Abstract
Objective To evaluate the accumulated information from genetic association studies investigating the impact of variants of the cytochrome P450 (CYP) 2C19 genotype on the clinical efficacy of clopidogrel.
Design Systematic review and meta-analysis with a structured search algorithm and prespecified eligibility criteria for retrieval of relevant studies; dominant genetic model assumptions and quantitative methods for calculating summary effect estimates from study level odds ratios; systematic assessment of bias within and between studies; and grading of the cumulative evidence by consensus criteria.
Data sources Medline, Embase, the Cochrane Library, online databases, contents pages and bibliographies of general medical, cardiovascular, pharmacological, and genetic journals.
Eligibility criteria for selecting studies Original full length reports assessing the cumulative incidence of major adverse cardiovascular events or stent thrombosis over a follow-up period of at least a month in association with carrier status for the loss of function or gain of function CYP2C19 allele in adult patients with coronary artery disease and a clinical presentation of acute coronary syndrome or stable angina pectoris who were taking clopidogrel.
Results 15 studies met the inclusion criteria. The random effects summary odds ratio for stent thrombosis in carriers of at least one CYP2C19 loss of function allele versus non-carriers combining nine studies was 1.77 (95% confidence interval 1.31 to 2.40; P<0.001). This nominally significant odds ratio was subject to considerable bias across the studies (small study effect bias and replication diversity). The adjustment for these quality modifiers tended to abolish the association. The corresponding random effects summary odds ratio of major adverse cardiovascular events for 12 studies combined was 1.11 (0.89 to 1.39; P=0.36). The random effects summary odds ratio of stent thrombosis in carriers versus non-carriers of at least one CYP2C19*17 gain of function allele for three studies combined was 0.99 (0.60 to 1.62; P=0.96), and the corresponding odds ratio of major adverse cardiovascular events in five studies was 0.93 (0.75 to 1.14; P=0.48). The overall quality of epidemiological evidence was graded as low, which excludes reliable clinical assessments.
Conclusions Accumulated information from genetic association studies does not indicate a substantial or consistent influence of CYP2C19 gene polymorphisms on the clinical efficacy of clopidogrel. The current evidence does not support the use of individualised antiplatelet regimens guided by CYP2C19 genotype.
Introduction
Along with aspirin, clopidogrel represents the mainstay of antiplatelet treatment for preventing thromboembolic events in patients with ischaemic cardiovascular diseases.1 It is the second biggest selling drug, with global sales of more than $9bn (£5.6bn, €6.3bn) a year.2 A major drawback to its use is its unpredictable clinical efficacy, with insufficient platelet inhibition in about 20% of patients,3 resulting in a higher incidence of atherothrombotic events.4 5 6
The primary source of variability in responsiveness to clopidogrel might lie in its pharmacokinetics. Clopidogrel is a prodrug that is transformed into its active metabolite through metabolic pathways involving cytochrome (CYP) P450 isoenzymes. CYP2C19 might be the crucial isoenzyme for the metabolic bioactivation of clopidogrel.7 Accordingly, polymorphisms of the CYP2C19 gene associated with loss of function have been associated with decreased antiplatelet responsiveness8 and are supposed to predict a large proportion of the variability in clinical response to clopidogrel.9 The quantitative relations between the presence of loss of function variants (specifically the most common *2 variant) and the risk of ischaemic cardiovascular events associated with use of clopidogrel have been summarised in five systematic reviews abstracting data from genetic association studies published up to 2009.10 11 12 13 14 The reported summary effect estimates consistently showed that carriers of a loss of function allele had about 1.5 to two times the risk of occurrence of a major adverse cardiovascular event or cardiovascular death compared with non-carriers. Particularly strong associations were found with the risk of stent thrombosis in patients who had undergone percutaneous coronary intervention. Patients who had at least one reduced function allele of CYP2C19 were about three or four times more likely to experience a stent thrombosis than non-carriers.10 11 12 13 14 Based on this evidence, CYP2C19 seems to be a major determinant of efficacy of clopidogrel. Therefore, determining a patient’s CYP2C19 genotype could be a straightforward diagnostic approach for optimising antiplatelet treatment and improving clinical outcome.9
In March 2010 the US Food and Drug Administration (FDA) issued a boxed warning on the clopidogrel label emphasising that individuals with a reduced function CYP2C19 genotype are at an increased risk of adverse cardiovascular outcomes and advocating strategies for adjustment of clopidogrel dose or the use of alternative antiplatelet agents in these high risk individuals.15 In accomplishing the strategy of genotype guided clopidogrel treatment, the FDA subsequently approved a validated genotyping assay for detecting variant genes of CYP2C19 (AutoGenomics CYP2C19 Assay, 510(k) clearance received October 2010).
Complementary to the associations between reduced function CYP2C19 alleles and increased cardiovascular risk, the common CYP2C19*17 gain of function allele has been associated with a better antiplatelet response to clopidogrel.16 17 Several studies have examined the possible linkage of CYP2C19*17 with clinical efficacy outcomes. Results might be expected to mirror the interaction with loss of function alleles, with lower risks of events in carriers of the *17 allele than in non-carriers.
Large genetic association studies published in 2010, however, failed to replicate the strong associations of the reduced function alleles with cardiovascular outcomes.18 19 Moreover, the results of a detailed biochemical investigation question a direct mechanistic involvement of CYP2C19 in the bioactivation of clopidogrel.20
We examined whether the new findings affect the strength of the association between CYP2C19 genotype and clinical efficacy of clopidogrel and whether the appraisal of the interaction between CYP2C19 and clopidogrel needs revision. We carried out a systematic review and meta-analysis of the accumulated information of association studies between loss of function or gain of function gene variants with clinical efficacy outcomes. We systematically assessed the quality of the individual studies and the risk of biases between the studies according to general epidemiological and specific genetic items. We used consensus criteria for rating the overall evidence of the meta-analyses to quantify the confidence of the effect estimates for guiding clinical decisions.
Methods
Our meta-analysis adhered to the MOOSE and PRISMA statements for reporting on systematic reviews and the STREGA recommendations for reporting of genetic association studies. We conducted the meta-analysis in accordance with the general guidelines of the Cochrane Handbook for Systematic Reviews of Interventions, version 5.0.2, and the specific recommendations of genetic meta-analysis of the HuGE Review Handbook, version 1.0.
Information sources and search strategy
We searched Medline (from 1966 to 31 December 2010), Embase (from 1974 to 31 December 2010), and the Cochrane Library (from 1980 to 31 December 2010) without language restrictions. The search algorithm combined the categories for “drug”, “gene”, and “outcome” by the Boolean operator “AND”. The search terms (medical subject headings and text words) in each category were combined with the operator “OR”. The algorithm was trimmed for maximum sensitivity by sequentially adding search items in each category until the total number of hits did not increase further. At each step we determined the explanatory power of the algorithm by testing that the removal of any item resulted in a lower number of total hits. We determined the specificity of the algorithm by testing that the combination of the three search categories yielded fewer hits than any combination of two categories. The following search strategy was applied: (clopidogrel OR plavix OR iscover OR thienop* OR P2Y12) AND (associat* OR cytochrome OR cyp OR polymorph* OR genetic* OR metabolis* OR enzyme) AND (cardiov* OR vascular OR coronar* OR stent* OR thrombos* OR myocar* OR heart OR infarct* OR death OR stroke OR ischem*).
In addition, we searched the online databases of general medical, cardiovascular, pharmacological, and genetic journals as indexed by ISI Web of Science to identify advance online publications. We hand searched the contents pages of the 2005-10 issues of these journals and the bibliographies of relevant articles to retrieve further potential publications.
We fixed 31 December 2010 as the cut-off date for inclusion of new studies, avoiding subjectivities in the choice of the time point when new information is reviewed and allowing calendar year based assessments.
Study selection and eligibility criteria
We included original peer reviewed reports of observational studies and clinical trials if published in full text or if we had full access to all original data and protocols. We excluded studies that were published only as abstracts or conference reports. We considered reports that evaluated the association of reduced function and increased function genetic variants of CYP2C19 with the occurrence of clinical outcomes in patients with established coronary artery disease who were treated with clopidogrel. Two researchers (DT and TB) independently retrieved studies. The strength of agreement was measured by Cohen’s κ coefficient with approximate standard errors,21 with κ=0.41-0.60 indicating moderate agreement, 0.61-0.80 good agreement, and ≥0.81 representing very good agreement.22 Disagreements were resolved by consensus.
Clinical eligibility criteria and outcome definitions
Eligible studies were conducted in unrelated men and women (aged ≥18) of any ethnicity with a clinical presentation of stable angina pectoris or acute coronary syndrome who were scheduled for administration of a loading dose of at least 300 mg clopidogrel and subsequent maintenance treatment with 75-150 mg clopidogrel a day for at least three months. For eligibility, studies must have reported follow-up data for at least 30 days after entry (inclusion or randomisation) of participants. We included studies if they provided absolute numbers of the cumulative incidence (first occurrence during follow-up) of the clinical efficacy end points: major adverse cardiovascular events or fatal or non-fatal stent thrombosis.
The primary definition of major adverse cardiovascular events was the composite of death from cardiovascular causes, non-fatal myocardial infarction, and non-fatal stroke or the composite of death from any cause, non-fatal myocardial infarction, and non-fatal stroke as evaluated by universal clinical guidelines. Other eligible definitions were death from cardiovascular causes and myocardial infarction; death from any cause and myocardial infarction; death from cardiovascular causes; and fatal and non-fatal myocardial infarction. We required that the components subsumed under the definition of major adverse cardiovascular events were sensitive and unbiased measurable events of the same underlying disease process. We excluded studies reporting only all cause mortality because of the high likelihood of bias by events without an underlying cardiovascular cause. Studies reporting only composite end points, including the clinician driven proxy outcomes of revascularisation or admission to hospital, were excluded because of uncertainties about precision, reproducibility, correlation with clinical end points, the use of non-standardised definitions, and a high risk of reporting bias.23 24
Studies of patients who underwent percutaneous coronary intervention and reporting definite stent thrombosis events were eligible if the stent thrombosis was evaluated according to the definition from the Academic Research Consortium.25 Probable stent thrombosis was considered only if definite and probable stent thrombosis were reported as a composite outcome. Possible stent thrombosis was not considered.
Genetic eligibility criteria
For unambiguous determination, polymorphisms of CYP2C19 needed to be designated by their NCBI dbSNP identifiers (“rs numbers”), their nucleotide exchange, or their common harmonised star allele nomenclature. We considered reports on the loss of function (reduced function) variants CYP2C19*2 (rs4244285), CYP2C19*3 (rs4986893), CYP2C19*4 (rs28399504), CYP2C19*5 (rs56337013), CYP2C19*6 (rs72552267), CYP2C19*7 (rs72558186), or CYP2C19*8 (rs41291556), and the gain of function variant CYP2C19*17 (rs12248560). The variant gene carrier status was required to be given as the distribution of genotypes among patients with and without the outcome event or as the number of individuals carrying at least one loss of function or gain of function allele. Studies reporting associations with a loss of function variant were eligible if they had genotyped at least the CYP2C19*2 allele because it accounts for more than 95% of the loss of function allele carrier status in white and black African populations and for more than 75% in Asian populations.26
Genetic model assumptions
Pharmacokinetic studies using different CYP2C19 substrates indicate an additive or dominant mode of inheritance of the loss of function metaboliser trait, showing either a per allele decrease in enzymatic activity27 or a similar drop of activity in carriers of one or two loss of function variants compared with non-carriers.28 29 Pharmacokinetic studies assessing the metabolic activity of CYP2C19 in relation to the gain of function CYP2C19*17 variant likewise indicate either additive inheritance with a per allele increase in activity between *1/*1, *1/*17, and *17/*17 carriers30 or dominant inheritance with a similar gain of the enzymatic activities in carriers of one or two *17 alleles compared with wild type *1/*1 carriers.31
From the available mechanistic evidence we inferred that the CYP2C19 metaboliser phenotype of clopidogrel is determined by additive or dominant genetic models of inheritance and that these models also apply to the associations of CYP2C19 with clinical events. We chose dominant genetic models to quantify the effect size in each study by comparing the genotype contrasts of carriers with one or two variant alleles with non-carriers of the variant alleles because dominant or additive models have a similar statistical power,32 genotype contrasts are more directly relevant at the individual and the clinical level compared with allele contrasts obtained from additive models, and allele contrasts could not be extracted from all studies.
Interaction between loss of function and gain of function polymorphisms
Except for one study,19 all association studies modelled loss of function and gain of function polymorphisms independently of each other, implying complete equilibrium of linkage and absence of functional interaction. An examination of the polymorphic loci of the CYP2C19 gene in white populations (HapMap-CEU database, Haploview, version 4.2 software, Broad Institute, Harvard, MA, USA) showed a low pairwise correlation coefficient (r2=0.047) between the most common loss of function polymorphism (*2) and the gain of function polymorphism *17. This excludes a substantial interaction between the single nucleotide polymorphisms at the haplotype level but entails the risk of interaction at the phenotype level. In single locus assessments of the CYP2C19*2 variant, about a quarter of all individuals classified as *2 allele carriers and about half of those classified as non-carriers were harbouring at least one *17 allele.18 In turn, in single locus analyses of the CYP2C19*17 variant, about 20% of people classified as carriers of the *17 allele and about 35% of those classified as non-carriers were harbouring at least one *2 allele.18 Pharmacokinetic analyses31 33 and pharmacodynamic platelet response studies17 suggest that the metaboliser phenotypes of mixed carriers of *2 and *17 alleles are comparable with individuals who are homozygous carriers of the wild type (*1) alleles at both loci. Therefore, the independently analysed associations of loss of function and gain of function single nucleotide polymorphisms with clinical outcomes are probably systematically distorted by the counteracting metaboliser trait. This does not necessarily invalidate the associations or increase the heterogeneity between studies, if it is assumed that a proportional bias applies to all studies. Because of higher frequencies of misclassified phenotypes among non-carriers of a certain variant allele, compared with the respective carriers, however, a trend towards overestimation of the single locus effect sizes is expected.
The rare loss of function variant loci CYP2C19*3-*8 are found within the same linkage disequilibrium block with the *2 variant. The pairwise correlation coefficients (r2≤0.019) indicate that their alleles assort independently of the *2 allele. Under the assumption of stochastic independence we pooled the data on loss of function alleles and analysed the combined values as data derived from a single bi-allelic loss of function locus.
Data collection process and extracted items
For standardisation of data extraction we adopted the Cochrane Consumers and Communication Review extraction template, modified it by the recommendations in the HuGE Review Handbook for abstracting genetic information, pilot tested five randomly selected included studies, and refined it accordingly. Two investigators independently extracted data from the included studies. Inconsistencies were resolved by consensus.
Extracted data included study identifiers (the first author’s name, year of publication, country or geographical origin of investigation, single or multicentre study); characteristics of study design (type of study, prospective or retrospective design, follow-up); characteristics of study participants (including diagnosis and procedural characteristics at study entry, demographic characteristics (number, sex, age, BMI (body mass index), and cardiovascular risk factors (smoking, hypertension, dyslipidaemia, diabetes mellitus)); characteristics of study intervention (loading dose of clopidogrel, duration of treatment, comedication with aspirin); outcome measures (type and number of events, carriers with loss of function or gain of function alleles) (table 1).
Table 1.
Characteristics of included studies of effect of variants of the cytochrome P450 (CYP) 2C19 genotype on clinical efficacy of clopidogrel
| Study | Design | Diagnosis at entry | Demographics (mean (SD) unless stated otherwise) | Cardiovascular risk factors | Clopidogrel loading dose, treatment duration; aspirin comedication | No (%) of events | No (%) of participants with ≥1 allele; MAF | |
|---|---|---|---|---|---|---|---|---|
| Loss of function CYP2C19 (at least *2) | Gain of function CYP2C19*17 | |||||||
| Trenk,63 2008, Germany, single centre | Cohort, retrospective, 12 months | Stable and unstable angina (ACS 27.4%, PCI 100%, DES 36.5%) | n=797, 78.0% men, age 66.4 (9.1) years, BMI 27.7 (3.9) | Smoking 10.9%, hypertension 82.3%, dyslipidaemia NR, diabetes 24.8% | 600 mg, median 1 (range 1-6) months (1 month for BMS, 6 months for DES); 100% aspirin | MACE (death, MI), 24 (3.0%) | 245 (30.7%), (*2); 16.4% | ND |
| Malek,64 2008, Poland, single centre | Cohort, prospective, 12 months | ACS (STEMI 81.9%, PCI 100%, DES NR) | n=105, 70.5% men, age 60.0 years, BMI 27.6 | Smoking 44.8%, hypertension 50.5%, dyslipidaemia 34.3%, diabetes 17.1% | 300 or 600 mg, NR; 100% aspirin | MACE (CV death, MI), 6 (5.7%) | 21 (20.0%) (*2); 10.5% | ND |
| Mega,34 2009; US, Europe, Oceania, Africa; multicentre | Substudy of RCT, retrospective, 15 months | ACS (STEMI 29.2%, PCI 95%, DES 47%†) | n=1459, 70.5% men, age 60.1 (11.1) years, median BMI 28† | Smoking 38.1%, hypertension 65.8%, dyslipidaemia 49.1%, diabetes 21.8% | 300 mg, ≤15 months (median 14.5 months)†; 99% aspirin† | MACE (CV death, MI, stroke) 129 (8.8%); ST (definite and probable) 17 (1.2%) | 395 (27.1%) (*2,*3,*4,*5,*8); 14.8% | ND |
| Simon,65 2009, France, multicentre | Cohort, retrospective, 12 months | Acute MI (STEMI 53.2%, PCI 69.5%, DES NR) | n=2208, 70.6% men, age 66.2 years, BMI 27.2 | Smoking 54.6%†, hypertension 58.0%, dyslipidaemia 49.3%, diabetes 31.6% | <300-900 (mean 300) mg, NR; 98.4% aspirin | MACE (death, MI, stroke) 294 (13.3%) | 635 (28.8%) (*2,*3,*4,*5), MAF: 15.7% | 774 (35.8%), MAF: 20.2% |
| Collet,66 2009, France, multicentre | Cohort, retrospective, mean: 34.6 (maximum 96) months | MI (STEMI 78.8%, PCI 73.0%, DES 32.0%) | n=259, 92.3% men, age 40.1 (5.1), BMI 25.7 (3.8) | Smoking 56.0%, hypertension 20.1%, dyslipidaemia 54.0%, diabetes 10.4% | NR, median 13.0 (IQR 3.4-36.0) months; 97.3% aspirin | MACE (CV death, MI), 19 (7.3%);ST (definite), 12 (5.4%) | 73 (28.2%) (*2,*3,*4,*5,*6); 15.8% | ND |
| Sibbing,67 2009, Germany, single centre | Substudy of RCTs, retrospective, 1 month | Stable and unstable angina and NSTEMI (ACS 34%, PCI 100%, DES 25.1%) | n=2485, 78.3% men, age 66.5 (10.2) years, BMI 27.2 (3.9) | Smoking 16.2%, hypertension 62.9%, dyslipidaemia 48.5%, diabetes 35.5% | 600 mg, ≥1 month; >95% aspirin | MACE (death, MI) 173 (7.0%); ST (definite) 17 (0.7%) | 680 (27.4%) (*2); 14.6% | ND |
| Giusti,68 2009, Italy, single centre | Cohort, retrospective, 6 months | Stable angina and ACS (ACS 65.7% PCI 100%, DES 100%) | n=772, 74.6% men, age 69 (11) years‡, BMI NR | Smoking 34.4%, hypertension 65.4%, dyslipidaemia 59.7%, diabetes 22.2% | 600 mg, ≥6 months; 100% aspirin | MACE (CV death), 18 (2.3%); ST (definite) 11 (1.4%) | 247 (32.0%) (*2); 17.7% | ND |
| Sibbing,69 2010, Germany, single centre | Cohort, retrospective, 1 month | Stable angina and ACS (ACS 33.1%§, PCI 100%, DES 98.0%§) | n=1524, 77.4% men, age 67.4 years, BMI 27.5 | Smoking 13.6%, 91. hypertension 3%, dyslipidaemia 70.1%, diabetes 28.2% | 600 mg, NR; 99.1% aspirin§ | MACE (MI) 50 (3.3%); ST (definite and probable) 14 (0.9%) | ND | 622 (40.8%), MAF: 22.9% |
| Tiroch,70 2010, Germany, single centre | Cohort, prospective, 12 months | Acute MI (STEMI NR, PCI 97.5%, DES >90%) | n=928, 74.8% men, age 64.8 years, BMI 27.0 | Smoking 36.5%, hypertension 74.5%, dyslipidaemia 51.9%, diabetes 24.1% | 600 mg, ≥6 months; 97.4% aspirin | MACE (death, MI, stroke) 82 (8.8%); ST (definite and probable) 10 (1.1%) | 248 (26.7%) (*2); 14.4% | 363 (39.1%), MAF: 22.5% |
| Wallentin,18 2010; America, Europe, Oceania, Asia; multicentre | Substudy of RCT, retrospective, 12 months | ACS (STEMI 38%¶, PCI 60.8%¶, DES 18.9%¶) | n=4904, 69.4% men, age 62.5 (11.0) years, median BMI 27¶ | Smoking 35.5%, hypertension 65.1%¶, dyslipidaemia 46.7%¶, diabetes 23.1% | 300-600 mg, median 9.2 (IQR 6-12) months; 96.1% aspirin | MACE (CV death, MI, stroke) 481 (9.8%); ST (definite) 56 (1.7%) | 1388 (28.3%) (*2,*3,*4,*5,*6,*7,*8); 15.4% | NR (no association with efficacy outcome reported) |
| Pare,19 2010; America, Europe, Oceania; multicentre | Substudy of RCT (CURE), retrospective, 12 months | Unstable angina and NSTEMI (STEMI 0%, PCI 15.5%, DES 0%) | n=2530, 58.8% men, age 63.8 (11.0) years, BMI 27.7 (4.2) | Smoking 23.1%, hypertension 59.9%**, dyslipidaemia NR, diabetes 20.7% | 300 mg, mean 9 (range 3-12) months; >94% aspirin | MACE (CV death, MI, stroke) 230 (9.1%) | 650 (25.7%) (*2,*3); 14.1% | 999 (39.1%); NA |
| Harmsze,71 2010, Netherlands, multicentre | Case-control, retrospective, 12 months | Stable angina and ACS (ACS 40.1%, PCI 100%, DES 42.6%) | n=596, 79.0% men, age 62.7 (10.3) years, BMI 27.3 | Smoking 15.1%, hypertension 48.7%, dyslipidaemia 51.0%, diabetes 16.8% | NR, ≥ 12 months; 100% aspirin | ST (definite) 176 (29.5%)†† | 193 (32.4%) (*2); 18.3% | ND |
| Sawada,72 2010, Japan, single centre | Cohort, prospective, mean 8.1 (range 0.23 to 18.2) months | Stable angina and ACS (ACS 9.0%, PCI 100%, DES 100%) | n=100, 85.0% men, age 69.6 years, BMI 23.7 | Smoking 41.0%, hypertension 81.0%, dyslipidaemia 69.0%, diabetes 42.0% | 300 mg, ≥follow-up period; 100% aspirin | MACE (death, MI) 4 (4.0%) | 42 (42.0%) (*2); NA | ND |
| Bouman,20 2010a; Germany, Netherlands; multicentre | Case-cohort, prospective, 18 months | Stable angina and ACS (ACS 50.9%, PCI 100%, DES 40.2%) | n=7719, 79.5% men, age 61.2 (8.5) years, BMI 27.0 (3.2) | Smoking 34.8%, hypertension 55.4%, dyslipidaemia 51.8%, diabetes 25.9% | 300-600 mg, median 12 (range 6-12) months; 91.2% aspirin | ST (definite) 41 (0.5%) | 2394 (31.0%)‡‡ (*2,*3,*4,*5); 16.9% | ND |
| Bouman,20 2010b; Germany, Netherlands; multicentre | Cohort, prospective, 12 months | ACS (STEMI 38.6% PCI 100%, DES 30.9%) | n=1982, 71.3% men, age 62.1 (10.2) years, BMI 27.1 (4.2) | Smoking 35.8%, hypertension 62.7%, dyslipidaemia 55.4%, diabetes 24.3% | 600 mg, ≥12 months; 100% aspirin | MACE (CV death, MI, stroke) 216 (10.9%); ST (definite) 44 (2.2%) | 678 (34.2%) (*2,*3,*4,*5,*6,*7,*8); 18.8% | 747 (37.7%); 20.9% |
ACS=acute coronary syndrome; BMI=body mass index; CV death=cardiovascular death; DES=drug eluting stent; IQR=interquartile range; MACE=major adverse cardiovascular event; MAF=minor allele frequency; MI=myocardial infarction; ND=not determined; NR=not reported; NA=not available from reported data; NSTEMI=non-ST segment elevation myocardial infarction; PCI=percutaneous coronary intervention; RCT=randomised controlled trial; SD=standard deviation; ST=stent thrombosis; STEMI=ST segment elevation myocardial infarction; BMS=bare metal stent.
†In 6795 patients assigned to clopidogrel treatment.79
‡In 804 patients.80
§In 1608 patients.81
¶In 9291 patients assigned to clopidogrel treatment.82
**In 6259 patients assigned to clopidogrel treatment.83
††Because of case-control design percentage does not reflect proportion of events in population.
‡‡Genotype distribution extrapolated to total cohort.
We included data from multiple published reports from the same study population only once. We used only the data from the initial report, and, in the case of overlapping samples, we used the data of the largest follow-up study. If a report referred to a previous publication for the description of study design, setting, and patients’ characteristics, we extracted these data. Missing data for one study34 were extracted from a subsequent meta-analysis14 conducted by the same corresponding author.
To avoid the risk of retrieval bias we did not contact the original investigators for more detailed information when we were unable to obtain complete data and protocols from all studies or to check accurateness and reliability of the obtained data.
Methods for assessing the risk of bias in individual studies
To explore the risk of bias in individual studies, we investigated indicators general to the quality of epidemiological studies and specific to the quality of genetic association studies.35 36 We extracted quality information on loss to follow-up, funding sources, comparability of groups (demographic and clinical homogeneity between groups with and without outcome, demographic and clinical homogeneity between carriers and non-carriers of the allele of interest, absence of population stratification (ethnic homogeneity)), reliability and validity of phenotype assessment (use of standardised definitions of disease phenotypes, blinding of clinical outcome assessors to patients’ genetic information), and reliability and validity of genotype assessment (consistency of observed genotype frequencies with the Hardy-Weinberg equilibrium, use of an appropriate genotyping method, high call rate, blinding of investigators who performed genotyping to clinical outcome).
Additionally, in adopting the guidelines of the Cochrane Handbook for Systematic Reviews of Interventions, we graded the methodological quality of the selected studies with a summary score using the Newcastle-Ottawa quality assessment scale.37 We applied a modified scale (adapted for genetic association studies), awarding a maximum score of 8, with one point each for representativeness of the exposed group (carriers of the genotype of interest) for the underlying population; selection of the unexposed group (non-carriers) from the same population as the exposed individuals; adequate measurement of exposure; adequate ascertainment of the absence of the outcome of interest at begin of the study; demographic and clinical comparability between groups (carriers and non-carriers of the genotype); appropriate measurement of outcome; adequate length of follow-up; and completeness of follow-up. Study quality was considered to be good when the score was ≥6 and poor to moderate when the score was <6. Two investigators independently scored quality, and the inter-rater agreement was determined by the κ statistic.
Consistency of the observed genotyping frequencies with the Hardy-Weinberg equilibrium provides an overall (albeit insensitive and non-specific) indication for the absence of any strong bias by the selection of patient groups, population stratification, or genotyping errors. We checked departure from Hardy-Weinberg equilibrium using Fisher’s exact test38 instead of the χ2 test reported in the individual studies as it yields increased statistical power. For the cohort studies and the genetic subgroup analyses, testing on Hardy-Weinberg equilibrium was performed in the whole population, and for the case-control study in the control group. We considered that significant departure from Hardy-Weinberg equilibrium (P<0.05) necessitated a correction of individual risk estimates.35
Summary effect measures and sample size estimation
We calculated crude unadjusted odds ratios and 95% confidence intervals for each study based on genotype contrasts of a dominant model comparing heterozygous and homozygous genotypes of the minor allele with homozygous genotypes of the major allele. As primary summary effect estimates we calculated summary odds ratios and 95% confidence intervals according to the DerSimonian and Laird random effects model, which utilises weights that incorporate variance within and between studies. In addition, we calculated the fixed effects summary estimates according to the Mantel-Haenszel method, which includes only variance within studies. Fixed effects meta-analysis assumes that the genetic effects are the same across all studied populations. Random effects calculations assume that the genetic effects might vary across populations because of genuine differences (such as population specific gene environment or gene-gene interactions) or differential biases (such as population stratification; genotyping error; phenotype misclassification; and population differences in correlation of clinical phenotypes, correlation between molecular and clinical phenotype, and linkage disequilibrium of gene variants). Anticipating heterogeneity between studies in meta-analyses of genetic association studies, random effects models are generally the preferred frequentist approach compared with fixed effect models.39 40 41 In the presence of high variance between studies, however, the random effects analysis has considerably less power to reject the null hypothesis of no association. By contrast, random effects models give relatively more weight to smaller studies, which involves the risk of generating higher summary estimates in the presence of bias from small study effects. Therefore, we have presented both random and fixed effects analysis. P<0.05 indicates a nominally significant overall association (according to the z test statistic for the null hypothesis of no association (odds ratio 1)).
To estimate the total sample size needed to be included in a meta-analysis to detect a significant association at low summary odds ratios, we performed Monte-Carlo simulations using PBAT software, version 3.61.42 For a dominant genotype contrast, to detect a summary odds ratio of 1.15 (the threshold for epidemiological credibility43) at a significance level α=0.05 with a power of 0.80 for cumulative event rates in the range of 1-10% and minor allele frequencies in the range of 15-25%, the required sample size was about 8000-9000, and to detect an odds ratio of 1.20 it was about 4500-5000 (based on 10 000 simulations each).
Heterogeneity measures
The presence of heterogeneity between studies was explored with the Cochran’s Q statistic, which is the weighted sum of squares of the deviations of individual study odds ratios from the Mantel-Haenszel summary odds ratio. The statistic follows a χ2 distribution with k−1 degrees of freedom (where k is the number of studies); P<0.10 indicated significant heterogeneity. The extent of variance between studies was estimated by the τ2 metric. The percentage of total variance attributable to heterogeneity between studies was quantified with the I2 metric and its approximate 95% confidence intervals (where I2=((Q−(k−1))/Q)×100%). I2 is independent of the number of studies and—in contrast with Q and τ2—allows comparison across different meta-analyses.44 Values of I2 <25%, ≥25%-<50%, and ≥50% were considered to represent low, modest, and large heterogeneity, respectively. Both Q and I2, however, have only low statistical power to detect heterogeneity with small numbers of studies45 and provide no information about the causes of heterogeneity. Hence, we performed additional prespecified heterogeneity and sensitivity analyses. Formal evaluations required that at least four studies were included in the meta-analysis.
Assessment of bias across studies
Inconsistency in replication is an important issue in genetic association. The first studies often suggest a stronger genetic effect than is found by subsequent studies.46 To assess the replication validity in the meta-analyses we compared the odds ratio of the first published studies with the random effects summary odds ratios without the first studies using the z test statistic, with P<0.05 indicating a significant inconsistency in replication.35 To explore the evolution and robustness of the summary effect estimates over time, we conducted cumulative and recursive cumulative meta-analyses. In cumulative meta-analysis, the random effects summary odds ratios are calculated with publication of each new study. In recursive cumulative meta-analysis, the ratio of the cumulative odds ratios in year n+1 to year n is calculated.47 These analyses facilitate the identification of early extreme contradictory estimates in genetic associations and of potential time lag bias resulting from a more rapid publication of studies with significant results compared with studies with non-significant results.35
To assess potential bias from small study effects we constructed funnel plots displaying the log odds ratios of individual studies on the horizontal axis and the standard errors of the log odds ratios (precision) on the vertical axis. Funnel plot asymmetry is a graphical means of indicating whether effect estimates of small studies differ from those in larger studies,48 but visual inspection is an unreliable method to detect bias. We carried out formal statistical assessment of funnel plot asymmetry with the Harbord-Egger regression test, which yields lower false positive and false negative rates when applied to dichotomous outcomes compared with the traditional Egger regression test or the Begg-Mazumdar rank correlation test.49 50 P<0.10 was assumed to indicate a significant difference of the precision in large versus small studies. For more specific evaluation of the presence and extent of publication bias we used the non-parametric trim and fill method according to Duval and Tweedie,51 which imputes missing studies in the funnel plot based on symmetry assumptions.
Sensitivity analyses
We performed a prespecified combinatorial exclusion sensitivity analysis to identify the individual studies or clusters of studies that provide the strongest contribution to the heterogeneity of the meta-analysis.52 Potential differences in the main characteristics of excluded studies in comparison with the remaining studies were investigated (considering the structured PICOS information of the Cochrane Handbook for Systematic Reviews, chapter 5).
Grading the evidence of meta-analyses
Reliable and valid instruments for evaluating the quality of the evidence derived from systematic reviews and meta-analyses are essential for developing transparent and unbiased clinical recommendations and avoiding implicit subjectivity. For judgment of the strength of the meta-analysed evidence for recommendations we applied the GRADE methods, proposed by WHO for producing practice guidelines.53 54 To evaluate the quality of evidence with specific regard to genetic topics, we used the Venice consensus criteria43 for rating the cumulative epidemiological evidence of meta-analyses of genetic association studies that yield significant (P<0.05) summary estimates.
Within the GRADE evaluation process observational studies are basically considered to present low quality of evidence. According to the GRADE handbook, version 3.2, and pragmatic instructions to guide the grading process54 judgments on the included studies are made with respect to the following five criteria that lower the quality of evidence: limitations of individual studies (risk of bias within a study); inconsistency (heterogeneity of results across studies); indirectness of evidence; imprecision (total number of events <300)55; and publication bias. Studies not downgraded for any reason are judged for three factors that increase the quality of evidence: dose-response gradient (gene-dose effect); large magnitude of effect (relative risk >2.0 or <0.5); and reduction of effect by all plausible biases present (potential underestimation of effect).
The Venice consensus criteria assign three levels for the amount of evidence, the consistency of replication, and the protection from bias. For amount of evidence, grade “A” is assigned when the total number of minor alleles of cases and controls combined in the meta-analyses exceeds 1000, “B” when it is between 100 and 1000, and “C” when it is less than 100. For replication and consistency, grade A is assigned for I2 <25%, B for I2 25-50%, and C for I2 >50%. For protection from bias, grade A implies that there is probably no bias that can affect the presence of the association, grade B that there is no demonstrable bias but important information is missing for its appraisal, and grade C that there is evidence for potential or clear bias that can invalidate the association. Specifically, whenever the summary odds ratio deviates less than 1.15-fold from null association (odds ratio=1), occult publication and selective reporting biases alone might invalidate the association, regardless of the presence or absence of other biases, and therefore a grade of C is assigned. When the summary odds ratio is ≥1.15-fold from null association, a grade of C is assigned when a significant modified regression test suggests the possibility of bias or when the association is no longer nominally significant on exclusion of the initial study or of studies violating the Hardy-Weinberg equilibrium. The composite epidemiological credibility is rated as “strong” if three A grades are assigned, “moderate” if at least one B grade but no C grades are assigned, and “weak” if a C grade in any of the three assessment criteria is assigned.
The grading was done independently by two investigators and repeated by a third investigator if disagreement occurred.
Statistical software programs
The statistical analyses were performed with PASW version 18.0.1 (SPSS, Il, US), Cochrane Review Manager 5.0 (Cochrane Library Software, Oxford, UK), and MIX version 1.7 (Department of Medical Informatics of Kitasato University, Japan).
Results
Study selection
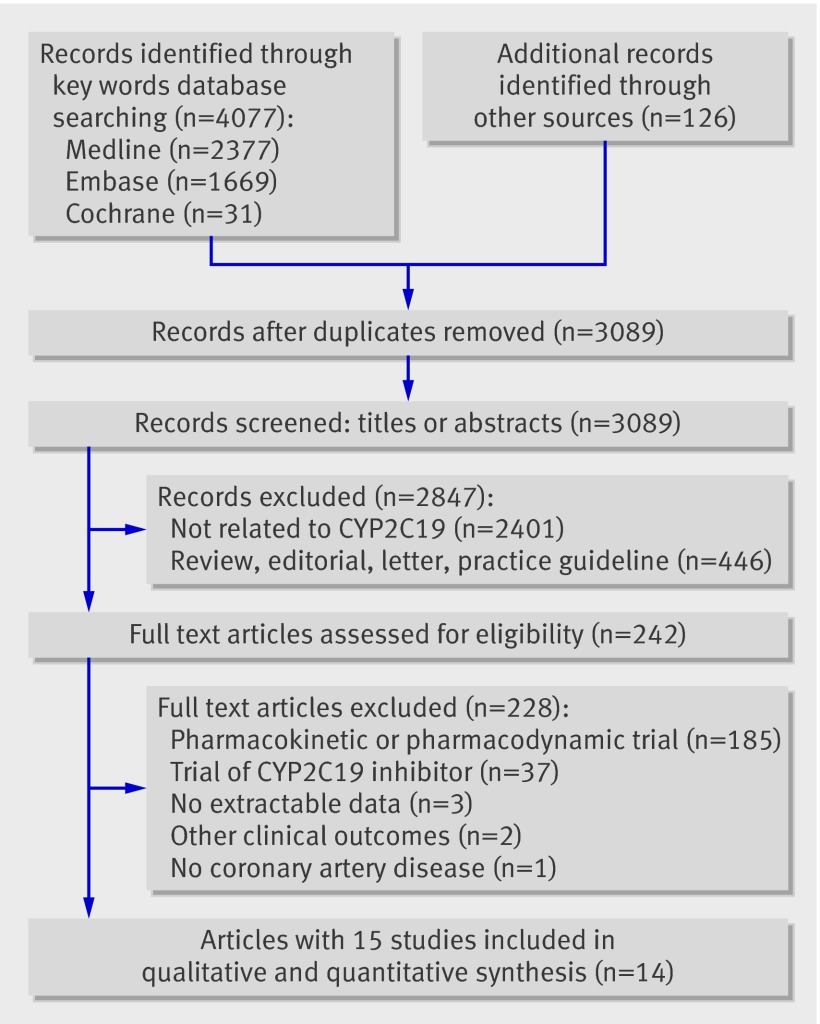
We identified a total of 4203 reports in the initial search (fig 1) and removed 1114 duplicate publications. We also excluded 2847 non-relevant records based on screens of titles or abstracts. Full text articles were retrieved for 242 publications and assessed for eligibility. We then excluded 228 articles, mainly because they reported only pharmacodynamic or pharmacokinetic associations with CYP2C19 or associations of inhibitors of CYP2C19 with clinical outcomes. The genetic substudy of the randomised controlled ACTIVE A trial was excluded because it was conducted in patients with atrial fibrillation and did not provide a genetic subgroup analysis for patients with additional coronary artery disease at baseline.19 Three studies were excluded because the outcome assessment did not meet the eligibility criteria; one study displayed only all cause mortality.56 One study reported only the combined end point of cardiovascular death, myocardial infarction, ischaemic stroke, stent thrombosis, revascularisation, and admission to hospital for coronary ischaemia.57 One study compared genotype frequencies in cases with untreated control subjects.58 We did not consider four potentially eligible studies that were published only in abstract form or as conference proceedings.59 60 61 62
Fig 1 Selection of studies of association of CYP2C19 variant genes with cardiovascular events in patients treated with clopidogrel
Overall, we identified 14 articles published in English involving 15 observational studies or genetic substudies from randomised controlled trials that met the inclusion criteria and included these in the systematic review.18 19 20 34 63 64 65 66 67 68 69 70 71 72
The κ statistic indicated a very good agreement between the abstracting investigators (κ 0.84; SE 0.07).
Characteristics of included studies
Twelve studies with a total of 18 529 participants examined the association of a reduced function variant of CYP2C19 with major adverse cardiovascular events,18 19 20 34 63 64 65 66 67 68 70 72 and nine studies combining 19 328 participants examined the association with stent thrombosis.18 20 34 66 67 68 70 71 Five studies with a total of 9128 participants investigated the association of the gain of function variant CYP2C19*17 with major adverse cardiovascular events,19 20 65 69 70 and three studies20 69 70 with 4434 participants examined the association with stent thrombosis. Table 1 presents the characteristics of the individual studies.
Design
Most studies (n=10) were retrospective genetic analyses of studies that were not primarily designed to evaluate genetic association.18 19 34 63 65 66 67 68 69 71 Nine studies were cohort studies,20 63 64 65 66 68 69 70 72 four were substudies of randomised controlled trials,18 19 34 67 one was a prospective case-cohort study,20 and one was a case-control study.71 Eight studies involved multiple centres.18 19 20 34 65 66 71
Participants
All studies included participants of both sexes; around 60-90% were men. In 14 studies the mean age at entry was over 60. In one study participants were younger (average age 40) and had a lower prevalence of hypertension and diabetes mellitus.66
Intervention
Most studies (n=13) noted that patients received a loading dose of clopidogrel (usually 300 or 600 mg) at occurrence of the index event or at the index procedure.18 19 20 34 63 64 65 67 68 69 70 72 The maintenance dose at discharge from hospital was 75 mg a day in all studies. More than 90% of patients in all studies were also treated with aspirin (75-325 mg a day). The follow-up periods ranged from 1 to 35 (median 12) months.
Outcomes
Six of 13 studies reporting the incidence of major adverse cardiovascular events met the primary outcome definition, constituting the composite of (cardiovascular or all cause) death, myocardial infarction, and stroke18 19 20 34 65 70; the other studies reported components of the primary definition. Seven of 10 studies that assessed stent thrombosis reported definite events18 20 66 67 68 71; three assessed the composite of definite and probable events.34 69 70 The cumulative incidence of major adverse cardiovascular events in the population based studies ranged from 3.0% to 13.3% and of stent thrombosis from 0.5% to 2.2%. The frequency of the *2 allele or of combinations of the loss of function alleles lay between 10.5% and 18.8%, and the allele frequency of the *17 variant was between 20.2% and 22.9%.
Risk of bias in individual studies
The risk of bias in the included studies was assessed with a standard approach with predefined criteria (table 2). The general methodological quality of the studies judged on the modified Newcastle-Ottawa scale was rated moderate to good (scores of 5-8). The agreement between raters in the awarded scores on the 8 point scale was considered good (12 agreements and three disagreements by 1 point; κ 0.69, SE 0.16). Potential risks of individual bias with respect to general epidemiological topics included missing information about loading dose (two studies66 71), no pre-fixed follow-up periods (two studies66 72), high rates of loss to follow-up (one study72), cessation of clopidogrel maintenance treatment before end of follow-up or missing information about duration of treatment (10 studies18 19 20 34 63 64 65 66 69 70), no explicit criteria for adherence to treatment (13 studies18 19 34 63 64 65 66 67 68 69 70 71 72), and funding by industry or healthcare providers (five studies18 19 34 65 69). One study assessed incident myocardial infarction in a sample of patients aged 18-45 who were not representative for a population of cardiovascular patients.66
Table 2.
Epidemiological and genetic quality assessment of included studies of effect of variants of the cytochrome P450 (CYP) 2C19 genotype on clinical efficacy of clopidogrel
| Study | Newcastle-Ottawa score (0-8); loss to follow-up | Funding | Demographic and clinical homogeneity between groups | Evidence of population stratification, ethnicity | Standardised definition and assessment of outcomes | Blinding: outcome assessment to genotypes†; genotyping to outcome status | Genotyping: errors; method (validation markers); call rate‡ | Hardy-Weinberg equilibrium§: loss of function alleles¶; CYP2C19*17; assumed genetic model | |
|---|---|---|---|---|---|---|---|---|---|
| With/without outcome | With/without exposure to genotype of interest | ||||||||
| Trenk63 (2008) | 8; 0.9% | Institutional grant | Unclear | Yes | Unclear, ethnicity NR | Yes | Yes; unclear | No; validated TaqMan PCR; 99.3% | Yes (P=0.30); ND; dominant |
| Malek64 (2008) | 6; 0% | Institutional grant | Unclear | Yes | Unclear, ethnicity NR | Unclear | Unclear; unclear | No; restriction fragment length PCR; 100% | Yes (P=1); ND; dominant |
| Mega34 (2009) | 8; 0.7%** | Industry | Unclear | Yes | No, 97.6% white (self reported) | Yes | Yes; unclear | No; Affymetrix DMET chip; 98.8% | Yes (P=0.48); ND; dominant |
| Simon65 (2009) | 7; 0.8% | Industry, state | No (patients with outcome event were older, had higher prevalence of CV risk factors and less often underwent PCI) | Unclear | Unclear, ethnicity NR | Yes | Yes; unclear | No; SNPlex oligoligation or TaqMan PCR; >98.0% | Yes (P=0.57); yes (P=0.12); codominant |
| Collet66 (2009) | 6; 0% | Institutional grant | No (hazard ratios of outcomes changed after adjustment for patients’ characteristics) | Yes | No, 78.0% white (method of assessment NR) | Yes | Unclear; yes | No; TaqMan PCR; 100% | Yes (P=0.24); ND; dominant |
| Sibbing67 (2009) | 8; 6.6% | Institutional grant | Unclear | Yes | Unclear, ethnicity NR | Yes | Yes; unclear | No; TaqMan PCR (quality checks); 100% | (Yes (P=0.38); ND; dominant and codominant models |
| Giusti68 (2009) | 7; 4% | Institutional grant | Unclear, NR | Unclear | Unclear, ethnicity NR | Yes | Yes; unclear | No; restriction fragment length PCR; 100% | Yes (P=0.62); ND; dominant and codominant models |
| Sibbing69 (2010) | 8; 5.2% | Industry and institutional grant | Unclear | Yes | Unclear, ethnicity NR | Yes | Yes; yes | No, TaqMan PCR (quality checks); 100% | ND; yes (P=0.61); dominant and co-dominant models |
| Tiroch70 (2010) | 7; 0% | Institutional grant | Yes | Yes | Unclear, ethnicity NR | Yes | Unclear; unclear | No; TaqMan PCR (quality checks); 100% | Yes (P=0.69); yes (P=0.19); dominant |
| Wallentin18 (2010) | 7; 4.7% | Industry | Unclear | Unclear | No, 98.3% white (self reported) | Yes | Yes; unclear | No; TaqMan PCR; ≥98.8% | Yes (P=0.35); unclear; dominant |
| Pare19 (2010) | 7; 0.1%†† | Industry | Unclear | Unclear | No, 86.2% white, 13.8% Hispanics (self reported) | Yes | Yes; unclear | No; TaqMan PCR; >98% | Yes (P=0.07); yes (P>0.05)‡‡; dominant and codominant models |
| Harmsze71 (2010) | 7; 5.4% | No funding | No (patients with outcome event had more ACS, were more often smokers, and fewer received DES) | Unclear | Unclear, ethnicity NR | Yes | Yes; unclear | No; TaqMan PCR; 100% | Yes (P=0.16); ND; dominant |
| Sawada72 (2010) | 5; 72.8% | No funding | Unclear | Yes | No, 100% East Asian (method of assessment NR) | Yes | Yes; unclear | No; TaqMan PCR; 100% | Unclear (genotype frequencies NR); ND; dominant |
| Bouman20 (2010a) | 7; 1.3% | No funding | Yes | Yes | No, 100% white (self reported, genomic control) | Yes | Yes; yes | No; direct sequencing (validation rules), 100% | Yes (P=0.90)§§; ND; codominant |
| Bouman20 (2010b) | 8; 8.4% | No funding | Yes | Yes | No, 100% white (self reported) | Yes | Yes; yes | No; direct sequencing (validation rules), 99.5% | Yes (P=0.88); yes (P=0.64); codominant |
ACS=acute coronary syndrome; CV=cardiovascular; PCI=percutaneous coronary intervention; DES=drug eluting stent; NR=not reported; ND=not determined; PCR= polymerase chain reaction.
†Outcome assessors classified as blinded to genotype if explicitly reported or genotyping conducted in subsequent study.
‡Fraction of complete and non-missing genotypes of number of total genotypes
§With Fisher’s exact test P value.
¶For testing deviation from Hardy-Weinberg equilibrium observed genotype frequencies of all loss of function alleles of CYP2C19 combined.
**From Mega et al.14
††In 6259 patients assigned to clopidogrel treatment.83
‡‡P value stated by authors (genotype distribution not presented).
§§P value derived from genotype distribution extrapolated to total cohort of 7719 patients (P=0.53, exact P value for genotype distribution of random subcohort).
The quality of the individual studies with respect to specific genetic indicators showed larger deficiencies: five studies did not assess the comparability of baseline demographic and clinical characteristics between carriers and non-carriers of the CYP2C19 genotype of interest,18 19 65 68 71 12 studies showed differences in characteristics between groups with and without outcome or did not perform group comparisons.18 19 34 63 64 65 66 67 68 69 71 72 This implies the possibility that the association with CYP2C19 could be confounded by variations in genetically or non-genetically determined factors that were associated with the outcome. Eight studies did not determine the ethnicity of participants, which introduces the risk of population stratification, although this is less likely to occur in studies conducted in Europe than in more heterogeneous American populations.63 64 65 67 68 69 70 71 All studies (except for one64) used standardised definitions for assessment of outcomes, and in most studies (except for three64 66 70) the outcome assessors were explicitly blinded or unaware (retrospective analysis) to the genotyping result of the individuals, which minimised the risk of misclassification of diseases. Only three studies, however, stated that the genotyping was done blinded to the outcome status of patients, which does not ensure an unbiased assessment.20 69 All studies reported high call rates of genotyping (that is, a high proportion of samples in which genotyping provided an unambiguous reading). This indicates a low extent of missing data on genotyping. Only five studies explicitly described the implementation of quality checks or validation rules to reduce genotyping errors.20 67 69 70 Such errors are not uncommon and can distort the robustness of association measures.73 Except for one study that did not provide genotype frequencies to allow testing,72 there was no significant departure from the Hardy-Weinberg equilibrium. All studies assumed dominant or codominant models for testing of genetic contrasts, but none gave a biological reason for the choice of the model and no adjustments for multiple comparisons were performed. Moreover, the studies reported different effect measures with or without adjustments for potential covariates. To yield consistency and comparability between effect estimates, we calculated the unadjusted odds ratios for each study.
Results of individual studies and synthesis of results
The forest plots show the odds ratios with 95% confidence intervals for each study. They also incorporate the proportions of events in specific groups and the weighting factors for the summary effect estimates and present measures of consistency between studies (Q, I2, τ2) and results of a significance test of the overall effect.
Associations with loss of function CYP2C19 variants
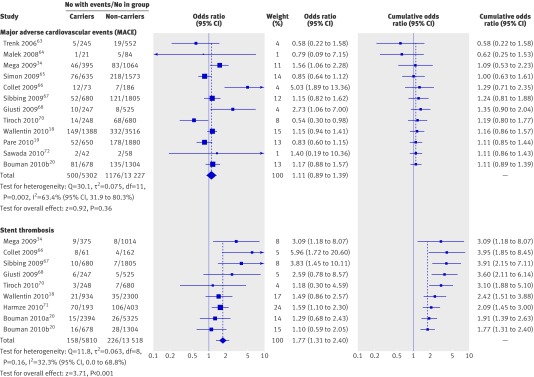
For comparison of carriers of at least one reduced function allele of CYP2C19 with non-carriers the unadjusted individual odds ratios of major adverse cardiovascular events were significantly higher in three of 12 studies,34 66 68 lower in one study,70 and not significantly different from null effect in eight (fig2).18 19 20 63 64 65 67 72 The odds ratios of stent thrombosis were positively associated with the reduced function CYP2C19 allele carrier status in four of nine studies,34 66 67 71 whereas five studies showed no significant association.18 20 68 70
Fig 2 Association between loss of function polymorphisms of CYP2C19 and major adverse cardiovascular events (MACE) or stent thrombosis in patients with coronary artery disease taking clopidogrel treatment. Odds ratios shown for individual studies for dominant model genotype contrasts (carriers of one or two loss of function alleles v non-carriers). Cumulative odds ratios shown for each additional information step obtained by stepwise inclusion of every new study into pooled estimate
The random effects summary odds ratio for major adverse cardiovascular events was 1.11 (95% confidence interval 0.89 to 1.39; P=0.36) and the corresponding fixed effects summary odds ratio was 1.07 (0.96 to 1.19; P=0.24), with heterogeneity measures: Q=30.1 (df=11, P=0.002); I2=63.4% (31.9% to 80.3%); τ2=0.075. The random effects summary odds ratio for stent thrombosis was 1.77 (1.31 to 2.40; P<0.001), and the fixed effects summary odds ratio was 1.67 (1.34 to 2.08; P<0.001) with Q=11.8 (df=8, P=0.16); I2=32.3% (0% to 68.8%); τ2=0.063.
Associations with gain of function CYP2C19*17 variant
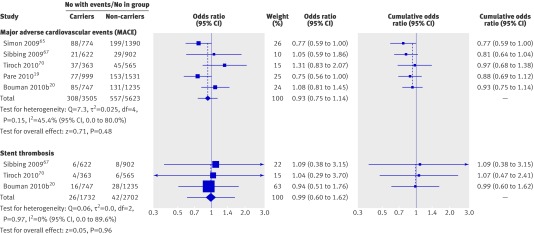
None of the individual studies showed a significant positive or negative association of major adverse cardiovascular events or stent thrombosis with CYP2C19*17 (fig 3). The random effects summary odds ratio for major adverse cardiovascular events was 0.93 (0.75 to 1.14; P=0.48), and the fixed effects summary odds ratio was 0.90 (0.78 to 1.04; P=0.15); heterogeneity statistics were: Q=7.3 (df=4, P=0.15); I2=45.4% (0% to 80.0%); τ2=0.025. The random and fixed effects summary odds ratios of stent thrombosis were identical: 0.99 (0.60 to 1.62; P=0.96), and indicators of heterogeneity were: Q=0.06 (df=2, P=0.97); I2 =0.0% (0% to 89.6%); τ2 =0.00.
Fig 3 Association between gain of function CYP2C19*17 polymorphisms with major adverse cardiovascular events (MACE) or stent thrombosis in patients with coronary artery disease receiving clopidogrel treatment. Odds ratios shown for individual studies for dominant model genotype contrasts (carriers of one or two gain of function alleles v non-carriers). Cumulative odds ratios shown for each additional information step obtained by stepwise inclusion of every new study into pooled estimate
The total sample size of the non-significant meta-analyses was lower compared with the meta-analysis showing a significant outcome. The simulations of the sample size needed to detect a given value of dominant contrast odds ratio, however, indicate that all meta-studies had sufficient power to identify clinically noteworthy associations with odds ratios ≤1.20.
Risk of bias across studies
Heterogeneity
The Q statistic and the point estimate ( I2) indicate the presence of high heterogeneity between studies in the meta-analysis of studies examining the association of CYP2C19 loss of function alleles with major adverse cardiovascular events, whereas the other meta-analyses did not exceed the conventional levels for significant heterogeneity (P<0.10 or I2≥50%). In all meta-analysis the upper 95% confidence interval of I2 exceeded the threshold of large heterogeneity (I2≥50%). We cannot therefore exclude the presence of considerable heterogeneity across the studies,45 which necessitates further assessments of heterogeneity. The meta-analysis of CYP2C19*17 and stent thrombosis included only three studies, which precluded the application of formal statistical methods for evaluation of heterogeneity.
Replication validity and evolution of summary estimates in time
The z statistic for comparison of the odds ratio of the first published study with the random effects pooled odds ratio of the remaining studies was not significant for the associations of major adverse cardiovascular events with loss of function variants (z=−1.28, P=0.20) or the gain of function variant (z=−1.38, P=0.17). There was, however, significant diversity within the associations of stent thrombosis with loss of function variants between the initial (almost simultaneously published) studies of Mega et al34 and Collet et al66 and the subsequent studies (z=2.39, P=0.017). Correspondingly, the cumulative meta-analysis for comparison of the loss of function variants with stent thrombosis showed a trend of decreasing strength of association as information accumulated. Recursive cumulative meta-analysis showed a significant relative change in the cumulative pooled odds ratio between information published in 2010 and information published in 2009 (0.395, z=−3.093, P=0.002). This suggests that the relation between the loss of function CYP2C19 genotypes and stent thrombosis is not sufficiently established by the accumulating evidence that the change in odds ratio could stabilise over time. No such time instabilities were observed in cumulative meta-analyses of the other associations. At no point in the acquisition of additional information was a significant cumulative odds ratio obtained. For the association of loss of function variants with major adverse cardiovascular events, the recursive cumulative comparison of odds ratios of studies published in 2010 with those published before showed a non-significant relative change of 0.722 (z=−1.337, P=0.181). It was not possible to apply recursive cumulative meta-analysis to the associations with CYP2C19*17 as all included studies (except for one65) were published in 2010.
Small study effects and publication bias
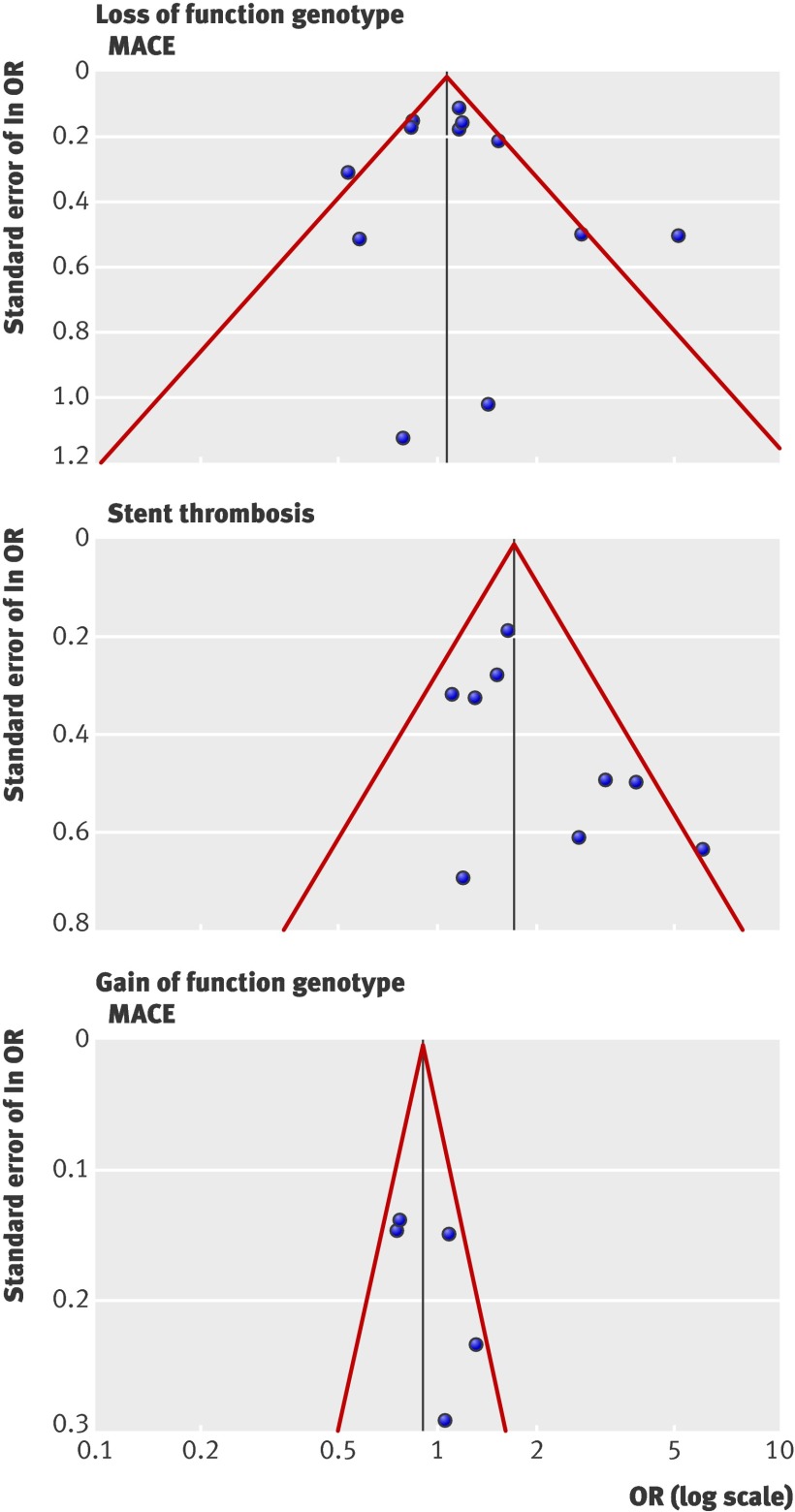
Figure 4 shows the funnel plots of the meta-analyses. In the funnel plot based on the results of 12 studies on the effect of loss of function variants on major adverse cardiovascular events, two studies66 70 were outside the expected 95% confidence limits. In the corresponding funnel plot of nine studies on the effect of loss of function variants on stent thrombosis, one study was outside the expected 95% confidence limits.66 This asymmetry indicates the possibility of heterogeneity by small studies or selection bias. The modified Egger regression test indicated a significant asymmetry across the association studies on loss of function variants with stent thrombosis (t=1.93, P=0.095), and the trim and fill analysis suggests that two studies were missing. The adjusted random effects summary odds ratio of 1.55 (1.10 to 2.20; P=0.014) obtained after symmetrically filling the funnel plot supposes that the strength of the association with stent thrombosis is overestimated (by 14%) because of unpublished studies. The modified Egger test was non-significant for the other associations yielding t=0.76 (P=0.46) for comparisons of loss of function variants with major adverse cardiovascular events and t=1.48 (P=0.24) for comparisons of the gain of function variant with major adverse cardiovascular events. Accordingly, the trim and fill analysis showed no missing studies.
Fig 4 Funnel plots for association studies of loss of function genotypes of CYP2C19 with major adverse cardiovascular events (MACE) or stent thrombosis and for association studies of the gain of function variant CYP2C19*17 with MACE. Solid vertical line represents summary effect estimate, derived by using fixed effects meta-analysis for displaying centre of plot in absence of bias.78 Red lines represent 95% confidence limits for expected distribution of studies in absence of heterogeneity between studies or of selection biases
Sensitivity analysis
Targeting for a minimum I2 below the 50% threshold in the meta-analysis of loss of function variants and major adverse cardiovascular events, the combinatorial algorithm led to exclusion of the studies of Collet et al66 and Tiroch et al70 (representing outliers in the funnel plot), giving I2=40.9% and a residual odds ratio of 1.09 (0.92 to 1.30; P=0.30). To achieve the minimal I2 value <25%, four studies34 65 66 70 had to be omitted, resulting in I2=18.8% and a virtually unchanged residual odds ratio of 1.09 (0.93 to 1.29; P=0.29). In the meta-analysis of loss of function variants and stent thrombosis, exclusion of the study of Collet et al66 decreased I2 below the requested 25% threshold, leading to the minimum final I2 of 8.6% and reducing the residual odds ratio (by 9%) to 1.61 (1.26 to 2.07; P<0.001). In the meta-analysis of CYP2C19*17 and major adverse cardiovascular events, targeting for minimum values of I2 and Q, the studies of Simon et al65 and Pare et al19 were excluded, yielding I2=0% (Q=0.6) and a residual odds ratio of 1.13 (0.90 to 1.41; P=0.29). In none of the meta-analyses did the nominal significance or non-significance of the summary effect estimates change on exclusion sensitivity analysis.
Grading of meta-analyses
Following the GRADE algorithm, we downgraded the quality of the evidence of studies investigating major adverse cardiovascular events in relation to loss of function genotypes from low to very low for heterogeneity between studies that was not explained by study design, differences between populations or interventions, or length of follow-up. We downgraded the meta-analysis of the associations of stent thrombosis with loss of function alleles for evidence of publication bias suggesting a systematic overestimation of the harmful effects of clopidogrel in loss of function allele carriers. The meta-analysis of the associations between stent thrombosis and CYP2C19*17 was downgraded because of imprecision (total number of events <300). By contrast, the meta-analysis of associations between major adverse cardiovascular events and CYP2C19*17 was not affected by confounders that were sufficient to downgrade the quality of evidence, but the overall judgment remained low as none of the criteria leading to upgrading in the quality of evidence were met. The grading of the quality of the epidemiological evidence as low or very low thus classifies any estimate of effect as uncertain or further research as likely to have an important impact on the confidence in the estimates.
When we applied the Venice criteria for the assessment of cumulative evidence in genetic association studies with nominally significant summary effect estimates, the meta-analysis of the association of stent thrombosis with loss of function variants was graded as A for criterion 1 (amount of evidence >1000 minor alleles in combined studies) and as B for criterion 2 (consistency of replication I2=25-50%). For criterion 3 (protection from bias) we assigned grade C because the modified Egger regression test suggested the presence of publication or small study effects bias. Based on the C grading, the overall epidemiological credibility of this meta-analysis was rated as “weak.” There were no disagreements between the two raters (DT and TB) in the judgments according to GRADE and Venice methods.
Discussion
Principal findings and relation to other studies
CYP2C19 gene polymorphisms do not influence the clinical efficacy of clopidogrel used to prevent thromboembolic events in patients with ischaemic cardiovascular diseases. We summarised the existing data on the association of the carrier status of variant alleles associated with loss and gain of function with the incidence of major adverse cardiovascular events or stent thrombosis in patients receiving clopidogrel antiplatelet treatment. We retrieved 15 studies that met the prespecified eligibility criteria using a systematic search strategy. The summary odds ratios derived by adopting a dominant genetic model indicate that carriers of at least one loss of function allele show an increased risk of stent thrombosis. The summary effect estimates were not significant for the association of major adverse cardiovascular events with carrier status for the loss of function allele and for the associations of major adverse cardiovascular events or stent thrombosis with carrier status for the gain of function CYP2C19*17 allele.
The systematic examination of potential sources of bias within the individual studies, the risk of misclassified phenotypes, and the assessment of inconsistencies between studies that used heterogeneity tests and sensitivity analyses indicate that all meta-analyses are affected by confounders that tend to weaken the strength or presence of the associations. Specifically, the association of stent thrombosis with loss of function genotypes was subject to bias from small study effects and to interaction with publication year. Adjustment for these quality modifiers tended to abolish the association. By using consensus grading systems to assess the cumulative evidence, all meta-analyses had low levels of epidemiological credibility or quality. We infer that, at the current state of accumulated information, there is no sufficiently robust and consistent evidence that CYP2C19 represents a strong susceptibility gene modifying the clinical efficacy of clopidogrel.
Our findings disagree with the results of previous meta-analyses, which reported a significant higher risk of major adverse cardiovascular events in carriers of the loss of function CYP2C19 gene variants and a considerably higher strength of the association between stent thrombosis and the loss of function variants. Two factors could explain the discrepancies. Firstly, previous meta-analyses included only those studies published before 2010 that generally show stronger effects compared with the subsequently published studies. Secondly, we extracted only the data of such prespecified clinical events that conformed to unbiased and standardised definitions, excluding clinician driven or less accurately defined events or outcomes with a high risk of aetiological confounding from genetic heterogeneity.
The lack of significance, inconsistency of results, or low epidemiological credibility do not rule out the presence of a true causal association with CYP2C19. A causal involvement of CYP2C19 in the bioactivation of clopidogrel and thus its clinical response, however, is not supported by stringent and unequivocal biological data. A recent metabolomic analysis,20 in the line with earlier studies,74 75 suggest that clopidogrel is metabolised primarily by CYP3A isoenzymes, but not by CYP2C19, to yield the intermediate product 2-oxo-clopidogrel. This is transformed into the active metabolite by hydrolysis not involving cytochrome P450 enzymes.
Limitations of the study
Our systematic meta-analyses had several limitations. Firstly, although we applied a highly sensitive search strategy for the retrieval of potentially eligible studies, we cannot rule out that some studies might have been overlooked. Secondly, we excluded results from association studies existing only as abstracts. This could have caused a disproportionate exclusion of significant or non-significant findings resulting in publication bias, but the incomplete data of abstracts entail uncertainties and did not allow us to perform validity assessments. Moreover, in line with our findings, the extractable data on major adverse cardiovascular events from the four potentially eligible studies that were available only as abstracts showed no significant association with carrier status for the CYP2C19 loss of function or gain of function alleles.59 60 61 62 Thirdly, differential genotype data for carrier status for the loss of function and gain of function alleles were missing in most studies. Thus, the single locus genotype contrasts were reported independently of each other, which could have led to a systematic overestimation of effects at the individual study level. Fourthly, unavailability of genotype for individual patients and demographic and clinical data in all studies meant we could not carry out haplotype based analyses, adjustment at the patient level for potentially confounding covariates, or time to event analyses. This might have removed the observed heterogeneity between studies by eliminating bias and genuine diversity at the individual level. A meta-analysis of the association between CYP2C19*2 and cardiovascular events combining individually adjusted hazard ratios, however, yielded a similar magnitude of effect estimates and a similar level of heterogeneity14 compared with meta-analyses that combined unadjusted odds ratios based on study level data.11 13 Fifthly, in contrast with stent thrombosis, the classification of major adverse cardiovascular events varied across studies and the components of the event cluster were aetiologically diverse and influenced by numerous covariates apart from platelet activity. The composite of death, myocardial infarction, and stroke, however, is a common primary end point in clinical trials of coronary artery disease and shows considerable practical relevance and high accurateness of diagnosis because of standardised definitions. Sixthly, our analyses did not consider cotreatment with CYP2C19 inhibitory drugs, specifically proton pump inhibitors, which could have confounded the genetic associations. None of the individual association studies that mentioned the use of proton pump inhibitors, however, observed significant interactions with CYP2C19 genotypes.18 20 65 66 68 69 71 72 Also, it has been suggested that effects of proton pump inhibitors on the platelet response to clopidogrel are independent of the CYP2C19 allele carrier status17 and that potential adverse cardiovascular effects of proton pump inhibitors are completely independent of clopidogrel use.76 77 Finally, the grading instruments we used for assessment of epidemiological and genetic quality of individual studies and of the overall epidemiological evidence of the meta-analyses are based on criteria that have been created by expert consensus but that need further prospective validation of their performance.
Implications for practice and future research
Summarised information from genetic association studies indicates that CYP2C19 has no major impact on the clinical efficacy of clopidogrel. There is no reliable and robust clinical justification to apply personalised treatment with clopidogrel tailored to CYP2C19 genotype.
Studies with a different design would be necessary to resolve the uncertainties and produce satisfactory answers to the role of CYP2C19 in clopidogrel activity. Specifically, the molecular mechanisms linking CYP2C19 activity to clinical effectiveness of clopidogrel should be explored in more detail. Moreover, results of large scale randomised controlled clinical trials that examine efficacy of clopidogrel in relation to the CYP2C19 allele carrier status would be less amenable to bias and could provide more reliable evidence.
What is already known on this topic
Genetic polymorphisms of the cytochrome P450 (CYP) 2C19 isoenzyme could affect the clinical efficacy of clopidogrel used for antiplatelet treatment
Current clinical practice and regulatory guidelines advise that tailored treatment with clopidogrel is guided by CYP2C19 genotype
What this study adds
Summary risk estimates from a systematic analysis indicate no effects or substantially biased effects of the CYP2C19 genotype on the occurrence of adverse cardiovascular events
The low overall epidemiological credibility of the associations makes all effect estimates and clinical inferences uncertain
There is no robust evidence to recommend individualised clopidogrel treatment driven by CYP2C19 genotype
Contributors: DT (principal investigator and guarantor) designed the study. TB and DT, with help from HJB, NFF, and JWvW, carried out literature searches, data entry, and data extraction. HJB, TB, and DT developed and wrote analysis scripts. All authors carried out the analyses. TB and DT wrote the manuscript, with contributions from HJB, NFF, and JWvW.
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Competing interests: All authors have completed the ICMJE uniform disclosure form at www.icmje.org/coi_disclosure.pdf (available on request from the corresponding author) and declare: no support from any organisation for the submitted work; JWvW has specified relationships with Accumetrics, Siemens, and The Medicines Company, and JMtB has specified relationships with Bristol-Myers Squibb, Eli Lilly, GlaxoSmithKline, Merck, Sanofi-Aventis, and Schering-Plough, that might have an interest in the submitted work in the previous three years; no other relationships or activities that could appear to have influenced the submitted work.
Ethical approval: Not required.
Data sharing: No additional data are available.
Cite this as: BMJ 2011;343:d4588
References
- 1.King SB 3rd, Smith SC Jr, Hirshfeld JW Jr, Jacobs AK, Morrison DA, Williams DO, et al. 2007 focused update of the ACC/AHA/SCAI 2005 guideline update for percutaneous coronary intervention: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines: 2007 writing group to review new evidence and update the ACC/AHA/SCAI 2005 guideline update for percutaneous coronary intervention, writing on behalf of the 2005 writing committee. Circulation 2008;117:261-95. [DOI] [PubMed] [Google Scholar]
- 2.Debnath B, Al-Mawsawi LQ, Neamati N. Are we living in the end of the blockbuster drug era? Drug News Perspect 2010;23:670-84. [DOI] [PubMed] [Google Scholar]
- 3.Snoep JD, Hovens MM, Eikenboom JC, van der Bom JG, Jukema JW, Huisman MV. Clopidogrel nonresponsiveness in patients undergoing percutaneous coronary intervention with stenting: a systematic review and meta-analysis. Am Heart J 2007;154:221-31. [DOI] [PubMed] [Google Scholar]
- 4.Sofi F, Marcucci R, Gori AM, Giusti B, Abbate R, Gensini GF. Clopidogrel non-responsiveness and risk of cardiovascular morbidity. An updated meta-analysis. Thromb Haemost 2010;103:841-8. [DOI] [PubMed] [Google Scholar]
- 5.Bonello L, Tantry US, Marcucci R, Blindt R, Angiolillo DJ, Becker R, et al. Consensus and future directions on the definition of high on-treatment platelet reactivity to adenosine diphosphate. J Am Coll Cardiol 2010;56:919-33. [DOI] [PubMed] [Google Scholar]
- 6.Breet NJ, van Werkum JW, Bouman HJ, Kelder JC, Ruven HJ, Bal ET, et al. Comparison of platelet function tests in predicting clinical outcome in patients undergoing coronary stent implantation. JAMA 2010;303:754-62. [DOI] [PubMed] [Google Scholar]
- 7.Kazui M, Nishiya Y, Ishizuka T, Hagihara K, Farid NA, Okazaki O, et al. Identification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab Dispos 2010;38:92-9. [DOI] [PubMed] [Google Scholar]
- 8.Hulot JS, Bura A, Villard E, Azizi M, Remones V, Goyenvalle C, et al. Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood 2006;108:2244-7. [DOI] [PubMed] [Google Scholar]
- 9.Damani SB, Topol EJ. The case for routine genotyping in dual-antiplatelet therapy. J Am Coll Cardiol 2010;56:109-11. [DOI] [PubMed] [Google Scholar]
- 10.Montalescot G, Hulot JS, Collet JP. Stent thrombosis: who’s guilty? Eur Heart J 2009;30:2685-8. [DOI] [PubMed] [Google Scholar]
- 11.Hulot J-S, Collet J-P, Silvain J, Pena A, Bellemain-Appaix A, Barthelemy O, et al. Cardiovascular risk in clopidogrel-treated patients according to cytochrome P450 2C19*2 loss-of-function allele or proton pump inhibitor coadministration: a systematic meta-analysis. J Am Coll Cardiol 2010;56:134-43. [DOI] [PubMed] [Google Scholar]
- 12.Sofi F, Giusti B, Marcucci R, Gori AM, Abbate R, Gensini GF. Cytochrome P450 2C19(*)2 polymorphism and cardiovascular recurrences in patients taking clopidogrel: a meta-analysis. Pharmacogenomics J 2011;11:199-206. [DOI] [PubMed] [Google Scholar]
- 13.Jin B, Ni HC, Shen W, Li J, Shi HM, Li Y. Cytochrome P450 2C19 polymorphism is associated with poor clinical outcomes in coronary artery disease patients treated with clopidogrel. Mol Biol Rep 2011;38:1697-702. [DOI] [PubMed] [Google Scholar]
- 14.Mega JL, Simon T, Collet JP, Anderson JL, Antman EM, Bliden K, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA 2010;304:1821-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Food and Drug Administration. FDA drug safety communication: reduced effectiveness of Plavix (clopidogrel) in patients who are poor metabolizers of the drug [safety announcement]. FDA, 2010.
- 16.Frere C, Cuisset T, Gaborit B, Alessi MC, Hulot JS. The CYP2C19*17 allele is associated with better platelet response to clopidogrel in patients admitted for non-ST acute coronary syndrome. J Thromb Haemost 2009;7:1409-11. [DOI] [PubMed] [Google Scholar]
- 17.Sibbing D, Gebhard D, Koch W, Braun S, Stegherr J, Morath T, et al. Isolated and interactive impact of common CYP2C19 genetic variants on the antiplatelet effect of chronic clopidogrel therapy. J Thromb Haemost 2010;8:1685-93. [DOI] [PubMed] [Google Scholar]
- 18.Wallentin L, James S, Storey RF, Armstrong M, Barratt BJ, Horrow J, et al. Effect of CYP2C19 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trial. Lancet 2010;376:1320-8. [DOI] [PubMed] [Google Scholar]
- 19.Pare G, Mehta SR, Yusuf S, Anand SS, Connolly SJ, Hirsh J, et al. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med 2010;363:1704-14. [DOI] [PubMed] [Google Scholar]
- 20.Bouman HJ, Schömig E, van Werkum JW, Velder J, Hackeng CM, Hirschhäuser C, et al. Paraoxonase-1 is a major determinant of clopidogrel efficacy. Nat Med 2011;17:110-6. [DOI] [PubMed] [Google Scholar]
- 21.Fleiss JL, Nee JCM, Landis JR. Large sample variance of kappa in the case of different sets of raters. Psychol Bull 1979;86:974-7. [Google Scholar]
- 22.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977;33:159-74. [PubMed] [Google Scholar]
- 23.Freemantle N, Calvert M, Wood J, Eastaugh J, Griffin C. Composite outcomes in randomized trials: greater precision but with greater uncertainty? JAMA 2003;289:2554-9. [DOI] [PubMed] [Google Scholar]
- 24.Cordoba G, Schwartz L, Woloshin S, Bae H, Gotzsche PC. Definition, reporting, and interpretation of composite outcomes in clinical trials: systematic review. BMJ 2010;341:c3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cutlip DE, Windecker S, Mehran R, Boam A, Cohen DJ, van Es GA, et al. Clinical end points in coronary stent trials: a case for standardized definitions. Circulation 2007;115:2344-51. [DOI] [PubMed] [Google Scholar]
- 26.Man M, Farmen M, Dumaual C, Teng CH, Moser B, Irie S, et al. Genetic variation in metabolizing enzyme and transporter genes. J Clin Pharmacol 2010;50:929-40. [DOI] [PubMed] [Google Scholar]
- 27.Yasui-Furukori N, Takahata T, Nakagami T, Yoshiya G, Inoue Y, Kaneko S, et al. Different inhibitory effect of fluvoxamine on omeprazole metabolism between CYP2C19 genotypes. Br J Clin Pharmacol 2004;57:487-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scholz I, Oberwittler H, Riedel KD, Burhenne J, Weiss J, Haefeli WE, et al. Pharmacokinetics, metabolism, and bioavailability of the triazole antifungal agent voriconazole in relation to CYP2C19 genotype. Br J Clin Pharmacol 2009;68:906-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoo HD, Cho HY, Lee YB. Population pharmacokinetic analysis of cilostazol in healthy subjects with genetic polymorphisms of CYP3A5, CYP2C19, and ABCB1. Br J Clin Pharmacol 2010;69:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sim SC, Risinger C, Dahl ML, Aklillu E, Christensen M, Bertilsson L, et al. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther 2006;79:103-13. [DOI] [PubMed] [Google Scholar]
- 31.Schenk PW, van Vliet M, Mathot RA, van Gelder T, Vulto AG, van Fessem MA, et al. The CYP2C19*17 genotype is associated with lower imipramine plasma concentrations in a large group of depressed patients. Pharmacogenomics J 2010;10:219-25. [DOI] [PubMed] [Google Scholar]
- 32.Lettre G, Lange C, Hirschhorn JN. Genetic model testing and statistical power in population-based association studies of quantitative traits. Genet Epidemiol 2007;31:358-62. [DOI] [PubMed] [Google Scholar]
- 33.Rudberg I, Mohebi B, Hermann M, Refsum H, Molden E. Impact of the ultrarapid CYP2C19*17 allele on serum concentration of escitalopram in psychiatric patients. Clin Pharmacol Ther 2008;83:322-7. [DOI] [PubMed] [Google Scholar]
- 34.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med 2009;360:354-62. [DOI] [PubMed] [Google Scholar]
- 35.Zintzaras E, Lau J. Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J Clin Epidemiol 2008;61:634-45. [DOI] [PubMed] [Google Scholar]
- 36.Attia J, Ioannidis JP, Thakkinstian A, McEvoy M, Scott RJ, Minelli C, et al. How to use an article about genetic association. B: are the results of the study valid? JAMA 2009;301:191-7. [DOI] [PubMed] [Google Scholar]
- 37.Wells GA, Shea B, O’Connell D, Peterson J, Welch V, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. 2011. www.ohri.ca/programs/clinical_epidemiology/oxford.htm.
- 38.Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 1992;48:361-72. [PubMed] [Google Scholar]
- 39.Ioannidis JP, Patsopoulos NA, Evangelou E. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One 2007;2:e841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kavvoura FK, Ioannidis JP. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Hum Genet 2008;123:1-14. [DOI] [PubMed] [Google Scholar]
- 41.Stephens M, Balding DJ. Bayesian statistical methods for genetic association studies. Nat Rev Genet 2009;10:681-90. [DOI] [PubMed] [Google Scholar]
- 42.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet 2004;74:367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ioannidis JP, Boffetta P, Little J, O’Brien TR, Uitterlinden AG, Vineis P, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol 2008;37:120-32. [DOI] [PubMed] [Google Scholar]
- 44.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539-58. [DOI] [PubMed] [Google Scholar]
- 45.Ioannidis JP, Patsopoulos NA, Evangelou E. Uncertainty in heterogeneity estimates in meta-analyses. BMJ 2007;335:914-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet 2001;29:306-9. [DOI] [PubMed] [Google Scholar]
- 47.Lau J, Antman EM, Jimenez-Silva J, Kupelnick B, Mosteller F, Chalmers TC. Cumulative meta-analysis of therapeutic trials for myocardial infarction. N Engl J Med 1992;327:248-54. [DOI] [PubMed] [Google Scholar]
- 48.Sterne JA, Egger M. Funnel plots for detecting bias in meta-analysis: guidelines on choice of axis. J Clin Epidemiol 2001;54:1046-55. [DOI] [PubMed] [Google Scholar]
- 49.Harbord RM, Egger M, Sterne JA. A modified test for small-study effects in meta-analyses of controlled trials with binary endpoints. Stat Med 2006;25:3443-57. [DOI] [PubMed] [Google Scholar]
- 50.Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA 2006;295:676-80. [DOI] [PubMed] [Google Scholar]
- 51.Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 2000;56:455-63. [DOI] [PubMed] [Google Scholar]
- 52.Patsopoulos NA, Evangelou E, Ioannidis JP. Sensitivity of between-study heterogeneity in meta-analysis: proposed metrics and empirical evaluation. Int J Epidemiol 2008;37:1148-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guyatt GH, Oxman AD, Kunz R, Vist GE, Falck-Ytter Y, Schunemann HJ. What is “quality of evidence” and why is it important to clinicians? BMJ 2008;336:995-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barbui C, Dua T, van Ommeren M, Yasamy MT, Fleischmann A, Clark N, et al. Challenges in developing evidence-based recommendations using the GRADE approach: the case of mental, neurological, and substance use disorders. PLoS Med 2010;7:pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mueller PS, Montori VM, Bassler D, Koenig BA, Guyatt GH. Ethical issues in stopping randomized trials early because of apparent benefit. Ann Intern Med 2007;146:878-81. [DOI] [PubMed] [Google Scholar]
- 56.Malek LA, Przyluski J, Spiewak M, Klopotowski M, Kostrzewa G, Kruk M, et al. Cytochrome P450 2C19 polymorphism, suboptimal reperfusion and all-cause mortality in patients with acute myocardial infarction. Cardiology 2010;117:81-7. [DOI] [PubMed] [Google Scholar]
- 57.Shuldiner AR, O’Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA 2009;302:849-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brackbill ML, Kidd RS, Abdoo AD, Warner JG Jr, Harralson AF. Frequency of CYP3A4, CYP3A5, CYP2C9, and CYP2C19 variant alleles in patients receiving clopidogrel that experience repeat acute coronary syndrome. Heart Vessels 2009;24:73-8. [DOI] [PubMed] [Google Scholar]
- 59.Mega JL, Thakuria JV, Cannon CP, Sabatine MS. Sequence variations in CYP metabolism genes and cardiovascular outcomes following treatment with clopidogrel: insights from the CLARITY-TIMI 28 genomic study (abstract 1017-69). J Am Coll Cardiol 2008;51(suppl A):A178-235. [Google Scholar]
- 60.Anderson JL, Mower CP, Home BD, Muhlestein JB, Park JJ, Bair TL, et al. Carriage of the CYP2C19*2 allele increases one-year risk of myocardial infarction among recipients of drug-eluting stents treated with clopidogrel (abstract 2903-8). J Am Coll Cardiol 2009;53(suppl A):A1-99. [Google Scholar]
- 61.Worrall A, Armesilla A, Norell M, Khogali S, Cusack M, Smallwood A, et al. The presence of the CYP p450 C19*2 allele is associated with impaired response to clopidogrel as measured by the verify now P2Y12 near-patient testing device in patients undergoing coronary angiography (abstract). Eur Heart J 2009;30(suppl):327. [Google Scholar]
- 62.Charisma Genomics Substudy. Evaluation of the CYP2C19 polymorphism in a prospective, randomized, placebo-controlled trial of chronic clopidogrel use for primary and secondary prevention. Annual meeting of transcatheter cardiovascular therapeutics, 24 September 2009, San Francisco, CA. www.cardiosource.org/news-media/meeting-coverage/tct/tct-2009.aspx.
- 63.Trenk D, Hochholzer W, Fromm MF, Chialda LE, Pahl A, Valina CM, et al. Cytochrome P450 2C19 681G. A polymorphism and high on-clopidogrel platelet reactivity associated with adverse 1-year clinical outcome of elective percutaneous coronary intervention with drug-eluting or bare-metal stents. J Am Coll Cardiol 2008;51:1925-34. [DOI] [PubMed] [Google Scholar]
- 64.Malek LA, Kisiel B, Spiewak M, Grabowski M, Filipiak KJ, Kostrzewa G, et al. Coexisting polymorphisms of P2Y12 and CYP2C19 genes as a risk factor for persistent platelet activation with clopidogrel. Circ J 2008;72:1165-9. [DOI] [PubMed] [Google Scholar]
- 65.Simon T, Verstuyft C, Mary-Krause M, Quteineh L, Drouet E, Meneveau N, et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med 2009;360:363-75. [DOI] [PubMed] [Google Scholar]
- 66.Collet JP, Hulot JS, Pena A, Villard E, Esteve JB, Silvain J, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet 2009;373:309-17. [DOI] [PubMed] [Google Scholar]
- 67.Sibbing D, Stegherr J, Latz W, Koch W, Mehilli J, Dorrler K, et al. Cytochrome P450 2C19 loss-of-function polymorphism and stent thrombosis following percutaneous coronary intervention. Eur Heart J 2009;30:916-22. [DOI] [PubMed] [Google Scholar]
- 68.Giusti B, Gori AM, Marcucci R, Saracini C, Sestini I, Paniccia R, et al. Relation of cytochrome P450 2C19 loss-of-function polymorphism to occurrence of drug-eluting coronary stent thrombosis. Am J Cardiol 2009;103:806-11. [DOI] [PubMed] [Google Scholar]
- 69.Sibbing D, Koch W, Gebhard D, Schuster T, Braun S, Stegherr J, et al. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation 2010;121:512-8. [DOI] [PubMed] [Google Scholar]
- 70.Tiroch KA, Sibbing D, Koch W, Roosen-Runge T, Mehilli J, Schomig A, et al. Protective effect of the CYP2C19 *17 polymorphism with increased activation of clopidogrel on cardiovascular events. Am Heart J 2010;160:506-12. [DOI] [PubMed] [Google Scholar]
- 71.Harmsze AM, van Werkum JW, Ten Berg JM, Zwart B, Bouman HJ, Breet NJ, et al. CYP2C19*2 and CYP2C9*3 alleles are associated with stent thrombosis: a case-control study. Eur Heart J 2010;31:3046-53. [DOI] [PubMed] [Google Scholar]
- 72.Sawada T, Shinke T, Shite J, Honjo T, Haraguchi Y, Nishio R, et al. Impact of cytochrome P450 2C19*2 polymorphism on intra-stent thrombus after drug-eluting stent implantation in Japanese patients receiving clopidogrel. Circ J 2010;75:99-105. [DOI] [PubMed] [Google Scholar]
- 73.Akey JM, Zhang K, Xiong M, Doris P, Jin L. The effect that genotyping errors have on the robustness of common linkage-disequilibrium measures. Am J Hum Genet 2001;68:1447-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Clarke TA, Waskell LA. The metabolism of clopidogrel is catalyzed by human cytochrome P450 3A and is inhibited by atorvastatin. Drug Metab Dispos 2003;31:53-9. [DOI] [PubMed] [Google Scholar]
- 75.Lau WC, Gurbel PA, Watkins PB, Neer CJ, Hopp AS, Carville DG, et al. Contribution of hepatic cytochrome P450 3A4 metabolic activity to the phenomenon of clopidogrel resistance. Circulation 2004;109:166-71. [DOI] [PubMed] [Google Scholar]
- 76.Charlot M, Ahlehoff O, Norgaard ML, Jorgensen CH, Sorensen R, Abildstrom SZ, et al. Proton-pump inhibitors are associated with increased cardiovascular risk independent of clopidogrel use: a nationwide cohort study. Ann Intern Med 2010;153:378-86. [DOI] [PubMed] [Google Scholar]
- 77.Bhatt DL, Cryer BL, Contant CF, Cohen M, Lanas A, Schnitzer TJ, et al. Clopidogrel with or without omeprazole in coronary artery disease. N Engl J Med 2010;363:1909-17. [DOI] [PubMed] [Google Scholar]
- 78.Poole C, Greenland S. Random-effects meta-analyses are not always conservative. Am J Epidemiol 1999;150:469-75. [DOI] [PubMed] [Google Scholar]
- 79.Wiviott SD, Braunwald E, McCabe CH, Montalescot G, Ruzyllo W, Gottlieb S, et al. Prasugrel versus clopidogrel in patients with acute coronary syndromes. N Engl J Med 2007;357:2001-15. [DOI] [PubMed] [Google Scholar]
- 80.Buonamici P, Marcucci R, Migliorini A, Gensini GF, Santini A, Paniccia R, et al. Impact of platelet reactivity after clopidogrel administration on drug-eluting stent thrombosis. J Am Coll Cardiol 2007;49:2312-7. [DOI] [PubMed] [Google Scholar]
- 81.Sibbing D, Braun S, Morath T, Mehilli J, Vogt W, Schomig A, et al. Platelet reactivity after clopidogrel treatment assessed with point-of-care analysis and early drug-eluting stent thrombosis. J Am Coll Cardiol 2009;53:849-56. [DOI] [PubMed] [Google Scholar]
- 82.Wallentin L, Becker RC, Budaj A, Cannon CP, Emanuelsson H, Held C, et al. Ticagrelor versus clopidogrel in patients with acute coronary syndromes. N Engl J Med 2009;361:1045-57. [DOI] [PubMed] [Google Scholar]
- 83.Yusuf S, Zhao F, Mehta SR, Chrolavicius S, Tognoni G, Fox KK. Effects of clopidogrel in addition to aspirin in patients with acute coronary syndromes without ST-segment elevation. N Engl J Med 2001;345:494-502. [DOI] [PubMed] [Google Scholar]