Abstract
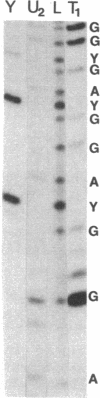
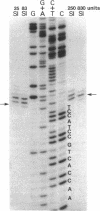
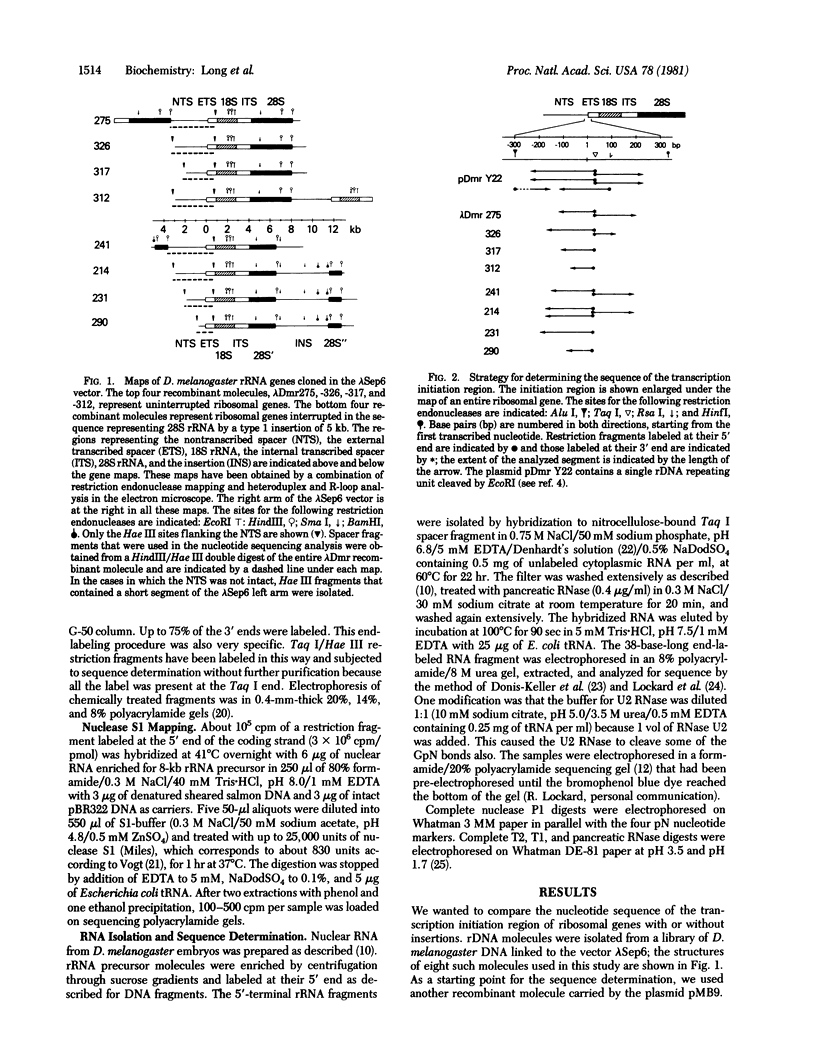
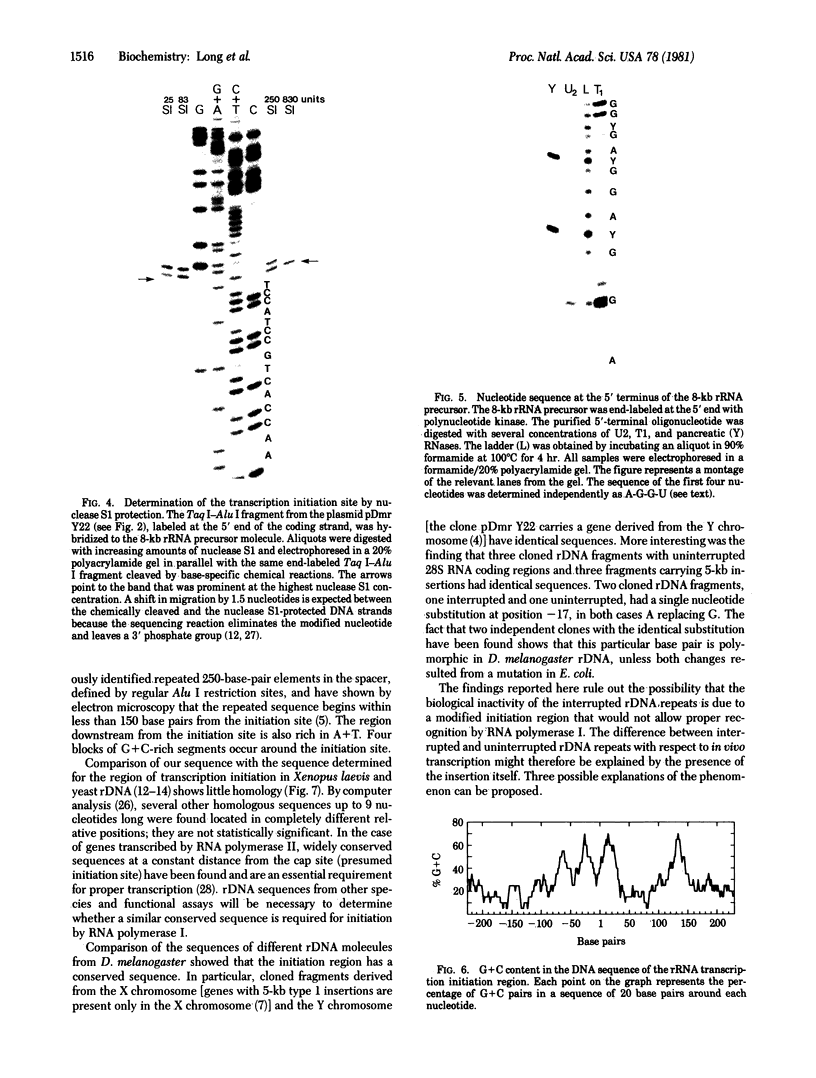
The sequence of 470 nucleotides surrounding the initiation site for rRNA transcription in Drosophila melanogaster has been determined. The precise initiation site was determined first by measuring the DNA fragment protected by the rRNA precursor against digestion by the single-strand specific nuclease S1 and second by direct sequence determination of the first 13 nucleotides of the rRNA precursor. Because greater than 80% od rRNA precursor molecules have been shown previously to bear pppA or ppA 5' termini, we assume that they represent the primary transcription product. Short sequence homologies exist with the initiation regions for rRNA transcription of Xenopus laevis and Saccharomyces cerevisiae. We have determined the nucleotide sequence of the initiation region in four cloned ribosomal genes from D. melanogaster which are not interrupted and in four cloned ribosomal genes in which the 28S rRNA coding region is interrupted by a 5-kilobase type 1 insertion. Three uninterrupted genes and three interrupted genes have identical sequences in the entire analyzed region. The remaining two genes have a single identical base substitution at position -17. We have shown previously that interrupted ribosomal genes in D. melanogaster are not effectively transcribed. Because the nucleotide sequences of the region where transcription initiates are identical in genes with or without insertions, we postulate that the presence of the insertion itself may be responsible for the inactivity of the interrupted genes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Dawid I. B., Wellauer P. K., Long E. O. Ribosomal DNA in Drosophila melanogaster. I. Isolation and characterization of cloned fragments. J Mol Biol. 1978 Dec 25;126(4):749–768. doi: 10.1016/0022-2836(78)90018-9. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glätzer K. H. Lengths of transcribed rDNA repeating units in spermatocytes of Drosophila hydei: only genes without an intervening sequence are expressed. Chromosoma. 1979 Nov;75(2):161–175. doi: 10.1007/BF00292205. [DOI] [PubMed] [Google Scholar]
- Klemenz R., Geiduschek E. P. The 5' terminus of the precursor ribosomal RNA of Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Jun 25;8(12):2679–2689. doi: 10.1093/nar/8.12.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Penman S. Processing steps and methylation in the formation of the ribosomal RNA of cultured Drosophila cells. J Mol Biol. 1978 May 15;121(2):219–238. doi: 10.1016/s0022-2836(78)80006-0. [DOI] [PubMed] [Google Scholar]
- Lockard R. E., Alzner-Deweerd B., Heckman J. E., MacGee J., Tabor M. W., RajBhandary U. L. Sequence analysis of 5'[32P] labeled mRNA and tRNA using polyacrylamide gel electrophoresis. Nucleic Acids Res. 1978 Jan;5(1):37–56. doi: 10.1093/nar/5.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Alternative pathways in the processing of ribosomal RNA precursor in Drosophila melanogaster. J Mol Biol. 1980 Apr 25;138(4):873–878. doi: 10.1016/0022-2836(80)90070-4. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Expression of ribosomal DNA insertions in Drosophila melanogaster. Cell. 1979 Dec;18(4):1185–1196. doi: 10.1016/0092-8674(79)90231-9. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Restriction analysis of spacers in ribosomal DNA of Drosophila melanogaster. Nucleic Acids Res. 1979 Sep 11;7(1):205–215. doi: 10.1093/nar/7.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Miller O. L., Jr Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell. 1976 Jun;8(2):305–319. doi: 10.1016/0092-8674(76)90014-3. [DOI] [PubMed] [Google Scholar]
- Pavlakis G. N., Jordan B. R., Wurst R. M., Vournakis J. N. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979 Dec 20;7(8):2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. P., Kelley D. E., Coleclough C., Seidman J. G., Leder P., Tonegawa S., Matthyssens G., Weigert M. Transcription of mouse kappa chain genes: implications for allelic exclusion. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1937–1941. doi: 10.1073/pnas.77.4.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C. L., Korn L. J. Computer analysis of nucleic acids and proteins. Methods Enzymol. 1980;65(1):595–609. doi: 10.1016/s0076-6879(80)65062-9. [DOI] [PubMed] [Google Scholar]
- Roiha H., Glover D. M. Chracterisation of complete type II insertions in cloned segments of ribosomal DNA from Drosophila melanogaster. J Mol Biol. 1980 Jun 25;140(2):341–355. doi: 10.1016/0022-2836(80)90110-2. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. I., Ohmori H., Bird R. E. Origin of replication of colicin E1 plasmid DNA. Proc Natl Acad Sci U S A. 1977 May;74(5):1865–1869. doi: 10.1073/pnas.74.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela P., Bell G. I., Venegas A., Sewell E. T., Masiarz F. R., DeGennaro L. J., Weinberg F., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. II. Physical map and nucleotide sequence of the 5 S ribosomal RNA gene and adjacent intergenic regions. J Biol Chem. 1977 Nov 25;252(22):8126–8135. [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Ribosomal DNA in Drosophila melanogaster. II. Heteroduplex mapping of cloned and uncloned rDNA. J Mol Biol. 1978 Dec 25;126(4):769–782. doi: 10.1016/0022-2836(78)90019-0. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B., Tartof K. D. X and Y chromosomal ribosomal DNA of Drosophila: comparison of spacers and insertions. Cell. 1978 Jun;14(2):269–278. doi: 10.1016/0092-8674(78)90113-7. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. The structural organization of ribosomal DNA in Drosophila melanogaster. Cell. 1977 Feb;10(2):193–212. doi: 10.1016/0092-8674(77)90214-8. [DOI] [PubMed] [Google Scholar]
- White R. L., Hogness D. S. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):177–192. doi: 10.1016/0092-8674(77)90213-6. [DOI] [PubMed] [Google Scholar]