1. Introduction
The discovery of the cannabinoid receptors CB1 and CB2 in 19881,2 and 1990,(3) respectively, and of the endogenous cannabinoid ligands (endocannabinoids) arachidonoylethanolamide (AEA) and 2-arachidonoylglycerol (2-AG) in 1992(4) and 1995,(5) respectively, represented major strides in the understanding of cannabinoid physiology and pharmacology. The realization that both endocannabinoids are derivatives of arachidonic acid (AA) also revealed a potential interrelationship between the endocannabinoid and eicosanoid signaling systems that is just beginning to be unraveled. In this review, we explore what is known about the interplay between the two lipid signaling networks and discuss the challenges and opportunities offered by this new field of inquiry.
1.1. Eicosanoid Biosynthetic Pathways
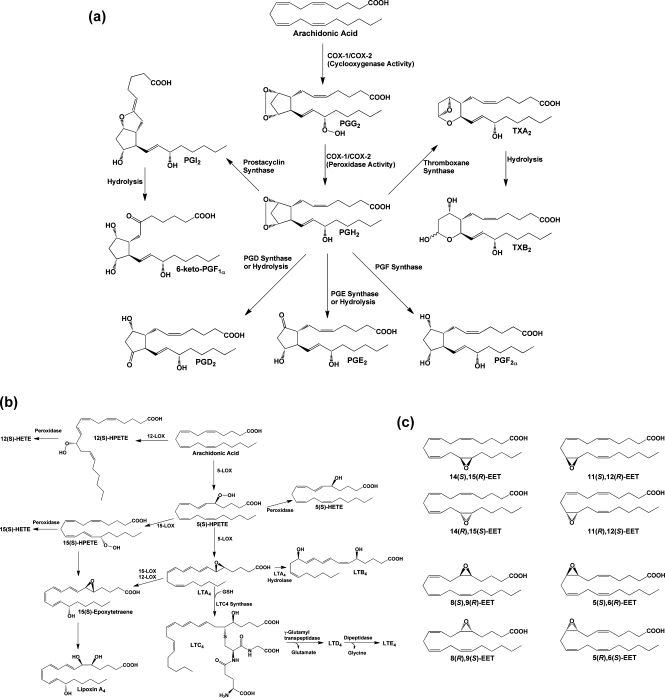
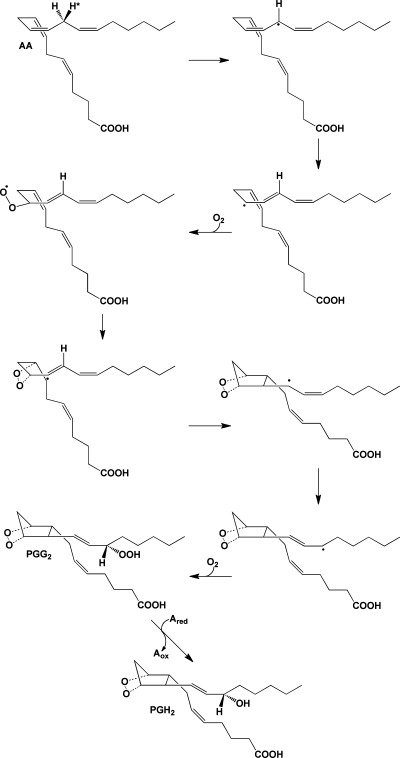
AA is an ω-6 tetraunsaturated fatty acid that is a component of mammalian cell membrane phospholipids, where it is predominantly esterified at the sn-2 position. AA’s role in eicosanoid signaling was first discovered in 1964, when Van Dorp et al. and Bergström et al. showed that incubation of the radiolabeled fatty acid with bull seminal vesicles led to the formation of prostaglandin E2 (PGE2).6,7 Since that time, we have come to appreciate that a wide range of stimuli (depending on cell type, tissue context, and physiologic state) can trigger the activation of cytosolic phospholipase A2 (cPLA2) and/or other phospholipases, leading to the release of free AA from phospholipid pools. The free fatty acid is then subject to oxidative metabolism by cyclooxygenase 1 and/or 2 (COX-1 and/or COX-2), leading to the formation of the endoperoxide PGH2. Tissue-specific metabolism of PGH2 by a group of PG synthases yields the biologically active PGs (PGE2, PGD2, PGF2α), prostacyclin (PGI2), and thromboxane A2 (TxA2) (Figure 1a).8,9 Alternatively, free AA may be metabolized by one of a variety of lipoxygenases (LOXs) that catalyze regio- and stereospecific oxygenation, yielding hydro-peroxyeicosatetraenoic acids (HPETEs). These compounds are enzymatically or chemically reduced to the corresponding hydroxy-eicosatetraenoic acids (HETEs) or undergo further metabolism. Multiple lipoxygenations produce the lipoxins,(10) or in the case of 5-HPETE, epoxidation followed by hydrolysis or glutathione adduction yields the leukotrienes (LTs)9,11 (Figure 1b). Finally, free AA may be oxidized at each of its double bonds or at the ω-terminus by cytochromes P450, leading to the epoxyeicosatrienoic acids (EETs) or HETEs(12) (Figure 1c). Members of each of these classes of compounds possess a unique range of biological activities.
Figure 1.
(a) Cyclooxygenase pathway of AA metabolism. AA is converted to PGG2 at the cyclooxygenase active site of COX-1 or COX-2 and is then reduced to PGH2 at the peroxidase active site. PGH2 spontaneously decomposes to yield PGE2 or PGD2, but these compounds, as well as PGF2α, PGI2, and TxA2, are also produced enzymatically by specific synthases. PGI2 and TXA2 are unstable and spontaneously decompose to yield 6-keto-PGF1α and TXB2, respectively. The ethanolamide and glyceryl ester of PGH2 produced from the metabolism of AEA and 2-AG, respectively, are converted to the same range of eicosanoid products as PGH2 with the exception of the TXA2 analogue. (b) Lipoxygenase pathway of AA metabolism. AA is converted by LOX enzymes to position-specific HPETEs, which are then reduced to the corresponding HETEs. 5-LOX also converts 5-HPETE to LTA4, which may then be metabolized to LTB4 or LTC4. The glutathionyl moiety of LTC4 is subject to enzymatic hydrolysis, yielding LTD4 and LTE4. The actions of multiple LOX enzymes lead to the formation of lipoxins. An example is provided for the synthesis of lipoxin A4 through multiple steps catalyzed by 5-LOX and either 15-LOX or 12-LOX. (c) Cytochromes P450 catalyze the epoxygenation of AA at each of the double bonds, producing the range of products shown.
1.2. Endocannabinoid Biosynthetic Pathways
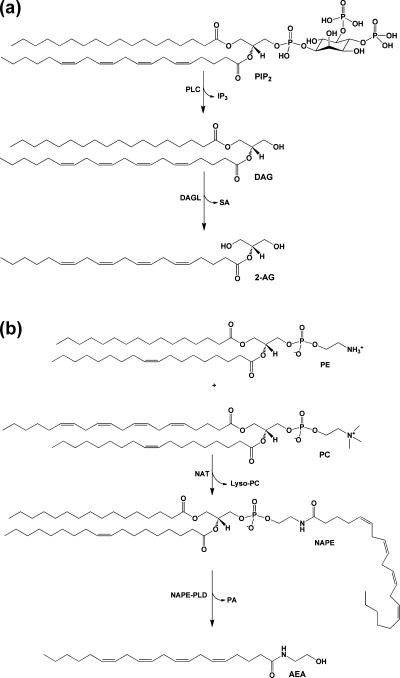
Endocannabinoids are also synthesized from AA-containing phospholipids, but by very different pathways. Activation of phospholipase C (PLC) leads to hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2), producing diacylglycerol (DAG). Because PIP2 is enriched in AA at the sn-2 position in most cells, and most PLCs favor substrates containing AA, a large proportion of the DAG formed in this reaction also contains AA. Hydrolysis by DAG lipases produces 2-AG (Figure 2a).13−15 2-AG can theoretically also be formed from the hydrolysis of DAGs produced in other reactions, such as the sequential hydrolysis of phosphatidylcholine (PC) by phospholipase D (PLD) and phosphatidic acid phosphatase. However, the importance of such alternative pathways in endocannabinoid lipid signaling is unclear.
Figure 2.
(a) Biosynthetic pathway for 2-AG. PIP2 containing AA at the sn-2 position is hydrolyzed by PLC to yield DAG and IP3. DAG is then further hydrolyzed by MAG lipase to 2-AG. (b) Biosynthetic pathway for AEA. An arachidonoyl moiety is transferred from the sn-1 position of a phospholipid, in this case PC, to the amino group of PE by NAT. The resulting product NAPE is then hydrolyzed by NAPE-PLD to yield AEA and PA.
Once generated, 2-AG is subject to hydrolysis, primarily by monoacylglycerol (MAG) lipases. The serine lipases α–β-hydrolase domains 6 and 12 (ABHD6 and ABHD12) have also been shown to play a role in 2-AG catabolism. On the basis of expression level and relative activity, MAG lipase, ABHD12, and ABHD6 are estimated to account for 85%, 9%, and 4% of 2-AG hydrolysis in mouse brain, respectively.16,17 Fatty acid amide hydrolase (FAAH), the enzyme primarily responsible for the hydrolysis of AEA, can also hydrolyze 2-AG; however, it plays only a minor role physiologically (approximately 1% in mouse brain).16,18 Under some circumstances, nonspecific esterases may catalyze 2-AG hydrolysis. For example, human carboxylesterases 1 and 2 (CES1 and CES2) metabolize 2-AG as efficiently as human and rat MAG lipase. Expression of CES1 accounts for 55% of 2-AG hydrolysis in the THP1 human monocytic leukemia cell line, suggesting that this may be a primary catabolic route in some leukocytes.(19) The presence of CES1 isoforms in rodent plasma could contribute to rapid 2-AG hydrolysis in those species; however, CES enzymes are not found in human plasma.20,21
It is generally agreed that the primary route to AEA begins with N-arachidonoylphosphatidylethanolamine (NAPE), which is synthesized by the transfer of AA from the sn-1 position of a donor phospholipid to phosphatidylethanolamine (PE) by a Ca2+-dependent N-acyltransferase (NAT). Hydrolysis of this precursor by an N-acylphosphatidylethanolamine-hydrolyzing PLD (NAPE-PLD) yields AEA13,14,22−26 (Figure 2b). Because NAT transfers a range of fatty acids from the sn-1 position of the donor phospholipid, NAPE biosynthesis is not specific for the incorporation of AA. Thus, this pathway produces a spectrum of fatty acyl ethanolamides, leading to a question of AEA specificity. Relevant to this question is the finding of Leung et al. that alternative pathways for the synthesis of AEA exist in mice bearing a targeted mutation of the known gene for NAPE-PLD.(27) As noted above, the major route of degradation of AEA and similar fatty acyl amides is hydrolysis by FAAH.(18)
Traditionally, the eicosanoid and endocannabinoid signaling systems have been investigated independently of one another, and it is conceivable that endocannabinoid signaling occurs in the absence of eicosanoid pathway activation and vice versa. However, the lipases that initiate both sets of pathways are responsive to some of the same second messengers (e.g., elevations in intracellular Ca2+). Therefore, it is likely that, in cells carrying the enzymatic machinery for both pathways, they will be activated together, presenting the potential for biochemical and pharmacologic cross-talk. As will be discussed below, the complexity of possible pathway interactions is increased by the fact that some enzymes of the eicosanoid biosynthetic pathways can metabolize endocannabinoids as well as AA. These considerations, combined with the sharing of common precursor lipid pools, guarantee multiple sites of interconnection. In this review, we will focus primarily on interactions that occur at the site of enzymatic reactions and receptor activation as outlined in the literature over the past 20 years. A considerable literature also exists on the effects of endocannabinoids on the expression of genes in the eicosanoid biosynthetic pathways and the converse. This topic will not be dealt with here.
2. Oxygenation of Endocannabinoids and Related Compounds by Eicosanoid Pathway Enzymes
Although the primary fate of endocannabinoids is inactivation through hydrolysis, increasing evidence indicates that these compounds are also subject to most of the oxidative metabolic pathways that lead to eicosanoid biosynthesis. Here we outline the specific enzymatic reactions shared by endocannabinoids and AA, compare their efficiency as substrates, and catalog the products formed.
2.1. Lipoxygenases: Studies with Purified or Partially Purified Proteins
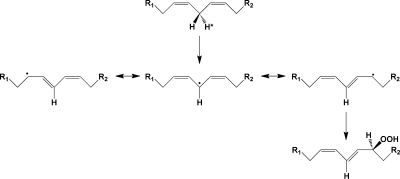
LOXs catalyze the abstraction of a hydrogen atom from a bisallylic carbon of a polyunsaturated fatty acid, followed by double bond migration and oxygen addition. The result is conversion of a 1,4-cis,cis-diene structure of the polyunsaturated fatty acid to a 1,3-cis,trans-5-peroxyl radical, which is reduced sequentially to a hydroperoxide and then the corresponding alcohol(28) (Figure 3). Most lipoxygenases will accept multiple fatty acids as substrates, but exhibit a high degree of regioselectivity and stereospecificity regarding the site and orientation of oxygen addition. The enzymes that metabolize AA are usually named by designating the number of the carbon atom where oxygen addition occurs. For most mammalian LOX enzymes, the antarafacial stereochemistry of oxygen addition leads to the formation of the (S)-hydroperoxide.(29)
Figure 3.
Mechanism of the lipoxygenase reaction. A hydrogen atom is abstracted from the bisallylic carbon of a polyunsaturated fatty acid, yielding a 1,5-pentadienyl radical. Addition of oxygen at the terminus of this radical yields a peroxyl radical, which is then reduced to the hydroperoxide.
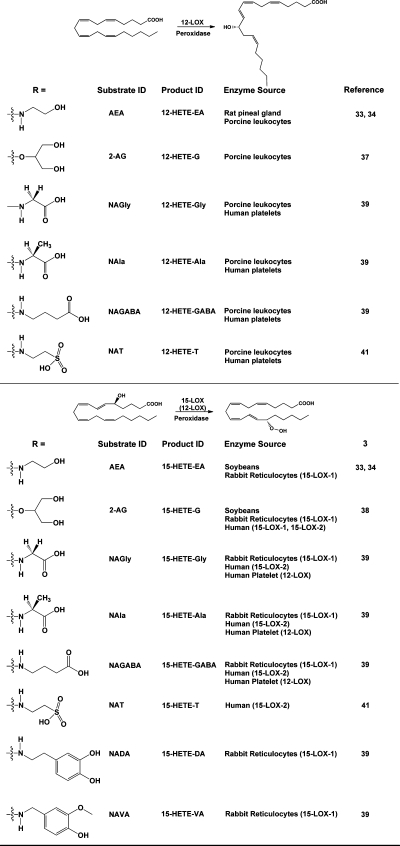
Although the majority of LOX enzymes display a preference for free fatty acid substrates, precedent exists for oxygenation of phospholipid-bound fatty acids and cholesteryl esters in the case of the rabbit reticulocyte 15-LOX, the human leukocyte 15-LOX-1, and the soybean 15-LOX-1.30−32 Thus, the possibility that ester or amide derivatives of AA could serve as LOX substrates was not unreasonable, and Hampson et al. and Ueda et al. were the first to test this hypothesis.33,34 Hampson et al. demonstrated metabolism of AEA by the 12-LOX enzymatic activity in an ammonium sulfate fraction of rat pineal gland. They identified the product of the reaction as the ethanolamide of 12-HETE (12-HETE-EA) following reduction of the hydroperoxide with NaBH4. This result indicated that the 12-LOX exhibited the same regioselectivity for AEA as it did for AA (Figure 4). Measurement of the total amount of product synthesized from AA and AEA suggested that the pineal gland 12-LOX metabolized the two substrates with equal efficiency. Hampson et al. went on to show that the purified 12-LOX from porcine leukocytes also catalyzed oxygenation of AEA to yield 12-HETE-EA and that the 15-LOX from soybeans produced predominantly 15-HETE-EA and minor amounts of 11-HETE-EA from AEA. In the case of the porcine 12-LOX, full kinetic studies yielded values for Km and Vmax that were statistically identical for AEA and AA, suggesting that the enzyme did not differentiate between the two substrates. In similar studies, Ueda et al. reported that the 12-LOX from porcine leukocytes, the 15-LOX-1 from rabbit reticulocytes, and the 15-LOX from soybeans could oxygenate AEA at rates roughly comparable to those for AA. In contrast, human platelet 12-LOX was only marginally active, and porcine leukocyte 5-LOX was inactive with AEA as the substrate. As for Hampson et al., characterization of the reaction products by Ueda et al. showed that the active enzymes exhibited the same regioselectivity for AEA as was observed for AA, producing the comparable ethanolamide product. Further characterization of the products of the porcine leukocyte 12-LOX and the soybean 15-LOX also confirmed that the stereospecificity of the reaction with AEA was identical to that of AA, with the major reduced products identified as 12(S)-HETE-EA and 15(S)-HETE-EA, respectively.
Figure 4.
Products of the action of lipoxygenases on endocannabinoids and related AA-derived compounds. The general reactions of 12-LOX and 15-LOX are shown, along with the structures of the endocannabinoid analogues that have been tested as substrates. The identities of the major products are indicated, along with the sources of the enzymes that have been shown to catalyze the reaction. Note that, in the case of the human platelet 12-LOX metabolizing arachidonoyl amino acids, both 12- and 15-hydroxylated products were identified. For this reason, 12-LOX is shown (in parentheses) as catalyzing the formation of this positional isomer.
Van der Stelt et al. carried out a structure–activity study, evaluating the capacity of the soybean 15-LOX to oxygenate linoleic acid and its amide, methylamide, dimethylamide, and ethanolamide derivatives.(35) The soybean enzyme oxygenated free linoleic acid at carbon 13, and the same regioselectivity was observed for all amides. Kinetic studies revealed similar Km values for the free acid, amide, and ethanolamide. Vmax values were similar for the free acid and ethanolamide, while the value for the amide was approximately 50% lower. Kinetic constants were not reported for the methylamide and dimethylamide. Zadelhoff et al. confirmed the ability of the soybean 15-LOX to efficiently metabolize AEA to the 15(S)-hydroperoxy product (Figure 4).(36) They also demonstrated that the 5-LOX enzymes from tomato and barley could metabolize AEA with efficiency equal to and better than, respectively, that of AA. However, these enzymes exhibited different regioselectivities for the two substrates, producing 11-HETE-EA, after reduction, from AEA in contrast to 5-HETE from AA.
Moody et al. extended the study of endocannabinoid lipoxygenation by demonstrating that the 12-LOX from porcine leukocytes, but not the enzyme from human platelets, could efficiently oxygenate 2-AG.(37) The reduced reaction product from the leukocyte enzyme was the glycerol ester of 12(S)-HETE (12(S)-HETE-G), indicating that the enzyme exhibited the same regio- and stereoselectivity with 2-AG as with AA (Figure 4). Kinetic studies with the porcine leukocyte 12-LOX revealed that the efficiency of 2-AG metabolism was approximately 40% as high as that of AA (as determined by kcat/Km), and a structure–activity relationship evaluation ranked a series of arachidonoyl esters as substrates from highest to lowest efficiency as 2-glyceryl ester (2-AG) > 1-glyceryl ester (1-AG) > hydroxyethyl ester > methoxyethyl ester > ethyl ester. This work was expanded by Kozak et al., who showed that soybean 15-LOX, rabbit reticulocyte 15-LOX-1, human 15-LOX-1, and human 15-LOX-2 all metabolized 2-AG efficiently, whereas potato and human leukocyte 5-LOXs showed no activity with this substrate.(38) Kinetic studies revealed that both human 15-LOX enzymes oxygenated 2-AG with efficiency equal or superior to that of AA, and structure–activity profiles for the soybean 15-LOX, rabbit reticulocyte 15-LOX-1, and human 15-LOX-2 were all similar to those observed for the porcine leukocyte 12-LOX as reported by Moody et al.(37)
The lipoamino acids are a class of compounds related to the endocannabinoids in that they are fatty acyl amides that may play a role in nociception and inflammation. Prusakiewicz et al. showed that the human platelet 12-LOX, porcine leukocyte 12-LOX, rabbit reticulocyte 15-LOX-1, and human 15-LOX-2 all metabolized the lipoamino acids N-arachidonoylglycine (NAGly), N-arachidonoylalanine (NAla), and N-arachidonoyl-γ-aminobutyric acid (NAGABA).(39) Following reduction, the products formed were the corresponding amino acid amides of 12- and 15-HETE, corresponding primarily to the regio-selectivity of each enzyme for AA. Efficiencies of lipoamino acid oxygenation based on kcat/Km values were between 42% and 105% of those of AA. This was the first report that the human platelet 12-LOX could metabolize a nonfree fatty acid substrate; however, the investigators noted that this enzyme displayed some loss of regioselectivity, as the amino acid amide derivatives of both 12-HETE and 15-HETE were produced (Figure 4). Prusakiewicz et al. also showed that reticulocyte 15-LOX-1 metabolized the vanilloid receptor ligands N-arachidon-oyldopamine (NADA) and N-arachidonoylvanillylamide (arvanil, NAVA) at 23–27% the rate of AA, while O-(3-methyl)-N-arachidonoyldopamine (OMDA) was not a substrate.(39) All of the vanilloids were poor substrates for the other LOX enzymes.
As noted above, the primary route of degradation of AEA is hydrolysis catalyzed by FAAH.(18) Saghatelian et al. have demonstrated that mice bearing a targeted deletion of the gene for FAAH exhibit an increase not only in fatty acyl ethanolamides, but also in N-acyltaurines, a previously unknown class of fatty acyl amides.(40) These compounds had no endocannabinoid activity, but were shown to activate multiple members of the transient receptor potential family of calcium channels. Turman et al. have reported that, like 2-AG and AEA, N-arachidon-oyltaurine (N-AT) is oxygenated by the human 15-LOX-2 and porcine leukocyte 12-LOX.(41) As in the case of the lipoamino acids, N-AT was also an efficient substrate for the human platelet 12-LOX. The human 15-LOX-2 and leukocyte 12-LOX exhibited the same regioselectivity for N-AT as for AA, but the platelet 12-LOX again showed some loss of regioselectivity, producing the taurine amides of both 12-HETE and 15-HETE (Figure 4).
In addition to AEA, the ethanolamides of other fatty acids have been detected in various tissues. The identification of docosahexaenoylethanolamide (DHEA) in the brain led Yang et al. to explore the lipoxygenase-dependent oxygenation of this endocannabinoid-related molecule.42,43 Incubation of human polymorphonuclear leukocytes (PMNs) or mouse brain homogenates with DHEA led to the formation of a number of oxygenated metabolites, including 17-hydroxy-DHEA, 10,17-, 14,17-, and 7,17-dihydroxy-DHEA, and (15-hydroxy-16(17)epoxydocosapentaenoyl)ethanolamide (15-HEDPEA). The production of 17-hydroxy-DHEA by incubation of DHEA with the 15-LOX from soybeans confirmed the LOX-dependent formation of this molecule. Furthermore, the presence of naturally occurring 17-hydroxy-DHEA in mouse brain homogenates suggests the possibility that lipoxygenation of DHEA may have physiological relevance.(43)
2.2. Cyclooxygenases: Studies with Purified or Partially Purified Proteins
The two COX isoforms catalyze the bisdioxygenation of AA, yielding the hydroperoxy endoperoxide PGG2 and the subsequent reduction of the hydroperoxide group of PGG2 to form PGH2 (Figure 5). The two enzymes exhibit 60% sequence identity and nearly overlapping three-dimensional structures. In vitro, their kinetics with AA as the substrate are very similar. Thus, research aimed at understanding the functional differences between the two isoforms has focused primarily on their differential expression. In most tissues, the gene for COX-1 is constitutively expressed, whereas COX-2 expression is inducible by stimuli such as growth factors, tumor promoters, and inflammatory agents. For this reason, it is generally believed that COX-1 produces PGs that regulate homeostatic functions, whereas COX-2 is responsible for PG formation in pathological states such as inflammation and tumorigenesis. Both COX isoforms are inhibited by nonsteroidal anti-inflammatory drugs (NSAIDs), such as aspirin, ibuprofen, and indomethacin, and this is believed to be the primary mechanism of action of these widely used pharmaceuticals. The association of COX-2 with the inflammatory response led to the development of COX-2-selective inhibitors (coxibs), with the expectation that such compounds would retain the anti-inflammatory activity of traditional NSAIDs, but with reduced side effects (e.g., gastrointestinal toxicity). Clinical experience with the coxibs that have reached the market (celecoxib, rofecoxib, valdecoxib) has supported this expectation; however, the recently discovered cardiovascular toxicity of these drugs has demonstrated that the relative roles of the two COX isoforms are not as clearly demarcated as was originally thought.44−47
Figure 5.
Mechanism of the cyclooxygenase reaction. The 13-pro-(S)-hydrogen of AA is removed by a radical at Y385 of the COX active site. The resulting radical migrates to position 11, which serves as the site of oxygen addition. Following the formation of the endoperoxide between carbons 11 and 9, a single bond links carbons 8 and 12 to form the prostanoid five-membered ring. The radical then migrates to carbon 15, which becomes the site of the second oxygen addition, forming a peroxyl radical, which is then reduced to a hydroperoxide (PGG2). Reduction of the 15-hydroperoxide using electrons from a coreductant (Ared) at the peroxidase active site yields PGH2.
COX-1, the first of the two isoforms to be discovered, has a strong requirement for a free carboxyl group in the substrate.48,49 Following the discovery of the endocannabinoids, Yu et al. challenged the assumption that this requirement also applies to COX-2.(48) Their discovery that purified COX-2, but not COX-1, could oxygenate AEA was the first demonstration of a substrate-based functional difference between the two isoforms. They showed that the product of the reaction of COX-2 with AEA was the ethanolamide of PGE2 (PGE2-EA) (Figure 6), but the efficiency of oxygenation of AEA was only about 18% as high as that of AA, on the basis of kcat/Km determinations. The major contributor to this difference in substrate preference was an approximately 4-fold higher Km for AEA as compared to AA. So et al. exploited this finding in their studies of the dynamics of catalysis and inhibition of the two COX isoforms.(50) They confirmed the results of Yu et al. and used AEA as a model substrate to explore the requirement of each enzyme for a free carboxyl group in the substrate.
Figure 6.
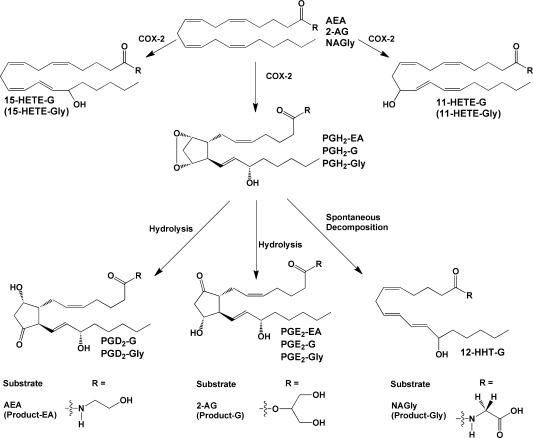
Products of the action of cyclooxygenase on endocannabinoids and related compounds. COX-2 produces PGH2 as the primary product, plus 11- and 15-HETE as minor products. In the absence of any additional metabolizing enzymes, PGH2 decomposes to form PGD2, PGE2, and HHT. Some or all of these compounds have been identified as products of the metabolism of endocannabinoid-like compounds as indicated above. For NAGly, a HETE-Gly derivative(s) was detected, but the exact position(s) of the hydroxyl group(s) was not determined. Endocannabinoid metabolism (2-AG and AEA) has been demonstrated for human COX-2; however, recombinant murine COX-2 has been used in the majority of these studies.
Kozak et al. performed structure–activity studies to explore the basis for AEA oxygenation using ovine COX-1 and murine COX-2.(51) For COX-2, the maximal rate of AEA oxygenation was 27% that of AA. For COX-1, this value was 11%, indicating that AEA is a preferred substrate for COX-2, but that COX-1 also has some capacity to oxygenate this substrate. Elimination of the terminal hydroxyl group of the ethanolamide moiety of AEA resulted in a marked reduction of oxygenation by both enzymes. Addition of a 1(S)-methyl group to the ethanolamide moiety of AEA increased catalytic activity approximately 2.4-fold for COX-2, but 3.6-fold for COX-1, reducing the COX-2:COX-1 selectivity ratio to 1.6:1 from 2.5:1 for AEA. In contrast, a 1(R)-methyl substituent increased the rate of COX-2 oxygenation by 1.2-fold, but decreased the rate for COX-1 by 75%, resulting in an increase in COX-2 selectivity to 12:1. The latter compound is (R)-methandamide, a metabolically stable AEA analogue that is frequently used in studies of AEA pharmacology. Dimethyl substitution at the 1-position of the ethanolamide resulted in activity similar to that of AEA for COX-2, but a 30% reduction in activity for COX-1 compared to that of AEA. Stereoselective effects were also observed with substitution at the 2-position of the ethanolamide, but the differences between the two COX isoforms were not as profound as those observed with 1-substitution. In most cases, replacement of the hydroxyl group of an analogue with a methoxy group decreased, but did not eliminate, oxygenation efficiency.(51)
Kozak et al. extended the study of COX-dependent endocannabinoid oxygenation to include 2-AG.(49) They showed that COX-2 metabolizes 2-AG with kcat and Km values similar to those of AA, while COX-1 utilizes this substrate poorly. The products of the reaction using purified COX-2 were the glyceryl esters of PGE2, PGD2, 11-HETE, 15-HETE, and 12-hydroxyheptadecatrienoic acid (HHT) (Figure 6). The relative quantities of these metabolites were not significantly different from those of the comparable free acid compounds formed when COX-2 metabolized AA. (Note that PGH2 spontaneously decomposes to form PGE2, PGD2, and HHT. 11- and 15-HETE are formed by monoxygenation of AA without cyclization.(44)) Structure–activity studies of various arachidonoyl esters revealed that 2-AG was utilized most efficiently followed by 1-AG and the hydroxyethyl ester. Esters that lack a free hydroxyl group were not substrates for COX-2, nor was cholesteryl ester or AA-containing DAG.
Prusakiewicz et al. demonstrated that substrate selectivity between the two COX isoforms is not restricted to 2-AG and AEA.(52) They reported that COX-2 but not COX-1 metabolizes the lipoamino acid NAGly with an efficiency approximately 10% that of AA and 23% that of 2-AG, but 8-fold higher than that of AEA, on the basis of kcat/Km values. The reaction yielded the glycine amides of products comparable to those obtained with AA (Figure 6). Expansion of these studies showed that COX-2 could also oxygenate NAla and NAGABA at 20–40% the maximal rate of AA, although the products of the reaction were not identified.(39) These are the first examples of charged substrates that are selective for COX-2. Neither COX isoform oxygenated N-AT or the vanilloids NADA, OMDA, or arvanil.39,41
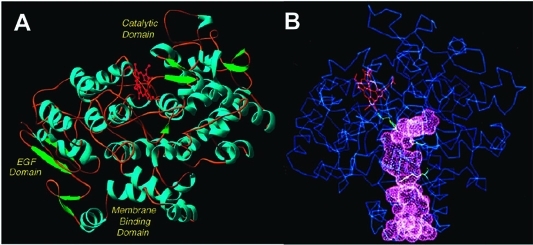
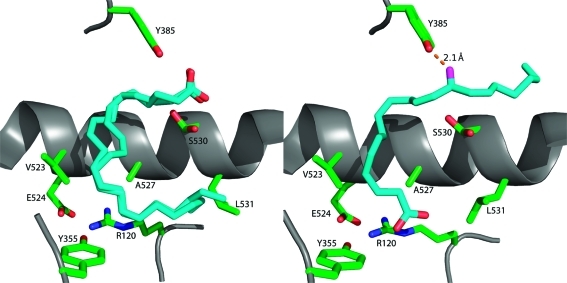
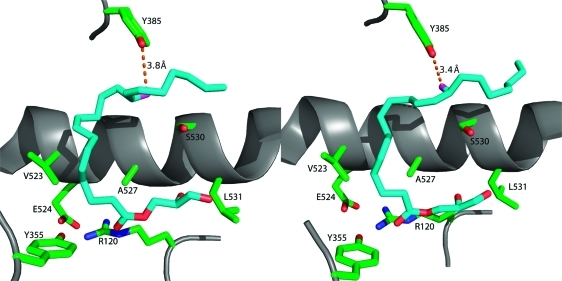
Recent crystal structures of murine COX-2 in complex with AA(53) or 1-AG(54) provide insight into the structural determinants of AA and 2-AG oxygenation. (Note that, under aqueous conditions, 2-AG undergoes rapid base-catalyzed acyl migration to form an equilibrium mixture of 1-AG and 2-AG at a ratio of approximately 8:2 (section 2.4).(55) Thus, use of 2-AG for crystallization studies is impractical.) Like COX-1, COX-2 is a homodimer, with each monomer consisting of an epidermal growth factor domain near the dimer interface, a membrane-binding domain, through which the enzyme interacts with one leaflet of the lipid bilayer, and a large catalytic domain. The active site of each monomer consists of a large “lobby” region that is bound by the four orthogonal α-helices of the membrane-binding domain. The roof of the lobby is demarcated by a “constriction site” formed by arginine-120, tyrosine-355, and glutamate-524. Above the constriction is a hydrophobic L-shaped channel that extends deep into the protein and forms the enzyme active site (Figure 7). In the muCOX-2:AA (mu = murine) structure, the orientation of substrate binding in this channel is distinctly different for the two monomers (Figure 8). In monomer B, AA is bound productively with its ω tail at the top of the channel and its carboxyl group forming a single hydrogen bond with tyrosine-355 at the constriction site. The cyclooxygenase reaction is initiated by a radical on tyrosine-385, which is located at the bend of the L-shaped channel (Figures 7 and 8). The binding orientation of AA in the active site places the tyrosyl radical in proximity to AA’s 13-pro-(S)-hydrogen, which is abstracted in the first step of the reaction mechanism (Figures 5 and 8). In monomer A, AA is bound unproductively in a reverse orientation with its carboxylate forming hydrogen bonds with tyrosine-385 and serine-530 at the bend of the active site channel. Movement of the side chain of leucine-531 provides room for AA’s ω tail to lie above arginine-120 at the constriction site (Figure 8).(53)
Figure 7.
(A) Domain structure of the COX enzymes. The N-terminus (not visible in the crystal structure) connects to the epidermal growth factor (EGF) domain. The EGF domain, in turn, connects to the membrane-binding domain, which comprises four α-helices (A–D). Helix D connects the membrane-binding domain to the large catalytic domain. α-helices are shown in blue, β-sheets are shown in green, and the heme prosthetic group is shown in red. (B) Drawing of the COX structure highlighting the hydrophobic channel of the cyclooxygenase active site. The large lobby region, which opens into the membrane-binding domain, is separated from the L-shaped channel above by a constriction. Key catalytic residues Tyr-385, Arg-120, and Ser-530 are shown in green. The heme prosthetic group is in red. These structural characteristics shown here for ovine COX-1 are representative of both isoforms. Panel B kindly provided by M. Garavito. Both figures reprinted from ref (44). Copyright 2003 American Chemical Society.
Figure 8.
Comparison of the conformations of AA in the active sites of the two monomers of murine COX-2. The left frame displays AA bound in monomer A of the COX-2 homodimer. It is bound in an inverted catalytically unproductive conformation in which its carboxyl group is H-bonded to Tyr-385 and Ser-530. The ω end of the fatty acid lies across Arg-120 and abuts Leu-531. Two molecules of AA are modeled in monomer A because of ambiguities in modeling from the electron density map. The right frame displays AA bound in monomer B of the homodimer in a productive conformation. The carboxylate is H-bonded to Arg-120, and the fatty acid chain projects up into the active site. The ω end of the fatty acid projects into an alcove above Ser-530, and the 13-pro-(S) hydrogen is located adjacent to the hydroxyl group of Tyr-385, which is converted to a tyrosyl radical during turnover.
The muCOX-2:1AG structure again reveals substrate binding in a productive conformation in monomer B and an unproductive conformation in monomer A; however, the differences in the two conformations are more subtle than those seen in the muCOX-2:AA structure (Figure 9). As for AA, the productive conformation of 1-AG in the active site of monomer B places the ω tail deep in the hydrophobic channel with tyrosine-385 oriented close to the 13-pro-(S)-hydrogen of 1-AG. Movement of the side chain of leucine-531 provides room for the 2,3-dihydroxypropyl moiety of 1-AG above the side chain of arginine-120. The only hydrophilic interaction between the substrate and the enzyme is a hydrogen bond between the phenolic hydroxyl of tyrosine-355 and the carboxyl oxygen of 1-AG. In monomer A, 1-AG is oriented as in monomer B, but the ω tail is not inserted deeply enough into the hydrophobic channel to bring the 13-pro-(S)-hydrogen into close proximity of tyrosine-385 (Figure 9). Hence, this conformation is unproductive.(54)
Figure 9.
Comparison of the conformations of 1-AG in the active sites of the two monomers of murine COX-2. The left frame displays 1-AG in an unproductive conformation in monomer A of the homodimer, whereas the right frame displays 1-AG bound in a productive conformation in monomer B of the homodimer. The two conformations are comparable, but the ω ends of the fatty acyl groups differ in conformation so that the 13-pro-(S) hydrogen is only close enough to Tyr-385 in monomer B to enable abstraction during the catalytic cycle.
In COX-1 an ionic interaction between the carboxylate of AA and arginine-120 is a key determinant of substrate binding. The absence of this interaction, confirmed by the muCOX-2:AA crystal structure, helps to explain COX-2’s ability to metabolize neutral ester and amide derivatives, which are poor substrates for COX-1. The data also suggest that the flexibility of leucine-531 is an important structural feature of COX-2 that allows it to accommodate the 2,3-dihydroxypropyl group of 1-AG; however, mutation of this residue to alanine, phenylalanine, or proline had minimal effect on the efficiency of oxygenation of AA, 1-AG, or 2-AG. For both AA and 1-AG, the only hydrophilic interaction with the enzyme was a hydrogen bond to tyrosine-355. Mutation of this residue to phenylalanine reduced the catalytic efficiency for metabolism of AA by approximately 80% (as determined by kcat/Km), while it increased the efficiency for metabolism of 2-AG by approximately 3-fold. The latter result was explained on the basis of elimination of hydrogen bonds between tyrosine-355 and other constriction site residues, giving 2-AG greater flexibility to attain an optimal binding position.(54)
A highly conserved difference between the COX isoforms is the presence of valine-523, arginine-513, and valine-434 in COX-2, as opposed to isoleucine-523, histidine-513, and isoleucine-434 in COX-1. The smaller side chains in COX-2 form a cavity, called the “side pocket”, which increases the volume of the active site. The side pocket has been exploited as a binding site for many highly effective COX-2-selective inhibitors. It is notable that the side pocket did not serve as a binding site for 1-AG in the muCOX-2:1AG structure, and mutation of arginine-513 to histidine had no effect on AA or 1-AG binding as observed in respective cocrystal structures obtained with the mutant enzyme. The R513H mutation also had no effect on the efficiency of 1-AG or 2-AG oxygenation (as determined by kcat/Km) in these studies.(54) Prior site-directed mutagenesis studies had suggested an important role for both the constriction site residues and the side pocket in the oxygenation of 2-AG, AEA, and lipoamino acids by COX-2.49,51,52,56 Thus, the near total absence of interaction between 1-AG and these active site residues as observed in the crystal structure was unexpected. However, both sets of studies agree that the overall binding orientation and reaction mechanism for endocannabinoid oxygenation are the same as for AA oxygenation.
2.3. Cytochromes P450: Studies with Purified or Partially Purified Proteins
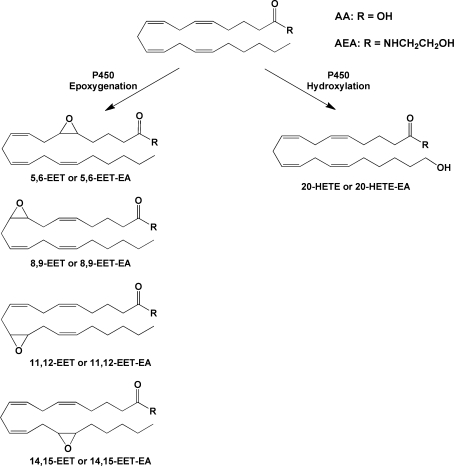
Cytochromes P450 are heme-containing monooxygenases that catalyze the NADPH-dependent biotransformation of a majority of clinically used drugs and xenobiotic toxicants as well as many endogenous substrates. There are 57 human P450 enzymes that display a wide range of substrate specificity, tissue distribution, and physiological function. P450 enzymes, primarily of the 2C and 2J families, catalyze the epoxygenation of AA at all four double bond positions, leading to the formation of 5,6-, 8,9-, 11,12-, and 14,15-EETs. P450s, primarily of the 4A and 4F families, catalyze terminal hydroxylation, leading to the formation of HETEs (Figure 10).(57)
Figure 10.
Examples of P450-catalyzed oxygenations of AA or AEA. Reactions shown are those that have been identified for AEA as well as AA. Additional reactions for AA have been reported.
Soon after the discovery of AEA, Bornheim et al. investigated its P450-mediated metabolism using the enzymatic activity in mouse tissue microsomes. They observed the formation of twenty and two metabolites using liver and brain microsomes, respectively. A combination of enzyme inducers and antibodies directed against specific P450s indicated that 3A family enzymes were primarily responsible for AEA metabolism by liver microsomes, but played a lesser role in the brain. These investigators did not identify the metabolites.(58)
More detailed studies of P450-mediated AEA metabolism did not occur until 2007, when Snider et al. reported the formation of the ethanolamides of EETs (EET-EAs) and HETE-EAs from the incubation of AEA with human kidney and liver microsomes. P450 4F2 was primarily responsible for AEA metabolism by kidney microsomes, and the only product formed was 20-HETE-EA. Human liver microsomes produced 5,6-, 8,9-, 11,12-, and 14,15-EET-EAs in addition to 20-HETE-EA. The major P450 responsible for EET-EA formation in liver microsomes was 3A4.(59) Soon thereafter, the same group reported that human recombinant P450 2D6 does not metabolize AA, but does convert AEA to 20-HETE-EA and all four EET-EAs. AEA was a high-affinity substrate for P450 2D6, and this enzyme accounted for most AEA oxygenation in brain mitochondrial preparations. Further studies demonstrated that a common genetic polymorphism of human P450 3A4 (I118 V) leads to a 60% reduction in EET-EA formation by this enzyme. One new monooxygenated and four new dioxygenated metabolites were formed by the I118V mutant enzyme as compared to products formed by the wild-type P450 3A4. These results suggest that P450 polymorphisms may be a source of variability in endocannabinoid metabolism and signaling.(60) Further support for this hypothesis came from studies of AEA metabolism by wild-type and polymorphic P450 2B6 and P450 2D6 enzymes.(61) Finally, the discovery that AEA is a high-affinity substrate for the orphan P450 4X1, with the primary product identified as 14,15-EET-EA, suggests that endocannabinoid metabolism may be an as yet unexplored function of additional P450 enzymes.(62)
Despite the considerable research on P450-mediated AEA oxygenation, little current evidence exists for direct metabolism of 2-AG. 2-AG was not a substrate for P450 4X1 and was not metabolized by microsomes from the livers and kidneys of rats, even following pretreatment by salt-loading or phenobarbital, which induces the expression of some P450s. Similarly, incubation of 2-AG with recombinant P450 2C8, 2C11, or 2C3, known AA epoxygenases, did not lead to product formation.62,63
2.4. Oxygenation of Endocannabinoids in Intact Cells and in Vivo
The capacity of some eicosanoid biosynthetic enzymes to metabolize endocannabinoids in vitro does not automatically imply that these reactions are of physiologic significance. Thus, investigators have explored this potential new biosynthetic pathway in intact cells and organisms. The first reported study of this nature was by Edgemond et al., who showed that human platelets convert exogenous AEA to 12(S)-HETE-EA and that human PMNs convert AEA to 15(S)-HETE-EA and to a lesser extent 12(S)-HETE-EA.(64) These results were generally consistent with those conducted in cell-free systems, although it is noteworthy that the finding with human platelets would not have been predicted from the poor efficiency of AEA oxygenation by the purified platelet 12-LOX in vitro (section 2.1).(34) Further studies of lipoxygenation of endocannabinoids in intact cells were reported by Moody et al. and Kozak et al., who showed that COS-7 cells transfected with porcine leukocyte 12-LOX or human 15-LOX-1 or 15-LOX-2 produced 12(S)-HETE-G or 15-HETE-G, respectively, from exogenous 2-AG.37,38 Kozak et al. also demonstrated conversion of 2-AG to 15-HETE-G by human keratinocytes, which constitutively express 15-LOX-2. Turman et al. incubated murine resident peritoneal macrophages (RPMs) with N-AT and identified 12-HETE-T along with minor amounts of 15-HETE-T as reaction products.(41)
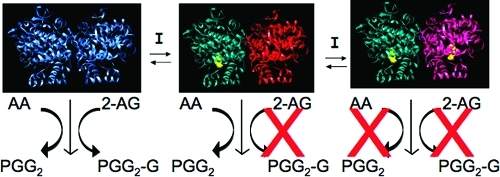
Treatment of the murine RAW264.7 macrophage-like cell line with bacterial lipopolysaccharide (LPS) and interferon-γ (IFN-γ) induces expression of COX-2. Kozak et al. showed that cells pretreated in this way synthesize the glyceryl ester of PGD2 (PGD2-G) from exogenous 2-AG and PGD2-EA from exogenous AEA.49,51 PGD2-G formation was also detected in the medium of LPS- and INF-γ-pretreated RAW264.7 cells exposed to ionomycin, which stimulates release of endogenous 2-AG. The finding that LPS/IFN-γ pretreatment was required and that synthesis was blocked by an isoform-selective COX-2 inhibitor verified that PGD2-G formation by RAW264.7 cells was COX-2-dependent in these experiments.
PGD2 is the major PG produced by RAW264.7 cells from AA. Therefore, the production of PGD2-G and PGD2-EA as the only COX-2-derived endocannabinoid products suggested that the PGD synthase in RAW264.7 cells is capable of efficiently metabolizing PGH2-G or PGH2-EA. PGH2 spontaneously decomposes to form PGD2 and PGE2 in ratios varying from 1:3 to 1:5, and PGH2-G and -EA suffer the same fate. Thus, in the absence of enzymatic conversion of the endoperoxide intermediates, one would expect to detect both PGE2 and PGD2 glyceryl ester or ethanolamide upon incubation of RAW264.7 cells with 2-AG or AEA, respectively, with PGE2 derivatives predominating. The preponderance of PGD2 derivatives suggests that PGH2-G and PGH2-EA are substrates for PGD syntheases. This discovery led Kozak et al. to explore the capacity of other PG synthases to metabolize PGH2-G and PGH2-EA.(51) They confirmed that purified hematopoietic PGD synthase, in the presence of COX-2, produced PGD2-G from 2-AG with an efficiency of about 50% compared to that of the conversion of AA to PGD2. Incubation of HCA-7 cells with 2-AG resulted in the formation of PGE2-G and PGF2α-G, while incubation with AEA resulted in the corresponding ethanolamides. These results suggested that the PGE and PGF synthases both accept PGH2-G and PGH2-EA as substrates, a conclusion that was confirmed for PGE synthase through incubation of the recombinant microsomal enzyme (mPGE synthase 1) with 2-AG in the presence of COX-2. PGE2-G was formed with efficiency approximately 60–75% of that of conversion of AA to PGE2. Similarly the reduction of PGD2-EA to PGF2α-EA by purified PGF synthase confirmed this enzyme’s ability to accept ethanolamide substrates.(65) Prostacyclin synthase in human platelet microsomes converted 2-AG and AEA to PGI2-G and PGI2-EA, respectively, in the presence of COX-2. The efficiency of PGI2-G and PGI2-EA synthesis was 70–80% of that of PGI2 synthesis from AA. Only human recombinant TX synthase showed poor ability to metabolize PGH2-G and PGH2-EA to the corresponding TXA2 analogues. Its ability to produce TXB2-G (the stable hydrolysis product of TXA2-G) from 2-AG in the presence of COX-2 was only 5% as high as the efficiency of TXB2 formation from AA.(51) Together, the results suggest that COX-2-dependent endocannabinoid oxygenation has the potential to produce a range of final products nearly as diverse as the products formed from AA oxygenation. The data also suggest that, with the exception of TX, the spectrum of products formed from 2-AG or AEA will be similar to that formed from AA in any given cell or tissue.
The studies discussed above clearly demonstrate that COX-2- and LOX-dependent endocannabinoid oxygenation can occur in the intracellular environment. However, most of these experiments were carried out with exogenous 2-AG or AEA, leaving unanswered the question of a cell’s ability to execute these biosynthetic pathways using substrates derived from endogenous lipid stores. To address this concern, Rouzer and Marnett investigated the biosynthesis of PG-Gs by murine RPMs in response to a zymosan stimulus.(66) Cells pretreated with LPS to induce COX-2 expression then exposed to a maximal zymosan stimulus synthesized approximately 16 pmol/107 cells of PG-Gs compared to 21 000 pmol/107 cells of PGs. The primary PG-Gs produced, PGE2-G and 6-ketoPGF1α-G (the stable breakdown product of PGI2-G), were consistent with the identity of the major PGs produced by these cells. Levels of free AA released in response to zymosan were approximately 10-fold higher than those of 2-AG, which partially accounted for the large differential in PG versus PG-G synthesis. However, even in the presence of 1 μM exogenous 2-AG, PGs were synthesized at higher levels (820 pmol/107 cells) than PG-Gs (78 pmol/107cells). Incubation of RPMs with exogenous PGE2-G indicated that the compound was stable, so degradation did not account for the comparatively low yield of PG-Gs in these cells. In contrast, exogenous 2-AG was rapidly hydrolyzed to AA, which accounted for PG synthesis upon addition of this substrate.
Murine RPMs constitutively express high levels of COX-1, so LPS-pretreated cells contain both isoforms of the enzyme. Rouzer and Marnett showed that selective inhibition of COX-2 by SC236 in these cells reduced zymosan-stimulated PG production by 17% and PG-G production by 49%.(66) This result suggested that the majority of PG formation and a substantial quantity of PG-G formation by the cells were COX-1-dependent. In contrast, when LPS-pretreated RPMs were exposed to exogenous 2-AG, SC236 reduced PG and PG-G synthesis by 76% and 88%, indicating a predominant role for COX-2 under these conditions. The apparent involvement of COX-1 in zymosan-stimulated PG-G synthesis was further explored using RPMs from mice bearing targeted deletions of the genes for COX-1 or COX-2.(67) These results confirmed a major role for COX-1 in zymosan-dependent PG and PG-G formation as indicated by the finding that COX-1 knockout markedly reduced the synthesis of both classes of product, whereas the effect of COX-2 knockout was not statistically significant. Knockout of either enzyme substantially reduced the synthesis of both PGs and PG-Gs from exogenous 2-AG. A somewhat different approach was taken by Yu et al., who replaced the gene for COX-2 with the gene for COX-1, generating a COX-1 > COX-2 “knockin” mouse.(68) They found that LPS-pretreated RPMs from these mice produced lower quantities of PG-Gs from exogenous 2-AG than RPMs from wild-type mice, again supporting a predominant role for COX-2 in the case of exogenously provided endocannabinoid. The substantial role for COX-1, particularly in zymosan-stimulated PG-G synthesis, was unexpected considering the fact that 2-AG is a relatively poor substrate for this enzyme. Kinetics studies suggested the possibility that COX-2 is rapidly inactivated in zymosan-stimulated cells. If this is true, it would also help to explain the low level of PG-G synthesis in response to zymosan, since COX-1-dependent oxygenation of 2-AG would be expected to be inefficient.
Attempts to detect oxygenation products of endocannabinoids in vivo have met with some success. Weber et al. detected low levels of PGE2-EA and PGD2-EA in the lungs and kidneys of wild-type mice following intravenous injection of AEA.(69) Higher levels of these compounds, in addition to PGF2α-EA, were detected in the lungs, kidneys, livers, and small intestines of mice bearing a targeted deletion of the gene for FAAH, but only after AEA injection. In these mice, knockout of FAAH reduced hydrolysis of AEA, providing higher levels of this substrate for oxygenation by COX-2. These results confirm that PG-EAs can be formed in vivo, but the conditions required for their detection in this study were not physiological.
Hu et al. provided convincing evidence of the presence of PGE2-G in homogenates of the hind paws of rats.(70) Quantities of PGE2-G detected were low (∼190 fmol/paw) compared to those of PGE2 (∼140 pmol/paw) and 2-AG (∼800 pmol/paw), and levels were undetectable in the brain and spinal cord. Higher quantities of PGE2-G were detected in the paws from rats pretreated with MAG lipase inhibitors, which prevented 2-AG breakdown, thus providing higher levels of substrate for PGE2-G formation. Lower levels of PGE2-G were detected in the paws of animals treated with COX inhibitors. However, no change in PGE2-G levels resulted from carageenan injection, which induces an inflammatory response in the paw accompanied by increased expression of COX-2. It should be noted that careful comparison of the published mass spectrum of the PGE2-G isolated from rat paw to that of the standard suggests the presence in the paw sample of a second compound of 2 units higher mass-to-charge ratio. It is quite possible that the material isolated from the paw was actually a mixture of PGE2-G and PGF2α-G, which would likely not have separated under the conditions used for chromatography in that study.
Although detection of PG-Gs in vivo has proved challenging, Chen et al. reported readily measurable quantities of the glyceryl esters of P450-derived EETs, including 2-(11,12-epoxyeicsatrienoyl)glycerol (2-11,12-EET-G) and 2-(14,15-epoxyeicosatrienoyl)glycerol (2-14,15-EET-G), in lipid extracts from rat spleen and kidney. 2-11,12-EET-G alone was found in the brain.(71) Quantities of the epoxygenase metabolites ranged from 0.2 to 1.5 ng/g of tissue as compared to those of 2-AG, which ranged from 5 to 11 ng/g of tissue. Therefore, these metabolites appear to be present in much higher quantity in vivo than PG-Gs. This discovery is notable in that these glyceryl epoxygenase metabolites are the only oxygenated endocannabinoids that were discovered in vivo prior to being generated enzymatically in vitro. However, the failure to demonstrate direct P450-dependent 2-AG epoxygenation (section 2.2) leaves the origin of these species unclear.
2.5. Metabolic Fate of PG-Gs and PG-EAs
To understand the production of oxygenated endocannabinoids detected in vivo requires knowledge of the chemical and metabolic fate of these compounds. 2-AG is initially formed containing AA in the sn-2 position of glycerol as a result of the fact that most AA in the parent phospholipid pool is esterified at this location. However, in aqueous solution, the arachidonoyl moiety of 2-AG undergoes acyl migration to the sn-1 position, yielding an equilibrium mixture of 1-AG and 2-AG at a ratio of roughly 8:2. This base-catalyzed first-order reaction occurs with a half-life of approximately 10 min at 37 °C and pH 7.4 in the absence of serum and 2.3 min in the presence of 10% serum.(55) Thus, it is unclear whether the substrate encountered by COX-2 in vivo is most likely to be 2-AG or 1-AG, although as noted above (section 2.2), both isomers are recognized by the enzyme. Similarly, PG-Gs and HETE-Gs synthesized from 2-AG are subject to acyl migration, so that these compounds might be present as either the 1- or 2-glyceryl esters in vivo.
Although 2-AG and PG-Gs are subject to acyl migration, these compounds and the fatty acyl ethanolamides are highly stable to chemical hydrolysis under physiological conditions. Thus, Kozak et al. explored their metabolic fate.(72) When injected intravenously into rats, PGE2-G disappeared from the circulation within 5 min, while PGE2-EA exhibited a plasma half-life of over 6 min and a large volume of distribution. Consistent with these findings, PG-Gs were rapidly hydrolyzed in rat plasma with a half-life of 14 s ex vivo, whereas PG-EAs were stable. In contrast, PG-Gs were much more stable in human plasma and whole blood (half-lives of >10 and 7 min, respectively), and no hydrolysis was observed in canine, bovine, or human cerebrospinal fluid. PG-EAs were stable to hydrolysis in all of these biological fluids.
Human 15-hydroxyprostaglandin dehydrogenase, the enzyme primarily responsible for inactivation of PGs, oxidized PGE2-G and PGE2-EA less efficiently than PGE2. Products of the reaction indicated that PGE2-G was oxidized only at carbon 15. The enzyme was nearly inactive with PGF2α-G. Together, the results suggest that PG-EAs are relatively stable metabolically to enzymatic hydrolysis and oxidation. Thus, failure to detect these compounds in vivo is not likely due to rapid oxidation to 15-keto derivatives. In contrast, PG-Gs are subject to rapid hydrolysis, especially in frequently used rodent models, a conclusion also supported by Hu et al., who observed rapid hydrolysis of PGE2-G upon injection into rat paw.(70) The product of PG-G hydrolysis is the corresponding PG. Thus, failure to detect these compounds in vivo may be due to rapid conversion to PGs, which are indistinguishable from PGs formed directly from AA.
The rapid hydrolysis of PG-Gs in vivo led investigators to explore the enzymes that might catalyze this reaction. Potential candidates include FAAH and MAG lipase, the enzymes primarily responsible for hydrolysis of AEA and 2-AG, respectively. However, Ross et al. showed that FAAH inhibitors had no effect on the binding of PGE2-EA to various membrane preparations.(73) Similarly, Fowler and Tiger showed that PGD2-G, PGE2-G, and PGF2α-G did not block the hydrolysis of AEA or 2-oleoylglycerol by cytosolic and membrane fractions from rat brain homogenates,(74) and Matias et al. showed that PGD2-EA, PGE2-EA, and PGF2α-EA did not block AEA hydrolytic activity in N18TG2 cell membranes, which are rich in FAAH.(75) These results suggest that PG-Gs and PG-EAs do not interact with FAAH or MAG lipase. This conclusion was further supported by Vila et al., who showed that PG-Gs are poor substrates for purified FAAH and MAG lipase and that specific inhibitors of these enzymes only partially blocked the hydrolysis of PGE2-G in RAW264.7 cells and dog brain homogenates.(76)
The discovery that human CES1 and CES2 can efficiently metabolize both PGE2-G and PGF2α-G, but not the corresponding ethanolamides, provides some insight into the mechanism of PG-G catabolism. CES1 was responsible for 80% and 97% of PGE2-G and PGF2α-G hydrolysis, respectively, in cultured human THP-1 monocytic leukemia cells.(19) The presence of CES enzymes in rodent, but not human, plasma could explain the striking species differences in PG-G half-lives in these body fluids.20,21,72 Thus, it appears likely that rapid hydrolysis of PG-Gs is catalyzed by an enzyme distinct from FAAH or MAG lipase, but the identity of the enzyme is unknown.
Biologically active free acid EETs are inactivated by epoxide hydrolases, which catalyze hydrolysis to the corresponding dihydroxy metabolites. Similarly, 5,6-EET-EA was subject to epoxide hydrolase-mediated hydrolysis, while exhibiting resistance to hydrolysis of the amide bond by FAAH. Resistance to FAAH rendered 5,6-EET-EA more stable in mouse brain homogenates than AEA.(77)
3. Cross-Talk between the Endocannabinoid and Eicosanoid Signaling Pathways
The common role of AA and the finding of oxygenation of endocannabinoids by some eicosanoid biosynthetic enzymes suggest a number of possible ways in which cross-talk between the endocannabinoid and eicosanoid pathways may occur. These include (1) hydrolysis of endocannabinoids to provide AA for eicosanoid biosynthesis, (2) production of oxygenated endocannabinods that are later hydrolyzed and act at eicosanoid receptors, (3) production of oxygenated endocannabinoids that act at eicosanoid or endocannabinoid receptors, (4) production of oxygenated endocannabinoids that act at distinct receptors, and (5) termination of endocannabinoid signaling by oxygenation of 2-AG or AEA. These possibilities have been explored to varying degrees, and results suggest that at least some of the potential cross-talk scenarios do, in fact, occur in cells and in vivo, whereas others are unlikely. Details of these investigations and the challenges that have arisen in these studies are outlined below.
3.1. Endocannabinoids as a Source of Free Acid Eicosanoids
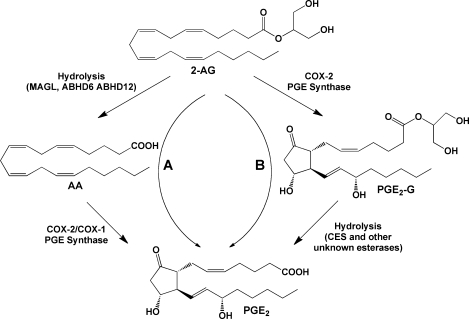
As noted above, 2-AG or AEA, produced from endogenous stores or provided exogenously, is subject to hydrolysis, yielding free AA, which may then be oxygenated by any eicosanoid biosynthetic enzymes that are present in the cell. This yields the corresponding free acid product (example for COX-dependent metabolism provided in Figure 11, pathway A). Alternatively, 2-AG or AEA could be oxygenated first, in which case the product eicosanoid glyceryl ester or ethanolamide is also subject to hydrolysis to produce the free acid eicosanoid (Figure 11, pathway B). The free acid eicosanoids produced by either pathway are indistinguishable from each other and from eicosanoids formed from AA that is released directly by PLA2-dependent pathways. This complicates the interpretation of data from experiments involving endocannabinoids in which free acid eicosanoid levels are measured or their pharmacologic effects are observed.
Figure 11.
Hydrolytic metabolism of 2-AG and PG-Gs. Pathway A: 2-AG may be hydrolyzed to AA, which is then subject to oxygenation by COX-2 or COX-1, yielding free acid PGs (illustrated here by PGE2). Pathway B: Oxygenation of 2-AG will produce PG-Gs (illustrated here by PGE2-G), hydrolysis of which will yield the corresponding free acid PG. The origin of the free acid PG product, through oxygenation of AA or 2-AG, cannot readily be distinguished.
A number of approaches help to distinguish the source of free acid eicosanoids in the complex cellular environment. Inhibitors of FAAH or MAG lipase block endocannabinoid hydrolysis, but not the hydrolysis of PG-Gs or PG-EAs. Thus, these inhibitors lower the level of eicosanoids formed from hydrolysis of endocannabinoids followed by oxygenation (Figure 11 pathway A), but not those formed by oxygenation followed by hydrolysis (Figure 11, pathway B). MAG lipase and FAAH inhibitors should also have no effect on eicosanoids synthesized from AA provided directly by PLA2-dependent phospholipid hydrolysis. When exogenous substrates are provided, the use of nonhydrolyzable endocannabinoid analogues, such as (R)-methandamide and 2-arachidonoyl glyceryl ether (noladin ether) in place of AEA and 2-AG, respectively, will prevent the formation of free acid eicosanoids by pathway A and will yield nonhydrolyzable eicosanoid amide or ester analogues by pathway B. COX and LOX inhibitors are frequently used to confirm the enzymatic origin of an oxygenated metabolite and may help to distinguish between free acid eicosanoids generated by pathway A versus pathway B. For example, nonselective COX inhibitors should block prostanoid synthesis occurring through either pathway, whereas a COX-2-selective inhibitor should completely block pathway B while having a variable effect on pathway A, depending on the isoforms involved. (However, see section 2.4 above regarding COX-1-dependent PG-G synthesis in RPMs.) Finally, when pharmacologic effects are being monitored, cannabinoid and prostanoid receptor antagonists help to distinguish between the pharmacologic activities of the parent endocannabinoid and its possible eicosanoid metabolites.
A substantial number of studies have used exogenously supplied AEA or 2-AG to investigate the pharmacology of these compounds. In some cases, exploration using inhibitors, receptor antagonists, and/or nonhydrolyzable endocannabinoid analogues has revealed that the observed physiological responses were likely due to hydrolysis of the endocannabinoid followed by COX- or LOX-dependent oxygenation of the resultant AA. A number of these studies focused on cardiovascular responses to endocannabinoids, such as contraction or relaxation of isolated vascular ring preparations78,79 and changes in arterial pressure in perfused lung.(80) However, such disparate responses as inhibition of prostate carcinoma cell invasiveness and cytotoxicity toward squamous cell carcinoma cultures have also been reported to be eicosanoid-mediated.81,82 These studies concluded that endocannabinoid hydrolysis produced AA for eicosanoid biosynthesis and identified the eicosanoid species likely responsible for the observed effect. However, a number of other studies have suggested eicosanoid-mediated effects of endocannabinoids without fully identifying the active species. Among the activities reported are relaxation of bovine arterial rings,(83) cerebral vasodilation in the rabbit,(84) induction of emesis in the shrew,(85) promotion of adipocyte differentiation,(86) inhibition of IL-2 secretion in splenocytes,(87) inhibition of 5-hydroxytryptamine receptor-induced head twitch in the mouse,(88) and non-CB receptor-dependent responses in a battery of cannabinoid responsiveness tests in mice.(89) Finally, some experiments have suggested that endocannabinoids can stimulate the release of AA from phospholipid stores, leading to eicosanoid synthesis that does not depend on endocannabinoid hydrolysis.90−92
Although the majority of these studies concluded that free acid eicosanoids were formed by endocannabinoid hydrolysis followed by oxygenation of the resultant free AA, the possibility of endocannabinoid oxygenation followed by hydrolysis of the glyceryl ester or ethanolamide product remained a viable option in some cases.82,88 As noted above, PG-EAs and PG-Gs are resistant to degradation by 15-hydroxyprostaglandin dehydrogenase when compared to their free acid counterparts. Thus, it is conceivable that these compounds could serve as a more metabolically stable pool of PGs that is transported to distant sites prior to hydrolysis and receptor binding. At present, there is relatively little direct evidence to support this as a significant PG signaling pathway.
Despite clear demonstrations that some endocannabinoid pharmacology is really eicosanoid pharmacology resulting from hydrolysis followed by oxygenation, there are also many compelling studies in which exogenously provided endocannabinoids act intact at CB receptors with no involvement of oxygenase enzymes. Excellent examples are seen in studies of the neurologic effects of endocannabinoids and in their modulation of membrane transporters. A major role of cannabinoids in the nervous system is seen in the phenomena of depolarization-induced suppressions of inhibition (DSI) and excitation (DSE), which occur in the hippocampus and cerebellum. These regions of the brain contain large neurons that are regulated by smaller inhibitory GABAergic or excitatory glutaminergic interneurons. Electrical or neuronal stimulation resulting in depolarization of the large neuron stimulates release of endocannabinoids, which travel to the smaller interneuron and suppress its activity through CB1-mediated signaling. Both DSI(93) and DSE(94) in hippocampal neurons were found to be due to the direct action of endocannabinoids without the influence of oxygenated metabolites. Similarly, inhibition of dopamine transport,(95) inhibition of Ca2+ efflux from T-tubule membranes,(96) stimulation of glucose uptake,(97) inhibition of endothelin 1-mediated Ca2+ mobilization,(98) and contraction of colonic smooth muscle(99) are not dependent on the activity of oxygenases. Finally, in contrast to the numerous studies showing that vascular effects of endocannabinoids were eicosanoid-mediated (see above), Gardiner et al. showed no effect of COX inhibition on the hemodynamic response of conscious rats to AEA.(100)
These studies vary tremendously with regard to how extensively the potential cross-talk between endocannabinoid and eicosanoid signaling was explored. In some cases, the simple inclusion of a COX inhibitor was used to show that an observed effect was not eicosanoid-mediated. In other cases, multiple inhibitors and receptor blockers were used, and the final eicosanoid product mediating an effect was identified. Thus, it is likely that further refinements of some of these observations will be made in the future. It is also important to note that the finding that exogenous endocannabinoids can serve as a source of eicosanoids does not necessarily mean that endogenously generated endocannabinoids are an important source of eicosanoids in vivo. The addition of exogenous endocannabinoids to cell culture media or a tissue bath provides a large absolute quantity of compound that can be hydrolyzed to free AA.(67) Such quantities of endocannabinoids are not available from endogenous sources. In fact, the levels of endocannabinoids produced in stimulated cells are usually at least an order of magnitude lower than the quantity of free AA released for eicosanoid biosynthesis. The majority of evidence suggests that the primary source of AA for stimulus-dependent eicosanoid biosynthesis in most cells/tissues is provided by PLA2-mediated phospholipid hydrolysis. However, in vivo studies in mice have shown that blockade of MAG lipase activity by organophosphorous nerve agents, a specific inhibitor (JZL184), or gene knockout results in increased 2-AG and decreased AA levels in brain tissue.101−103 Similarly, mice bearing a targeted deletion of the gene for DAG lipase-α exhibit decreased brain levels of both 2-AG and AA when compared to wild-type controls.(104) These results suggest that 2-AG is a source of steady-state AA in mouse brain. In addition, the finding of substantial PG biosynthesis in zymosan-stimulated macrophages from cPLA2–/– mice suggests the possibility that endogenously generated AEA or 2-AG could serve as a source of AA for stimulus-dependent eicosanoid synthesis under some circumstances.(67)
3.2. Oxygenated Endocannabinoids as Receptor Ligands
3.2.1. Activity at Known Receptors
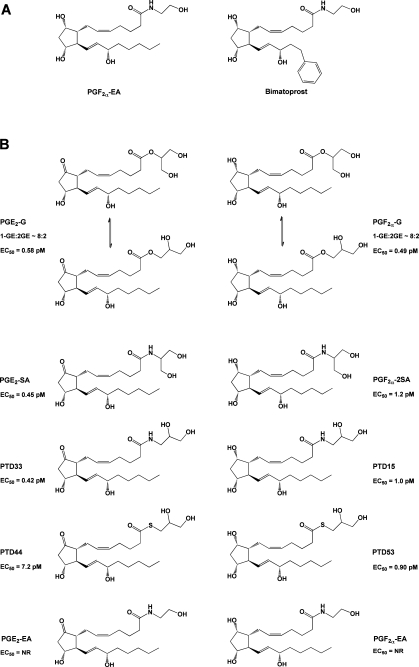
Definitive studies have shown that COX-derived oxygenated endocannabinoids are not ligands for the traditional eicosanoid or endocannabinoid receptors. Pinto et al. showed that the ethanolamides of PGE2, PGA2, PGB1, and PGB2 do not bind to the CB1 receptor, and Ross et al. showed that PGE2-EA’s affinity for the four EP receptors was at least 2 orders of magnitude lower than that of PGE2.73,105 Similarly, Nirodi et al. reported that the binding affinity of PGE2-G was at least 2 orders of magnitude lower than that of PGE2 at all four EP receptors and that the compound was completely inactive at the FP, DP, TP, and IP receptors.(106) An analogue of PGF2α-EA, bimatoprost (Figure 12A), is used clinically for the treatment of glaucoma. Hence, there is an extensive literature on the pharmacology of PGF2α-EA, indicating that it does not interact with the FP receptor.75,107 Considering the fact that oxygenated endocannabinoids are usually present at much lower levels than free acid eicosanoids in vivo, it is highly unlikely that these compounds can successfully compete for binding to the traditional prostanoid receptors. Thus, there is no current evidence that PG-Gs or PG-EAs act as endocannabinoids or free acid prostanoids or that they serve as antagonists for these compounds through direct receptor interactions.
Figure 12.
(A) Structures of PGF2α-EA and bimatoprost. (B) Structures of compounds tested for the ability to mobilize Ca2+ in RAW264.7 cells and H1819 cells.(115) Note that, in aqueous solution, PG-Gs rapidly equilibrate to form a mixture of the 1- and 2-glyceryl esters, with the 1-glyceryl ester predominating at a ratio of approximately 8:2. In contrast, the stereochemistry of the corresponding amide analogues remains fixed. EC50 values are given for Ca2+ mobilization in H1819 cells. NR indicates no response.
In contrast, Edgemond et al. and Van der Stelt et al. showed that the 12(S)-, 12(R)-, and 20-HETE-EAs have nearly the same affinity for the CB1 receptor as AEA.64,108 12(S)- and 20-HETE-EA were also comparable in affinity to AEA in binding to CB2. 15(S)-HETE-EA exhibited poor affinity for both receptors. Similarly, the l5-LOX product of linoleoyl amide and linoleoyl-EA demonstrated no affinity for CB1.(109) These results were supported by Hampson et al.,(33) who showed that 12-HETE-EA was more active than 15-HETE-EA in eliciting cannabinoid-dependent contraction of mouse vas deferens and in blocking forskolin-mediated cAMP production. In contrast, Ueda et al.(34) found higher activity for 15-HETE-EA than for 12-HETE-EA in the vas deferens assay. Thus, it appears that at least some LOX-derived metabolites of AEA have the potential to act as endocannabinoids.
Yang et al. have reported that the DHEA-derived lipoxygenase metabolites 10,17-dihydroxy-DHEA and 15-HEDPEA have endocannabinoid activity. Both of these compounds showed potency comparable to that of AEA and superior to that of DHEA at the CB2 receptor. They were also active at the CB1 receptor, but required considerably higher concentrations than AEA. In addition to CB receptor binding, 10,17-dihydroxy-DHEA and 15-HEDPEA inhibited chemotaxis of human PMN, blocked leukocyte–platelet aggregate formation, and exhibited protective activity in a mouse model of reperfusion second organ injury. It is unclear, however, whether these effects are mediated by the activity of these compounds at CB receptors or as yet unidentified receptors.(43)
Snider et al. showed that the P450-dependent metabolite 5,6-EET-EA has a higher affinity for the CB2 receptor than its parent AEA. Increased biosynthesis of this compound was observed concomitantly with augmented CB2 expression in IFN-γ-stimulated microglia, suggesting that this pathway may play a role in inflammatory signaling in these cells. Chen et al. showed that the P450 epoxygenase metabolites 2-11,12-EET-G and 2-14,15-EET-G have affinity for and pharmacologic activity at CB1 and CB2.(71) These compounds were detected in sizable quantities in vivo, suggesting that they could play a significant role in endocannabinoid signaling.
Some evidence has been presented that oxygenated eicosanoids may act at peroxisome proliferator-activated receptors (PPARs). Kozak et al. reported that 15-HETE-G, but not 15-HETE, is an agonist at PPAR-α in NIH 3T3 cells expressing a PPAR-α-dependent luciferase reporter gene.(38) Ghosh et al. demonstrated that 2-AG activates PPAR-δ in human vascular endothelial cells by a process that requires COX-2 and prostacyclin synthase.(110) PPAR-δ activation was also observed with AEA and the nonhydrolyzable analogue of 2-AG, noladin ether, but not with AA. These results suggest that 2-AG is converted to PGI2-G, which then serves as a PPAR-δ agonist. However, 6-keto-PGF1α-G, the stable breakdown product of PGI2-G, was not detectable in the cells. Similarly, Rockwell et al. demonstrated that 2-AG, AEA, and noladin ether inhibit IL-2 secretion in activated Jurkat T cells and primary splenocytes.(111) The effect was blocked by selective inhibitors of COX-2 and a PPAR-γ antagonist. The results suggest that the effect was due to a COX-2-dependent metabolite of 2-AG; however, the finding that the same effect could be observed upon addition of AA suggests that a free acid PG may be the active agent.(87)
3.2.2. Activity at Novel Receptors
A number of reports suggest that PG-Gs and/or PG-EAs have biological activities distinct from those of their free acid counterparts and may act at novel receptors. The most extensive studies of this nature have focused on the biological activity of PGF2α-EA because, as noted above, an analogue of this compound is used clinically in the treatment of glaucoma. In the eye, PGF2α-EA and its clinical counterpart bimatoprost (Figure 12A) have effects similar to those of PGF2α on ocular tension. However, extensive pharmacologic data indicate that these compounds do not act at the FP receptor.75,107 The discovery of antagonists that block the action of PGF2α-EA and bimatoprost but not PGF2α in the eye further supports the conclusion that there are distinct sites of action for these two compounds.112,113 Efforts to characterize a specific PGF2α-EA receptor led Liang et al. to identify six splice variants of the FP receptor in human ocular tissues.(114) They showed that HEK293/EBNA cells coexpressing the wild-type FP and the altFP4 splice variant responded to both PGF2α and PGF2α-EA binding with distinct patterns of Ca2+ mobilization. The response to PGF2α-EA but not PGF2α was blocked by antagonists to bimatoprost. Only PGF2α mobilized Ca2+ in HEK298/EBNA cells expressing the wild-type FP receptor alone. The FP receptor exists as a homodimer. Liang et al. showed that cells expressing both wild-type FP and altFP4 form heterodimers of the two receptor gene products. They propose that it is this heterodimeric receptor that responds to PGF2α-EA and bimatoprost. It will be interesting to see if this paradigm applies to other biologically active ester and amide derivatives of prostanoids.
Although not as advanced as the pharmacology of PGF2α-EA, some progress has been made on characterizing distinct biological activities of PG-Gs. Nirodi et al. showed that PGE2-G, but not PGD2-G, or PGF2α-G induced Ca2+ mobilization in RAW264.7 cells.(106) The EC50 for this response was 1 pM, as compared to 15 nM for PGF2α. Nirodi et al. demonstrated that no significant hydrolysis of PGE2-G occurred within the time frame of the response, and PGE2 did not elicit Ca2+ mobilization in these cells. PGE2-G induced a transient increase in inositol 1,4,5-trisphosphate (IP3) levels and the membrane association of protein kinase C (PKC). An increase in levels of the phosphorylated forms of the mitogen-activated protein kinases (MAPKs) and extracellular signal regulated kinases (ERKs) 1 and 2 also occurred, and this response was attenuated by pharmacologic blockade of the IP3 receptor and inhibitors of PKC and PLCβ. The investigators concluded that PGE2-G, likely through binding to an as yet unknown receptor, stimulates activation of PLCβ, leading to IP3 production, Ca2+ mobilization, PKC activation, and ultimately MAPK phosphorylation and activation. Follow-up studies by Richie-Janetta et al. showed that the human non-small-cell lung cancer cell line H1819 also responds to PGE2-G with Ca2+ mobilization and that the maximal response in these cells (2.5–6-fold) was greater than that of RAW264.7 cells (1.5–3.5-fold).(115) Structure–activity relationship experiments showed that the cells responded similarly to PGE2-G and its analogue PGE2-serinol amide (Figure 12B). PGE2-serinol amide is more stable to ester hydrolysis than PGE2-G and does not undergo acyl migration, allowing the investigators to show that both RAW264.7 cells and H1819 cells responded similarly regardless of whether the PGE2 moiety was linked at the sn-1 or sn-2 position of the glycerol. In contrast, replacing the ester linkage with a thioester resulted in a marked reduction in potency. In these studies, PGF2α-G and its amide analogues were found to have activity similar to that of PGE2-G. The ethanolamides of both PGE2 and PGF2α were inactive. These results suggest that the putative receptor recognizes key features of the glyceryl headgroup but is not highly specific for the substituents on the prostanoid ring.
As noted above (section 3.1), in the hippocampus 2-AG invokes DSI, which can be measured electrophysiologically as decreases in miniature inhibitory postsynaptic currents (mIPSCs). Sang et al. showed that PGE2-G, PGD2-G, PGF2α-G, and PGD2-EA but not PGE2-EA or PGF2α-EA have an effect opposite that of 2-AG, invoking increases in mIPSCs in mouse hippocampal neurons.(116) Since PGE2 and PGD2 had an effect opposite that of their corresponding esters or amides, and PGF2α had no effect, Sang et al. concluded that the effects of the glyceryl esters and amides were not mediated by prostanoid receptors. The effect of PGE2-G was not inhibited by a CB1 antagonist, but it was blocked by an IP3 receptor antagonist and a MAPK inhibitor. A second major effect of endocannabinoids in the hippocampus is the suppression of long-term potentiation. Yang et al. found that increased COX-2 expression enhances basal synaptic transmission and augments long-term potentiation in the mouse hippocampus.(117) This response was blocked by COX-2 inhibitors, and COX-2 gene knockout had an effect opposite that of overexpression. As in the case of increased COX-2 expression, addition of the glyceryl ester or ethanolamide of PGD2, PGE2, or PGF2α to hippocampal slices increased basal synaptic transmission and long-term potentiation, and these effects were attenuated by IP3 receptor blockade and MAPK inhibition. Together, the results of Sang et al. and Yang et al. support the conclusions of Nirodi et al. and Richie-Janetta et al. that the prostanoid esters and ethanolamides act at one or more as yet unknown receptors via IP3-dependent Ca2+ mobilization and activation of MAPK. However, it should be noted that the concentrations of ligand used in the hippocampus studies were quite high (10–30 μM), and no dose–response experiments to determine the potencies of the various prostanoid esters and amides were reported. Thus, the sensitivity and specificity of the putative receptor(s) remain unclear.
A number of additional studies have suggested biological activities for PG-Gs and PG-EAs. For example, Hu et al. showed that PGE2-G induces hyperalgesia and modulates NF-κB activity in carageenan-induced inflammation in the rat paw.(70) The pharmacology of this response was distinct from that of PGE2, suggesting that it was not mediated by EP receptors, although hydrolysis of PGE2-G to PGE2 was demonstrated in the paw. Patsos et al. showed that AEA induced cell death in an apoptosis-resistant colon cancer cell line by a mechanism requiring COX-2 activity.(118) They also showed that PGE2-EA and PGD2-EA induce apoptosis in colorectal cancer cells, whereas PGE2 was reported to induce proliferation in these cells.(119) Correa et al. demonstrated that, like AEA, PGE2-EA suppresses the expression of interleukin 12 (IL-12) and IL-23 in microglial cells.(120) However, these effects were partially blocked by an EP2 receptor blocker, so it is possible that the active compound was PGE2 formed after hydrolysis of PGE2-EA and/or following hydrolysis and oxygenation of AEA. Van Dross et al. demonstrated that PGD2-EA is cytotoxic to squamous cell carconima cells, but the finding that PGD2 has the same effect suggests that this may be the active compound.(82)
3.3. Oxygenation as a Termination of Endocannabinoid Signaling
A growing number of studies suggest that COX- or LOX-dependent oxygenation serves as a mechanism to terminate endocannabinoid signaling by removing the active ligand. As noted above, available data indicate that PG-EAs, and likely PG-Gs, do not bind to the CB receptors, so COX-dependent endocannabinoid oxygenation results in inactivation. The majority of studies supporting this hypothesis depend on the use of nonselective, COX-2-selective, or LOX inhibitors. If oxygenation is involved in termination of endocannabinoid signaling, blockade of the relevant COX or LOX enzymes will lead to facilitation of signaling by increasing the levels of the endocannabinoids. However, a potentially confounding aspect of using NSAIDs as COX inhibitors in studies of endocannabinoid-eicosanoid cross-talk arises from reports that some of these compounds also inhibit FAAH.(121) In a rank order of decreasing potency, FAAH inhibitory activity was reported for suprofen > ibuprofen > fenoprofen > naproxen > ketoprofen > diclofenac > sulindac. Isobutyric acid, hydrocinnamic acid, acetylsalicylic acid, and acetaminophen were inactive. Later work showed ketorolac and flurbiprofen to be more potent than ibuprofen and revealed stereoselectivity for the (R)-isomers of ibuprofen and ketorolac.122,123 It is notable that the active enantiomer for COX inhibition is (S) for ibuprofen and ketorolac, thus differentiating between the FAAH and COX inhibitory activities. The IC50 values for FAAH inhibition by (S)-ketorolac and (R,S)-flubiprofen, in the range of 50 μM are clinically relevant, since similar concentrations may be reached in the plasma of patients taking these compounds as anti-inflammatory drugs. Holt et al. have proposed that the combination of COX and FAAH inhibition provides the basis for an improved anti-inflammatory agent, which would not only block the synthesis of pro-inflammatory prostanoids, but also prolong the anti-inflammatory and antinociceptive action of endocannabinoids.(124) They conducted structure–activity relationship studies that yielded the 6-methylpyridin-2-yl derivative of ibuprofen. This compound exhibited substantially higher FAAH inhibitory potency (IC50 ≈ 8 μM) than ibuprofen (IC50 ≈ 100 μM), while retaining ibuprofen’s COX inhibitory activity. The clinical value of this approach remains to be evaluated as well as the utility of the compounds as probes for FAAH inhibition. An IC50 value of 50 μM is much higher than the IC50 values of most NSAIDs for inhibition of COX activity in intact cells.
The antinociceptive activity of NSAIDs has traditionally been attributed to their inhibition of free acid PG synthesis by COX enzymes. This mechanism is well supported in the case of pain signals arising at the site of peripheral inflammation, but has been questioned with regard to central pain transmission at the level of the spinal cord. For example, Ates et al.(125) showed that the NSAID flurbiprofen is antinociceptive in the formalin-induced nociception model in the rat. However, intrathecal injection of PGE2 is also antinociceptive in this model. Thus, blocking PGE2 formation by COX inhibition cannot be the mechanism of the antinociceptive action of intrathecal flurbiprofen. Ates et al. went on to show that flurbiprofen’s antinociceptive action was blocked by a CB1 antagonist but not by adding PGE2, suggesting that it was endocannabinoid-mediated. Support for this conclusion comes from the work of Gühring et al.,(126) who demonstrated that CB1 receptor knockout or a CB1 antagonist, but not PGE2, blocked the antinociceptive action of the NSAID indomethacin in the formalin pain model. Seidel et al. showed that tetrahydrocannabinol and flurbiprofen inhibit capsaicin-induced calcitonin gene related peptide release from the spinal cord, another model of central nociceptive nerve transmission.(127) As in the reports of Ates et al. and Gühring et al., this effect was blocked by a CB1 antagonist but not by PGE2. In all of these cases, the investigators concluded that flurbiprofen increased endocannabiniod tone by blocking COX-mediated oxygenation of AA, thereby increasing the pool of AA available for AEA synthesis. They argued that this effect, combined with inhibition of FAAH (in the case of flurbiprofen), accounted for the NSAID-mediated increased endocannabinoid tone. However, they did not consider the possibility that the NSAIDs acted by inhibiting the direct COX-dependent oxygenation of AEA or 2-AG, which is not excluded by their data. In contrast, Bishay et al. showed that (R)-flubiprofen reduces pain transmission in a sciatic nerve injury model by reducing glutamate release in the dorsal horn of the spinal cord. This effect was mediated by increased levels of endocannabinoids. Since (R)-flurbiprofen is the inactive isomer with regard to COX inhibition, Bishay et al. argued that increased endocannabinoid levels in this model resulted from (R)-flurbiprofen-mediated FAAH inhibition and a reduction in the expression of NAPE-PLD.(128)
Despite these potential points of confusion, a series of additional studies argue strongly that COX-2 plays a role in regulation of signaling by endocannabinoids. Kim et al. showed that COX-2 inhibitors prolong endocannabinoid-mediated DSI in hippocampal slices.(129) FAAH inhibitors did not have the same effect, and the COX inhibitors used, nimesulide and meloxicam, do not have FAAH inhibitory activity. Thus, Kim et al. attributed the effects of the COX-2 inhibitors to blockade of 2-AG oxygenation. Further evidence that COX-2 plays a role in modulating endocannabinoid signaling in the hippocampus comes from Straiker et al., who characterized murine hippocampal neurons with regard to their temporal response to activation of endocannabinoid signaling by direct depolarization.(94) They found two populations of neurons that responded to endocannabinoid activation with DSI. One of these populations exhibited rapid recovery from this suppression, while the other population recovered much more slowly. The rapidly recovering cells were sensitive to COX-2 inhibition, which caused a prolongation of the endocannabinoid effect. In a subsequent study, Straiker et al. showed that overexpression of COX-2 in cultured excitatory autaptic hippocampal neurons results in a more rapid recovery from DSE.(130) Together, these findings suggest that COX-2-dependent metabolism of endocannabinoids is responsible for a termination of endocannabinoid signaling that results in rapid desuppression in these cells.
As noted above, in addition to mediating DSI or DSE, endocannabinoids also act to inhibit long-term potentiation in the hippocampus. Slanina et al. showed that COX-2-selective inhibitors, but not COX-1-selective inhibitors, blocked the development of long-term potentiation in rat hippocampal slices in an endocannabinoid-dependent manner.(131) Endocannabinoids also exert a tonic suppression of synaptic responses evoked upon stimulation of Schaffer collaterals in the hippocampus.(132) COX-2 inhibitors increased the suppression of excitatory transmission in these cells. The investigators concluded that, in both of these models, COX-2 inhibitors may block oxygenation of endocannabinoids, resulting in increased endocannabinoid tone and signaling.
The studies described above all relied on pharmacology to dissect the role of COX-2 in endocannabinoid regulation. However, a number of investigators have actually measured endocannabinoid levels and shown that COX-2 inhibition results in an increase in those levels. Wang et al. reported that AEA and 2-AG levels are increased in COX-2 knockout mice.(133) Telleria-Diaz used a model of inflammation in the rat knee joint that is characterized by spinal neuron hyperexcitability.(134) In this model, COX-2 inhibitors reversed hyperexcitability after inflammation was established, and this effect was accompanied by an increase in 2-AG levels. The finding that the effects of COX-2 inhibitors were partially blocked by a CB1 antagonist led Telleria-Diaz et al. to conclude that one mechanism by which COX-2 inhibitors suppress hyperexcitability is through facilitation of endocannabinoid signaling. Jhaveri et al. reported that the COX-2 inhibitor nimesulide increased levels of AEA in the paws of rats treated with carageenan to induce inflammation.(135) This result suggests that COX-2 inhibition prevents oxygenation of AEA, leading to higher levels. However, the discovery that nimesulide also leads to increased levels of palmitoylethanolamide, which is not a COX-2 substrate, calls this interpretation of the data into question. Furthermore, although Staniaszek et al. found that CB1 receptor blockade inhibited the antinociceptive action of intrathecal nimesulide in a model of mechanical allodynia, the NSAID treatment had no effect on 2-AG levels and actually decreased levels of AEA in the spinal cords of treated animals.(136)
Glaser et al. reported a novel, albeit indirect, approach to monitor COX-2-mediated endocannabinoid metabolism. Their method was based on the knowledge that, following AEA or 2-AG hydrolysis, the free AA is rapidly incorporated into cellular membrane lipids. Thus, mice injected intravenously with [3H]AEA carrying the label in the AA moiety will exhibit the incorporated label upon tissue autoradiography within 15 min. In contrast, the products of COX-2-dependent oxygenation are not incorporated into lipids, so the presence of this pathway would lead to a reduction in the amount of radiolabel detected by autoradiography. Glaser et al. pretreated mice with the COX-2-selective inhibitor nimesulide, or vehicle, prior to an intravenous injection of [3H]AEA. Subsequent autoradiography of brain tissue from the mice revealed increased label in the nimesulide-treated mice as compared to the control. The investigators concluded that the increase in label in the presence of the COX-2-selective inhibitor represented the amount of AEA that is oxygenated by COX-2 as opposed to inactivated by FAAH-mediated hydrolysis.(137)
Most of the data discussed above are consistent with the hypothesis that COX-2 modulates endocannabinoid tone by converting 2-AG and AEA into oxygenated derivatives that are not active at the cannabinoid receptors. However, it is important that we do not develop tunnel vision in interpreting these data. Clearly, other mechanisms may explain the ability of COX-2 inhibitors to increase apparent endocannabinoid signaling. As noted above, some investigators have proposed that free AA not used for PG synthesis is diverted to endocannabinoid formation in NSAID-treated animals. It is also possible that free acid prostanoids produced by COX-2 suppress endocannabinoid release or responses. Finally, in the case of parecoxib and valdecoxib, a direct interaction of the drug with the CB1 receptor has been reported.(138) Clearly, further work is required to develop a full understanding of the role of COX-2-dependent oxygenation on the modulation of endocannabinoid signaling.
3.4. Substrate-Selective COX-2 Inhibition
NSAIDs are among the oldest, most widely used, and thoroughly studied drugs in the modern pharmacopeia. As such, they have proven extremely useful in determining the role of COX enzymes in physiology and pathophysiology. The newer COX-2-selective coxibs have served a similar function for the elucidation of the specific role of the COX-2 isoform in conditions such as inflammation and cancer. However, when used at concentrations that block AA oxygenation by COX-2, both NSAIDs and coxibs also block endocannabinoid oxygenation. Consequently, they have been of limited value for the selective study of COX-2-dependent endocannabinoid metabolism in vivo.
Recently, Prusakiewicz et al. reported that weak, competitive inhibitors of AA oxygenation by COX-2, such as ibuprofen and mefenamic acid, are potent, time-dependent inhibitors of 2-AG oxygenation.(139) The differences in potency for the two activities are reflected in the Ki values for inhibition of AA versus 2-AG oxygenation. The reported values were 60 μM versus 1.2 μM for ibuprofen and 10 μM versus 4 nM for mefenamic acid, respectively. The differences in kinetic behavior and binding constants observed for the two substrates strongly suggest distinct inhibitory mechanisms. This led Prusakiewicz et al. to propose that the two subunits of the COX-2 homodimer act differently with regard to inhibitor interactions (Figure 13). In the case of inhibitors such as ibuprofen and mefenamic acid, the first molecule binds to one subunit of COX-2 with high affinity. This induces a conformational change in the second subunit that effectively blocks oxygenation of 2-AG, but not AA, at that subunit. To inhibit AA oxygenation, a second molecule of inhibitor must bind in the remaining subunit’s active site, but this interaction occurs with lower affinity. Thus, 2-AG oxygenation is blocked by high-affinity inhibitor binding to the first subunit in a noncompetitive fashion, while AA oxygenation is blocked by lower affinity, competitive binding to the second subunit.
Figure 13.
Mechanism of substrate-selective inhibition of endocannabinoid oxygenation by rapid, reversible inhibitors of COX-2. Inhibitor binding in one subunit of the homodimer induces a conformational change in the second subunit, which blocks 2-AG and AEA oxygenation but not AA oxygenation. Another molecule of inhibitor must bind in the second subunit to inhibit AA oxygenation. Reprinted from ref (134). Copyright 2009 American Chemical Society.
Substrate-selective inhibition was not observed for potent, time-dependent COX inhibitors such as indomethacin. For these compounds, Prusakiewicz et al. proposed that tight binding of a single molecule of inhibitor in one subunit is sufficient to induce a conformational change that blocks oxygenation of both AA and 2-AG. The mechanism proposed for substrate-selective inhibition is consistent with reports from the Smith laboratory. They have shown that binding of a fatty acid molecule to one subunit of COX induces a conformational change that alters the capacity of the second subunit to catalyze the oxygenation reaction.(140) They have also shown that binding of a molecule of celecoxib to one subunit of COX-2 induces a conformational change that inhibits binding of aspirin in the second subunit.(141) Thus, growing evidence supports the hypothesis that the two subunits of COX homodimers act as functional heterodimers.
The discovery of substrate-selective inhibition provides a mechanism by which 2-AG oxygenation may be pharmacologically distinguished from AA oxygenation in vivo. However, further work will be required to refine the conditions needed to achieve this goal.
4. Future Challenges
The fact that the endocannabinoids AEA and 2-AG are metabolites of AA ensures that there must be cross-talk between the endocannabinoid and eicosanoid signaling systems. However, the complexity of the possible interrelationships was not fully appreciated until the first reports that some LOX enzymes and COX-2 can oxygenate both AEA and 2-AG as well as AA. Evidence is mounting that these biochemical conversions are more than a test tube curiosity. Both LOX- and COX-2-derived endocannabinoid metabolites have biological activities distinct from those of their free acid counterparts, and considerable data support the hypothesis that COX-2-dependent oxygenation modulates endocannabinoid tone. Despite these advances, questions remain regarding the importance of these pathways in health and disease. Challenges for future research include the following:
-
(1)
Quantifying endocannabinoid oxygenation in vivo. As noted above, rapid hydrolysis of endocannabinoid-derived oxygenated products yields free acid eicosanoids that are indistinguishable from those produced by direct metabolism of AA. Thus, despite the availability of excellent mass-spectrometry-based assays for oxygenated endocannabinoids,(142) determination of the actual levels of these compounds produced in vivo remains elusive. It should be noted that a precedent exists for the generation of short-lived biologically active eicosanoids. Both PGI2 and TXA2 are chemically unstable and undergo rapid hydrolysis in vivo. However, in the case of these two compounds, a distinctive decomposition product allows estimation of the amount of eicosanoid originally produced. This is not the case for enzymatic hydrolysis of endocannabinoid-derived oxygenation products. A possible solution to this dilemma is to identify the hydrolytic enzymes so that chemical or genetic inhibition can be used to block the degradation pathway. In the absence of hydrolysis, the full quantity of oxygenated endocannabinoids may then be assessed.
-
(2)
As noted above, considerable evidence supports the hypothesis that oxygenated endocannabinoids exert biological activities through distinct receptors. However, with the exception of the prostamides (section 3.2.2), no specific receptors for these ligands have been characterized. Clearly, receptor identification is a major goal in clarifying the role that these pathways play in vivo. The existence of specific receptors would lend support to the hypothesis that endocannabinoid oxygenation has a defined physiological function, and receptor blockade or knockout can be used to interrogate that function under various physiological or pathological conditions.
-
(3)
Evidence is mounting to support the hypothesis that oxygenation serves as a mechanism to modulate endocannabinoid tone. This scenario is particularly appealing in the central nervous system, where the effects of NSAIDs appear to be endocannabinoid-dependent, and under conditions of inflammation that would lead to increased expression of COX-2. Indeed, reduction in levels of anti-inflammatory endocannabinoids may be one mechanism by which COX-2 exerts its pro-inflammatory effects. The recent discovery of substrate-selective inhibition may provide an approach for determining the role of COX-2-dependent oxygenation in modulation of endocannabinoid tone.
Acknowledgments
Work in the Marnett laboratory was supported by research grants from the National Institutes of Health (CA89450 and GM15431) and the National Foundation for Cancer Research. We are grateful to M. Windsor for assistance with the molecular graphics.
Glossary
Abbreviations
- CB1
cannabinoid receptor 1
- CB2
cannabinoid receptor 2
- AEA
arachidonoylethanolamide
- 2-AG
2-arachidonoylglycerol
- AA
arachidonic acid
- PG
prostaglandin
- cPLA2
cytosolic phospholipase A2
- COX
cyclooxygenase
- PGI2
prostacyclin
- TX
thromboxane
- LOX
lipoxygenase
- HPETE
hydroperoxyeicosatetraenoic acid
- HETE
hydroxyeicosatetraenoic acid
- LT
leukotriene
- EET
epoxyeicosatrienoic acid
- PLC
phospholipase C
- PIP2
phosphatidylinositol 4,5-bisphosphate
- DAG
diacylglycerol
- PC
phosphatidylcholine
- PLD
phospholipase D
- MAG
monoacylglycerol
- ABHD
α,β-hydrolase domain
- FAAH
fatty acid amide hydrolase
- CES
carboxylesterase
- NAPE
N-arachidonoylphosphatidylethanolamine
- PE
phosphatidylethanolamine
- NAT
N-acyltransferase
- NAPE-PLD
N-acylphosphatidylethanolamine-hydrolyzing PLD
- HETE-EA
hydroxyeicosatetraenoic acid ethanolamide
- HETE-G
hydroxyeicosatetraenoic acid glyceryl ester
- NAGly
N-arachidonoylglycine
- NAla
N-arachidonoylalanine
- NAGABA
N-arachidonoyl-γ-aminobutyric acid
- NADA
N-arachidonoyldopamine
- arvanil
N-arachidonoylvanillylamide
- OMDA
O-(3-methyl)-N-arachidonoyldopamine
- N-AT
N-arachidonoyltaurine
- DHEA
docosahexaenoylethanolamide
- PMN
polymorphonuclear leukocyte
- 15-HEDPEA
(15-hydroxy-16(17)-epoxydocosapentaenoyl)ethanolamide
- NSAID
nonsteroidal anti-inflammatory drug
- coxib
COX-2-selective inhibitor
- PG-EA
prostaglandin ethanolamide
- HHT
12-hydroxyheptadecatrienoic acid
- EET-EA
epoxyeicosatrienoic acid ethanolamide
- RPM
resident peritoneal macrophage
- LPS
lipopolysaccharide
- IFN-γ
interferon-γ
- PG-G
prostaglandin glyceryl ester
- DSI
depolarization-induced suppression of inhibition
- DSE
depolarization-induced suppression of excitation
- IP3
inositol trisphosphate
- PKC
protein kinase C
- MAPK
mitogen-activated protein kinase
- ERK
extracellular signal regulated kinase
- mIPSC
miniature inhibitory postsynaptic current
- PPAR
peroxisome proliferator-activated receptor
Biographies

Carol A. Rouzer received her B.A. in Chemistry from McDaniel College, her Ph.D. in Biomedical Sciences from The Rockefeller University (1982), and her M.D. from the Weill Medical College of Cornell University (1983). After postdoctoral work at the Karolinska Institute, she accepted a position as Research Fellow at the Merck Frosst Center for Therapeutic Research. She moved to McDaniel College in 1989, where she was engaged primarily in undergraduate education until 2000, when she joined the Biochemistry Faculty of Vanderbilt University as Research Professor. Her main research interests have focused on the total metabolism of arachidonic acid in inflammatory and malignant cells. Currently, she is the Managing Editor of Chemical Research in Toxicology and Communications Director of the Vanderbilt Institute of Chemical Biology. She is the author of over 50 publications.

Lawrence J. Marnett received his B.S. in Chemistry from Rockhurst College (1969) and his Ph.D. in Chemistry from Duke University (1973). After postdoctoral training at Karolinska Institute and Wayne State University, he joined the faculty in Chemistry at Wayne in 1975. He moved to Vanderbilt University in 1989 as Mary Geddes Stahlman Professor of Cancer Research and Professor of Biochemistry and Chemistry. His research interests are in the chemical biology of oxygenation of polyunsaturated fatty acids. Dr. Marnett is Editor-in-Chief of Chemical Research in Toxicology and Director of the Vanderbilt Institute of Chemical Biology. He is the author of over 400 publications.
Funding Statement
National Institutes of Health, United States
References
- Devane W. A.; Dysarz F. A. 3rd; Johnson M. R.; Melvin L. S.; Howlett A. C. Mol. Pharmacol. 1988, 34, 605. [PubMed] [Google Scholar]
- Matsuda L. A.; Lolait S. J.; Brownstein M. J.; Young A. C.; Bonner T. I. Nature 1990, 346, 561. [DOI] [PubMed] [Google Scholar]
- Munro S.; Thomas K. L.; Abu-Shaar M. Nature 1993, 365, 61. [DOI] [PubMed] [Google Scholar]
- Devane W. A.; Hanus L.; Breuer A.; Pertwee R. G.; Stevenson L. A.; Griffin G.; Gibson D.; Mandelbaum A.; Etinger A.; Mechoulam R. Science 1992, 258, 1946. [DOI] [PubMed] [Google Scholar]
- Sugiura T.; Kondo S.; Sukagawa A.; Nakane S.; Shinoda A.; Itoh K.; Yamashita A.; Waku K. Biochem. Biophys. Res. Commun. 1995, 215, 89. [DOI] [PubMed] [Google Scholar]
- Van Dorp D. A.; Beerthuis R. K.; Nugeteren D. H.; Vonkeman H. Biochim. Biophys. Acta 1964, 90, 204. [DOI] [PubMed] [Google Scholar]
- Bergström S.; Danielsson H.; Samuelsson B. Biochim. Biophys. Acta 1964, 90, 207. [DOI] [PubMed] [Google Scholar]
- Samuelsson B.; Goldyne M.; Granström E.; Hamberg M.; Hammarström S.; Malmsten C. Annu. Rev. Biochem. 1978, 47, 997. [DOI] [PubMed] [Google Scholar]
- Funk C. D. Science 2001, 294, 1871. [DOI] [PubMed] [Google Scholar]
- Serhan C. N.; Chiang N.; Van Dyke T. E. Nat. Rev. Immunol. 2008, 8, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R. C.; Gijón M. A. Biochem. J. 2007, 405, 379. [DOI] [PubMed] [Google Scholar]
- Spector A. A. J. Lipid Res. 2009, 50 (Suppl.), S52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Ueda N. Prostaglandins Other Lipid Mediators 2009, 89, 112. [DOI] [PubMed] [Google Scholar]
- Di Marzo V. Rev. Physiol. Biochem. Pharmacol. 2008, 160, 1. [DOI] [PubMed] [Google Scholar]
- Bisogno T.; Howell F.; Williams G.; Minassi A.; Cascio M. G.; Ligresti A.; Matias I.; Schiano-Moriello A.; Paul P.; Williams E.-J.; Gangadharan U.; Hobbs C.; Di Marzo V.; Doherty P. J. Cell Biol. 2003, 163, 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankman J. L.; Simon G. M.; Cravatt B. F. Chem. Biol. 2007, 14, 1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs W. R.; Blankman J. L.; Horne E. A.; Thomazeau A.; Lin Y. H.; Coy J.; Bodor A. L.; Muccioli G. G.; Hu S. S.; Woodruff G.; Fung S.; Lafourcade M.; Alexander J. P.; Long J. Z.; Li W.; Xu C.; Moller T.; Mackie K.; Manzoni O. J.; Cravatt B. F.; Stella N. Nat. Neurosci. 2010, 13, 951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney M. K.; Cravatt B. F. Annu. Rev. Biochem. 2005, 74, 411. [DOI] [PubMed] [Google Scholar]
- Xie S.; Borazjani A.; Hatfield M. J.; Edwards C. C.; Potter P. M.; Ross M. K. Chem. Res. Toxicol. 2010, 23, 1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B.; Yang D.; Bullock P.; Parkinson A. J. Biol. Chem. 1995, 270, 19128. [DOI] [PubMed] [Google Scholar]
- Li B.; Sedlacek M.; Manoharan I.; Boopathy R.; Duysen E. G.; Masson P.; Lockridge O. Biochem. Pharmacol. 2005, 70, 1673. [DOI] [PubMed] [Google Scholar]
- Reddy P. V.; Schmid P. C.; Natarajan V.; Muramatsu T.; Schmid H. H. Biochim. Biophys. Acta 1984, 795, 130. [DOI] [PubMed] [Google Scholar]
- Sugiura T.; Kondo S.; Sukagawa A.; Tonegawa T.; Nakane S.; Yamashita A.; Ishima Y.; Waku K. Eur. J. Biochem. 1996, 240, 53. [DOI] [PubMed] [Google Scholar]
- Cadas H.; di Tomaso E.; Piomelli D. J. Neurosci. Res. 1997, 17, 1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marzo V.; Fontana A.; Cadas H.; Schinelli S.; Cimino G.; Schwartz J. C.; Piomelli D. Nature 1994, 372, 686. [DOI] [PubMed] [Google Scholar]
- Okamoto Y.; Morishita J.; Tsuboi K.; Tonai T.; Ueda N. J. Biol. Chem. 2004, 279, 5298. [DOI] [PubMed] [Google Scholar]
- Leung D.; Saghatelian A.; Simon G. M.; Cravatt B. F. Biochemistry 2006, 45, 4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge S.; Boyington J.; Faig M.; Doctor K.; Gaffney B.; Amzel L. Biochimie 1997, 79, 629. [DOI] [PubMed] [Google Scholar]
- Lehmann W. D. Free Radical Biol. Med. 1994, 16, 241. [DOI] [PubMed] [Google Scholar]
- Schewe T.; Halangk W.; Hiebsch C.; Rapoport S. M. FEBS Lett. 1975, 60, 149. [DOI] [PubMed] [Google Scholar]
- Jung G.; Yang D. C.; Nakao A. Biochem. Biophys. Res. Commun. 1985, 130, 559. [DOI] [PubMed] [Google Scholar]
- Schewe T. Biol. Chem. 2002, 383, 365. [DOI] [PubMed] [Google Scholar]
- Hampson A. J.; Hill W. A.; Zan-Phillips M.; Makriyannis A.; Leung E.; Eglen R. M.; Bornheim L. M. Biochim. Biophys. Acta 1995, 1259, 173. [DOI] [PubMed] [Google Scholar]
- Ueda N.; Yamamoto K.; Yamamoto S.; Tokunaga T.; Shirakawa E.; Shinkai H.; Ogawa M.; Sato T.; Kudo I.; Inoue K. Biochim. Biophys. Acta 1995, 1254, 127. [DOI] [PubMed] [Google Scholar]
- van der Stelt M.; Nieuwenhuizen W. F.; Veldink G. A.; Vliegenthart J. F. FEBS Lett. 1997, 411, 287. [DOI] [PubMed] [Google Scholar]
- van Zadelhoff G.; Veldink G. A.; Vliegenthart J. F. Biochem. Biophys. Res. Commun. 1998, 248, 33. [DOI] [PubMed] [Google Scholar]
- Moody J. S.; Kozak K. R.; Ji C.; Marnett L. J. Biochemistry 2001, 40, 861. [DOI] [PubMed] [Google Scholar]
- Kozak K. R.; Gupta R. A.; Moody J. S.; Ji C.; Boeglin W. E.; DuBois R. N.; Brash A. R.; Marnett L. J. J. Biol. Chem. 2002, 277, 23278. [DOI] [PubMed] [Google Scholar]
- Prusakiewicz J. J.; Turman M. V.; Vila A.; Ball H. L.; Al-Mestarihi A. H.; Di Marzo V.; Marnett L. J. Arch. Biochem. Biophys. 2007, 464, 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghatelian A.; McKinney M. K.; Bandell M.; Patapoutian A.; Cravatt B. F. Biochemistry 2006, 45, 9007. [DOI] [PubMed] [Google Scholar]
- Turman M. V.; Kingsley P. J.; Rouzer C. A.; Cravatt B. F.; Marnett L. J. Biochemistry 2008, 47, 3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A.; Crozier G.; Bisogno T.; Cavaliere P.; Innis S.; Di Marzo V. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R.; Fredman G.; Krishnamoorthy S.; Agrawal N.; Irimia D.; Piomelli D.; Serhan C. N. J. Biol. Chem. 2011, 286, 31532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzer C. A.; Marnett L. J. Chem. Rev. 2003, 103, 2239. [DOI] [PubMed] [Google Scholar]
- Rouzer C. A.; Marnett L. J. J. Lipid Res. 2009, 50 (Suppl.), S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W. L.; DeWitt D. L.; Garavito R. M. Annu. Rev. Biochem. 2000, 69, 145. [DOI] [PubMed] [Google Scholar]
- Marnett L. J. Annu. Rev. Pharmacol. Toxicol. 2009, 49, 265. [DOI] [PubMed] [Google Scholar]
- Yu M.; Ives D.; Ramesha C. S. J. Biol. Chem. 1997, 272, 21181. [DOI] [PubMed] [Google Scholar]
- Kozak K. R.; Rowlinson S. W.; Marnett L. J. J. Biol. Chem. 2000, 275, 33744. [DOI] [PubMed] [Google Scholar]
- So O. Y.; Scarafia L. E.; Mak A. Y.; Callan O. H.; Swinney D. C. J. Biol. Chem. 1998, 273, 5801. [DOI] [PubMed] [Google Scholar]
- Kozak K. R.; Crews B. C.; Morrow J. D.; Wang L.-H.; Ma Y. H.; Weinander R.; Jakobsson P.-J.; Marnett L. J. J. Biol. Chem. 2002, 277, 44877. [DOI] [PubMed] [Google Scholar]
- Prusakiewicz J. J.; Kingsley P. J.; Kozak K. R.; Marnett L. J. Biochem. Biophys. Res. Commun. 2002, 296, 612. [DOI] [PubMed] [Google Scholar]
- Vecchio A. J.; Simmons D. M.; Malkowski M. G. J. Biol. Chem. 2010, 285, 22152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchio A. J.; Malkowski M. G. J. Biol. Chem. 2011, 286, 20736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzer C. A.; Ghebreselasie K.; Marnett L. J. Chem. Phys. Lipids 2002, 119, 69. [DOI] [PubMed] [Google Scholar]
- Kozak K. R.; Prusakiewicz J. J.; Rowlinson S. W.; Schneider C.; Marnett L. J. J. Biol. Chem. 2001, 276, 30072. [DOI] [PubMed] [Google Scholar]
- Capdevila J. H.; Falck J. R. Prostaglandins Other Lipid Mediators 2002, 68–69, 325. [DOI] [PubMed] [Google Scholar]
- Bornheim L. M.; Kim K. Y.; Chen B.; Correia M. A. Biochem. Pharmacol. 1995, 50, 677. [DOI] [PubMed] [Google Scholar]
- Snider N. T.; Kornilov A. M.; Kent U. M.; Hollenberg P. F. J. Pharmacol. Exp. Ther. 2007, 321, 590. [DOI] [PubMed] [Google Scholar]
- Pratt-Hyatt M.; Zhang H.; Snider N. T.; Hollenberg P. F. Drug Metab. Dispos. 2010, 38, 2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridar C.; Snider N. T.; Hollenberg P. F. Drug Metab. Dispos. 2011, 39, 782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark K.; Dostalek M.; Guengerich F. P. FEBS J. 2008, 275, 3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Chen J.-K.; Falck J. R.; Guthi J. S.; Anjaiah S.; Capdevila J. H.; Harris R. C. Mol. Cell. Biol. 2007, 27, 3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgemond W. S.; Hillard C. J.; Falck J. R.; Kearn C. S.; Campbell W. B. Mol. Pharmacol. 1998, 54, 180. [DOI] [PubMed] [Google Scholar]
- Koda N.; Tsutsui Y.; Niwa H.; Ito S.; Woodward D. F.; Watanabe K. Arch. Biochem. Biophys. 2004, 424, 128. [DOI] [PubMed] [Google Scholar]
- Rouzer C.; Marnett L. J. Biol. Chem. 2005, 280, 26690. [DOI] [PubMed] [Google Scholar]
- Rouzer C. A.; Tranguch S.; Wang H.; Zhang H.; Dey S. K.; Marnett L. J. Biochem. J. 2006, 399, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.; Fan J.; Hui Y.; Rouzer C. A.; Marnett L. J.; Klein-Szanto A. J.; Fitzgerald G. A.; Funk C. D. J. Biol. Chem. 2006, 282, 1498. [DOI] [PubMed] [Google Scholar]
- Weber A.; Ni J.; Ling K.-H. J.; Acheampong A.; Tang-Liu D. D.-S.; Burk R.; Cravatt B. F.; Woodward D. J. Lipid Res. 2004, 45, 757. [DOI] [PubMed] [Google Scholar]
- Hu S. S.-J.; Bradshaw H. B.; Chen J. S.-C.; Tan B.; Walker J. M. Br. J. Pharmacol. 2008, 153, 1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-K.; Chen J.; Imig J. D.; Wei S.; Hachey D. L.; Guthi J. S.; Falck J. R.; Capdevila J. H.; Harris R. C. J. Biol. Chem. 2008, 283, 24514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak K. R.; Crews B. C.; Ray J. L.; Tai H. H.; Morrow J. D.; Marnett L. J. J. Biol. Chem. 2001, 276, 36993. [DOI] [PubMed] [Google Scholar]
- Ross R. A.; Craib S. J.; Stevenson L. A.; Pertwee R. G.; Henderson A.; Toole J.; Ellington H. C. J. Pharmacol. Exp. Ther. 2002, 301, 900. [DOI] [PubMed] [Google Scholar]
- Fowler C. J.; Tiger G. Biochem. Pharmacol. 2005, 69, 1241. [DOI] [PubMed] [Google Scholar]
- Matias I.; Chen J.; De Petrocellis L.; Bisogno T.; Ligresti A.; Fezza F.; Krauss A. H.-P.; Shi L.; Protzman C. E.; Li C.; Liang Y.; Nieves A. L.; Kedzie K. M.; Burk R. M.; Di Marzo V.; Woodward D. F. J. Pharmacol. Exp. Ther. 2004, 309, 745. [DOI] [PubMed] [Google Scholar]
- Vila A.; Rosengarth A.; Piomelli D.; Cravatt B.; Marnett L. J. Biochemistry 2007, 46, 9578. [DOI] [PubMed] [Google Scholar]
- Snider N. T.; Nast J. A.; Tesmer L. A.; Hollenberg P. F. Mol. Pharmacol. 2009, 75, 965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke-Labesque F.; Mallaret M.; Lefebvre B.; Hardy G.; Caron F.; Bessard G. Cardiovasc. Res. 2004, 63, 155. [DOI] [PubMed] [Google Scholar]
- Herradón E.; Martín M. I.; López-Miranda V. Br. J. Pharmacol. 2007, 152, 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahn H.; Wolf J.; Kram F.; Frantz S.; Wagner J. A. Am. J. Physiol.: Heart Circ. Physiol. 2005, 289, H2491. [DOI] [PubMed] [Google Scholar]
- Endsley M. P.; Aggarwal N.; Isbell M. A.; Wheelock C. E.; Hammock B. D.; Falck J. R.; Campbell W. B.; Nithipatikom K. Int. J. Cancer 2007, 121, 984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dross R. T. Mol. Carcinog. 2009, 48, 724. [DOI] [PubMed] [Google Scholar]
- Gauthier K. M.; Baewer D. V.; Hittner S.; Hillard C. J.; Nithipatikom K.; Reddy D. S.; Falck J. R.; Campbell W. B. Am. J. Physiol.: Heart Circ. Physiol. 2005, 288, H1344. [DOI] [PubMed] [Google Scholar]
- Ellis E. F.; Moore S. F.; Willoughby K. A. Am. J. Physiol. 1995, 269, H1859. [DOI] [PubMed] [Google Scholar]
- Darmani N. A. J. Pharmacol. Exp. Ther. 2002, 300, 34. [DOI] [PubMed] [Google Scholar]
- Karaliota S.; Siafaka-Kapadai A.; Gontinou C.; Psarra K.; Mavri-Vavayanni M. Obesity (Silver Spring) 2009, 17, 1830. [DOI] [PubMed] [Google Scholar]
- Rockwell C. E.; Kaminski N. E. J. Pharmacol. Exp. Ther. 2004, 311, 683. [DOI] [PubMed] [Google Scholar]
- Egashira N.; Mishima K.; Uchida T.; Hasebe N.; Nagai H.; Mizuki A.; Iwasaki K.; Ishii H.; Nishimura R.; Shoyama Y.; Fujiwara M. Psychopharmacology (Berlin) 2004, 171, 382. [DOI] [PubMed] [Google Scholar]
- Wiley J. L.; Razdan R. K.; Martin B. R. Life Sci. 2006, 80, 24. [DOI] [PubMed] [Google Scholar]
- Pestonjamasp V. K.; Burstein S. H. Biochim. Biophys. Acta 1998, 1394, 249. [DOI] [PubMed] [Google Scholar]
- Baldassarri S.; Bertoni A.; Bagarotti A.; Sarasso C.; Zanfa M.; Catani M. V.; Avigliano L.; Maccarrone M.; Torti M.; Sinigaglia F. J. Thromb. Haemostasis 2008, 6, 1772. [DOI] [PubMed] [Google Scholar]
- Maccarrone M.; Lorenzon T.; Bari M.; Melino G.; Finazzi-Agro A. J. Biol. Chem. 2000, 275, 31938. [DOI] [PubMed] [Google Scholar]
- Hashimotodani Y.; Ohno-Shosaku T.; Kano M. J. Neurosci. 2007, 27, 1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straiker A.; Mackie K. Neuroscience 2009, 163, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oz M.; Jaligam V.; Galadari S.; Petroianu G.; Shuba Y. M.; Shippenberg T. S. J. Neurochem. 2010, 112, 1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oz M.; Tchugunova Y. B.; Dunn S. M. Eur. J. Pharmacol. 2000, 404, 13. [DOI] [PubMed] [Google Scholar]
- Gasperi V.; Fezza F.; Pasquariello N.; Bari M.; Oddi S.; Agrò A. F.; Maccarrone M. Cell. Mol. Life Sci. 2007, 64, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; McCarron R. M.; Ohara Y.; Bembry J.; Azzam N.; Lenz F. A.; Shohami E.; Mechoulam R.; Spatz M. Circ. Res. 2000, 87, 323. [DOI] [PubMed] [Google Scholar]
- Kojima S.-i.; Sugiura T.; Waku K.; Kamikawa Y. Eur. J. Pharmacol. 2002, 444, 203. [DOI] [PubMed] [Google Scholar]
- Gardiner S. M.; March J. E.; Kemp P. A.; Bennett T. Br. J. Pharmacol. 2009, 158, 1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosburg J. E.; Blankman J. L.; Long J. Z.; Nomura D. K.; Pan B.; Kinsey S. G.; Nguyen P. T.; Ramesh D.; Booker L.; Burston J. J.; Thomas E. A.; Selley D. E.; Sim-Selley L. J.; Liu Q. S.; Lichtman A. H.; Cravatt B. F. Nat. Neurosci. 2010, 13, 1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J. Z.; Li W.; Booker L.; Burston J. J.; Kinsey S. G.; Schlosburg J. E.; Pavon F. J.; Serrano A. M.; Selley D. E.; Parsons L. H.; Lichtman A. H.; Cravatt B. F. Nat. Chem. Biol. 2009, 5, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura D. K.; Blankman J. L.; Simon G. M.; Fujioka K.; Issa R. S.; Ward A. M.; Cravatt B. F.; Casida J. E. Nat. Chem. Biol. 2008, 4, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Vasilyev D. V.; Goncalves M. B.; Howell F. V.; Hobbs C.; Reisenberg M.; Shen R.; Zhang M. Y.; Strassle B. W.; Lu P.; Mark L.; Piesla M. J.; Deng K.; Kouranova E. V.; Ring R. H.; Whiteside G. T.; Bates B.; Walsh F. S.; Williams G.; Pangalos M. N.; Samad T. A.; Doherty P. J. Neurosci. 2010, 30, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto J. C.; Potié F.; Rice K. C.; Boring D.; Johnson M. R.; Evans D. M.; Wilken G. H.; Cantrell C. H.; Howlett A. C. Mol. Pharmacol. 1994, 46, 516. [PubMed] [Google Scholar]
- Nirodi C. S.; Crews B. C.; Kozak K. R.; Morrow J. D.; Marnett L. J. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward D. F.; Krauss A. H.-P.; Chen J.; Liang Y.; Li C.; Protzman C. E.; Bogardus A.; Chen R.; Kedzie K. M.; Krauss H. A.; Gil D. W.; Kharlamb A.; Wheeler L. A.; Babusis D.; Welty D.; Tang-Liu D. D.-S.; Cherukury M.; Andrews S. W.; Burk R. M.; Garst M. E. J. Pharmacol. Exp. Ther. 2003, 305, 772. [DOI] [PubMed] [Google Scholar]
- van der Stelt M.; van Kuik J. A.; Bari M.; van Zadelhoff G.; Leeflang B. R.; Veldink G. A.; Finazzi-Agrò A.; Vliegenthart J. F. G.; Maccarrone M. J. Med. Chem. 2002, 45, 3709. [DOI] [PubMed] [Google Scholar]
- van der Stelt M.; Paoletti A. M.; Maccarrone M.; Nieuwenhuizen W. F.; Bagetta G.; Veldink G. A.; Finazzi Agrò A.; Vliegenthart J. F. FEBS Lett. 1997, 415, 313. [DOI] [PubMed] [Google Scholar]
- Ghosh M.; Wang H.; Ai Y.; Romeo E.; Luyendyk J. P.; Peters J. M.; Mackman N.; Dey S. K.; Hla T. J. Exp. Med. 2007, 204, 2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell C. E.; Raman P.; Kaplan B. L. F.; Kaminski N. E. Biochem. Pharmacol. 2008, 76, 353. [DOI] [PubMed] [Google Scholar]
- Woodward D. F.; Krauss A. H.; Wang J. W.; Protzman C. E.; Nieves A. L.; Liang Y.; Donde Y.; Burk R. M.; Landsverk K.; Struble C. Br. J. Pharmacol. 2007, 150, 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward D. F.; Carling R. W. C.; Cornell C. L.; Fliri H. G.; Martos J. L.; Pettit S. N.; Liang Y.; Wang J. W. Pharmacol. Ther. 2008, 120, 71. [DOI] [PubMed] [Google Scholar]
- Liang Y.; Woodward D. F.; Guzman V. M.; Li C.; Scott D. F.; Wang J. W.; Wheeler L. A.; Garst M. E.; Landsverk K.; Sachs G.; Krauss A. H.-P.; Cornell C.; Martos J.; Pettit S.; Fliri H. Br. J. Pharmacol. 2008, 154, 1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richie-Jannetta R.; Nirodi C. S.; Crews B. C.; Woodward D. F.; Wang J. W.; Duff P. T.; Marnett L. J. Prostaglandins Other Lipid Mediators 2010, 92, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang N.; Zhang J.; Chen C. J. Physiol. (London) 2006, 572, 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.; Zhang J.; Andreasson K.; Chen C. Mol. Cell. Neurosci. 2008, 37, 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsos H. A.; Greenhough A.; Hicks D. J.; Al Kharusi M.; Collard T. J.; Lane J. D.; Paraskeva C.; Williams A. C. Int. J. Oncol. 2010, 37, 187. [DOI] [PubMed] [Google Scholar]
- Patsos H. A.; Hicks D. J.; Dobson R. R. H.; Greenhough A.; Woodman N.; Lane J. D.; Williams A. C.; Paraskeva C. Gut 2005, 54, 1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa F.; Docagne F.; Clemente D.; Mestre L.; Becker C.; Guaza C. Biochem. J. 2008, 409, 761. [DOI] [PubMed] [Google Scholar]
- Fowler C. J.; Stenström A.; Tiger G. Pharmacol. Toxicol. 1997, 80, 103. [DOI] [PubMed] [Google Scholar]
- Fowler C. J.; Janson U.; Johnson R. M.; Wahlström G.; Stenström A.; Norström K.; Tiger G. Arch. Biochem. Biophys. 1999, 362, 191. [DOI] [PubMed] [Google Scholar]
- Tiger G.; Stenström A.; Fowler C. J. Biochem. Pharmacol. 2000, 59, 647. [DOI] [PubMed] [Google Scholar]
- Holt S.; Paylor B.; Boldrup L.; Alajakku K.; Vandevoorde S.; Sundström A.; Cocco M. T.; Onnis V.; Fowler C. J. Eur. J. Pharmacol. 2007, 565, 26. [DOI] [PubMed] [Google Scholar]
- Ates M.; Hamza M.; Seidel K.; Kotalla C. E.; Ledent C.; Gühring H. Eur. J. Neurosci. 2003, 17, 597. [DOI] [PubMed] [Google Scholar]
- Gühring H.; Hamza M.; Sergejeva M.; Ates M.; Kotalla C. E.; Ledent C.; Brune K. Eur. J. Pharmacol. 2002, 454, 153. [DOI] [PubMed] [Google Scholar]
- Seidel K.; Hamza M.; Ates M.; Gühring H. Neurosci. Lett. 2003, 338, 99. [DOI] [PubMed] [Google Scholar]
- Bishay P.; Schmidt H.; Marian C.; Häussler A.; Wijnvoord N.; Ziebell S.; Metzner J.; Koch M.; Myrczek T.; Bechmann I.; Kuner R.; Costigan M.; Dehghani F.; Geisslinger G.; Tegeder I. PLoS ONE 2010, 5, e10628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.; Alger B. E. Nat. Neurosci. 2004, 7, 697. [DOI] [PubMed] [Google Scholar]
- Straiker A.; Wager-Miller J.; Hu S. S.; Blankman J. L.; Cravatt B. F.; Mackie K.. Br. J. Pharmacol. 2011, in press. [DOI] [PMC free article] [PubMed]
- Slanina K. A.; Schweitzer P. Neuropharmacology 2005, 49, 653. [DOI] [PubMed] [Google Scholar]
- Slanina K. A.; Roberto M.; Schweitzer P. Neuropharmacology 2005, 49, 660. [DOI] [PubMed] [Google Scholar]
- Wang H.; Xie H.; Sun X.; Kingsley P. J.; Marnett L. J.; Cravatt B. F.; Dey S. K. Prostaglandins Other Lipid Mediators 2007, 83, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telleria-Diaz A.; Schmidt M.; Kreusch S.; Neubert A.-K.; Schache F.; Vazquez E.; Vanegas H.; Schaible H.-G.; Ebersberger A. Pain 2010, 148, 26. [DOI] [PubMed] [Google Scholar]
- Jhaveri M. D.; Richardson D.; Robinson I.; Garle M. J.; Patel A.; Sun Y.; Sagar D. R.; Bennett A. J.; Alexander S. P. H.; Kendall D. A.; Barrett D. A.; Chapman V. Neuropharmacology 2008, 55, 85. [DOI] [PubMed] [Google Scholar]
- Staniaszek L. E.; Norris L. M.; Kendall D. A.; Barrett D. A.; Chapman V. Br. J. Pharmacol. 2010, 160, 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser S. T.; Kaczocha M. J. Pharmacol. Exp. Ther. 2010, 335, 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder H.; Höllt V.; Becker A. Neurochem. Int. 2011, 58, 9. [DOI] [PubMed] [Google Scholar]
- Prusakiewicz J. J.; Duggan K. C.; Rouzer C. A.; Marnett L. J. Biochemistry 2009, 48, 7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C.; Sidhu R. S.; Kuklev D. V.; Kado Y.; Wada M.; Song I.; Smith W. L. J. Biol. Chem. 2009, 284, 10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimon G.; Sidhu R. S.; Lauver D. A.; Lee J. Y.; Sharma N. P.; Yuan C.; Frieler R. A.; Trievel R. C.; Lucchesi B. R.; Smith W. L. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley P. J.; Marnett L. J. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2009, 877, 2746. [DOI] [PMC free article] [PubMed] [Google Scholar]