Abstract
Here we report and validate a simple method for measuring intracellular activities of glial glutamine synthetase (GS) and glutaminase (GLNase) in intact glial cells. These enzymes are responsible for glutamate and glutamine recycling in the brain, where glutamate and glutamine transport from the blood stream is strongly limited by the blood-brain barrier. The intracellular levels of glutamate and glutamine are dependent on activities of numerous enzymatic processes, including 1) cytosolic production of glutamine from glutamate by GS, 2) production of glutamate from glutamine by GLNase that is primarily localized between mitochondrial membranes, and 3) mitochondrial conversion of glutamate to the tricarboxylic cycle intermediate α-ketoglutarate in the reactions of oxidative deamination and transamination. We measured intracellular activities of GS and GLNase by quantifying enzymatic interconversions of l-[3H]glutamate and l-[3H]glutamine in cultured rat astrocytes. The intracellular substrate and the products of enzymatic reactions were separated in one step using commercially available anion exchange columns and quantified using a scintillation counter. The involvement of GS and GLNase in the conversion of 3H-labeled substrates was verified using irreversible pharmacological inhibitors for each of the enzymes and additionally validated by measuring intracellular amino acid levels using an HPLC. Overall, this paper describes optimized conditions and pharmacological controls for measuring GS and GLNase activities in intact glial cells.
Keywords: glutamate-glutamine cycle, metabolism, astrocytes
l-glutamate and l-glutamine are two major intermediates of nitrogen metabolism. These two amino acids are present at high micromolar (∼30–500 μM) levels in the circulatory system. They diffuse freely from capillaries to the interstitial liquids and extracellular space and are taken inside animal cells by several types of amino acid transporters (24). In cytosol and mitochondrial matrix, concentrations of glutamate and glutamine are in the millimolar range. These two amino acids exist in equilibrium because they are consumed and produced in numerous interrelated biochemical reactions. The abundance and the generic roles of glutamate and glutamine in metabolism and protein synthesis interfere with the signaling properties of glutamate in the central nervous system (CNS). In the brain, glutamate serves as the main excitatory neurotransmitter. Therefore, extracellular levels of glutamate have to be tightly regulated and kept at low levels (reviewed in Refs. 9 and 14). This is possible due to two major factors: 1) limited transport of glutamate and glutamine via the endothelial blood-brain barrier (33), and 2) continuous recycling of glutamate and glutamine in the so-called glutamate-glutamine cycle as originally proposed in Refs. 3 and 42 and reviewed in Refs. 2, 5, and 20.
Under normal physiological conditions, neuronal release from synaptic vesicles serves as the major source of extracellular glutamate in the CNS. Once released in the extracellular space, glutamate activates post- and presynaptic glutamate receptors. After neuronal stimulation, glutamate levels rapidly decline due to diffusion from synaptic cleft and fast removal by the Na+-K+-dependent glutamate transporters, predominantly by the astrocytic transporters, glutamate transporter 1 (GLT1)/EAAT2 and glutamate aspartate transporter (GLAST)/EAAT1 (9). In astrocytes, glutamate is converted to glutamine by the ATP-dependent enzyme glutamine synthetase (GS) (19, 20). During the normal neuronal signaling, up to 80–90% of synaptically released glutamate is recycled via the GS activity (17). Astrocytic glutamine is released predominantly by the amino acid transporter SNAT3/SN1 but also by SNAT5/SN2 and antiport via LAT2; extracellular glutamine is then taken into neuronal cells, predominantly by the SNAT1/ATA1 at the level of nerve terminals, and SNAT2 in neuronal bodies (5, 8, 10). In neurons, glutamine is hydrolyzed back to glutamate by glutaminase (GLNase), mainly by its phosphate-activated “kidney” isoform (2, 18). The resulting glutamate is repackaged into synaptic vesicles, hence completing the glutamate-glutamine cycle, or alternatively diverted to biosynthetic and metabolic reactions (2, 9, 20).
The activities of GS and GLNase and consequently glutamate-glutamine recycling in the brain tissue can be strongly modulated in a number of neural pathologies. For example, in stroke and ischemia the GS activity is strongly reduced due to the drop in intracellular ATP levels but also via oxidative damage and proteolytic degradation (23, 28). This may indirectly contribute to the dramatic increases in extracellular glutamate levels seen in the ischemic brain (4, 13, 44). At least some forms of human epilepsy and animal epilepsy models are associated with loss of the GS protein in hippocampus that may contribute to increased extracellular glutamate levels and hyperexcitability (12, 15). It has been reported that pharmacological inhibition of GS in the rat hippocampus produces recurrent seizures resembling epilepsy (11). In hyperammonemia and hepatic encephalopathy, excessive flux of ammonia from circulation to the CNS leads to markedly increased glutamine production by astrocytes, astroglial swelling, and brain edema, potentially leading to coma and death (22, 37, 41). Systemic supplementation of the GS inhibitor methionine sulfoximine (MSO) strongly reduces mortality in animal hyperammonemia models (45), likely via attenuation of cerebral edema (38). Oxidative damage and reduced activity of GS have also been reported in the aging brain (see for example Refs. 6 and 26).
Growing understanding of the pathological impact of impaired glutamate and glutamine metabolism increased interest in mechanistic studies measuring individual activities of the enzymes involved in metabolism and recycling of these amino acids, particularly GS and GLNase. Yet, studying individual enzymatic reactions in cultured cells or more complex model systems is quite challenging because of rapid intracellular turnover of glutamate and glutamine and intimate connection of these amino acids to bioenergetic and biosynthetic processes. In complex systems, including in vivo, enzymatic activities can be estimated via quantifying metabolic conversions of 13C-labeled compounds by a nuclear magnetic resonance spectroscopy (see for example Refs. 21, 34, and 43). However, such an approach requires sophisticated equipment and data processing. More accessible methodology for accurate measurements of GS and GLNase activities involves quantification of the enzymatic conversion of 3H- or 14C-labeled amino acids (25, 27). Yet, the existing radiolabeled amino acid assays were developed for purified proteins or tissue homogenates. In the present paper, we identified experimental conditions that are suitable for determining the GS and GLNase activities in intact cultured astrocytes and proposed a number of solutions to pitfalls associated with the whole cell assays. For instance, GS and GLNase inhibitors strongly interfere with glutamine transport and intracellular glutamine levels but can be used in the preincubation media because they act irreversibly. We performed our study in astrocytes because these cells nearly exclusively express GS in the brain (19). However, a technique developed by us may be readily adapted to study modulation of GS and GLNase activities in other cell types and perhaps more complex tissue preparations.
MATERIALS AND METHODS
Materials.
l-Glutamate, l-glutamine, 6-diazo-5-oxo-l-norleucine (DON), MSO, 2-deoxy-d-glucose, and rotenone were purchased from Sigma-Aldrich (St. Louis, MO). l-[3H]glutamate and l-[3H]glutamine were acquired from PerkinElmer-New England Nuclear (Waltham, MA). All cell culture media and sera were obtained from Invitrogen (Carlsbad, CA). All salts, buffers, solvents, and other reagents were from Sigma-Aldrich unless otherwise specified.
Preparation of primary astrocyte cultures.
Confluent primary astrocyte cultures were prepared from brain cortical tissue of 1- to 2-day-old Sprague-Dawley rats. All animal procedures were approved by the Albany Medical Center Institutional Animal Use and Care Committee. Pups were euthanized by rapid decapitation. The cerebral cortices were dissected from the meninges, hippocampi, and basal ganglia and transferred to ice-cold OptiMEM medium (Invitrogen). Cortical tissue collected from four animals was minced and transferred into 10 ml of a solution of the recombinant protease TrypLE (Invitrogen), which was diluted 1:1 (vol:vol) with OptiMEM. Cells were extracted at 37°C using three 10-min incubations with TrypLE additionally supplemented with bovine pancreatic DNAse I (Sigma). The first extraction was discarded, while the second and the third extractions were combined with minimal essential medium (MEM) containing 10% heat-inactivated horse serum (HIHS) and 50 U/ml penicillin plus 50 μg/ml streptomycin (Pen-Strep). After each extraction cells were sedimented by a brief centrifugation (1,000 g for 1.5 min) and then resuspended in MEM-HIHS. Dissociated cells were seeded on poly-l-lysine-coated T75 culture flasks (Techno Plastic Products, TPP, Trasadingen, Switzerland) at the density of 250,000 cells/flask. Cultures were grown for 2–3 wk in MEM-HIHS + Pen-Strep at 37°C in a humidified atmosphere of 5% CO2-95% air. The culture medium was changed twice a week. Culture purity was periodically verified by staining with antibody recognizing specific astrocytic marker, glial fibrillary acidic protein (GTAP, Sigma); ≥98% of cells were GFAP positive. Confluent cells were replated as necessary on 6- or 12-well tissue culture plates (TPP) or 18-mm square coverslips (Carolina Biological, Burlington, NC).
Assay of glutamine synthetase activity.
The activity of GS was quantified as intracellular conversion of l-[3H]glutamate to l-[3H]glutamine. Because the GS and the subsequent GLNase assays are the subject of the present methodological paper, we describe them in a step-by-step manner with brief comments on the significance of each step.
Astrocytes grown in six-well plates were washed from the culture media three times with HEPES-buffered basal solution of the following composition (in mM): 135 NaCl, 3.8 KCl, 1.2 MgSO4, 1.3 CaCl2, 1.2 KH2PO4, 10 d-glucose, 10 HEPES (pH = 7.4). This was necessary to remove extracellular amino acids, particularly 2 mM glutamine that is present in cell culture media. All the subsequent steps were performed at 37°C in an air atmosphere in a water-jacketed incubator.
Cells were preincubated at 37°C in basal medium for 40 min with the irreversible GLNase inhibitor 1 mM DON (46). At this concentration and duration of treatment DON irreversibly blocked GLNase activity by >75% and prevented reverse conversion of glutamine to glutamate. As seen in results, this degree of inhibition was sufficient for specific measurements of GS activity. DON could not be present in the subsequent steps because it strongly interferes with transport of amino acids (see results).
Cells were washed from DON two times with 2 ml of basal solution and transferred into 2 ml of the GS reaction medium that was prepared on the basis of basal with addition of 2 μCi/ml of l-[3H]glutamate (final concentration adjusted to 2 μM with unlabeled l-glutamate) and 100 μM of (NH4)2SO4 ([NH4+/NH3] = 200 μM). Ammonium sulfate was added to provide sufficient NH4+ levels for the GS reaction. Cells were incubated in this reaction mix for 30 min at 37°C.
The reaction was terminated and extracellular isotope was removed by three consecutive washes with 2 ml of ice-cold basal solution.
One milliliter of milliQ H2O was added to each well to lyse astrocytes; cells were scraped and sonicated for 4 min using Branson 200 Ultrasonic Cleaner. Lysates were clarified by rapid centrifugation (4 min × 12,100 g, room temperature).
Each cell lysate (1 ml) was added onto activated AG 1-X8 Polyprep column (Bio-Rad, Hercules, CA) for anion exchange separation of l-[3H]glutamate and l-[3H]glutamine. Column content was eluted with three 2-ml volumes of milliQ H2O, followed by three 2-ml volumes of 0.1 M HCl. Water elution removed uncharged l-[3H]glutamine, while subsequent acid elution extracted negatively charged l-[3H]glutamate as well as its metabolites [such as α-ketoglutarate and other intermediates of the mitochondrial tricarboxylic acid cycle (TCA)].
Eluent fractions were collected into scintillation vials and 4 ml of scintillation cocktail (Ecoscint A, National Diagnostics, Atlanta, GA) were added to each vial. 3H content was measured in a Tri-Carb 1900TR Liquid Scintillation Analyzer. The GS activity was calculated as percent conversion of l-[3H]glutamate into l-[3H]glutamine, which was normalized to the total 3H recovered from each sample. This was done using a simple formula: % conversion = [(dpms in “glutamine” fractions nos. 1–3)/(dpms in “glutamine” fractions nos. 1–3) + (dpms in “glutamate” fractions no. 4–6)] × 100%.
Assay of glutaminase activity.
The activity of GLNase was quantified as the intracellular conversion of l-[3H]glutamine to l-[3H]glutamate.
Confluent astrocyte cultures grown in six-well plates were initially washed from culture media three times with HEPES-buffered basal solution. All the subsequent steps were performed at 37°C in an air atmosphere in a water-jacketed incubator.
Cells were then preincubated at 37°C in basal medium for 40 min with the irreversible inhibitor of GS 1 mM MSO (30). At this concentration and duration of treatment, MSO irreversibly blocked GLNase activity by >95% and prevented reverse conversion of glutamate to glutamine. This is a particularly critical treatment because of the very high GS activity in glial cells. MSO could not be present in the subsequent steps because it interferes with transport of amino acids (see results).
Cells were washed from MSO two times with 2 ml of basal solution and transferred into 2 ml of the GLNase reaction medium that was prepared on the basis of basal with addition 4 μCi/ml of l-[3H]glutamine (final concentration adjusted to 2 μM with unlabeled l-glutamine). Cells were incubated with this reaction mix for 30 min at 37°C.
The reaction was terminated and extracellular isotope was removed by three consecutive washes with 2 ml of ice-cold basal solution.
One milliter of milliQ H2O was added to each well to lyse astrocytes; cells were scraped and sonicated for 4 min. Lysates were clarified by rapid centrifugation (4 min × 12,100 g at room temperature).
Each cell lysate (1 ml) was added onto AG 1-X8 Polyprep column, and l-[3H]glutamate was separated from l-[3H]glutamine by subsequent H2O and 0.1 M HCl elutions as described above.
Eluent fractions were collected into scintillation vials, and 3H content was determined as described in the GS assay section. The GLNase activity was calculated as percent conversion of l-[3H]glutamine to l-[3H]glutamate, which was normalized to the total 3H recovered from each sample. This was done using the following formula: % conversion = [(dpms in “glutamate” fractions nos. 4–6)/(dpms in “glutamine” fractions nos. 1–3 + dpms in “glutamate” fractions nos. 4–6)] × 100%.
HPLC assay of intracellular amino acid content.
For determination of intracellular amino acid content, cells were treated under identical conditions as in the enzymatic assay experiments with the exception of 3H-labeled compounds. Confluent cell cultures grown in six-well plates were preincubated in basal HEPES-buffered medium for 40 min with or without inhibitors of GS and GLNase, as indicated in figure legends. They were then washed from inhibitors two times with 2 ml basal medium and transferred to media containing 2 μM glutamate plus 100 μM (NH4)2SO4 or 2 μM glutamine to mimic enzymatic assay conditions for GS and GLNase, respectively. After 30 min incubation at 37°C, experimental media were aspirated, cells were washed three times from extracellular amino acids, and 1 ml of solution containing 5 mM HEPES and 1 mM EDTA was added to each well. Cells were scraped and sonicated for 4 min at room temperature. Aliquots (100 μl) of cell lysates were taken for protein assays, and the remaining lysates were clarified by rapid centrifugation (4 min × 12,100 g, room temperature). Amino acid levels in each supernatant were determined by a reverse-phase HPLC using an Agilent 1200 HPLC setup and Eclipse XDB-C18 column (4.6 × 150 mm, 5 μm particle diameter). Precolumn derivatization of amino acids was performed with freshly prepared mix of o-phthaldialdehyde and 2-mercaptoethanol in 0.4 M sodium tetraborate buffer (pH = 9.5). The amino acid derivatives were eluted with solvent containing 30 mM NaH2PO4, 1% tetrahydrofuran, 30 mM sodium acetate, 0.05% sodium azide, and increasing concentration of HPLC grade methanol (10–30%). Fluorescence signals were registered using a programmable 1200 series fluorescence detector (Agilent). Amino acid standards (l-alanine, l-aspartate, l-glutamate, l-glutamine, taurine), which were processed in the same fashion, were used to locate amino acid peaks and calculate concentrations of individual amino acids in the samples.
3H-labeled amino acid uptake assay.
To determine rates of amino acids accumulation, confluent astrocyte cultures grown in 12-well plates were washed three times from serum-containing media with 1 ml of HEPES-buffered basal medium and incubated for 5–40 min at 37°C with basal medium containing additionally 2 μCi/ml of l-[3H]glutamine plus 2 μM of unlabeled l-glutamine or 1 μCi/ml of l-[3H]glutamate plus 2 μM of unlabeled l-glutamate. Amino acid uptake was terminated by three washes with 1 ml of ice-cold basal medium. Cells were then lysed in 1 ml of 2% SDS plus 8 mM EDTA. Lysates were transferred into scintillation vials and 3H content was determined as described above. Rates of amino acid accumulation were then normalized to specific activity of each radiolabeled compound and average protein content. Protein content was determined using bicinchoninic acid (BCA) assay kit (Thermo Scientific-Pierce, Rockford, IL) according to the manufacturer's instructions. Optical density was read at 590 nm using an ELx800 plate reader (Bio-Tek Instruments, Winooski, VT).
l-[3H]glutamine release assay.
To measure l-[3H]glutamine release, cultured astrocytes grown on 18-mm square coverslips were preincubated for 40 min in basal medium with the GLNase inhibitor 1 mM DON to minimize conversion of l-[3H]glutamine to l-[3H]glutamate. Coverslips were then washed from DON and placed into fresh portion of basal medium containing additionally 4 μCi/ml of l-[3H]glutamine and 2 μM of unlabeled l-glutamine. After 30 min loading, cells were washed from extracellular isotope and transferred to a Lucite perfusion chamber. This chamber had a depression precisely cut in the bottom to accommodate the coverslip and a Teflon screw top leaving space above the cells of around 150–200 μm in height. Cells were superfused at a flow rate of 1.2 ml/min in an incubator set at 37°C with basal medium as indicated in figures. One-minute perfusate fractions were collected and analyzed for 3H content. At the end of each experiment, astrocytes on coverslips were lysed with 1 ml of 2% sodium dodecyl sulfate (SDS) plus 8 mM EDTA to calculate remaining isotope content. Fractional isotope release for each time point was calculated by dividing radioactivity released in each 1-min interval by the radioactivity left in the cells.
Statistical analyses.
All data in this study are presented as mean values ± SE. Statistical differences between groups were determined using t-test, one-way ANOVA, or repeated measures ANOVA as appropriate. Post hoc Neman-Keuls test was performed for multiple group comparisons. Origin 8.1 (Origin Labs, Northampton, MA) and Prism 5 (GraphPad Software, San Diego, CA) were used for analyses.
RESULTS
Effects of the inhibitor of glutamine synthetase MSO and the inhibitor of glutaminase DON on l-[3H]glutamate and l-[3H]glutamine transport in primary rat astrocytes.
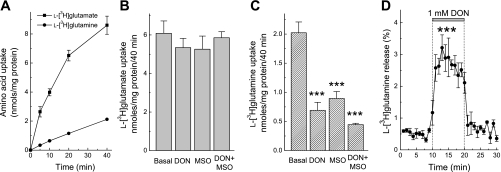
The inhibitors of GS and GLNase are known to interfere with transport of amino acids, particularly with l-glutamine transporters. DON has been shown to be a competitive inhibitor of the Na+-dependent l-glutamine uptake in Xenopus oocytes (39), whereas MSO was reported to inhibit l-glutamine uptake in brain cortical slices (48). Such nonspecific actions can seriously complicate whole cell l-[3H]glutamine metabolism experiments. To quantify to what degree DON and MSO interfere with glutamate and glutamine transport under our experimental conditions, we first tested their effects on amino acid transport in cultured astrocytes. Both 1 mM DON and 1 mM MSO potently reduced l-[3H]glutamine accumulation, and their combination showed a trend for additive action (Fig. 1C). Neither of these two compounds affected uptake of l-[3H]glutamate (Fig. 1B). In addition, 1 mM DON triggered a three- to fourfold increase in the release of preloaded l-[3H]glutamine likely mediated by heteroexchange via one of l-glutamine transporters (Fig. 1D). These data resemble the previously reported effects of MSO, which also induced intracellular l-glutamine release in cultured rat astrocytes (1). Therefore, in the whole cell assays MSO and DON can only be used in preincubation media.
Fig. 1.
The glutaminase (GLNase) inhibitor 6-diazo-5-oxo-l-norleucine (DON) and the glutamine synthetase (GS) inhibitor l-methionine sulfoximine (MSO) interfere with uptake of l-[3H]glutamine but not l-[3H]glutamate in primary rat astrocytes. A: comparative kinetics of l-[3H]glutamine and l-[3H]glutamate uptake by cultured astrocytes. Total concentration of both amino acids was adjusted to 2 μM. The results are mean values ± SE of 6 independent determinations. When not shown, SE values were less than symbol. B: effects of 1 mM DON and 1 mM MSO on cumulative astrocytic l-[3H]glutamate uptake measured for 40 min. The results are the mean values ± SE of 6 independent determinations in 2 experiments. C: effects of 1 mM DON and 1 mM MSO on cumulative astrocytic l-[3H]glutamine uptake measured for 40 min. The results are the mean values ± SE of 6 independent determinations in 2 experiments. ***P < 0.001 vs. basal uptake values. D: 1 mM DON triggers efflux of intracellular l-[3H]glutamine. The results are the mean values ± SE of 4 experiments. ***P < 0.001 DON vs. basal release values, repeated measures ANOVA.
Optimization and validation of conditions for the GS assay.
We quantified the activity of GS as enzymatic conversion of radiolabeled l-glutamate to l-glutamine in the intact astrocytes. This assay was designed based on the method proposed by Prusiner and Milner (27) that involves separation of radiolabeled amino acids on Dowex anion exchange columns. However, the original method was designed for use with isolated enzymes and did not take into account the possibility that both the substrate and the product of the GS reaction may be consumed in alternative enzymatic processes. For example, newly formed l-[3H]glutamine may be hydrolyzed back to l-[3H]glutamate by GLNase. We, therefore, made a number of changes to adapt our assay for measuring enzymatic activities in intact cells and to make it easily reproducible in other laboratories. Briefly, main modifications can be summarized as follows. First, to protect the newly formed l-[3H]glutamine from hydrolysis by intracellular GLNase, we preincubated cells with the irreversible GLNase inhibitor DON (46). Second, instead of l-[14C]glutamate, we utilized less expensive l-[3H]glutamate. Finally, we used commercially available Dowex AG 1-X8 columns that allow for rapid and reproducible filtration.
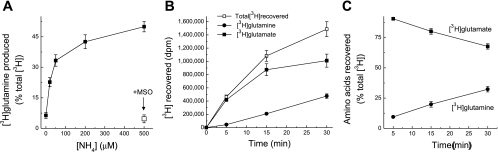
We first determined optimal substrate concentrations for measuring the GS reaction. The concentration of extracellular l-[3H]glutamate was selected empirically to allow for robust labeling of intracellular amino acid pool. We used 2 μCi/ml of l-[3H]glutamate and adjusted the total concentration of this amino acid to 2 μM by adding unlabeled l-glutamate. We then tested the dependence of the GS reaction on presence of extracellular ammonia. Under physiological conditions, ammonia exists in an equilibrium of charged (ammonium, NH4+) and uncharged (ammonia, NH3) forms. In the blood stream, levels of NH4+/NH3 are in the range of 20–80 μM, whereas in the CNS, due to local oxidative deamination and equilibrating diffusion from the blood, NH4+/NH3 concentrations are estimated to be up to 100–300 μM (7). In the culture dish, rapid diffusion to the extracellular space does not allow cells to sustain physiological levels of intracellular ammonia. As seen in Fig. 2A, adding extracellular (NH4+)2SO4 drastically increased conversion of l-[3H]glutamate to l-[3H]glutamine with saturation at the ammonia concentrations of ≥500 μM. These observations are consistent with the previously reported Michaelis constant of 150–160 μM for this enzyme (40). For the subsequent experiments we selected to use [NH4+/NH3] = 200 μM, which produces nearly maximal GS activity. Higher concentrations of ammonia can interfere with multiple cellular processes (22).
Fig. 2.
Kinetics and efficacy of enzymatic conversion of l-[3H]glutamate into l-[3H]glutamine in rat astrocyte cultures. A: dependence of the GS activity on extracellular ammonia levels. l-[3H]glutamate (2 μM) and indicated concentrations of ammonia were added to the reaction medium and intracellular l-[3H]glutamine production was determined as described in materials and methods. Activity values were calculated as production of intercellular l-[3H]glutamine in relationship to the total isotope content. Arrow points to the GS activity values in cells preincubated with the GS inhibitor 1 mM MSO. The data are the mean values ± SE from 3 experiments. B: time course of total l-[3H]glutamate accumulation and intracellular l-[3H]glutamate to l-[3H]glutamine conversion in cultured astrocytes. Data are the mean values ± SE of 5 experiments. C: relative efficacy of l-[3H]glutamate to l-[3H]glutamine conversion at different incubation times. These data were derived from those presented in B, the conversion values were normalized to the total isotope content at each time point.
In the next experiments, we established suitable reaction times for measuring the GS activity. Inside astrocytes, radiolabeled glutamate is consumed by GS but also in the reactions of transamination and oxidative deamination. The product of transamination and oxidative deamination α-ketoglutarate is utilized in the TCA. Therefore, with longer times of measuring the GS activity, a substantial fraction of l-[3H]glutamate taken a into cells may be metabolized and completely oxidized in the TCA cycle. However, because exogenous l-[3H]glutamate and unlabeled endogenous l-glutamate need to equilibrate, shorter reaction times may not be sufficient for synthesis of measurable quantities of l-[3H]glutamine. As seen in Fig. 2B, l-[3H]glutamate is progressively converted to l-[3H]glutamine inside cultured astrocytes. The relative fraction of newly produced l-[3H]glutamine constantly increased during the 30-min incubation period (Fig. 2C). Based on these results we selected 30 min incubation as time for measuring the GS activity.
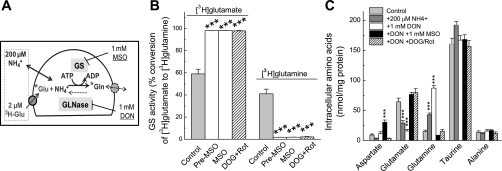
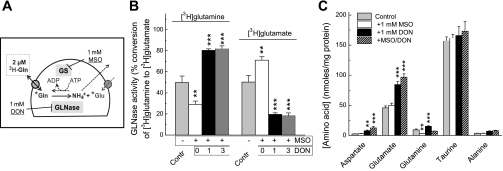
With the reaction time and substrate concentrations selected by us, which are schematically summarized in Fig. 3A, we tested whether the assay designed by us specifically measures the GS activity. Over a 30-min reaction period, astrocytes, which were preincubated with 1 mM DON, converted ∼40% of accumulated l-[3H]glutamate into l-[3H]glutamine (Fig. 3B). Addition of MSO, which irreversibly binds to the catalytic center of GS (31), reduced the conversion rate by 98% (Fig. 3B). These results suggest that the conditions chosen by us allow for selective measurements of the GS activity. Importantly, MSO was equally potent when added to preincubation media only or in both the preincubation and the reaction media, confirming that MSO effect is irreversible. This is particularly critical for the subsequent measurements of the GLNase activity, because MSO blocks l-[3H]glutamine uptake, if present in the reaction media (see Fig. 1C). Since GS is the ATP-dependent enzyme, we additionally tested the effect of metabolic inhibition. We used combinations of 10 mM 2-deoxy-d-glucose (nonmetabolized glucose analog) and 10 μM rotenone (blocker of the mitochondrial complex I), which in our previous study was shown to cause rapid and complete depletion of intracellular ATP in astrocyte cultures (32). In the present experiments, these two agents inhibited conversion of l-[3H]glutamate to l-[3H]glutamine by 97%, further supporting the notion that this process was catalyzed by GS (Fig. 3C).
Fig. 3.
Validation of the method measuring GS activity in intact astrocytes based on quantification of the enzymatic conversion of l-[3H]glutamate into l-[3H]glutamine. A: schematic summary of experimental design. B: confirmation of the specificity of GS assay using the GS inhibitor 1 mM MSO or the metabolic inhibitors 10 mM 2-deoxy-d-glucose plus 10 μM rotenone (DOG + Rot). Intracellular l-[3H]glutamate and l-[3H]glutamine content was determined as described in materials and methods and normalized to the total isotope content (100%). Cells were preincubated with MSO and DOG + Rot for 40 min before and throughout the enzymatic assay. Alternatively, MSO was added to the preincubation media only (pre-MSO). The results are the mean values ± SE of 6–8 independent experiments performed in 4 different culture preparations. ***P < 0.001 vs. control. C: effects of the inhibitors of GS and glutaminase (GLNase) on the intracellular content of endogenous amino acids. All assay conditions and concentration of inhibitors including the GLNase inhibitor 1 mM DON were the same as in enzymatic assays presented in B. Intracellular content of l-aspartate (Asp), l-glutamate (Glu), l-glutamine (Gln), taurine (Tau), and l-alanine (Ala) were determined using an HPLC assay as described in materials and methods. The data are the mean values ± SE of 4–8 independent experiments performed in 3 different culture preparations. ***P < 0.001 compared with Control.
To additionally validate our assay and explore to what extent changes in l-[3H]glutamate to l-[3H]glutamine conversion reflect changes in concentrations of endogenous l-glutamate and l-glutamine, we measured intracellular amino acid levels using an HPLC approach. These experiments were performed under conditions identical to those employed in the enzymatic GS assays, with 2 μM l-glutamate and 200 μM ammonia present in extracellular media. Addition of ammonia decreased endogenous l-glutamate and increased endogenous l-glutamine levels inside the cells (Fig. 3C), clearly demonstrating that ammonia concentration is a rate-limiting factor for the GS reaction in cultured cells. Treatment with MSO or metabolic inhibition with 2-deoxy-d-glucose plus rotenone, both strongly reduced intracellular glutamine levels and significantly increased intracellular glutamate levels (Fig. 3C). These results validate the specificity of assay conditions and their suitability for measuring the GS activity in astrocytes.
Optimization and validation of conditions for the GLNase assay.
In our pilot experiments, we found that commercial lots of l-[3H]glutamine may contain up to 8% of 3H label incorporated into negatively charged compound(s), likely l-[3H]glutamate. Therefore, it is important to test the purity of l-[3H]glutamine lots, and, if necessary, to additionally purify radiolabeled amino acid using AG 1-X8 Polyprep columns.
We measured GLNase activity as the conversion of l-[3H]glutamine to l-[3H]glutamate, in a similar fashion as in the GS assay. Because the GS activity in astrocyte is very high, to preserve the GLNase-derived l-[3H]glutamate, astrocytes were pretreated with the irreversible GS inhibitor MSO in all experiments, unless stated otherwise. The efficacy of GS inhibition with MSO was validated in the experiments presented in Fig. 3B. Concentration and specific activity of the GLNase substrate l-[3H]glutamine was selected empirically as 4 μCi/ml of l-[3H]glutamine, adjusted to 2 μM with unlabeled l-glutamine. These loading conditions allowed for high levels of l-[3H]glutamine labeling of the intracellular amino acid pool.
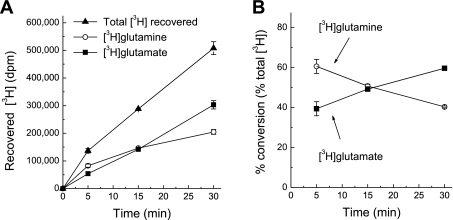
As in the case of the GS assay, we started with determining the suitable time for measuring GLNase activity. Incubation of cells with l-[3H]glutamine led to a time-dependent accumulation of l-[3H]glutamate (Fig. 4A). The relative conversion rates of l-[3H]glutamine to l-[3H]glutamate steadily increased over a 30-min incubation period (Fig. 4B). Somewhat surprisingly, we found very high relative conversion rates (>35%) within 5 min after addition of extracellular l-[3H]glutamine (Fig. 4B). That was in contrast to much lower conversion of l-[3H]glutamate to l-[3H]glutamine by GS measured at the same time period (∼8–9%, as seen in Fig. 2C). Possible reasons for differences in relative conversion rates are discussed in the following sections. To yield high intracellular levels of l-[3H]glutamine and l-[3H]glutamate and to make the GLNase assay comparable to the GS assay, we selected 30 min time for all the following experiments.
Fig. 4.
Kinetics of l-[3H]glutamine accumulation and efficacy of l-[3H]glutamine to l-[3H]glutamate conversion in astrocyte cultures. A: time course of total l-[3H]glutamine accumulation and intracellular l-[3H]glutamine to l-[3H]glutamate conversion. Data are the mean values ± SE of 5 experiments. B: relative efficacy of l-[3H]glutamine to l-[3H]glutamate conversion at different incubation times. These data were derived from those presented in A, the conversion values were normalized to the total isotope content at each time point.
The conditions of the GLNase assay are schematically summarized in Fig. 5A. To validate specificity of our assay conditions, we performed pharmacological controls. Without MSO pretreatment, the l-[3H]glutamate to l-[3H]glutamine conversion rate was ∼50% after 30 min (Fig. 5B). Treatment with MSO increased the conversion rate to ∼70% (Fig. 5B) indicating that substantial fraction of newly formed l-[3H]glutamate is converted back to l-[3H]glutamine by the GS. The irreversible GLNase inhibitor DON blocked l-[3H]glutamine to l-[3H]glutamate conversion by ∼75%, when cells were preincubated with this agent at 1 mM concentration (Fig. 5B). We checked if the greater inhibition values can be achieved by increasing concentration of DON but found essentially the same degree of inhibition with 3 mM DON (Fig. 5B). The incomplete inhibition with DON may be due to non-GLNase-mediated l-glutamine conversions. This amino acid may be involved in other biosynthetic reactions, in which amide nitrogen is transferred from glutamine to acceptor substrates to produce other amino acids, purine and pyrimidine bases, and amino sugars (47). Based on our results, at least 75% of l-[3H]glutamine-to-l-[3H]glutamate conversion should be attributed to the GLNase activity. However, as discussed below, this may be underestimation.
Fig. 5.
Validation of measurements of GLNase activity in intact astrocytes based on quantification of the enzymatic conversion of l-[3H]glutamine into l-[3H]glutamate. A: schematic summary of experimental design. B: confirmation of the specificity of GLNase assay with the irreversible GLNase inhibitor DON (1 or 3 mM). Intracellular l-[3H]glutamate and l-[3H]glutamine content was determined as described in materials and methods and normalized to the total isotope content (100%). The results are the mean values ± SE of 6 independent experiments performed in 2 different culture preparations. **P < 0.01; ***P < 0.001 vs. control. C: effects of inhibitors of GLNase and GS on the intracellular content of endogenous amino acids. The design of this experiment was identical to that presented in B. The data are the mean values ± SE of 6–10 experiments. **P < 0.01; ***P < 0.001 compared with Control.
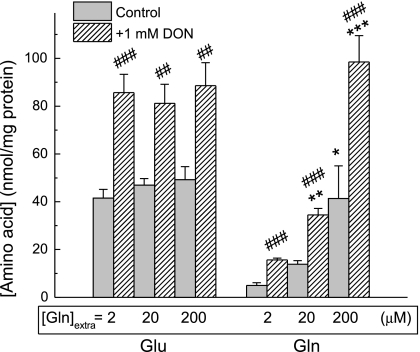
To test whether changes in l-[3H]glutamine-to-l-[3H]glutamate conversion rates translate to the changes in total intracellular amino acid levels, we performed additional HPLC experiments in the presence of 2 μM l-glutamine in extracellular media to mirror conditions of the GLNase assays. Surprisingly, we found that GLNase inhibition with DON produced very modest elevation in the intracellular l-glutamine and increased, rather than decreased, l-glutamate (Fig. 5C). These results contrasted with strong and predictable changes in l-glutamine levels found when the same experiments were performed under the “GS assay” conditions (with extracellular glutamate and ammonium added to the media, see Fig. 3C). To resolve this paradox we elevated extracellular glutamine from 2 to 20 to 200 μM, bringing it close to the concentrations seen in vivo, which are estimated to be in the range of 300–600 μM. With elevated extracellular l-glutamine concentrations, 1 mM DON produced very strong net increases in the intracellular glutamine levels (Fig. 6). Interestingly, there was no relationship between intracellular glutamine and intracellular glutamate.
Fig. 6.
The effects of 1 mM DON on intracellular amino acid levels as a function of increasing concentrations of extracellular l-glutamine. Cells were pretreated with DON for 40 min and then transferred for 30 min into medium containing 2, 20, or 200 μM l-glutamine. Intracellular amino acid levels were determined using an HPLC assay. For clarity only Glu and Gln levels are shown. The results are the mean values ± SE of 3 experiments. *P < 0.05; **P < 0.01; ***P < 0.001 vs. value measured with 2 μM extracellular glutamine. ##P < 0.01; ###P < 0.001, t-test, samples without or with DON measured with the same concentrations of extracellular glutamine.
DISCUSSION
The new GS and GLNase assays presented here provide an important advantage of monitoring changes of enzymatic activities in intact cultured cells. GS and GLNase are two critical enzymes of nitrogen metabolism that are regulated on numerous levels. The activity of cytosolic GS depends on the intracellular levels of glutamate, ammonium, and ATP and is a subject to cumulative feedback inhibition by more than 40 biosynthetic products of nucleotide synthesis and by adenylation (35). GLNase, which is predominantly localized between outer and inner mitochondrial membranes, is regulated by local glutamine availability but also by the levels of intracellular inorganic phosphate N-acetyl-aspartate, taurine, histidine, lysine, and intermediates of TCA (29, 36). The previously developed methods were optimized for isolated enzymes or homogenates of cells and tissues (25, 27). However, using isolated enzymes removes the intrinsic regulation of GS and GLNase in a “native” environment. For example, metabolic inhibition causes strong reductions in the GS activity in the whole cell assays (see Fig. 3), but no such changes could be observed in cell homogenates when all the substrates and ATP are included in the assay media.
However, the whole cell assays have certain inherent limitations. Enzymatic substrates (l-[3H]glutamate and ammonium for GS, and l-[3H]glutamine for GLNase) that are added to the extracellular media have to be transported across plasma membrane. Once in the intracellular compartment, the radiolabeled compounds are diluted by the millimolar pool of the endogenous amino acids. As a result, it is extremely difficult to accurately determine, or even estimate, the specific activities and the precise concentrations of substrates and products of relevant enzymatic reactions. Therefore, obtained activity values are calculated as relative conversion values, which are normalized to the total isotope loading, rather than in standard units of enzymatic activity per minute per milligram of protein. Furthermore, because radiolabeled amino acids have numerous metabolic fates, the rates of enzymatic reactions may be somewhat underestimated because substrates are metabolized in alternative processes. For example, l-[3H]glutamate could be converted to l-[3H]glutamine by the GS but also to α-[3H]ketoglutarate by mitochondrial glutamate dehydrogenase and aminotransferases (20). Under our experimental conditions, this seems to not be the case, because l-[3H]glutamate conversion was inhibited by ≥97% by the GS blocker MSO and by metabolic inhibition.
For the GLNase-mediated reaction, part of the newly synthesized l-[3H]glutamate is diverted to the mitochondrial metabolism by glutamate dehydrogenase. This should not be a substantial problem for the whole cell assays because resulting α-[3H]ketoglutarate and TCA metabolites are acid substances, which are eluted from Dowex columns together with negatively charged l-[3H]glutamate. Besides GLNase, glutamine is a subject for other metabolic reactions, in which amide nitrogen is transferred to various acceptor substrates to produce other amino acids, purine and pyrimidine bases, and amino sugars (47). Therefore, it is critical to have proper pharmacological controls. In our assays, the GLNase inhibitor DON reduced the l-[3H]glutamine-to-“l-[3H]glutamate” (3H-labeled acid substances) conversion by 75% only, when added at 1 or 3 mM levels. This may be explained by the non-GLNase reactions or, alternatively, by an incomplete inhibition of GLNase. Willis and Seegmiller (46) reported that in cellular assays, DON required 2.5-h preincubation to exert a complete inhibitory action. Although the 40 min preincubation may not be sufficient to completely block the GLNase with DON, very long preincubation periods seemed unpractical in our case. As previously shown and reproduced in our control experiments, DON strongly interferes with l-[3H]glutamine transport (39, 48) and can rapidly deplete intracellular glutamine via DONout/l-[3H]glutaminein heteroexchange.
In conclusion, we developed and validated simple enzymatic assays, which can be useful for studying regulation of GS and GLNase without disruption of cell integrity. In our recently published work we successfully used similar approach to discover that cellular swelling inhibits GS but not GLNase in cultured astrocytes (16). Present method may be utilized for cultured glial cells to model physiological and pathological changes in glutamate and glutamine metabolism and otherwise adapted for neuronal cells or cell cultures prepared from other tissues.
GRANTS
This work was funded by the National Institute for Neurological Disorders and Stroke Grant NS-061953 (to A. A. Mongin).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
ACKNOWLEDGMENTS
We thank Drs. Paul J. Feustel, Iskandar F. Abdullaev, and Preeti Dohare for helpful discussions of this work.
REFERENCES
- 1. Albrecht J, Norenberg MD. l-methionine-DL-sulfoximine induces massive efflux of glutamine from cortical astrocytes in primary culture. Eur J Pharmacol 182: 587–589, 1990 [DOI] [PubMed] [Google Scholar]
- 2. Albrecht J, Sonnewald U, Waagepetersen HS, Schousboe A. Glutamine in the central nervous system: function and dysfunction. Front Biosci 12: 332–343, 2007 [DOI] [PubMed] [Google Scholar]
- 3. Balazs R, Machiyama Y, Hammond BJ, Julian T, Richter D. The operation of the gamma-aminobutyrate bypath of the tricarboxylic acid cycle in brain tissue in vitro. Biochem J 116: 445–461, 1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benveniste H, Drejer J, Schousboe A, Diemer NH. Elevation of the extracellular concentrations of glutamate and aspartate in rat hippocampus during transient cerebral ischemia monitored by intracerebral microdialysis. J Neurochem 43: 1369–1374, 1984 [DOI] [PubMed] [Google Scholar]
- 5. Broer S, Brookes N. Transfer of glutamine between astrocytes and neurons. J Neurochem 77: 705–719, 2001 [DOI] [PubMed] [Google Scholar]
- 6. Butterfield DA, Howard BJ, Yatin S, Allen KL, Carney JM. Free radical oxidation of brain proteins in accelerated senescence and its modulation by N-tert-butyl-alpha-phenylnitrone. Proc Natl Acad Sci USA 94: 674–678, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cooper AJL, Plum F. Biochemistry and physiology of brain ammonia. Physiol Rev 67: 440–519, 1987 [DOI] [PubMed] [Google Scholar]
- 8. Cubelos B, Gonzalez-Gonzalez IM, Gimenez C, Zafra F. Amino acid transporter SNAT5 localizes to glial cells in the rat brain. Glia 49: 230–244, 2005 [DOI] [PubMed] [Google Scholar]
- 9. Danbolt NC. Glutamate uptake. Prog Neurobiol 65: 1–105, 2001 [DOI] [PubMed] [Google Scholar]
- 10. Deitmer JW, Broer A, Broer S. Glutamine efflux from astrocytes is mediated by multiple pathways. J Neurochem 87: 127–135, 2003 [DOI] [PubMed] [Google Scholar]
- 11. Eid T, Ghosh A, Wang Y, Beckstrom H, Zaveri HP, Lee TS, Lai JC, Malthankar-Phatak GH, de Lanerolle NC. Recurrent seizures and brain pathology after inhibition of glutamine synthetase in the hippocampus in rats. Brain 131: 2061–2070, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eid T, Thomas MJ, Spencer DD, Runden-Pran E, Lai JC, Malthankar GV, Kim JH, Danbolt NC, Ottersen OP, de Lanerolle NC. Loss of glutamine synthetase in the human epileptogenic hippocampus: possible mechanism for raised extracellular glutamate in mesial temporal lobe epilepsy. Lancet 363: 28–37, 2004 [DOI] [PubMed] [Google Scholar]
- 13. Feustel PJ, Jin Y, Kimelberg HK. Volume-regulated anion channels are the predominant contributors to release of excitatory amino acids in the ischemic cortical penumbra. Stroke 35: 1164–1168, 2004 [DOI] [PubMed] [Google Scholar]
- 14. Fonnum F. Glutamate: a neurotransmitter in mammalian brain. J Neurochem 42: 1–11, 1984 [DOI] [PubMed] [Google Scholar]
- 15. Hammer J, Alvestad S, Osen KK, Skare O, Sonnewald U, Ottersen OP. Expression of glutamine synthetase and glutamate dehydrogenase in the latent phase and chronic phase in the kainate model of temporal lobe epilepsy. Glia 56: 856–868, 2008 [DOI] [PubMed] [Google Scholar]
- 16. Hyzinski-Garcia MC, Vincent MY, Haskew-Layton RE, Dohare P, Keller RW, Jr, Mongin AA. Hypoosmotic swelling modifies glutamate-glutamine cycle in the cerebral cortex and in astrocyte cultures. J Neurochem 118: 140–152, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kanamori K, Ross BD, Kondrat RW. Glial uptake of neurotransmitter glutamate from the extracellular fluid studied in vivo by microdialysis and (13)C NMR. J Neurochem 83: 682–695, 2002 [DOI] [PubMed] [Google Scholar]
- 18. Laake JH, Takumi Y, Eidet J, Torgner IA, Roberg B, Kvamme E, Ottersen OP. Postembedding immunogold labelling reveals subcellular localization and pathway-specific enrichment of phosphate activated glutaminase in rat cerebellum. Neuroscience 88: 1137–1151, 1999 [DOI] [PubMed] [Google Scholar]
- 19. Martinez-Hernandez A, Bell KP, Norenberg MD. Glutamine synthetase: glial localization in brain. Science 195: 1356–1358, 1977 [DOI] [PubMed] [Google Scholar]
- 20. McKenna MC. The glutamate-glutamine cycle is not stoichiometric: fates of glutamate in brain. J Neurosci Res 85: 3347–3358, 2007 [DOI] [PubMed] [Google Scholar]
- 21. McKenna MC, Sonnewald U, Huang X, Stevenson J, Zielke HR. Exogenous glutamate concentration regulates the metabolic fate of glutamate in astrocytes. J Neurochem 66: 386–393, 1996 [DOI] [PubMed] [Google Scholar]
- 22. Norenberg MD, Jayakumar AR, Rama Rao KV, Panickar KS. New concepts in the mechanism of ammonia-induced astrocyte swelling. Metab Brain Dis 22: 219–234, 2007 [DOI] [PubMed] [Google Scholar]
- 23. Oliver CN, Starke-Reed PE, Stadtman ER, Liu GJ, Carney JM, Floyd RA. Oxidative damage to brain proteins, loss of glutamine synthetase activity, and production of free radicals during ischemia/reperfusion-induced injury to gerbil brain. Proc Natl Acad Sci USA 87: 5144–5147, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Palacin M, Estevez R, Bertran J, Zorzano A. Molecular biology of mammalian plasma membrane amino acid transporters. Physiol Rev 78: 969–1054, 1998 [DOI] [PubMed] [Google Scholar]
- 25. Pishak MR, Phillips AT. A modified radioisotopic assay for measuring glutamine synthetase activity in tissue extracts. Anal Biochem 94: 82–88, 1979 [DOI] [PubMed] [Google Scholar]
- 26. Poon HF, Calabrese V, Calvani M, Butterfield DA. Proteomics analyses of specific protein oxidation and protein expression in aged rat brain and its modulation by l-acetylcarnitine: insights into the mechanisms of action of this proposed therapeutic agent for CNS disorders associated with oxidative stress. Antioxid Redox Signal 8: 381–394, 2006 [DOI] [PubMed] [Google Scholar]
- 27. Prusiner S, Milner L. A rapid radioactive assay for glutamine synthetase, glutaminase, asparagine synthetase, and asparaginase. Anal Biochem 37: 429–438, 1970 [DOI] [PubMed] [Google Scholar]
- 28. Rivett AJ. Preferential degradation of the oxidatively modified form of glutamine synthetase by intracellular mammalian proteases. J Biol Chem 260: 300–305, 1985 [PubMed] [Google Scholar]
- 29. Roberg B, Torgner IA, Kvamme E. Inhibition of glutamine transport in rat brain mitochondria by some amino acids and tricarboxylic acid cycle intermediates. Neurochem Res 24: 809–814, 1999 [DOI] [PubMed] [Google Scholar]
- 30. Ronzio RA, Meister A. Phosphorylation of methionine sulfoximine by glutamine synthetase. Proc Natl Acad Sci USA 59: 164–170, 1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ronzio RA, Rowe WB, Meister A. Studies on the mechanism of inhibition of glutamine synthetase by methionine sulfoximine. Biochemistry 8: 1066–1075, 1969 [DOI] [PubMed] [Google Scholar]
- 32. Rutledge EM, Mongin AA, Kimelberg HK. Intracellular ATP depletion inhibits swelling-induced d-[3H]aspartate release from primary astrocyte cultures. Brain Res 842: 39–45, 1999 [DOI] [PubMed] [Google Scholar]
- 33. Smith QR. Transport of glutamate and other amino acids at the blood-brain barrier. J Nutr 130: 1016S–1022S, 2000 [DOI] [PubMed] [Google Scholar]
- 34. Sonnewald U, Westergaard N, Petersen SB, Unsgard G, Schousboe A. Metabolism of [U-13C]glutamate in astrocytes studied by 13C NMR spectroscopy: incorporation of more label into lactate than into glutamine demonstrates the importance of the tricarboxylic acid cycle. J Neurochem 61: 1179–1182, 1993 [DOI] [PubMed] [Google Scholar]
- 35. Stadtman ER. The story of glutamine synthetase regulation. J Biol Chem 276: 44357–44364, 2001 [DOI] [PubMed] [Google Scholar]
- 36. Svenneby G, Tveit B, Kvamme E. Glutaminase from pig renal cortex. II. Activation by inorganic and organic anions. J Biol Chem 245: 1878–1882, 1970 [PubMed] [Google Scholar]
- 37. Swain M, Butterworth RF, Blei AT. Ammonia and related amino acids in the pathogenesis of brain edema in acute ischemic liver failure in rats. Hepatology 15: 449–453, 1992 [DOI] [PubMed] [Google Scholar]
- 38. Takahashi H, Koehler RC, Brusilow SW, Traystman RJ. Inhibition of brain glutamine accumulation prevents cerebral edema in hyperammonemic rats. Am J Physiol Heart Circ Physiol 261: H825–H829, 1991 [DOI] [PubMed] [Google Scholar]
- 39. Taylor PM, Mackenzie B, Hundal HS, Robertson E, Rennie MJ. Transport and membrane binding of the glutamine analogue 6-diazo-5-oxo-L-norleucine (DON) in Xenopus laevis oocytes. J Membr Biol 128: 181–191, 1992 [DOI] [PubMed] [Google Scholar]
- 40. Tiemeier DC, Milman G. Chinese hamster liver glutamine synthetase. Purification, physical and biochemical properties. J Biol Chem 247: 2272–2277, 1972 [PubMed] [Google Scholar]
- 41. Tofteng F, Hauerberg J, Hansen BA, Pedersen CB, Jorgensen L, Larsen FS. Persistent arterial hyperammonemia increases the concentration of glutamine and alanine in the brain and correlates with intracranial pressure in patients with fulminant hepatic failure. J Cereb Blood Flow Metab 26: 21–27, 2006 [DOI] [PubMed] [Google Scholar]
- 42. van den Berg CJ, Garfinkel D. A stimulation study of brain compartments. Metabolism of glutamate and related substances in mouse brain. Biochem J 123: 211–218, 1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Waagepetersen HS, Sonnewald U, Larsson OM, Schousboe A. Multiple compartments with different metabolic characteristics are involved in biosynthesis of intracellular and released glutamine and citrate in astrocytes. Glia 35: 246–252, 2001 [DOI] [PubMed] [Google Scholar]
- 44. Wahl F, Obrenovitch TP, Hardy AM, Plotkine M, Boulu R, Symon L. Extracellular glutamate during focal cerebral ischaemia in rats: time course and calcium dependency. J Neurochem 63: 1003–1011, 1994 [DOI] [PubMed] [Google Scholar]
- 45. Warren KS, Schenker S. Effect of an inhibitor og glutamine synthesis (methionine sulfoximine) on ammonia toxicity and metabolism. J Lab Clin Med 64: 442–449, 1964 [PubMed] [Google Scholar]
- 46. Willis RC, Seegmiller JE. The inhibition by 6-diazo-5-oxo-l-norleucine of glutamine catabolism of the cultured human lymphoblast. J Cell Physiol 93: 375–382, 1977 [DOI] [PubMed] [Google Scholar]
- 47. Zalkin H, Smith JL. Enzymes utilizing glutamine as an amide donor. Adv Enzymol Relat Areas Mol Biol 72: 87–144, 1998 [DOI] [PubMed] [Google Scholar]
- 48. Zielinska M, Stafiej A, Law RO, Albrecht J. Effects of methionine sulfoximine on the glutamine and glutamate content and cell volume in rat cerebral cortical slices: involvement of mechanisms not related to inhibition of glutamine synthesis. Neurotoxicology 25: 443–449, 2004 [DOI] [PubMed] [Google Scholar]