Abstract
Oxygen provides a crucial energy source in eukaryotic cells. Hence, eukaryotes ranging from yeast to humans have developed sophisticated mechanisms to respond to changes in oxygen levels. Regulation of protein localization, like protein modifications, can be an effective mechanism to control protein function and activity. However, the contribution of protein localization in oxygen signaling has not been examined on a genomewide scale. Here, we examine how hypoxia affects protein distribution on a genomewide scale in the model eukaryote, the yeast Saccharomyces cerevisiae. We demonstrate, by live cell imaging, that hypoxia alters the cellular distribution of 203 proteins in yeast. These hypoxia-redistributed proteins include an array of proteins with important functions in various organelles. Many of them are nuclear and are components of key regulatory complexes, such as transcriptional regulatory and chromatin remodeling complexes. Under hypoxia, these proteins are synthesized and retained in the cytosol. Upon reoxygenation, they relocalize effectively to their normal cellular compartments, such as the nucleus, mitochondria, endoplasmic reticulum, and cell periphery. The resumption of the normal cellular locations of many proteins can occur even when protein synthesis is inhibited. Furthermore, we show that the changes in protein distribution induced by hypoxia follow a slower trajectory than those induced by reoxygenation. These results show that the regulation of protein localization is a common and potentially dominant mechanism underlying oxygen signaling and regulation. These results may have broad implications in understanding oxygen signaling and hypoxia responses in higher eukaryotes such as humans.
Keywords: oxygen signaling, live cell imaging, nuclear import
most eukaryotic cells rely on aerobic respiration to generate ATP for cellular energy supply. Hence, living organisms ranging from yeast to humans need to respond effectively to the changes in oxygen levels in the environment (6, 48). In humans, hypoxia is a widely occurring condition associated with an array of disease states, such as cancer and stroke (15, 39, 48). Hypoxia elicits numerous acute and adaptive responses at the molecular and cellular levels, such as changes in gene expression (6, 48). To gain a global understanding of hypoxia-induced molecular events, many labs have applied genomewide approaches. Particularly, several groups, including ours, have performed a series of detailed studies to characterize hypoxia-induced genomewide changes in gene expression by combining microarray gene expression profiling with computational analyses (24, 25, 29, 31, 40). Such studies have provided useful insights into transcriptional regulatory programs. Likewise, a number of proteomic studies have been performed to identify changes in protein levels induced by altered oxygen levels in various organisms (5, 10, 11, 17, 30, 37, 47). However, such studies are limited by the sensitivity of the techniques used, mainly shotgun proteomic or even less sensitive methods like two-dimensional differential gel electrophoresis (2D DIGE) (1). They often identify changes in abundant enzymes, structural proteins, and chaperones, but they rarely identify low-level regulatory proteins such as nuclear proteins and signaling proteins (5, 10, 11, 17, 30, 37, 47). Hence, more sensitive methods that can directly monitor nuclear and signaling proteins should enable us to glean important regulatory and signaling events.
Crucially, the distribution of proteins in different cell organelles is a major factor in determining the functions of many proteins (4, 34, 42, 43). Particularly, the regulated distribution of many important proteins between the nucleus and cytosol is known to be critical for development, differentiation, and transformation. A variety of signals including hormones and growth factors, cytokines, cell-cycle signals, and stress can elicit changes in nucleocytoplasmic distribution (34, 42, 43). Such changes in turn alter the activities of a wide array of signaling and regulatory proteins. These regulated proteins include tumor suppressors (e.g., BRCA1 and p53), signaling transducers (e.g., Pho4, FOXO1, and APC), steroid receptors (e.g., androgen receptor and glucocorticoid receptor), cell cycle regulators (e.g., Swi5, Swi6, and cyclin D), developmental regulators (e.g., Dorsal and SRY), and immune regulators (e.g., NF-AT, NF-κB p65, and NF-κB p105) (34, 42, 43). Furthermore, emerging evidence shows that protein localization to other organelles can be regulated (16). For example, endoplasmic reticulum (ER) stress attenuates the translocation of secretory and membrane proteins in a signal sequence-selective manner (22). Clearly, regulation of protein distribution in cell organelles can be a major mechanism controlling a wide array of physiological and pathological processes.
Hypoxia is known to induce broad changes in gene expression in eukaryotes. For example, it can change the transcript levels of over 1,000 genes in yeast and mammalian cells (24, 25, 29, 31, 40). It is conceivable that hypoxia may induce changes in protein distribution, and such changes may contribute to the changes in gene expression and other processes. Many years ago, the potential effect of hypoxia on the Golgi apparatus was noted by Cajal (8). Hence, we decided to identify hypoxia-induced changes in protein distribution on a genomewide scale. We took advantage of a comprehensive collection of over 4,000 green fluorescent protein (GFP)-tagged yeast strains generated by Huh and colleagues (19). Each of the strains expresses a yeast protein tagged with GFP at the COOH terminus, under the control of their natural promoters. By using fluorescent microscopy, we monitored the distribution of all expressed proteins in live normoxic and hypoxic cells. We found that hypoxia selectively altered the cellular distribution patterns of 203 proteins. Among them, 121 are nuclear proteins. Many are components of important regulatory protein complexes, such as the SAGA, Swi/Snf, and transcription factor II H (TFIIH) complexes. Over 30 mitochondrial proteins also exhibited different cellular distribution patterns in hypoxic cells. Other hypoxia-relocalized proteins include those in ER, cell periphery, and vacuoles. Evidently, in hypoxic cells, the proteins accumulated in the cytosol. In response to increased oxygen levels, they localize to their normal cellular compartments. These findings not only reveal novel mechanisms governing oxygen signaling and regulation, but also provide a global, genomewide picture of the extent of protein redistribution elicited by a major nutritional and environmental signal, oxygen.
MATERIALS AND METHODS
Yeast strains and antibodies.
The yeast GFP clone collection of 4,159 strains expressing GFP-tagged proteins (19) was purchased from Invitrogen. Red fluorescent protein (RFP)-tagged haploid marker strains were provided by Dr. Erin K. O'Shea's lab (Harvard University, Cambridge, MA). The anti-GFP monoclonal antibody was purchased from Santa Cruz Biotechnology.
The creation of hypoxic growth conditions.
Hypoxic [∼10 parts per billion (ppb) O2] growth condition was created by using a hypoxia chamber (Coy Laboratory,) and by filling the chamber with a mixture of 5% H2 and 95% N2 in the presence of palladium catalyst (18). The oxygen level in the chamber was monitored by using the model 10 gas analyzer (Coy Laboratory Products). The precise level of oxygen was also estimated by using a CHEMetrics rhodazine oxygen detection kit (K-7511) with the minimum detection limit at 1 ppb, and a range of 0–20 ppb. The hypoxic state was further confirmed by measuring oxygen-controlled promoter activities, including UAS1/CYC1, ANB1, and OLE1 (18, 20, 28).
Fluorescent live cell imaging and screening.
The screen to identify proteins whose cellular distribution was altered by hypoxia was performed in three stages. The initial screen of 4,159 strains was performed by visually viewing and comparing the GFP fluorescent images of live cells grown in air vs. hypoxia (in agar plates containing synthetic complete medium lacking histidine), using a multichannel Zeiss Axio Observer.Z1 fluorescent microscope with a Zeiss ×100 oil immersion lens (numerical aperture 0.55, working distance 0.26 mm). Briefly, GFP-tagged cells on 96-well plates were transferred to a new set of plates by using a 96-well pin tool. Colonies were grown large enough, in air or in a hypoxia chamber, to allow a sufficient number of cells to be picked for microscopic viewing. Positive strains which showed different locations in hypoxic vs. normoxic cells were selected. Then, new stocks containing the initial positive strains were made on 96-well plates.
Subsequently, we captured fluorescent live cell (hypoxic and normoxic) images of the initially identified positive strains by using a high-speed AxioCam MRm Rev3 monochrome camera. The software used was Carl Zeiss AxioVision Rel 4.8.1, set to the multidimensional mode that allows automatic filter changes and image capturing. The image resolution is 1,388 × 1,040 pixels without binning and with fast acquisition. All images acquired from each channel have the maximum image size of 2.8 MB. The captured images were viewed to assess the effect of hypoxia on the locations of GFP-tagged proteins. In the follow-up confirmation screen, we stained live cells briefly with 4,6-diamidino-2-phenylindole (DAPI), before collecting them and capturing GFP and DAPI fluorescent images. For hypoxic cells, the staining was performed in a hypoxia chamber. The cells were quenched on ice before being taken out of the chamber, and imaged by using the aforementioned fluorescent microscope and camera. Normoxic cells were grown in air, stained, and then imaged in the same way as hypoxic cells. Three researchers examined these images to confirm those GFP-tagged proteins that exhibited altered locations in hypoxic and normoxic cells. For each condition, at least 25 cells were examined and counted to determine the percentage of cells expressing GFP-tagged proteins in various cellular compartments, and three sets of cells were counted independently. Owing to the large number of strains, we selected only those yeast strains that repeatedly showed different patterns of GFP fluorescence in hypoxic vs. normoxic cells. Cells with excessive bright or weak fluorescence or with aberrant morphology were not scored. None of the proteins that show cell-cell variability (19) were selected in our screen, although we did not deliberately rule them out. This approach should minimize false positives, but it is possible that some proteins that may have changed locations in response to hypoxia were missed.
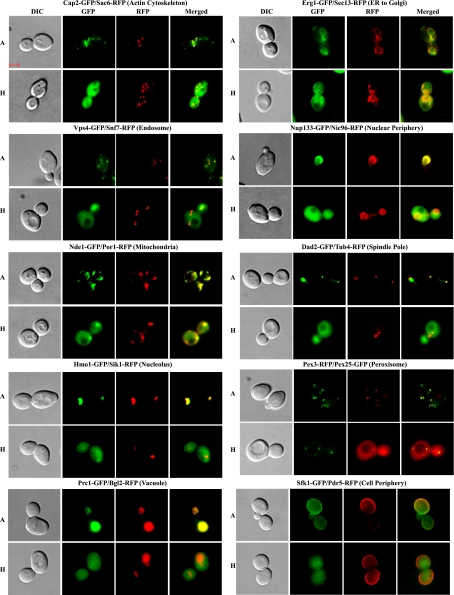
To further verify the subcellular localization changes in hypoxic vs. normoxic cells, we performed colocalization experiments. Briefly, we generated diploid or haploid yeast cells that express a GFP-tagged protein whose location was found to be altered in hypoxic cells and a RFP-tagged protein whose location was unaffected by hypoxia. Diploid strains were generated by mating a GFP-tagged yeast strain with a RFP-tagged marker strain obtained from the O'Shea lab. For mitochondria, spindle pole, cell periphery, and vacuole, there was no available marker strain expressing a RFP-tagged protein whose localization was not affected by hypoxia. In these cases, we constructed new strains that express a GFP-tagged protein whose cellular localization was affected by hypoxia and a reference RFP-tagged protein whose cellular localization was not affected by hypoxia. For peroxisome, the tags were reversed (see Fig. 5). These strains were generated by transforming GFP-tagged strains with PCR products with gene-specific cassettes containing a COOH-terminally positioned RFP tag followed by the KanMX6 described previously (19). The GFP tag may affect localization of some proteins; but the vast majority of the proteins appeared to be not affected (19). We think that the effect of hypoxia on the majority of the proteins should be biologically relevant.
Fig. 5.
DIC, GFP, red fluorescent protein (RFP), and merged fluorescent images of cells expressing a hypoxia-redistributed and an unaffected protein in various organelles. Cells expressing a pair of GFP- and RFP-tagged proteins were grown in air (A) or under hypoxia (H), and the images were captured and are shown here. Except for the proteins showing a peroxisome location in air, the hypoxia-redistributed proteins were tagged with GFP, while the ones unaffected by hypoxia were tagged with RFP. For peroxisome, the RFP-tagged protein was affected by hypoxia, while the GFP-tagged protein was not. The scale bar represents 2 μm. The cellular locations of proteins in normoxic cells are also indicated at top.
For a quantitative characterization of the time courses of nuclear protein localization in response to hypoxia or reoxygenation, we used a previously defined nuclear protein import assay in yeast (13, 26, 41). Briefly, cells at various time points of hypoxia or reoxygenation treatment were collected, and images were acquired. At least 25 cells were counted at each time point, and three sets of cells were counted. A particular cell was counted as having the GFP-tagged protein in the nucleus if the nucleus was much brighter than the surrounding cytoplasm and a clear nuclear-cytoplasmic boundary was visible. For a comparison, we also monitored and plotted the time course of the effect of 200 ng/ml rapamycin on Sfp1 nuclear localization. Cells with excessive bright or weak fluorescence or with aberrant morphology were not scored.
Cycloheximide treatment.
A total of 50 μg/ml cycloheximide was added to yeast cells grown in air or in a hypoxic chamber.
Preparation of yeast cell extracts and Western blotting.
Yeast cells expressing various GFP-tagged proteins were grown to an optical density (OD600) of ∼0.8. Cells were harvested and resuspended in three packed cell volumes of buffer (20 mM Tris, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 0.3 M NaCl, 1 mM phenylmethylsulfonyl fluoride, 1 mg of pepstatin per ml, 1 mg of leupeptin per ml). Cells were then permeabilized by agitation with four packed cell volumes of glass beads, and extracts were collected as described previously (49). Protein concentrations were determined by the bicinchoninic acid (BCA) protein assay kit (Pierce).
For Western blotting, ∼100 μg of whole cell extracts were first separated on sodium dodecyl sulfate (SDS)-8% polyacrylamide gels and then transferred to polyvinylidene difluoride or nitrocellulose membranes (Bio-Rad). GFP-tagged proteins were detected by using a monoclonal antibody against GFP and a chemiluminescence Western blotting kit (Roche Diagnostics). The signals were detected by using a Kodak image station 4000MM Pro with the molecular imaging software (version 4.5).
Protein gene ontology analysis and construction of the network map.
The analysis of functional categories of relocalized proteins was performed on Funspec (http://funspec.med.utoronto.ca/). P values were calculated using the hypergeometric distribution, with Bonferroni correction. For constructing the protein network map, the Cytoscape application program (http://www.cytoscape.org/) was used. The 203 hypoxia-relocalized proteins were mapped according to their gene ontology (GO) terms. They were obtained by using the Saccharomyces Genome Database (SGD) Gene Ontology Slim Mapper Web Tool set to “Macromolecular Complex terms: Components” on the SGD website. The mapped output file was reformatted into a Cytoscape-compatible network file, and the network map was created. The network map was further graphically refined by using the Canvas application program. The subcellular compartments of these proteins in normoxic cells were designated based on data from the O'Shea lab (19).
RESULTS
Hypoxia selectively alters the cellular distribution of 203 proteins in diverse cell organelles.
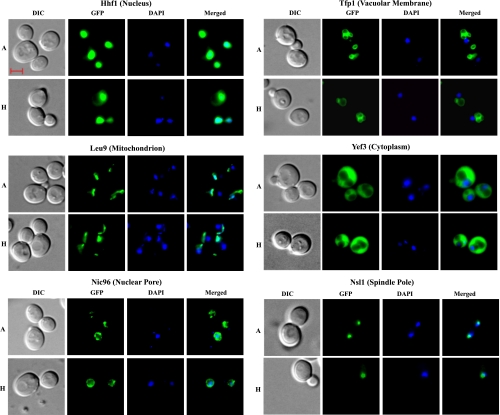
To identify proteins whose cellular distribution was altered in hypoxic cells, we used the comprehensive collection of over 4,159 yeast GFP-tagged strains generated previously by Huh et al. (19). Each of the strains expresses a GFP-tagged protein from its natural promoter and chromosomal location. We used fluorescent microscopy to monitor the distribution of proteins in normoxic and hypoxic cells. To capture the protein distribution patterns in hypoxic cells, we used a multichannel Zeiss fluorescent microscope with an AxioCam camera (see materials and methods). We screened for the proteins that exhibited different cellular distribution patterns in hypoxic and normoxic cells in three stages (see materials and methods). In the final stage, cells were stained briefly with DAPI, before imaging. Then, the differential interference contrast (DIC), DAPI, and GFP fluorescent images of normoxic and hypoxic live cells were captured. We found that the vast majority of proteins did not show changes in their cellular distribution in hypoxic cells, compared with normoxic cells. Figure 1 shows several examples of such proteins in different organelles, including the nucleus, nuclear pore, cytosol, mitochondria, and vacuolar membrane. Clearly, hypoxia did not cause dramatic changes in the morphology of these organelles or nonspecific changes in protein distribution in general.
Fig. 1.
Examples of differential interference contrast (DIC), green fluorescent protein (GFP), 4,6-diamidino-2-phenylindole (DAPI), and merged fluorescent images of cells in which the distribution patterns of GFP-tagged proteins are not affected by hypoxia. Cells were grown in air (A) or under hypoxia (H), and the images were captured. The scale bar represents 2 μm. The cellular locations of proteins in normoxic cells are also indicated at top.
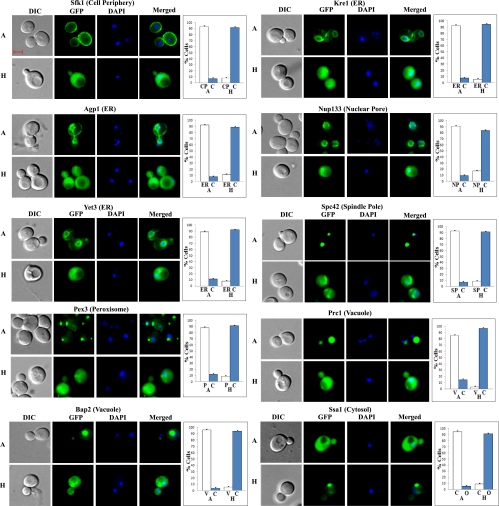
However, the distribution of 203 proteins did clearly change in hypoxic cells, compared with normoxic cells. Figure 2 shows several examples of nuclear proteins that redistributed to the cytosol in hypoxic cells. Figure 3 shows several examples of mitochondrial proteins that redistributed in response to hypoxia, while Fig. 4 shows several examples of redistributed proteins from other organelles. Furthermore, we show that RFP-tagged marker proteins, Sac6 (actin cytoskeleton), Pdr5 (cell periphery), Sec13 (ER to Golgi), Snf7 (endosome), Por1 (mitochondria), Nic96 (nuclear periphery), Sik1 (nucleolus), Tub4 (spindle pole), and Bgl2 (vacuole) in different organelles did not alter their distribution in hypoxic cells, in contrast to the hypoxia-redistributed proteins tagged with GFP (Fig. 5). For peroxisome, we show that Pex3-RFP localization was affected by hypoxia, while Pex25-GFP was not affected.
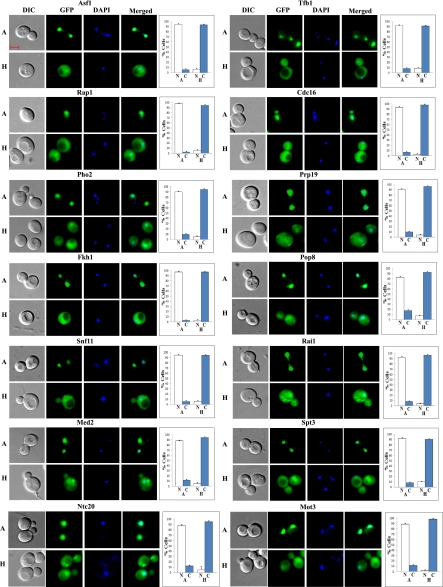
Fig. 2.
Examples of DIC, GFP, DAPI, and merged fluorescent images of cells expressing hypoxia-redistributed proteins that are nuclear in normoxic cells. Cells were grown in air (A) or under hypoxia (H), and the images were captured. The number of cells showing GFP-tagged proteins in the nucleus (N) or cytosol (C) was counted and the percentage was calculated and is plotted. The scale bar represents 2 μm.
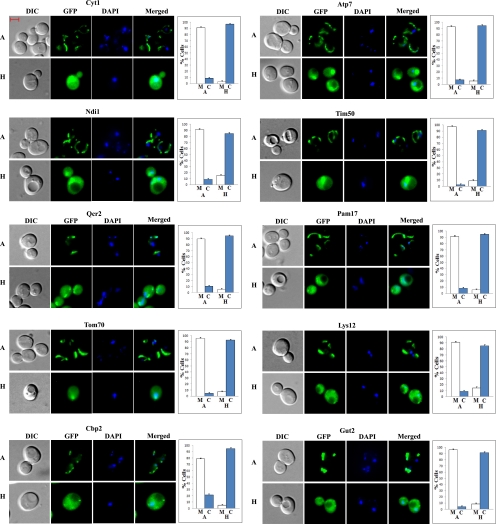
Fig. 3.
Examples of DIC, GFP, DAPI, and merged fluorescent images of cells expressing hypoxia-redistributed proteins that are mitochondrial in normoxic cells. Cells were grown in air (A) or under hypoxia (H), and the images were captured. The number of cells showing GFP-tagged proteins in the mitochondria (M) or cytosol (C) was counted and the percentage was calculated and is plotted. The scale bar represents 2 μm.
Fig. 4.
Examples of DIC, GFP, DAPI, and merged fluorescent images of cells expressing hypoxia-redistributed proteins that are in organelles other than the nucleus or mitochondria in normoxic cells. Cells were grown in air (A) or under hypoxia (H), and the images were captured. The number of cells showing GFP-tagged proteins in the cell periphery (CP), cytosol (C), endoplasmic reticulum (ER), nuclear pore (NP), peroxisome (P), spindle pole (SP), vacuole (V), or other cellular locations (O) was counted and the percentage was calculated and is plotted. The scale bar represents 2 μm. The cellular locations of proteins in normoxic cells are also indicated at top.
Besides several predominantly cytosolic heat shock proteins, Ssa1, Ssa2, and Hsp104 (see Table S1; Supplemental Material for this article is available online at the Journal website), the dominant pattern of protein redistribution appeared to be as follows: under hypoxic conditions, proteins in the nucleus, mitochondria, and other organelles generally did not localize to their presumed cellular compartments but accumulated in the cytosol (see Figs. 2–5). This observation raises the possibility that the change may be attributable to the degradation of proteins and the resulting accumulation of GFP fragments in the cytosol in hypoxic cells. Although this possibility seems unlikely since GFP-tagged proteins are often very stable, we tested it by using Western blotting with anti-GFP antibodies. We found that all detected GFP-tagged proteins appeared to be predominantly in their complete, full-length forms in hypoxic cells (see Fig. S1 in the supplemental material), even after undergoing the rigorous disruptive procedure for preparing extracts.
Hypoxia-redistributed proteins are predominantly involved in nuclear and mitochondrial functions.
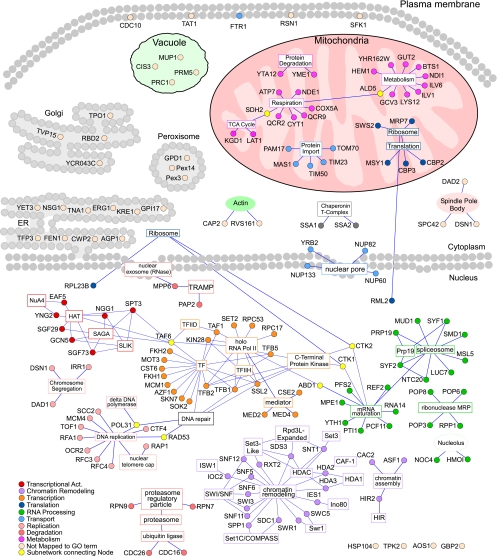
To gain insights into the global molecular events underlying hypoxia responses, we performed GO and network analyses of the hypoxia-redistributed proteins. As shown in Table 1, more than half (121) of the redistributed proteins are nuclear in normoxic cells. About half of those are involved in transcriptional control (Supplemental Table S2). Many of these nuclear proteins are components of previously characterized regulatory complexes mediating the regulation of diverse genes, such as the SAGA complex, the Swi/Snf complex, the NuA4 histone acetyltransferase complex, the SRB mediator complex, the polyadenylation complex, and the related ribonuclease MRP complex (2, 3, 7, 9, 12, 14, 27, 35, 36, 44, 45). The clustering of the hypoxia-redistributed proteins into several major functional groups becomes even clearer when we plotted their network map (Fig. 6). The nuclear proteins form four clusters: those involved in chromatin remodeling, in DNA replication and repair, in transcriptional regulatory and Pol II complexes, and in mRNA maturation, RNA processing, and spliceosome. The second largest class of hypoxia-redistributed proteins is mitochondrial proteins (Table 1 and Fig. 6). Three functional clusters of proteins appear to be selectively affected by hypoxia: those involved in the generation of cellular energy, including enzymes of TCA cycle and respiratory metabolism, those involved in mitochondrial protein import, and those involved in ribosome and translation (Fig. 6).
Table 1.
Summary of the location and functional categories of hypoxia-relocalized proteins
|
Location | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cyto | Cyto + Nuc | ER | Mito | Nuc | Nuc Peri | Spin Pole | Vac | Cell Peri | Others |
| 3 | 5 | 12 | 32 | 121 | 4 | 3 | 4 | 6 | 13 |
|
Functional Category | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Essen | Trans | DNA binding | Chrom | DNA repair | Splic | mRNA | rRNA | Protein localiz | Perm | Respir |
| 55 | 65 | 38 | 16 | 14 | 16 | 25 | 24 | 10 | 4 | 12 |
Listed are the numbers of relocalized proteins in each location or functional category.
Cyto, cytoplasm; Nuc, nucleus; ER, endoplasmic reticulum; Mito, mitochondrion; Nuc Peri, nuclear periphery; Spin pole, spindle pole; Vac, vacuole; Cell Peri, cell periphery; Essen, essential; Trans, transcription control; Chrom, chromatin modification, Splic, splicing; mRNA, mRNA cleavage and processing; rRNA, rRNA metabolic process; Protein localiz, protein localization; Perm, amino acid permeases; Respir, respiration.
P value <0.05, see materials and methods.
Fig. 6.
Graphical representation of protein-protein interaction networks for hypoxia-redistributed proteins. The information on the biochemical interactions and complex formation of the 203 hypoxia-redistributed proteins were downloaded from the Saccharomyces Genome Database (SGD) database, and then imported to Cytoscape for network construction. The proteins (shown as round nodes in different colors based on their cellular functions) are placed in their respective cellular compartments in normoxic cells. The gene ontology (GO) terms for protein complexes or functional categorizations are indicated and are shown in square nodes. Nodes of the same subnetworks are colored similarly, and a key for the coloring of the nodes is shown. Lines represent an association of the protein with a particular complex or functional GO term.
Half of the redistributed proteins recover their normoxic distribution in response to reoxygenation even in the absence of protein synthesis.
To further probe the mechanisms by which proteins redistribute in response to changes in oxygen levels, we monitored protein distribution following the shift of cells from hypoxic to normoxic growth conditions, and vice versa. Proteins likely sense oxygen by binding to oxygen, and the increase of oxygen levels can be readily controlled, whereas oxygen depletion is more difficult to control. We therefore reasoned that monitoring changes during the shift of hypoxic to normoxic growth conditions may be more reflective of mechanisms of oxygen sensing and regulation. This is indeed the case, as our results showed. We found that 184 of 203 redistributed proteins recovered their normoxic distribution or locations 2 h (<1 generation) after cells were shifted from hypoxic to normoxic growth conditions. More than half of these proteins (103 out of 184) recovered their normoxic distribution, even when protein synthesis was inhibited by the potent protein synthesis inhibitor cycloheximide (38).
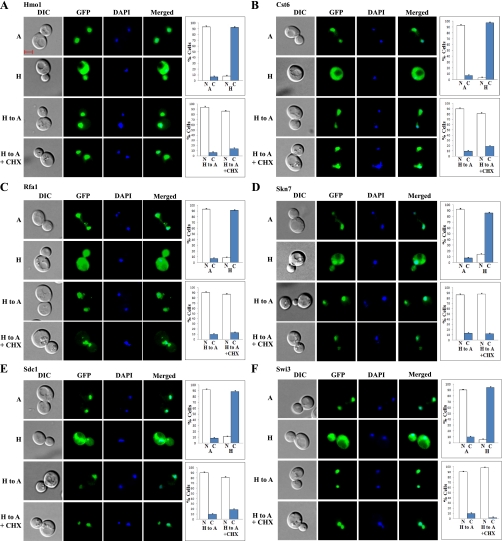
Figure 7 shows six examples of the redistributed proteins, Hmo1, Cst6, Rfa1, Skn7, Sdc1, and Swi3, which recovered their normoxic distribution in the absence of protein synthesis. These proteins were nuclear in normoxic cells, as shown in the first panel (marked “A”) of images in Fig. 7, A–F. They were retained in the cytosol in hypoxic cells, as shown in the second panel (marked “H”) of images in Fig. 7, A–F. When cells were shifted from hypoxic to normoxic growth conditions, the proteins recovered their normoxic, nuclear distribution within 2 h after the shift, as shown in the third panel (marked “H to A”) of images in Fig. 7, A–F. This recovery of normoxic distribution occurred even in the presence of cycloheximide, as shown in the fourth panel (marked “H to A + CHX”) of images in Fig. 7, A–F. Evidently, cycloheximide had very little, if any, effect on the recovery of nuclear localization of these proteins, in response to reoxygenation. These results demonstrate that the retained proteins in the cytosol in hypoxic cells can be imported to the nucleus readily upon reoxygenation. They also show that redistributed proteins in the cytosol in hypoxic cells were not degraded. For controls and comparison, we showed six examples of redistributed proteins Asf1, Fkh1, Mot3, Snf5, Dsn1, and Gcn5, which required new protein synthesis to recover their normoxic distribution (Supplemental Fig. S2, A–F). Like those shown in Fig. 7, reoxygenation caused the recovery of nuclear localization of these proteins. However, these proteins cannot be reimported to the nucleus without newly synthesized proteins, as shown in the fourth panel (marked “H to A + CHX”) of images in Supplemental Fig. S2, A–F.
Fig. 7.
Examples of images of cells expressing redistributed nuclear proteins that recovered their normoxic nuclear locations in response to increased oxygen levels even in the presence of cycloheximide. Cells expressing GFP-tagged Hmo1 (A), Cst6 (B), Rfa1 (C), Skn7 (D), Sdc1 (E), and Swi3 (F), respectively, were grown under hypoxic conditions and then shifted to normoxic conditions for 2 h in the presence or absence of cycloheximide (CHX). The images of cells grown under different conditions were captured. A, images of cells grown in air; H, images of cells grown in a hypoxia chamber; H to A, images of cells grown in a hypoxia chamber and then shifted to air for 2 h; H to A + CHX, images of cells grown in a hypoxia chamber and then shifted to air for 2 h in the presence of cycloheximide. The number of cells showing GFP-tagged proteins in the nucleus (N) or cytosol (C) was counted and the percentage was calculated and is plotted. The scale bar represents 2 μm.
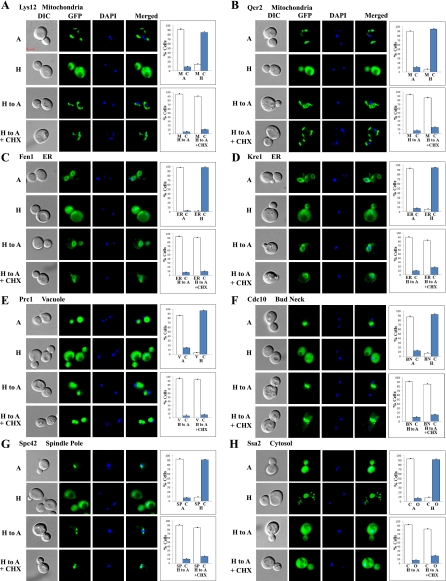
The vast majority of nonnuclear proteins recovered their normoxic locations 2 h after shifting from hypoxic to normoxic growth conditions, and they did so even in the presence of cycloheximide. All but three redistributed mitochondrial proteins, Ndi1, Gut2, and Cbp2 (Supplemental Table S1), recovered their normoxic distribution even in the absence of protein synthesis; see Lys12 and Qcr2 in Fig. 8, A and B, for examples. Remarkably, a majority of redistributed ER, cell periphery, and other proteins recovered their normoxic distribution patterns even in the presence of cycloheximide. The images of some proteins are shown in Fig. 8, C–H. Two proteins in ER, Fen1 and Kre1 (Fig. 8, C and D), recovered the ringlike ER pattern in the presence of cycloheximide. Likewise, vacuolar protein Prc1 (Fig. 8E), bud neck protein Cdc10 (Fig. 8F), spindle pole protein Spc42 (Fig. 8G), and cytosolic protein Ssa2 (Fig. 8H) recovered their normoxic distribution patterns after shifting from hypoxic to normoxic growth conditions in the presence of cycloheximide. These results demonstrate that redistributed proteins of diverse cell organelles can readily recover their cellular distribution or locations upon reoxygenation.
Fig. 8.
Examples of images of cells expressing redistributed nonnuclear proteins that recovered their normoxic nuclear locations in response to increased oxygen levels even in the presence of cycloheximide. Cells expressing GFP-tagged mitochondrial proteins Lys12 (A) and Qcr2 (B), ER proteins Fen1 (C) and Kre1 (D), vacuolar protein Prc1 (E), bud neck protein Cdc10 (F), spindle pole protein Spc42 (G), and cytosolic protein Ssa2 (H), respectively, were grown under hypoxic conditions and then shifted to normoxic conditions for 2 h in the presence or absence of cycloheximide. The images of cells grown under different conditions were captured. A, images of cells grown in air; H, images of cells grown in a hypoxia chamber; H to A, images of cells grown in a hypoxia chamber and then shifted to air for 2 h; H to A + CHX, images of cells grown in a hypoxia chamber and then shifted to air for 2 h in the presence of cycloheximide. The number of cells showing GFP-tagged proteins in the cytosol (C), mitochondria (M), vacuole (V), ER, bud neck (BN), spindle pole (SP), or other cellular locations (O) was counted and the percentage was calculated and is plotted. The scale bar represents 2 μm.
Molecular events induced by hypoxia are not reciprocal to those induced by reoxygenation.
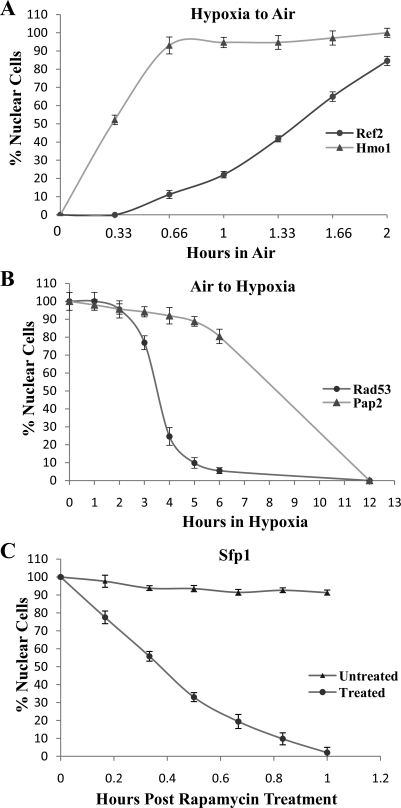
In living organisms in the wilderness, cells invariably experience fluctuations in oxygen levels and need to respond to both hypoxia and reoxygenation. Thus, it is interesting to examine cellular responses to both increasing oxygen levels and hypoxia. We therefore followed the time course of protein redistribution when cells were shifted from normoxic to hypoxic growth conditions. Notably, changes in protein distribution were significantly slower when cells were shifted from normoxic to hypoxic conditions, compared with the hypoxic to normoxic shift, likely because of the lag time required to exhaust bound oxygen in cells. When cells were shifted from hypoxic to normoxic growth conditions, proteins recovered their normoxic cellular locations within 2 h. Using a previously developed nuclear protein import assay (13, 26, 41), we performed a quantitative analysis of the time courses of those nuclear proteins in response to hypoxia or reoxygenation. Figure 9A shows two representative examples. One class of proteins, including Hmo1 (shown in Fig. 9A) and 10 other proteins, recovered their nuclear locations in <1 h. The rest of the proteins, exemplified by Ref2 (Fig. 9A), recovered their nuclear locations in 2 h. In contrast, longer times were required for the proteins to adopt their cytosolic locations in response to hypoxia. Figure 9B shows two examples of the time courses of protein accumulation in the cytosol in response to hypoxia. Only partial hypoxic patterns were observed 3 h after shifting for all proteins. One class of proteins, including Rad53 and 23 other proteins, adopted cytosolic locations within 6 h after shifting of cells from normoxic to hypoxic growth conditions. The rest of the proteins, exemplified by Pap2 shown in Fig. 9B, adopted mostly cytosolic locations after shifting for 12 h.
Fig. 9.
Time course characteristics of protein relocalization elicited by hypoxia, reoxygenation, or rapamycin. A: time courses of relocalization of Ref2 and Hmo1 in response to reoxygenation. Cells expressing Ref2-GFP or Hmo1-GFP were grown under hypoxia and then shifted to normoxic growth conditions. At various time points, cells were imaged, and the number of cells showing GFP-tagged proteins in the nucleus (N) or cytosol (C) was counted. The percentage of cells showing nuclear locations is plotted. B: time courses of relocalization of Rad53 and Pap2 in response to hypoxia. Cells expressing Rad53-GFP or Pap2-GFP were grown in air and then shifted to hypoxic growth conditions. At various time points, cells were imaged, and the number of cells showing GFP-tagged proteins in the nucleus (N) or cytosol (C) was counted. The percentage of cells showing nuclear locations was calculated and is plotted. C: time course of Sfp1 protein relocalization in response to rapamycin. Cells expressing Sfp1-GFP were grown in the absence or presence of 200 ng/ml rapamycin for various times. Cells were imaged, and the number of cells showing GFP-tagged proteins in the nucleus (N) or cytosol (C) was counted. The percentage of cells showing nuclear locations is plotted here.
For a comparison, we examined the time course of the effect of rapamycin on Sfp1 localization. Figure 9C shows that rapamycin caused Sfp1 to relocalize within 1 h of treatment. These results show that oxygen can act, at a rate comparable to rapamycin, to induce changes in protein localization. This result, showing that changes induced by reoxygenation occur much more quickly than those induced by hypoxia, is consistent with a previous time course experiment of the transcriptome response to hypoxia and subsequent reoxygenation (25). Supplemental Figure S3 shows the time courses of transcriptome changes in response to hypoxia and reoxygenation based on principal component analysis, which suggests that changes induced by air occur much more quickly than changes induced by hypoxia.
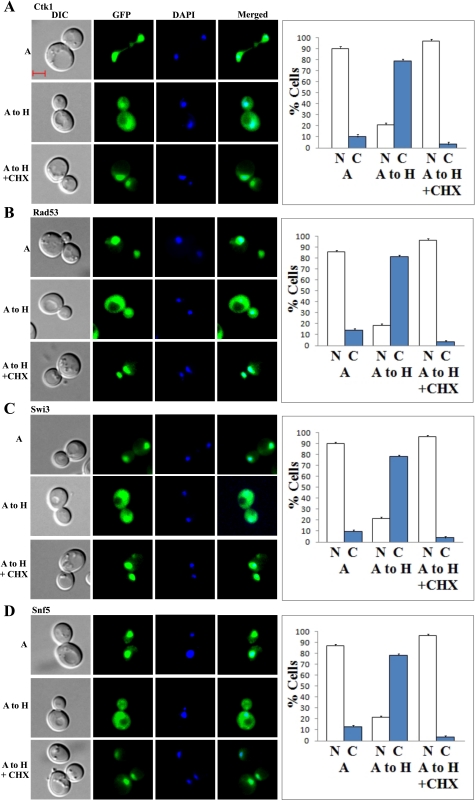
To further compare the effects of hypoxia and reoxygenation, we also examined the effect of cycloheximide on protein localization changes induced by hypoxia. Figure 10, A–D, shows four examples of proteins that exhibited complete hypoxic distribution patterns 6 h after shifting from normoxic to hypoxic growth conditions. Strikingly, the hypoxia-induced redistribution of Ctk1, Rad53, Snf5, and Swi3 was inhibited by cycloheximide. In fact, the hypoxia-induced redistribution of most proteins was inhibited by cycloheximide. Even partial redistribution of proteins was inhibited by cycloheximide. This is a stark contrast to the effect of shifting cells from hypoxic to normoxic growth conditions, where the redistribution of a majority of proteins (103 out of 183 in 2 h) was not inhibited by cycloheximide (see Figs. 7 and 8). It appeared that all presynthesized nuclear proteins in air (Figs. 10, A–D) remained in the nucleus under hypoxia. In the presence of cycloheximide, cell division was likely slowed down. Thus, nuclear proteins would not be diluted by cell division as much as in the absence of cycloheximide, and can be observed. This result also unequivocally supports the conclusion that proteins do not redistribute to the cytosol under hypoxia due to protein degradation and diffusion of degraded proteins to the cytosol. Together, these results show that changes induced by hypoxia and reoxygenation are asymmetric, and they follow different paths.
Fig. 10.
A–D: examples of images of cells expressing redistributed nuclear proteins when shifted from normoxic to hypoxic growth conditions. Cells expressing GFP-tagged Ctk1 (A), Rad53 (B), Swi3 (C), and Snf5 (D), respectively, were grown under normoxic conditions and then shifted to hypoxic conditions for 6 h in the presence or absence of cycloheximide. A, images of cells grown in air; A to H, images of cells grown in air and then shifted to a hypoxia chamber for 6 h; A to H + CHX, images of cells grown in air and then shifted to a hypoxia chamber for 6 h in the presence of cycloheximide. The number of cells showing GFP-tagged proteins in the nucleus (N) or cytosol (C) was counted and the percentage was calculated and is plotted here. The scale bar represents 2 μm.
DISCUSSION
In this report, we describe a genomewide study to examine the effect of oxygen on protein distribution in live yeast cells. We discovered that 203 proteins in various cellular compartments redistributed in response to hypoxia. Evidently, when most of these proteins were synthesized in hypoxic cells, they were not transported to their presumed organelles. Rather, they were retained in the cytosol. In response to oxygen, they were then localized to their presumed normoxic locations. These results suggest novel mechanisms of oxygen signaling and protein localization in eukaryotes.
Previous studies have shown that hypoxia causes broad molecular and cellular changes in diverse eukaryotic cells ranging from yeast to mammalian cells. Particularly, hypoxia causes changes in the expression of a wide array of genes. For example, nearly 20% of yeast genes change their transcript levels in response to changes in oxygen levels (24). In the human arterial endothelial cells, >8% of all genes alter their transcript levels by at least 1.5-fold in response to hypoxia (29). In primary human astrocytes, >5% of the genes alter their transcript levels by at least twofold in response to hypoxia (31). What is the molecular basis underlying hypoxia-induced changes in thousands of genes in diverse eukaryotic cells? Our results presented here may shed some light on this question.
The most prominent hypoxia-induced change in protein localization is that over 100 nuclear proteins relocalize to the cytosol in hypoxic cells. The results suggest that hypoxia and reoxygenation may cause changes in the organization and thereby the functional property of important macromolecular complexes controlling gene expression and other important processes. For example, in hypoxic cells, it appears that a number of components of the Swi/Snf and SAGA complexes (Fig. 6), which play global roles in the regulation of many genes, are retained in the cytosol. The lack of certain components of these complexes in the nucleus would alter their composition and organization. Consequently, this would cause a change in the functional properties of these complexes and affect the target genes which they regulate. Consistent with this idea, the hypoxia-induced changes in protein localization appear to precede the changes in transcriptome (25). In response to air or reoxygenation, changes in protein localization are completed in less than one generation (2 h, Fig. 9B), while changes in transcriptome are largely completed in two generations (25). Likewise, in response to hypoxia, changes in protein localization are completed in about three generations (12 h, Fig. 9A), while changes in transcriptome are completed in six generations (25). Very likely, changes induced by hypoxia or reoxygenation in protein localization contribute, at least partially, to the broad changes induced in gene expression.
Another significant change in hypoxia-induced protein relocalization is that many mitochondrial proteins were synthesized but were retained in the cytosol in hypoxic cells. In response to reoxygenation, all but two of the proteins relocalized to mitochondria in 2 h, as the nuclear proteins shown in Fig. 9B. These proteins include many directly involved in energy generation, such as enzymes in TCA cycle and oxidative phosphorylation, as well as those involved in mitochondrial protein import (Fig. 6). The mitochondrion is an organelle dedicated to cellular energy production by using oxygen. When oxygen is limiting, as in hypoxic cells, there is reduced need for mitochondrial enzymes. Hence, these mitochondrial proteins may not need to be imported, and are retained in the cytosol. Furthermore, the presynthesis and accumulation of these proteins/enzymes in the cytosol and rapid import into the mitochondria would ensure that cells can readily recover and generate respiratory energy upon reoxygenation. This mechanism allows cells to respond to changes in oxygen levels effectively, and maximize the capacity of cells to utilize oxygen.
Besides nuclear and mitochondrial proteins, the experiments here show that proteins can be translocated from the cytosol into other organelles, posttranslationally and directly. It is well documented that protein transport to the nucleus, mitochondria, and peroxisomes often occurs posttranslationally and directly from the cytosol (32, 46). However, protein transport to other organelles, specifically ER and cell periphery, occurs generally cotranslationally (33, 46). Our results here unexpectedly identified an array of proteins that can be localized posttranslationally from the cytosol to diverse organelles, in response to oxygen. As listed in Supplemental Table S3, four cell periphery proteins (Cdc10, Ftr1, Sfk1, and Tat1), one endosomal protein (Vps4), nine ER proteins (Agp1, Cwp2, Erg1, Fen1, Kre1, Tna1, Yet3, Tpo1, and Erg25), one Golgi protein (YCR043C), and three vacuolar proteins (Mup1, Prc1, and Prm5) can be transported from the cytosol (their location in hypoxic cells) directly to their corresponding organelles, in response to reoxygenation. Our results also support the emerging idea that the regulation of protein localization can be an important regulatory mechanism (16), and identify a series of proteins whose activity can be regulated by modulating their localization. The regulation of protein localization can be an efficient mechanism to respond and adapt to changes in oxygen levels, as well as other environmental and cellular cues.
This mechanism of rapid response and adaptation may have general implications in higher eukaryotes. Notably, of the 203 identified yeast proteins that are found to be relocalized in response to hypoxia, 121 have conserved human homologues (Supplemental Table S4). It is plausible that some of these conserved human proteins can also change their cellular locations in response to hypoxia. Consistent with this idea, recent evidence shows that the Swi/Snf complex, which is highly conserved from yeast to humans, plays an important role in mediating oxygen signaling and hypoxia responses in mammalian cells (21, 23). Our results here may provide some clues about whether and how the mammalian homologues of hypoxia-relocalized proteins may contribute to oxygen signaling and regulation. It is likely that various groups of mammalian proteins, like those identified yeast proteins, can relocalize and alter their properties and functions, in response to hypoxia and reoxygenation.
GRANTS
This work was supported by National Institutes of Health Grant GM62246 (to L. Zhang).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dr. Erin K. O'Shea's lab for providing the RFP-tagged marker strains. We are indebted to Mr. Steve Lianoglou and Dr. Christina Leslie (Memorial Sloan-Kettering Cancer Center) for providing computational assistance in this project, for doing the transcriptome analysis in Supplemental Fig. S3, and for helpful comments on the manuscript.
REFERENCES
- 1. Aggarwal K, Choe LH, Lee KH. Shotgun proteomics using the iTRAQ isobaric tags. Brief Funct Genomic Proteomic 5: 112–120, 2006 [DOI] [PubMed] [Google Scholar]
- 2. Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J 18: 5108–5119, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ansari SA, He Q, Morse RH. Mediator complex association with constitutively transcribed genes in yeast. Proc Natl Acad Sci USA 106: 16734–16739, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borgese N, Driessen AJ, Rapaport D, Robinson C. The quest for a better resolution of protein-translocation processes. Conference on the Control, Co-ordination and Regulation of Protein Targeting and Translocation. EMBO Rep 10: 337–342, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bruckmann A, Hensbergen PJ, Balog CI, Deelder AM, Brandt R, Snoek IS, Steensma HY, van Heusden GP. Proteome analysis of aerobically and anaerobically grown Saccharomyces cerevisiae cells. J Proteomics 71: 662–669, 2009 [DOI] [PubMed] [Google Scholar]
- 6. Bunn HF, Poyton RO. Oxygen sensing and molecular adaptation to hypoxia. Physiol Rev 76: 839–885, 1996 [DOI] [PubMed] [Google Scholar]
- 7. Cairns BR, Kim YJ, Sayre MH, Laurent BC, Kornberg RD. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA 91: 1950–1954, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cajal SR. Algunas variaciones fisiologicas y patologicas del aparato reticular de Golgi. In: Trabajos del Laboratorio de investigaciones Biolo'gicas de la Universidad de Madrid. Madrid: 1914, p. 127–227 [Google Scholar]
- 9. Chamberlain JR, Lee Y, Lane WS, Engelke DR. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev 12: 1678–1690, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Datta A, Park JE, Li X, Zhang H, Ho ZS, Heese K, Lim SK, Tam JP, Sze SK. Phenotyping of an in vitro model of ischemic penumbra by iTRAQ-based shotgun quantitative proteomics. J Proteome Res 9: 472–484, 2010 [DOI] [PubMed] [Google Scholar]
- 11. De Groot MJ, Daran-Lapujade P, van Breukelen B, Knijnenburg TA, de Hulster EA, Reinders MJ, Pronk JT, Heck AJ, Slijper M. Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveals post-transcriptional regulation of key cellular processes. Microbiology 153: 3864–3878, 2007 [DOI] [PubMed] [Google Scholar]
- 12. Esakova O, Krasilnikov AS. Of proteins and RNA: the RNase P/MRP family. RNA 16: 1725–1747, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fahrenkrog B, Hubner W, Mandinova A, Pante N, Keller W, Aebi U. The yeast nucleoporin Nup53p specifically interacts with Nic96p and is directly involved in nuclear protein import. Mol Biol Cell 11: 3885–3896, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636, 2006 [DOI] [PubMed] [Google Scholar]
- 15. Giaccia AJ, Simon MC, Johnson R. The biology of hypoxia: the role of oxygen sensing in development, normal function, and disease. Genes Dev 18: 2183–2194, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hegde RS, Kang SW. The concept of translocational regulation. J Cell Biol 182: 225–232, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Helbig AO, de Groot MJ, van Gestel RA, Mohammed S, de Hulster EA, Luttik MA, Daran-Lapujade P, Pronk JT, Heck AJ, Slijper M. A three-way proteomics strategy allows differential analysis of yeast mitochondrial membrane protein complexes under anaerobic and aerobic conditions. Proteomics 9: 4787–4798, 2009 [DOI] [PubMed] [Google Scholar]
- 18. Hon T, Dodd A, Dirmeier R, Gorman N, Sinclair PR, Zhang L, Poyton RO. A mechanism of oxygen sensing in yeast: multiple oxygen-responsive steps in the heme biosynthetic pathway affect Hap1 activity. J Biol Chem 278: 50771–50780, 2003 [DOI] [PubMed] [Google Scholar]
- 19. Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature 425: 686–691, 2003 [DOI] [PubMed] [Google Scholar]
- 20. Jiang Y, Vasconcelles MJ, Wretzel S, Light A, Gilooly L, McDaid K, Oh CS, Martin CE, Goldberg MA. Mga2p processing by hypoxia and unsaturated fatty acids in Saccharomyces cerevisiae: impact on LORE-dependent gene expression. Eukaryot Cell 1: 481–490, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson AB, Barton MC. Hypoxia-induced and stress-specific changes in chromatin structure and function. Mutat Res 618: 149–162, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kang SW, Rane NS, Kim SJ, Garrison JL, Taunton J, Hegde RS. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell 127: 999–1013, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kenneth NS, Mudie S, van Uden P, Rocha S. SWI/SNF regulates the cellular response to hypoxia. J Biol Chem 284: 4123–4131, 2009 [DOI] [PubMed] [Google Scholar]
- 24. Kundaje A, Xin X, Lan C, Lianoglou S, Zhou M, Zhang L, Leslie C. A predictive model of the oxygen and heme regulatory network in yeast. PLoS Comput Biol 4: e1000224, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lai LC, Kosorukoff AL, Burke PV, Kwast KE. Metabolic-state-dependent remodeling of the transcriptome in response to anoxia and subsequent reoxygenation in Saccharomyces cerevisiae. Eukaryot Cell 5: 1468–1489, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leslie DM, Timney B, Rout MP, Aitchison JD. Studying nuclear protein import in yeast. Methods 39: 291–308, 2006 [DOI] [PubMed] [Google Scholar]
- 27. Li Y, Bjorklund S, Jiang YW, Kim YJ, Lane WS, Stillman DJ, Kornberg RD. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc Natl Acad Sci USA 92: 10864–10868, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lowry CV, Zitomer RS. ROX1 encodes a heme-induced repression factor regulating ANB1 and CYC7 of Saccharomyces cerevisiae. Mol Cell Biol 8: 4651–4658, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 105: 659–669, 2005 [DOI] [PubMed] [Google Scholar]
- 30. Mary J, Rogniaux H, Rees JF, Zal F. Response of Alvinella pompejana to variable oxygen stress: a proteomic approach. Proteomics 10: 2250–2258, 2010 [DOI] [PubMed] [Google Scholar]
- 31. Mense SM, Sengupta A, Zhou M, Lan C, Bentsman G, Volsky DJ, Zhang L. Gene expression profiling reveals the profound upregulation of hypoxia-responsive genes in primary human astrocytes. Physiol Genomics 25: 435–449, 2006 [DOI] [PubMed] [Google Scholar]
- 32. Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem 76: 723–749, 2007 [DOI] [PubMed] [Google Scholar]
- 33. Nickel W, Seedorf M. Unconventional mechanisms of protein transport to the cell surface of eukaryotic cells. Annu Rev Cell Dev Biol 24: 287–308, 2008 [DOI] [PubMed] [Google Scholar]
- 34. Poon IK, Jans DA. Regulation of nuclear transport: central role in development and transformation? Traffic 6: 173–186, 2005 [DOI] [PubMed] [Google Scholar]
- 35. Rino J, Carmo-Fonseca M. The spliceosome: a self-organized macromolecular machine in the nucleus? Trends Cell Biol 19: 375–384, 2009 [DOI] [PubMed] [Google Scholar]
- 36. Rodriguez-Navarro S. Insights into SAGA function during gene expression. EMBO Rep 10: 843–850, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ruiz-Romero C, Calamia V, Rocha B, Mateos J, Fernandez-Puente P, Blanco FJ. Hypoxia conditions differentially modulate human normal and osteoarthritic chondrocyte proteomes. J Proteome Res 9: 3035–3045, 2010 [DOI] [PubMed] [Google Scholar]
- 38. Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol 6: 209–217, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Semenza GL. Hypoxia-inducible factor 1 and cancer pathogenesis. IUBMB Life 60: 591–597, 2008 [DOI] [PubMed] [Google Scholar]
- 40. Seta KA, Spicer Z, Yuan Y, Lu G, Millhorn DE. Responding to hypoxia: lessons from a model cell line. SciSTKE 2002: re11, 2002 [DOI] [PubMed] [Google Scholar]
- 41. Shulga N, Roberts P, Gu Z, Spitz L, Tabb MM, Nomura M, Goldfarb DS. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J Cell Biol 135: 329–339, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sorokin AV, Kim ER, Ovchinnikov LP. Nucleocytoplasmic transport of proteins. Biochemistry (Mosc) 72: 1439–1457, 2007 [DOI] [PubMed] [Google Scholar]
- 43. Thompson ME. BRCA1 16 years later: nuclear import and export processes. FEBS J 277: 3072–3078, 2010 [DOI] [PubMed] [Google Scholar]
- 44. Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718, 2009 [DOI] [PubMed] [Google Scholar]
- 45. Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev 10: 2117–2130, 1996 [DOI] [PubMed] [Google Scholar]
- 46. Wickner W, Schekman R. Protein translocation across biological membranes. Science 310: 1452–1456, 2005 [DOI] [PubMed] [Google Scholar]
- 47. Yang HY, Kwon J, Cho EJ, Choi HI, Park C, Park HR, Park SH, Chung KJ, Ryoo ZY, Cho KO, Lee TH. Proteomic analysis of protein expression affected by peroxiredoxin V knock-down in hypoxic kidney. J Proteome Res 9: 4003–4015, 2010 [DOI] [PubMed] [Google Scholar]
- 48. Zhang H, Semenza GL. The expanding universe of hypoxia. J Mol Med 86: 739–746, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang L, Hach A, Wang C. Molecular mechanism governing heme signaling in yeast: a higher-order complex mediates heme regulation of the transcriptional activator HAP1. Mol Cell Biol 18: 3819–3828, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.