Abstract
SunS is a novel S-glycosyltransferase involved in the biosynthesis of the antimicrobial peptide sublancin. It selectively modifies Cys22 in a 56 amino acid peptide substrate SunA and can accept a variety of NDP sugars. This study reports the substrate selectivity with regard to the peptide substrate and the antimicrobial activity of the resulting sublancin analogues. The results suggest that SunS recognizes an α-helix N-terminal of the Cys to be glycosylated, which is present in a flexible linker. Interestingly, when Cys22 is mutated, sugar attachment is not required for sublancin antimicrobial activity. Furthermore, the sublancin-producing strain Bacillus subtilis 168 also becomes susceptible to such mutants. These data suggest that S-glycosylation may be important for self-resistance.
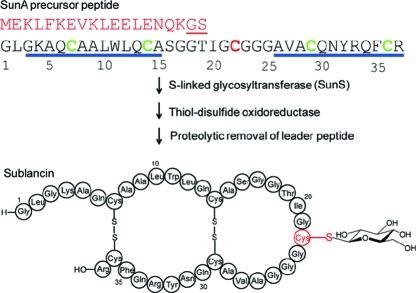
N-Linked and O-linked glycosylation of proteins are common post-translational modifications.1−4 Examples of S-linked glycopeptides are rare5−10 and until recently have not been well characterized. In the past year, two examples of bacterial S-linked glycopeptides that display antimicrobial activities were reported: sublancin produced by Bacillus subtilis 168 and glycocin F produced by Lactobacillus plantarum KW30.11,12 The novel S-glycosyltransferase SunS was demonstrated to glucosylate Cys22 during the biosynthesis of sublancin (Figure 1). Its enzymatic activity was reconstituted in vitro, and investigation of its substrate specificity showed that SunS can accept a variety of nucleotide sugars as sugar donors.(11) In this study, we explored the peptide substrate specificity of SunS and its potential application in generating sublancin analogues. These studies suggest that SunS recognizes an α-helical segment of its substrate and then glycosylates cysteine residues in a flexible loop following this helix. Interestingly, when Cys22 in sublancin was mutated to Ser or Ala, the nonglycosylated mutant retained antimicrobial activity against sublancin-sensitive strains and also gained activity against the producer organism. These findings, combined with the previous observation that the type of sugar attached to Cys22 is not important for bioactivity,(11) suggest that glycosylation at Cys22 may constitute an unusual type of self-resistance.
Figure 1.
Post-translational modifications during sublancin biosynthesis. The leader peptide of the SunA precursor peptide is shown in red, and the double-glycine-type proteolytic cleavage site is underlined. The predicted helix-forming regions are underlined with blue bars. The cysteine residues that are not glycosylated are shown in green, and the glycosylated residue Cys22 is shown in red. The order of glycosylation and disulfide-bond formation in vivo is unknown and is arbitrarily shown with glycosylation occurring first. Residue numbering is based on the core peptide of SunA.(13)
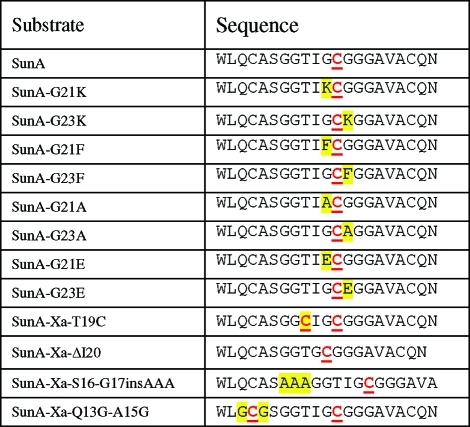
Under the conditions of the in vitro assays, all five Cys residues in the SunA substrate peptide are reduced and available for glycosylation, yet only Cys22 is modified. As noted previously,(11) the four Cys residues in SunA that are not glycosylated by SunS are flanked by Gln, Ala, Arg, and Phe, whereas Cys22 that is glycosylated is flanked by two Gly amino acids (Figure 1). Therefore, the importance of the flanking residues of Cys22 for the regioselectivity displayed by SunS was first investigated. The two glycine residues flanking Cys22 were mutated to Ala, Lys, Glu, and Phe (Table 1). Interestingly, all of these SunA mutants were accepted as substrates by SunS in the presence of uridine diphosphate glucose (UDP-glucose), as demonstrated by MALDI-TOF-MS (Figure S1 in the Supporting Information), and all were glucosylated on Cys22. Therefore, the flanking residues do not provide the basis for the observed regiochemistry. However, the flanking residues do impact the efficiency of catalysis, as demonstrated by the time dependence of substrate conversion monitored by HPLC. At a final concentration of 25 μM, wild type (wt) SunA, SunA-G23A, and SunA-G23F were fully converted to their glycopeptide products in 15 min in the presence of 150 nM SunS. Under the same conditions, conversion of SunA-G23E took 3 h, whereas modification of SunA-G23K was not observed. However, SunA-G23K was glucosylated when a longer incubation time and higher enzyme concentration (5 μM) were used (Figure S1g). These results indicate that a charged flanking residue of Cys22 slows down glycosylation. The size of the flanking residues appears to be less important than their charge state.
Table 1. Partial Sequences of SunA Peptides (Residues 11–30)a.
Mutations are highlighted in yellow. Cysteines that were glucosylated by SunS are shown in red underlined font.
To investigate whether the selectivity for Cys22 is based on its position within the SunA peptide, two mutant peptides were produced to investigate the effect of a “change in register” (Table 1). In one peptide, SunA-Xa-ΔI20, Ile20 was deleted, bringing Cys22 one residue closer to the N-terminus. In a second peptide, SunA-Xa-(S16-G17insAAA), three Ala residues were inserted between Ser16 and Gly17 to position the Cys three residues further away from the N-terminus. In both SunA mutant peptides, a Factor Xa cleavage site was engineered to allow removal of the leader peptide and evaluation of the bioactivities of the sublancin analogues. Incubation of these substrate analogues with SunS resulted in glucosylation of both peptides at the third Cys residue from the N-terminus (equivalent to Cys22 in wt SunA), as determined by ESI-MSMS. Thus, SunS tolerates changes in the position of the targeted Cys residue toward either the N- or C-terminus without compromising its regio- and chemoselectivity (Figure S2).
The observation that SunS can selectively modify Cys residues at positions other than the 22nd amino acid of the core peptide encouraged exploration of its capability to conjugate multiple sugar moieties. First, the mutant peptide SunA-Xa-T19C was produced to introduce an additional Cys in the stretch of amino acids between Cys14 and Cys29 that encompasses Cys22 (Table 1). After incubation with SunS, only a monoglycosylated peptide was observed by MALDI-TOF-MS. However, ESI-MSMS analysis revealed that the glucose was present on either Cys19 or Cys22 (Figure S3). These results indicate that SunS can modify Cys residues at either position but not both. Although the MSMS data could not quantify the selectivity of SunS for Cys19 or Cys22 in this mutant, the intensities of the fragment ions showed that the enzyme does not display a strong preference for either.
Interestingly, incubation of SunS with a SunA mutant in which the flanking residues of Cys14 (one of the residues not modified in wt SunA) were mutated to glycines (SunA-Xa-Q13G-A15G, Table 1) resulted in a mixture of mono- and bisglycosylated products (Figure S4a). Subsequent ESI-MSMS analysis showed that both the Cys14 and Cys22 residues were modified (Figure S4b,c). The observation that SunS can modify two Cys residues in SunA-Xa-Q13G-A15G but not in SunA-Xa-T19C indicates that a minimal distance between two Cys residues is required in order to accommodate the initially formed monoglycosylated peptide into the active site for a subsequent second glycosylation.
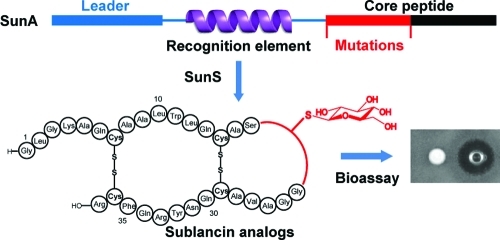
Whereas it is clear that SunS has substantial tolerance toward mutations in its peptide substrate, the basis of its substrate recognition mechanism is still largely unknown. Previous studies showed that the leader peptide of SunA is not required for enzymatic processing by SunS in vitro.(11) Therefore, the core peptide must provide sufficient binding affinity for substrate recognition and processing. Given the outcome of our mutagenesis studies, we wondered whether perhaps secondary structure elements in SunA would be recognized by SunS. Structure prediction tools (PSIPRED(14)) anticipated two α-helical regions in the reduced core peptide spanning residues 3–15 and 26–36 (Figure 1). Circular dichroism experiments support the existence of α-helical character in both reduced (no disulfides) and native sublancin and in the reduced, linear His6-SunA peptide (data not shown). In addition, the recently solved NMR structure of the S-linked glycopeptide bacteriocin glycocin F, which shares structural similarity with sublancin (Figure S5), consists of two α-helices held together by a pair of disulfide bonds.(15) The two α-helices in glycocin F project well onto regions 7–14 and 29–36 of SunA, although the two peptides have no sequence identity except for the four cysteines (Figure S5). If such a helical structure provides the basis for enzyme–substrate recognition, it offers a possible explanation for the observed regioselectivity of SunS. The four Cys residues in SunA that are not modified by SunS are all inside the proposed helical regions, whereas the Cys that is modified (Cys22) is in a flexible coil region connecting the helices. This hypothesis is supported by the double glycosylation observed in SunA-Xa-Q13G-A15G. The PSIPRED structure prediction program(14) suggests that because of the helix-breaking residues Gly13 and Gly15, Cys14 is no longer within the N-terminal α-helix in this mutant, possibly explaining why Cys14 was glucosylated by SunS.
To investigate further the importance of each of the two proposed helices in the core peptide of SunA for substrate recognition by SunS, two peptides spanning residues 1–27 (SunA-1-27) and 16–26 (SunA-16-26) were synthesized by solid-phase peptide synthesis (Table S2). A third peptide, SunA-17-37, was obtained by expression of SunA-S16E in Escherichia coli, digestion of the peptide with endoproteinase GluC, and HPLC purification. Incubation of these three peptides with SunS under the standard assay conditions resulted in glycosylated SunA-1-27, whereas SunA-16-26 and SunA-17-37 were not recognized as substrates. These results suggest that the presence of the N-terminal helical region (residues 3–15) of the core peptide is required for substrate recognition whereas the C-terminal helix (residues 26–36) is dispensable and the flexible linker connecting the helices (residues 16–26) is not sufficient. As a further test of the hypothesis that the N-terminal helix in the core peptide of SunS is important for substrate recognition, the helix-breaking residue Pro was introduced at position 11 of SunA by site-directed mutagenesis. The peptide fragments SunA-(5-33)-W11P and SunA-(5-33) were then generated by trypsin digestion of SunA-W11P and wt SunA, respectively (Table S2). Incubation of these peptides with SunS showed that the proline mutant SunA-(5-33)-W11P peptide was not a substrate whereas SunA-(5-33) was still glycosylated (Figure S6). Therefore, the N-terminal helix appears to be essential for substrate recognition by SunS.
The relaxed substrate specificity of SunS observed herein also provided a convenient tool for generating a series of sublancin analogues. The leader peptides were removed from glucosylated SunA-Xa-G23E, SunA-Xa-G23F, SunA-Xa-ΔI20, and SunA-Xa-(S16-G17insAAA) using Factor Xa. Subsequently, these peptides were subjected to the previously reported oxidative folding procedure to facilitate the formation of the correct disulfides.(11) Interestingly, all of the reconstituted sublancin analogues exhibited similar growth inhibitory activity against B. subtilis 6633 as reconstituted wt sublancin (Figure 2), suggesting that neither the residues flanking the glucosylated Cys nor the precise position of glucose attachment are critical for its antimicrobial potency.
Figure 2.
Antimicrobial assays of sublancin analogues against B. subtilis 6633. (1) sublancin analogue G23E; (2) sublancin analogue G23F; (3) sublancin analogue S16G17insAAA; (4) sublancin analogue ΔI20; (5) reconstituted wt sublancin; (6) oxidative folding buffer (50 mM Tris, pH 7.5, 2 mM oxidized glutathione, 2 mM reduced glutathione, 0.1 mM EDTA). All of the peptides were reconstituted from their precursor peptides.
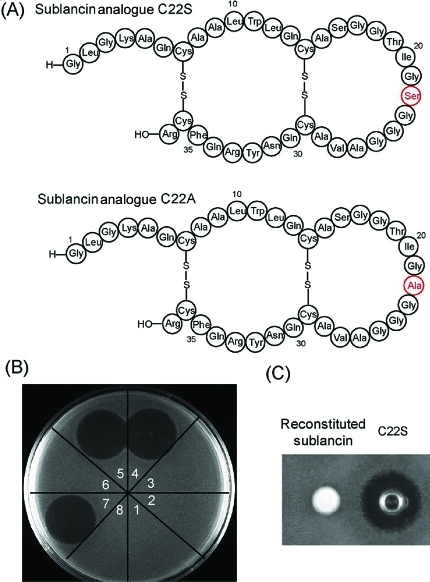
In our previous study, we reported that glycosylation of SunA is essential for antimicrobial activity of sublancin.(11) Further investigation revealed that without glycosylation, the correct disulfide bonds of sublancin are not formed during the oxidative folding process (Figure S7). Presumably, the free thiol of unmodified Cys22 disrupts the formation of the correct disulfide bridges by thiol–disulfide exchange.(16) Hence, the importance of the actual glycosylation for antimicrobial activity has not been tested. To provide additional information regarding this question, the mutants SunA-Xa-C22S and SunA-Xa-C22A were generated. Surprisingly, after proteolytic removal of the leader peptide and subsequent oxidative folding, both sublancin-C22S and sublancin-C22A exhibited similar growth inhibitory activity against B. subtilis 6633 as reconstituted wt sublancin (Figure 3B). Similar to the observations for sublancin, removal of the leader peptide was required for the bioactivity of both sublancin analogues. These results indicate that the presence of sugar moieties at position 22 is not required for the antimicrobial activity of sublancin. The observation that the glucose is not important for bioactivity also explains why sublancin analogues with a variety of different sugars attached to Cys22 all exhibit similar antimicrobial activity(11) and why analogues with the glucosylated Cys at different positions in the loop connecting the two α-helices are also all active (Figure 2). It therefore appears that the two helices held together by the disulfide bonds are most important for biological activity. To rule out the possibility that the sublancin C22S mutant acts by a different mechanism of action, we tested the compound against a B. subtilis 6633 mutant strain that acquired resistance upon exposure to wt sublancin. Neither wt sublancin nor the C22S mutant displayed antimicrobial activity against this mutant strain at concentrations of up to 50 μM, suggesting that the C22S mutant acts by the same mechanism of action as wt sublancin.
Figure 3.
Bioactivity assays of sublancin analogues C22S and C22A. (A) Structures of sublancin analogues C22S and C22A. (B) Agar-well diffusion assay of sublancin and sublancin analogues C22S and C22A against B. subtilis 6633: (1) negative control; (2) SunA-Xa after glucosylation and oxidative folding; (3) SunA-Xa after oxidative folding and proteolytic removal of the leader peptide; (4) SunA-Xa after glucosylation, proteolytic removal of the leader peptide, and oxidative folding (reconstituted sublancin); (5) SunA-Xa C22S after proteolytic removal of the leader peptide and oxidative folding; (6) product of SunA-Xa-C22S after oxidative folding; (7) SunA-Xa C22A after proteolytic removal of the leader peptide and oxidative folding; (8) SunA-Xa-C22A after oxidative folding. (C) Activities of reconstituted sublancin and its analogue C22S (35 μM) against B. subtilis 168.
We also tested the activity of sublancin-C22S against the sublancin producing strain B. subtilis 168. As expected, neither authentic nor reconstituted sublancin showed inhibitory activity against B. subtilis 168 (Figure 3C), presumably because of the self-resistance mechanisms in the producer organism mediated by SunI.(17) Surprisingly, however, B. subtilis 168 was susceptible to sublancin-C22S (Figure 3C), suggesting the possibility that glycosylation is related to a self-resistance mechanism, possibly in combination with SunI. Future studies will focus on further evaluation of this possibility.
In summary, we have demonstrated that a novel S-linked glycosyl transferase, SunS, exhibits high promiscuity in regard to its peptide substrates without losing its chemo- and regioselectivity. The data presented here suggest that the enzyme recognizes an α-helix that spans approximately residues 3–15. A variety of sublancin analogues were generated by utilizing SunS to modify mutant substrates. These mutants show that glucose attachment is not required for the antimicrobial activity of sublancin.
Acknowledgments
The authors thank Trent Oman for assistance in the initial stages of this study and Chantal Garcia de Gonzalo for generating sublancin-resistant B. subtilis mutants. This work was supported by the U.S. National Institutes of Health (GM58822 to W.A.v.d.D.).
Supporting Information Available
Molecular biology procedures, protein purifications, and supporting figures. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Spiro R. G. Glycobiology 2002, 12, 43R–56R. [DOI] [PubMed] [Google Scholar]
- Nothaft H.; Szymanski C. M. Nat. Rev. Microbiol. 2010, 8, 765–778. [DOI] [PubMed] [Google Scholar]
- Hart G. W.; Slawson C.; Ramirez-Correa G.; Lagerlof O. Annu. Rev. Biochem. 2011, 80, 825–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E.; Imperiali B. Glycobiology 2006, 16, 91R–101R. [DOI] [PubMed] [Google Scholar]
- Lote C. J.; Weiss J. B. Biochem. J. 1971, 123, 25P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J. B.; Lote C. J.; Bobinski H. Nature, New Biol. 1971, 234, 25–26. [DOI] [PubMed] [Google Scholar]
- Lote C. J.; Weiss J. B. FEBS Lett. 1971, 16, 81–85. [DOI] [PubMed] [Google Scholar]
- Elsayed S.; Bennich H. Scand. J. Immunol. 1975, 4, 203–208. [DOI] [PubMed] [Google Scholar]
- Olsen E. H.; Rahbek-Nielsen H.; Thogersen I. B.; Roepstorff P.; Enghild J. J. Biochemistry 1998, 37, 408–416. [DOI] [PubMed] [Google Scholar]
- Garavelli J. S. Proteomics 2004, 4, 1527–1533. [DOI] [PubMed] [Google Scholar]
- Oman T. J.; Boettcher J. M.; Wang H.; Okalibe X. N.; van der Donk W. A. Nat. Chem. Biol. 2011, 7, 78–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepper J.; Shastri S.; Loo T. S.; Preston J. C.; Novak P.; Man P.; Moore C. H.; Havlicek V.; Patchett M. L.; Norris G. E. FEBS Lett. 2011, 585, 645–650. [DOI] [PubMed] [Google Scholar]
- Oman T. J.; van der Donk W. A. Nat. Chem. Biol. 2010, 6, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin L. J.; Bryson K.; Jones D. T. Bioinformatics 2000, 16, 404–405. [DOI] [PubMed] [Google Scholar]
- Venugopal H.; Edwards P. J.; Schwalbe M.; Claridge J. K.; Libich D. S.; Stepper J.; Loo T.; Patchett M. L.; Norris G. E.; Pascal S. M. Biochemistry 2011, 50, 2748–2755. [DOI] [PubMed] [Google Scholar]
- Alternatively, the presence of the sugar may facilitate folding of wt sublancin. We attempted to address this question by following the rate of folding of glycosylated wt SunA and SunA-C22S, but because of overlapping peaks in the HPLC, we were unable to distinguish between these two possibilities.
- Dubois J. Y.; Kouwen T. R.; Schurich A. K.; Reis C. R.; Ensing H. T.; Trip E. N.; Zweers J. C.; van Dijl J. M. Antimicrob. Agents Chemother. 2009, 53, 651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.