levels of protein detected on Western blots were quantified by a PhosphorImager, subtracting background density and normalizing the results to the amount of protein loaded. t-Tests and analysis of variance were used to analyze the in vitro data. P < 0.05 was considered significant. All values shown are means ± SE. The numbers of independent experiments are indicated in the figure legends.
RESULTS
PPAR-α transcriptional activity is reduced by AMPK activators and increased by the AMPK inhibitor compound C.
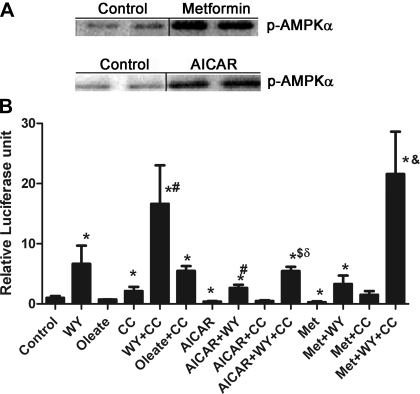
H4IIEC3 cells are a well-differentiated hepatoma cell line that expresses alcohol dehydrogenase, metabolizes ethanol, and was used to demonstrate the effect of ethanol on AMPK and PPAR-α. The expression of RXR in these cells is high, but that of the PPARs is relatively low (14, 31). Therefore, to test the effect of AMPK activity on PPAR-α activity, H4IIEC3 cells were transfected with a PPAR-α expression plasmid, a tk-Renilla luciferase plasmid to control for transfection efficiency, and a PPRE-luciferase reporter plasmid. Cells were then treated with either the AMPK inhibitor compound C or with AICAR or metformin, both well-characterized AMPK activators. Compound C is a potent reversible inhibitor of AMPK, which is competitive with ATP (Ki = 109 ± 16 nM in the absence of AMP) (44). AICAR is an adenosine analog that is taken up into the cells by the adenosine transporter and phosphorylated by adenosine kinase to 5-aminoimidazole-4-carboxamide ribonucleotide, an AMP analog (16). It may also activate Ataxia Telangiectasia mutated, which is reported to be an AMPK kinase (33). The effect of metformin on AMPK was originally proposed to occur through inhibition of mitochondrial complex I, but in the range of 0.5–2.0 mM it activates AMPK in rat hepatocytes with no detectable change in the AMP/ATP ratio (44). Thus metformin can activate AMPK through a number of mechanisms. Western blotting confirmed increased phosphorylation of AMPK in the presence of metformin or AICAR in H4IIEC3 cells (Fig. 1A), which correlated with increased AMPK activity as determined using a standard AMPK kinase assay (data not shown). Compound C does not affect Thr172 phosphorylation, so this could not be used to follow AMPK activity in the compound C-treated cells.
Fig. 1.
A: treatment of H4IIEC3 cells with AMP-activated protein kinase (AMPK) activators results in increased phosphorylation of AMPK. H4IIEC3 cells were treated with metformin or 5-aminoimidazole-4-carboxamide-1-β-d-ribofuranoside (AICAR) for 24 h before harvesting and Western blotting for phospho-AMPK. Bars between columns signify noncontiguous lanes on the same gel. B: H4IIEC3 cells were transfected with a proliferator-activated receptor (PPAR)-α expression plasmid and a peroxisome-proliferator-response element (PPRE) luciferase reporter plasmid in addition to a tk-Renilla luciferase plasmid to control for transfection efficiency. Cells were treated for 24 h as described in the text before harvesting and measuring luciferase activity. Concentrations of the treatments were as follows: 1 μM WY-14,643 (WY), 100 μM oleic acid, 40 μM compound C (CC), 500 μM AICAR, 1 mM metformin (Met). N = 4. *P < 0.05 compared with control; #P < 0.05 compared with WY; $P < 0.05 compared with WY + AICAR; &P < 0.05 compared with metformin + WY; δP < 0.05 compared with WY + CC.
Expression of the reporter plasmid was low in the absence of ligand, but treatment with 1 μM WY resulted in a sixfold increase in PPRE reporter activity (Fig. 1B). Oleate, reported to be a weak but physiological PPAR-α agonist, had no significant effect on PPAR-α activity in this experiment. The length of exposure to or the concentration of oleate may have been inadequate to induce measurable changes in PPAR-α activity. Thus WY was used in subsequent experiments to allow for detection of changes in PPAR-α activity. Compound C treatment increased basal PPAR-α activity and more than doubled the effect of WY. Compound C also increased PPRE reporter activity in cells treated with oleate, but this was not significantly different from cells treated with compound C alone. AICAR treatment inhibited basal PPAR-α activity by 60% and reversed the stimulatory effect of WY. Compound C reversed the effects of AICAR on basal PPRE reporter activity and partially restored the effect of WY on AICAR-treated cells. Metformin had similar effects except that compound C was able to completely overcome the effect of metformin and return luciferase activity to the level of WY plus compound C. These results suggest that activation of AMPK inhibits PPAR-α activity.
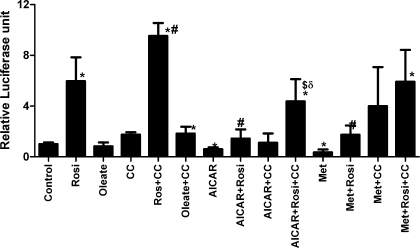
PPAR-γ transcriptional activity is also inhibited by AMPK activators and stimulated by inhibition of AMPK.
To test whether the effect of the AMPK activators and inhibitor was unique to PPAR-α, parallel experiments were performed with PPAR-γ expression plasmids. PPAR-γ is a nuclear receptor closely related to PPAR-α, which also heterodimerizes with RXR and activates transcription when exposed to its ligands. Treatment of transfected cells with the PPAR-γ agonist rosiglitazone resulted in a sixfold increase in PPRE reporter activity (Fig. 2). Oleate did not stimulate PPAR-γ activity, nor did it synergize with compound C. Inhibition of AMPK with compound C doubled the activity of PPAR-γ and potentiated the effect of rosiglitazone. AICAR and metformin inhibited basal PPAR-γ activity by 40–65% and inhibited rosiglitazone-stimulated PPAR-γ activity by 75%. Compound C partially reversed the effect of AICAR and metformin on basal- and rosiglitazone-stimulated PPAR-γ activity, restoring luciferase activity to that seen with rosiglitazone plus compound C in metformin-treated cells, but not in AICAR-treated cells.
Fig. 2.
H4IIEC3 cells were transfected with PPAR-γ expression plasmids, along with PPRE reporter plasmid and tk-Renilla luciferase plasmid. Twenty-four hours before harvest, cells were exposed to the treatments listed above. Concentrations of the compounds were as described in Fig. 1; 30 μM rosiglitazone (Rosi) was used. N = 3. *P < 0.05 compared with control; #P < 0.05 compared with rosiglitazone alone, $P = 0.05 compared with AICAR + rosiglitazone; &P = 0.05 compared with metformin + rosiglitazone, δP < 0.05 compared with CC + rosiglitazone.
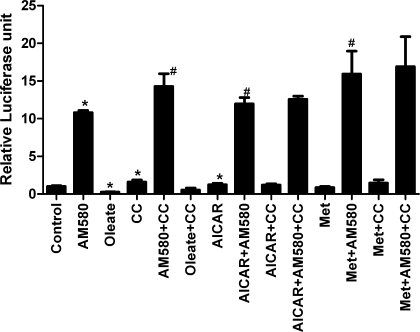
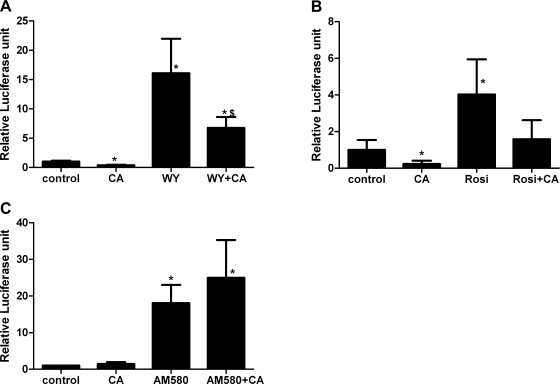
AMPK activators do not inhibit RAR/RXR transcriptional activity.
Given that both PPAR-α and PPAR-γ heterodimerize with RXR, it was possible that the actions of AMPK activators were mediated by effects on RXR, and not on the PPARs themselves. We obtained a luciferase reporter plasmid for RAR-α, which also heterodimerizes with RXR. Cells transfected with the RAR luciferase reporter plasmid and the tk-Renilla luciferase plasmid were treated with AM580, an RAR agonist. The reporter plasmid was highly responsive to this ligand (Fig. 3), demonstrating the presence of RAR in the cells. Oleate significantly inhibited RAR reporter activity by 75%, which has not been previously reported. Neither AMPK activator inhibited the reporter activity. This indicates that the drugs are not globally reducing transcription or protein synthesis and make it unlikely that they are affecting PPAR-α or -γ activity through a modification of the RXR component. However, compound C increased basal and AM580-stimulated RAR activity by about 40%.
Fig. 3.
H4IIEC3 cells were transfected with a retinoic acid receptor (RAR) luciferase reporter plasmid and tk-Renilla-luciferase plasmid to control for transfection efficiency. Treatments were for 24 h, and concentrations of the compounds were the same as described in Fig. 1; 50 nM AM580 was used. N = 4. *P < 0.05 compared with control; #P < 0.05 compared with AM580.
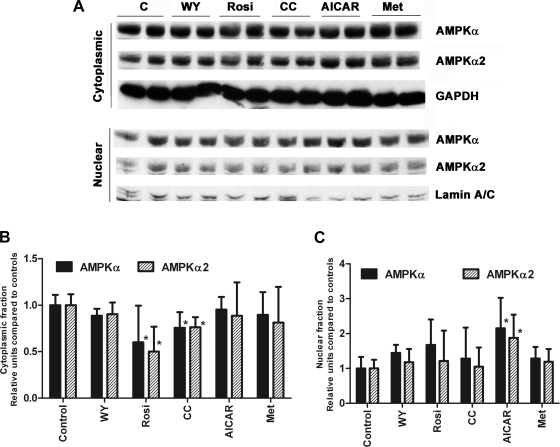
AICAR increases nuclear AMPK-α, whereas metformin has no effect.
If AMPK affects PPAR activity through a direct association, one might expect changes in its cellular localization during the manipulations of AMPK activity. This was tested by Western blotting cytoplasmic and nuclear fractions from cells treated with the AMPK activators and inhibitor for total AMPK-α subunit and -α2 subunits (Fig. 4). The α2-isoform has a propensity to localize to the nucleus and has been previously reported to exit the nucleus with AICAR treatment (3, 17). The blots are consistent with the notion that the α2-subunit is the more abundant subunit in the nuclear fraction. Rosiglitazone and compound C treatment significantly decreased cytoplasmic total AMPK-α and -α2 but did not significantly change nuclear levels. AICAR treatment had no detectable effect on cytoplasmic levels of AMPK-α or -α2 but increased the nuclear levels of AMPK-α and -α2. Metformin had no effect on cytoplasmic or nuclear levels of AMPK-α or -α2, indicating that the effect of AICAR is probably independent of its ability to activate AMPK.
Fig. 4.
A: H4IIEC3 cells were subjected to the treatments as described for 24 h before harvesting and cellular fractionation. Nuclear and cytoplasmic fractions (10 μg) were subjected to Western blotting for AMPK-α and -α2. B: cytoplasmic levels of total AMPK-α and -α2. Protein levels were normalized to GAPDH. *P < 0.05 compared with control. C: nuclear levels of total AMPK-α and α-2. Protein levels were normalized to the levels of lamin. *P < 0.05 compared with control. N = 3.
AMPK activators do not impair PPAR-α DNA binding.
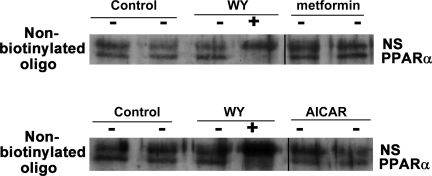
We next sought to determine whether the inhibition of PPAR-α by AMPK activators was attributable to impaired DNA binding. H4IIEC3 cells were transfected with PPAR-α and treated with the different compounds for 24 h before harvesting. PPAR-α DNA binding was determined by incubating the lysate overnight with biotinylated PPRE oligonucleotides, which were then pulled down with streptavidin beads. The bound proteins were then subjected to Western blotting for PPAR-α. PPAR-α present in the extracts bound the oligonucleotides in the presence or absence of ligand (Fig. 5). The lower band was competed with unbiotinylated oligonucleotides, demonstrating that it represents specifically bound PPAR-α. Although the amount of PPAR pulled down appears less than the control in this figure, there was no consistent reduction with four replications of this experiment. Thus neither AMPK activator affected the ability of PPAR-α to bind DNA.
Fig. 5.
H4IIEC3 cells were transfected with PPAR-α and treated with the AMPK activators for 24 h as labeled. Lysates were then incubated with biotinylated double-stranded PPRE oligonucleotides overnight, before using streptavidin beads to pull down the biotinylated oligonucleotide and any protein bound to it. After being boiled with SDS-SB, samples were subjected to Western blotting for PPAR-α. The presence of nonbiotinylated PPRE oligonucleotide prevented the pull down of the lower, immunoreactive PPAR-α band, demonstrating that the upper band was attributable to a nonspecific interaction. NS, nonspecific band. Bars between columns signify noncontiguous lanes on the same gel. There was no consistent effect of AICAR, WY, or metformin on binding of PPAR-α to its consensus binding site in cellular extracts from 4 independent experiments.
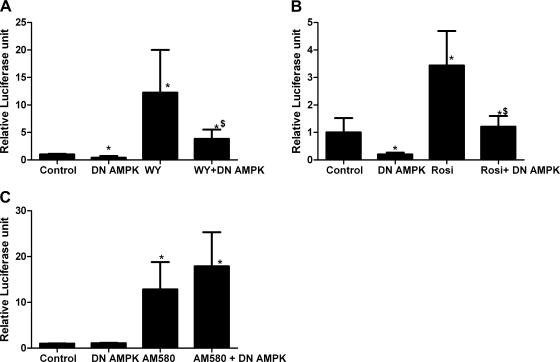
Activity of PPAR-α and -γ is inhibited by expression of a constitutively active AMPK-α1 and a dominant negative mutant AMPK-α1 subunit.
The effect of AMPK activators and compound C suggested that the kinase activity of AMPK was correlated with its ability to modify PPAR transcriptional activity. To further explore this, we transfected H4IIEC3 cells with a constitutively active AMPK-α1 (truncated at residue 312) (AMPK-α1312) expression plasmid. Truncation of the constitutively active AMPK-α mutant at residue 312 allows for activity of the kinase independent of its association with the β- and γ-subunit, and mutation of the threonine at residue 172 to an aspartic acid mimics phosphorylation, further increasing its activity (8, 40). Consistent with the effect of metformin and AICAR, AMPK-α1312 inhibited baseline and WY-stimulated PPAR-α activity by 60% (Fig. 6A). Similarly, AMPK-α1312 inhibited basal PPAR-γ activity by 75% (Fig. 6B). Rosiglitazone-stimulated PPAR-γ activity was also inhibited by AMPK-α1312 by 60%, approaching significance (P = 0.06) compared with rosiglitazone stimulation, reaching a level of activity not significantly different from the control. AMPK-α1312 had no effect on basal or AM580-stimulated RAR reporter activity (Fig. 6C).
Fig. 6.
H4IIEC3 cells were transfected with the reporter plasmids and PPAR-α (A) or -γ (B), and the AMPK-α1312 expression plasmid as noted by CA (constitutively active). The cells were treated with WY, rosiglitazone, and AM580 at the same concentrations noted in Figs. 1–3 for 24 h. A: PPAR-α, *P < 0.05 compared with control; $P < 0.05 compared with WY-stimulated cells. B: PPAR-γ, *P < 0.05 compared with control. C: RAR. *P < 0.05 compared with control.
We therefore predicted that a dominant negative AMPK-α subunit would activate PPAR transcriptional activity similar to the effect of compound C. Dominant negative AMPK-α1 (α1-DN AMPK) contains a mutation of aspartate 157 to alanine in the ATP binding site of the kinase domain. This blocks kinase activity while preserving the ability of the subunit to bind the β- and γ-subunits. This displaces the wild-type α-subunit (which is degraded) and reduces cellular AMPK activity (40).
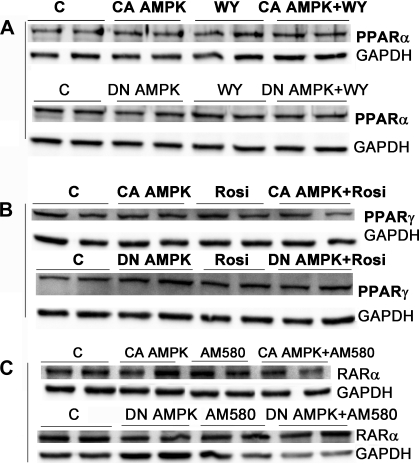
Surprisingly, transfection of α1-DN AMPK inhibited basal and WY-stimulated PPAR-α reporter activity by 60% and 70%, respectively (Fig. 7A). It also inhibited basal and rosiglitazone-stimulated PPAR-γ activity by 80% and 100%, respectively (Fig. 7B). α1-DN AMPK expression had no effect on RAR reporter activity (Fig. 7C). It was possible that the inhibitory effect of transfecting the cells with the expression plasmids for the AMPK variants reduced the expression of the nuclear receptors. Therefore we examined the levels of PPAR-α and -γ, and RAR-α in transfected cells. Western blotting confirmed that transfection of the AMPK-α1312 and α1-DN AMPK constructs did not reduce expression of the PPAR-α, PPAR-γ, or RAR-α proteins in the absence or presence of receptor ligands (Fig. 8).
Fig. 7.
H4IIEC3 cells were transfected with the reporter plasmid and PPAR-α or -γ as labeled. Dominant negative (α1-DN) AMPK was transfected at the same time as the reporter and expression plasmids. Concentrations of WY, rosiglitazone, and AM580 were as described in Figs. 1–3, and treatments were for 24 h. A: PPAR-α, *P < 0.05 compared with control; $P < 0.05 compared with WY-stimulated cells. B: PPAR-γ, *P < 0.05 compared with control; $P < 0.05 compared with rosiglitazone-stimulated cells. C: RAR, *P < 0.05 compared with control.
Fig. 8.
H4IIEC3 cells were transfected with the expression plasmids for PPAR-α or -γ (A and B) and with the constitutively active (CA AMPK) or dominant negative (DN AMPK) expression plasmids. Where noted, the cells were treated with WY, rosiglitazone, or AM580 as described in Figs. 1–3. The cells were harvested after 24 h and the cellular extracts blotted for PPAR-α, -γ, or RAR-α. GAPDH was used as the loading control. There was no effect of the transfection of the AMPK variants or treatment with receptor ligands on the expression of the nuclear receptors. C, control.
DISCUSSION
We have demonstrated that the AMPK activators AICAR and metformin decreased transcriptional activity of PPAR-α/RXR and PPAR-γ/RXR but not of RAR-α/RXR in the rat hepatoma cell line H4IIEC3. This suggests that the effect of the activators did not involve alteration in the properties of RXR or of the general transcriptional machinery. There was no effect of the activators on the ability of PPAR-α/RXR in the cellular extracts to bind DNA, and there was no correlation between the effects of the compounds tested and the nuclear-cytosolic shuttling of AMPK-α subunits. The effect of the activators was partially reversed by the presence of the AMPK inhibitor compound C. However, transfection of either constitutively active or dominant negative AMPK-α1 expression plasmids resulted in inhibition of the PPARs but not RAR-α/RXR. These data appear to exclude several explanations for the interaction between AMPK and PPAR-α or -γ. Neither the enzymatic activity of AMPK nor its ability to bind ATP at the catalytic site were required for the inhibitory effect, as evidenced by the ability of the α1-DN subunit to inhibit the PPARs. Association of the α-subunit with the β- and γ-subunits was not required, as shown by the ability of the AMPK-α1312 mutant to inhibit PPAR-α. We cannot state whether an acidic group (phosphoryl-threonine or an aspartoyl residue in the AMPK-α1312 mutant) is required because all of the AMPK subunits we tested could be phosphorylated at that site (compound C did not prevent phosphorylation, not shown) or contained a T172D mutation. These results suggest that a protein-protein interaction between the NH2-terminal domain of AMPK-α and PPAR mediates the effects observed.
Recent structural studies of human AMPK have demonstrated an important role for the autoinhibitory domain (AID) located in the COOH-terminal region of the α-subunit in the regulation of AMPK activity; when it interacts with the NH2-terminal kinase domain, it markedly reduces catalytic activity and increases the Km for substrates (6). This interaction is independent of phosphorylation of Thr172. AMP was suggested to activate the enzyme by triggering a conformational change that alters the interaction of the AID with the kinase domain. We would suggest that an active conformation of the AMPK-α subunit is required for the inhibition of PPAR-α. This is consistent with the ability of metformin and AICAR or transfection of the AMPK-α1312 subunit to inhibit PPAR. This model requires that the presence of compound C prevents the subunit from assuming the active conformation. The mechanism of inhibition of AMPK by compound C has not been rigorously studied. It inhibits AMPK in partially purified extracts competitively with ATP. This suggests that it competes for ATP binding at the kinase domain ATP binding site, but it might also bind the ATP/AMP binding site of the γ-subunit. Perhaps the presence of compound C bound to the active site prevents the active conformation, or an interaction with the γ-subunit maintains the AID in contact with the kinase domain. In fact, the results with compound C suggest that, when AMPK is not in the active conformation, it can activate PPAR. The stimulatory effect of compound C seen with the RAR reporter plasmid is likely an off target, AMPK-independent effect, as α1-DN AMPK had no significant effect on the RAR reporter plasmid. To date, only inhibitory effects of compound C on general transcriptional processes have been reported (10), which would not explain the increase in activity of the reporters we observed.
How could these results be reconciled with the results reported by Bronner et al. (3)? Their data showed that transfection of expression plasmids for a variety of AMPK subunits increased PPAR-α transcriptional activity. This was associated with a physical interaction of AMPK with the ligand-binding domain of PPAR-α that was mediated through regions of the COOH-terminal regulatory domain with PPAR-α but also requiring a contribution of the NH2-terminal catalytic domain for the activation. The α-subunits tested and shown to activate PPAR-α included native α1- and α2-subunits, a kinase-deficient subunit (Thr172Ala), and two kinaseless subunits (Asp157Ala and Asp139Ala). We suggest that mere transfection of expression plasmids for the subunits would not necessarily result in the activation of AMPK, and thus each of the constructs tested may have been in an inactive conformation, except when AICAR was present and the endogenous and transfected AMPK were activated. We propose that AMPK in the active conformation inhibits PPARs, whereas AMPK in the inactive conformation activates them. The ability of the AMPK-α1312 mutant, which lacks the COOH-terminal regulatory domain, to inhibit PPAR activity also suggests that, in the active conformation, the interaction domain reported in the NH2 terminus of the subunit is sufficient to mediate the inhibition, whereas, in the inactive conformation, additional sites in the regulatory domain are required. Alternatively, there may be interactions with other domains of the PPAR-α outside the ligand-binding domain tested.
An inconsistency with this hypothesis was the ability of the α1-DN (D157A) mutant to activate PPAR-α in Bronner's hands and to inactivate it in our experiments. The α1-DN AMPK construct has impaired MgATP binding to the α-subunit. Thus it competes with kinase-active AMPK for binding to the β- and γ-subunits by the two subunits, regardless of kinase activity. However, we have no way of knowing whether the transfected DN-α1 mutant was in an active conformation because its catalytic activity is disabled. A simple explanation for the difference is that different cell lines were used by Bronner et al. (3) (293, Hela and COS-7 cells) and our studies (H4IIEC3). Furthermore, AMPK activity is very sensitive to cellular stress, and thus it is plausible that subtle differences in experimental conditions account for the differences in results. Alternatives for which we have no direct evidence include effects of phosphorylation of additional sites in the α- and β-subunits or additional interactions of AMPK with other cellular components that alter the active conformation of the enzyme, such as glycogen interaction with the β-subunit. These, too, might differ between the cell lines tested by Bronner and our data and modify the effects of interaction of AMPK with PPARs.
This model provides an attractive physiological signaling mechanism. AMPK activity is altered by a diverse set of cellular stresses, not all of which demand an increase in the long-term ability of the cells to oxidize fat (e.g., the induction of PPAR-α), and, conversely, AMPK initially must signal increased energy generation, without committing ATP to transcription and translation of PPAR-induced genes. When AMPK is in the activated conformation, the interaction of AMPK with PPAR-α or -γ results in inhibition of the transcriptional activity of the PPARs; this would delay the expression of the PPAR-regulated proteins until the cell had restored its energy stores. Conversely, when the energy stores are adequate, AMPK will be in the inactive conformation, and its interaction with PPARs would be activating. This would serve to amplify the effectiveness of signaling via molecules like AMP, which reflect energy stores. In a sense, AMPK could be seen as a coactivator of PPAR that is switched off by cellular stresses.
We initially became interested in the interactions between AMPK and PPAR-α because both are inhibited in the livers of ethanol-fed animals despite increased levels of free fatty acids in the liver. We conclude that the inhibitory effect of ethanol on AMPK in H4IIEC3 cells does not account for the inhibition of PPAR-α because the presence of inactive AMPK in ethanol-treated cells would, on the basis of our data, be expected to activate PPAR-α.
In summary, our studies demonstrated an inhibitory effect of AMPK on PPAR-α and -γ when AMPK was in a putative activated conformation but was independent of kinase activity, effects of AMPK on RXR, or on DNA-binding activity of the PPAR. Clearly, the interactions between PPAR-α and AMPK are more complex than previously thought. Further studies are required to better understand the nature of the activated conformation of AMPK and the physiological role of and cellular context within which AMPK modulates PPAR-α and -γ activity.
GRANTS
This research was supported by P60AA 07611 (D. Crabb), R01 AA15070 (D. Crabb), K08 AA016570 (S. Liangpunsakul), and F32 AA017800 (M. Sozio).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
REFERENCES
- 1.Barger PM, Brandt JM, Leone TC, Weinheimer CJ, Kelly DP. Deactivation of peroxisome proliferator-activated receptor-alpha during cardiac hypertrophic growth. J Clin Invest 105: 1723–1730, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barger PM, Browning AC, Garner AN, Kelly DP. p38 mitogen-activated protein kinase activates peroxisome proliferator-activated receptor alpha: a potential role in the cardiac metabolic stress response. J Biol Chem 276: 44495–44501, 2001 [DOI] [PubMed] [Google Scholar]
- 3.Bronner M, Hertz R, Bar-Tana J. Kinase-independent transcriptional co-activation of peroxisome proliferator-activated receptor alpha by AMP-activated protein kinase. Biochem J 384: 295–305, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns KA, Vanden Heuvel JP. Modulation of PPAR activity via phosphorylation. Biochim Biophys Acta 1771: 952–960, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carey AL, Steinberg GR, Macaulay SL, Thomas WG, Holmes AG, Ramm G, Prelovsek O, Hohnen-Behrens C, Watt MJ, James DE, Kemp BE, Pedersen BK, Febbraio MA. Interleukin-6 increases insulin-stimulated glucose disposal in humans and glucose uptake and fatty acid oxidation in vitro via AMP-activated protein kinase. Diabetes 55: 2688–2697, 2006 [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Jiao ZH, Zheng LS, Zhang YY, Xie ST, Wang ZX, Wu JW. Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature 459: 1146–1149, 2009 [DOI] [PubMed] [Google Scholar]
- 7.Corton JM, Gillespie JG, Hardie DG. Role of the AMP-activated protein kinase in the cellular stress response. Curr Biol 4: 315–324, 1994 [DOI] [PubMed] [Google Scholar]
- 8.Crute BE, Seefeld K, Gamble J, Kemp BE, Witters LA. Functional domains of the alpha1 catalytic subunit of the AMP-activated protein kinase. J Biol Chem 273: 35347–35354, 1998 [DOI] [PubMed] [Google Scholar]
- 9.Dobrzyn P, Dobrzyn A, Miyazaki M, Cohen P, Asilmaz E, Hardie DG, Friedman JM, Ntambi JM. Stearoyl-CoA desaturase 1 deficiency increases fatty acid oxidation by activating AMP-activated protein kinase in liver. Proc Natl Acad Sci USA 101: 6409–6414, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emerling BM, Viollet B, Tormos KV, Chandel NS. Compound C inhibits hypoxic activation of HIF-1 independent of AMPK. FEBS Lett 581: 5727–5731, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett L, Galli A, Crabb D. The role of hepatic peroxisome proliferator-activated receptors (PPARs) in health and disease. Liver 20: 191–199, 2000 [DOI] [PubMed] [Google Scholar]
- 12.Fischer M, You M, Matsumoto M, Crabb DW. Peroxisome proliferator-activated receptor alpha (PPARalpha) agonist treatment reverses PPARalpha dysfunction and abnormalities in hepatic lipid metabolism in ethanol-fed mice. J Biol Chem 278: 27997–28004, 2003 [DOI] [PubMed] [Google Scholar]
- 13.Fujii N, Hayashi T, Hirshman MF, Smith JT, Habinowski SA, Kaijser L, Mu J, Ljungqvist O, Birnbaum MJ, Witters LA, Thorell A, Goodyear LJ. Exercise induces isoform-specific increase in 5′AMP-activated protein kinase activity in human skeletal muscle. Biochem Biophys Res Commun 273: 1150–1155, 2000 [DOI] [PubMed] [Google Scholar]
- 14.Galli A, Stewart M, Dorris R, Crabb D. High-level expression of RXRalpha and the presence of endogenous ligands contribute to expression of a peroxisome proliferator-activated receptor-responsive gene in hepatoma cells. Arch Biochem Biophys 354: 288–294, 1998 [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Villafranca J, Guillen A, Castro J. Ethanol consumption impairs regulation of fatty acid metabolism by decreasing the activity of AMP-activated protein kinase in rat liver. Biochimie 90: 460–466, 2008 [DOI] [PubMed] [Google Scholar]
- 16.Giri S, Nath N, Smith B, Viollet B, Singh AK, Singh I. 5-aminoimidazole-4-carboxamide-1-beta-4-ribofuranoside inhibits proinflammatory response in glial cells: a possible role of AMP-activated protein kinase. J Neurosci 24: 479–487, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardie DG. Roles of the AMP-activated/SNF1 protein kinase family in the response to cellular stress. Biochem Soc Symp 64: 13–27, 1999 [PubMed] [Google Scholar]
- 18.Hardie DG, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem 67: 821–855, 1998 [DOI] [PubMed] [Google Scholar]
- 19.Hawley SA, Davison M, Woods A, Davies SP, Beri RK, Carling D, Hardie DG. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J Biol Chem 271: 27879–27887, 1996 [DOI] [PubMed] [Google Scholar]
- 20.Horman S, Browne G, Krause U, Patel J, Vertommen D, Bertrand L, Lavoinne A, Hue L, Proud C, Rider M. Activation of AMP-activated protein kinase leads to the phosphorylation of elongation factor 2 and an inhibition of protein synthesis. Curr Biol 12: 1419–1423, 2002 [DOI] [PubMed] [Google Scholar]
- 21.Hua S, Kittler R, White KP. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell 137: 1259–1271, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kliewer SA, Forman BM, Blumberg B, Ong ES, Borgmeyer U, Mangelsdorf DJ, Umesono K, Evans RM. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci USA 91: 7355–7359, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature 358: 771–774, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kudo N, Barr AJ, Barr RL, Desai S, Lopaschuk GD. High rates of fatty acid oxidation during reperfusion of ischemic hearts are associated with a decrease in malonyl-CoA levels due to an increase in 5′-AMP-activated protein kinase inhibition of acetyl-CoA carboxylase. J Biol Chem 270: 17513–17520, 1995 [DOI] [PubMed] [Google Scholar]
- 25.LeBrasseur NK, Kelly M, Tsao TS, Farmer SR, Saha AK, Ruderman NB, Tomas E. Thiazolidinediones can rapidly activate AMP-activated protein kinase in mammalian tissues. Am J Physiol Endocrinol Metab 291: E175–E181, 2006 [DOI] [PubMed] [Google Scholar]
- 26.Lee WJ, Kim M, Park HS, Kim HS, Jeon MJ, Oh KS, Koh EH, Won JC, Kim MS, Oh GT, Yoon M, Lee KU, Park JY. AMPK activation increases fatty acid oxidation in skeletal muscle by activating PPARalpha and PGC-1. Biochem Biophys Res Commun 340: 291–295, 2006 [DOI] [PubMed] [Google Scholar]
- 27.Leff T. AMP-activated protein kinase regulates gene expression by direct phosphorylation of nuclear proteins. Biochem Soc Trans 31: 224–227, 2003 [DOI] [PubMed] [Google Scholar]
- 28.Mao X, Bravo IG, Cheng H, Alonso A. Multiple independent kinase cascades are targeted by hyperosmotic stress but only one activates stress kinase p38. Exp Cell Res 292: 304–311, 2004 [DOI] [PubMed] [Google Scholar]
- 29.Marsin AS, Bertrand L, Rider MH, Deprez J, Beauloye C, Vincent MF, Van den Berghe G, Carling D, Hue L. Phosphorylation and activation of heart PFK-2 by AMPK has a role in the stimulation of glycolysis during ischaemia. Curr Biol 10: 1247–1255, 2000 [DOI] [PubMed] [Google Scholar]
- 30.Muoio DM, Seefeld K, Witters LA, Coleman RA. AMP-activated kinase reciprocally regulates triacylglycerol synthesis and fatty acid oxidation in liver and muscle: evidence that sn-glycerol-3-phosphate acyltransferase is a novel target. Biochem J 338: 783–791, 1999 [PMC free article] [PubMed] [Google Scholar]
- 31.Pinaire J, Chou WY, Morton M, You M, Zeng Y, Cho WK, Galli A, Everett L, Breen H, Dumaual N, Smith JR, Crabb D. Identification of a retinoid receptor response element in the human aldehyde dehydrogenase-2 promoter. Alcohol Clin Exp Res 27: 1860–1866, 2003 [DOI] [PubMed] [Google Scholar]
- 32.Schmidt MC, McCartney RR. beta-subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J 19: 4936–4943, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun Y, Connors KE, Yang DQ. AICAR induces phosphorylation of AMPK in an ATM-dependent, LKB1-independent manner. Mol Cell Biochem 306: 239–245, 2007 [DOI] [PubMed] [Google Scholar]
- 34.Suzuki A, Kusakai G, Kishimoto A, Shimojo Y, Ogura T, Lavin MF, Esumi H. IGF-1 phosphorylates AMPK-alpha subunit in ATM-dependent and LKB1-independent manner. Biochem Biophys Res Commun 324: 986–992, 2004 [DOI] [PubMed] [Google Scholar]
- 35.Suzuki A, Kusakai G, Shimojo Y, Chen J, Ogura T, Kobayashi M, Esumi H. Involvement of transforming growth factor-beta 1 signaling in hypoxia-induced tolerance to glucose starvation. J Biol Chem 280: 31557–31563, 2005 [DOI] [PubMed] [Google Scholar]
- 36.Takeda Y, Arii S, Kaido T, Niwano M, Moriga T, Mori A, Hanaki K, Gorrin-Rivas MJ, Ishii T, Sato M, Imamura M. Morphologic alteration of hepatocytes and sinusoidal endothelial cells in rat fatty liver during cold preservation and the protective effect of hepatocyte growth factor. Transplantation 67: 820–828, 1999 [DOI] [PubMed] [Google Scholar]
- 37.Tomita K, Tamiya G, Ando S, Kitamura N, Koizumi H, Kato S, Horie Y, Kaneko T, Azuma T, Nagata H, Ishii H, Hibi T. AICAR, an AMPK activator, has protective effects on alcohol-induced fatty liver in rats. Alcohol Clin Exp Res 29: 240S-–245S., 2005 [DOI] [PubMed] [Google Scholar]
- 38.Towler MC, Hardie DG. AMP-activated protein kinase in metabolic control and insulin signaling. Circ Res 100: 328–341, 2007 [DOI] [PubMed] [Google Scholar]
- 39.Winder WW, Hardie DG. Inactivation of acetyl-CoA carboxylase and activation of AMP-activated protein kinase in muscle during exercise. Am J Physiol Endocrinol Metab 270: E299–E304, 1996 [DOI] [PubMed] [Google Scholar]
- 40.Woods A, Azzout-Marniche D, Foretz M, Stein SC, Lemarchand P, Ferre P, Foufelle F, Carling D. Characterization of the role of AMP-activated protein kinase in the regulation of glucose-activated gene expression using constitutively active and dominant negative forms of the kinase. Mol Cell Biol 20: 6704–6711, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamauchi T, Kamon J, Minokoshi Y, Ito Y, Waki H, Uchida S, Yamashita S, Noda M, Kita S, Ueki K, Eto K, Akanuma Y, Froguel P, Foufelle F, Ferre P, Carling D, Kimura S, Nagai R, Kahn BB, Kadowaki T. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat Med 8: 1288–1295, 2002 [DOI] [PubMed] [Google Scholar]
- 42.Yoon MJ, Lee GY, Chung JJ, Ahn YH, Hong SH, Kim JB. Adiponectin increases fatty acid oxidation in skeletal muscle cells by sequential activation of AMP-activated protein kinase, p38 mitogen-activated protein kinase, and peroxisome proliferator-activated receptor alpha. Diabetes 55: 2562–2570, 2006 [DOI] [PubMed] [Google Scholar]
- 43.You M, Matsumoto M, Pacold CM, Cho WK, Crabb DW. The role of AMP-activated protein kinase in the action of ethanol in the liver. Gastroenterology 127: 1798–1808, 2004 [DOI] [PubMed] [Google Scholar]
- 44.Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, Musi N, Hirshman MF, Goodyear LJ, Moller DE. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest 108: 1167–1174, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]