Abstract
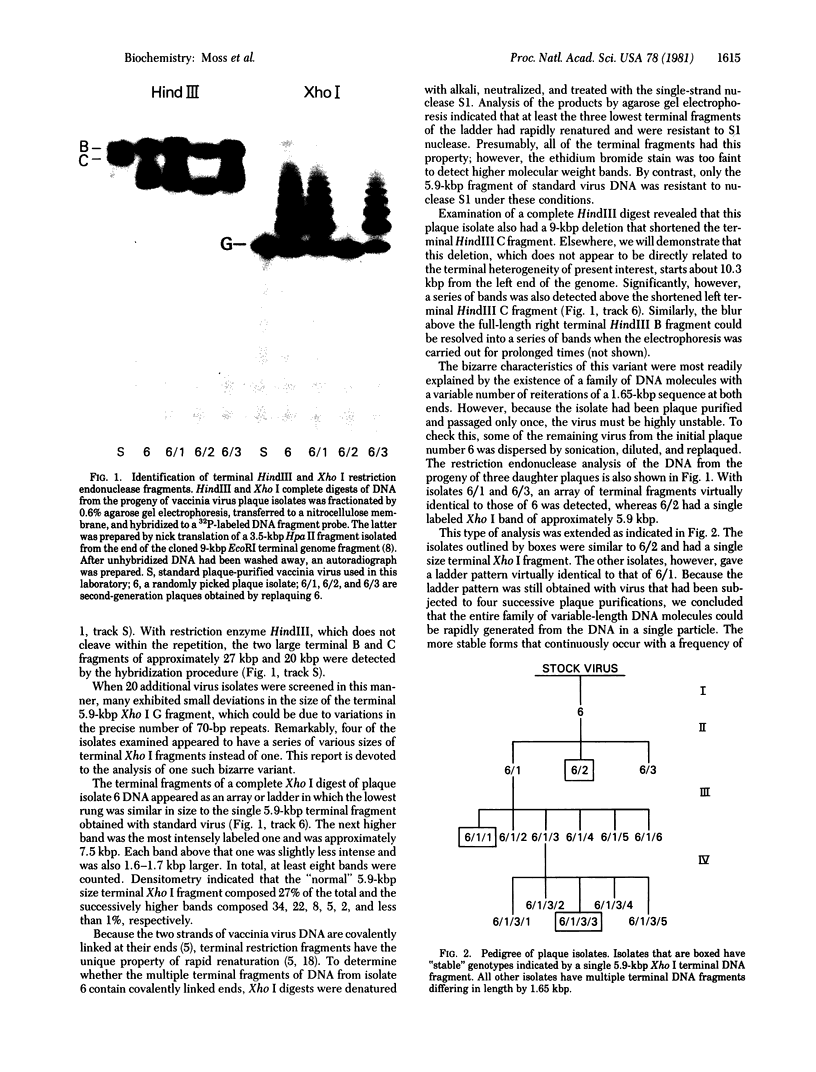
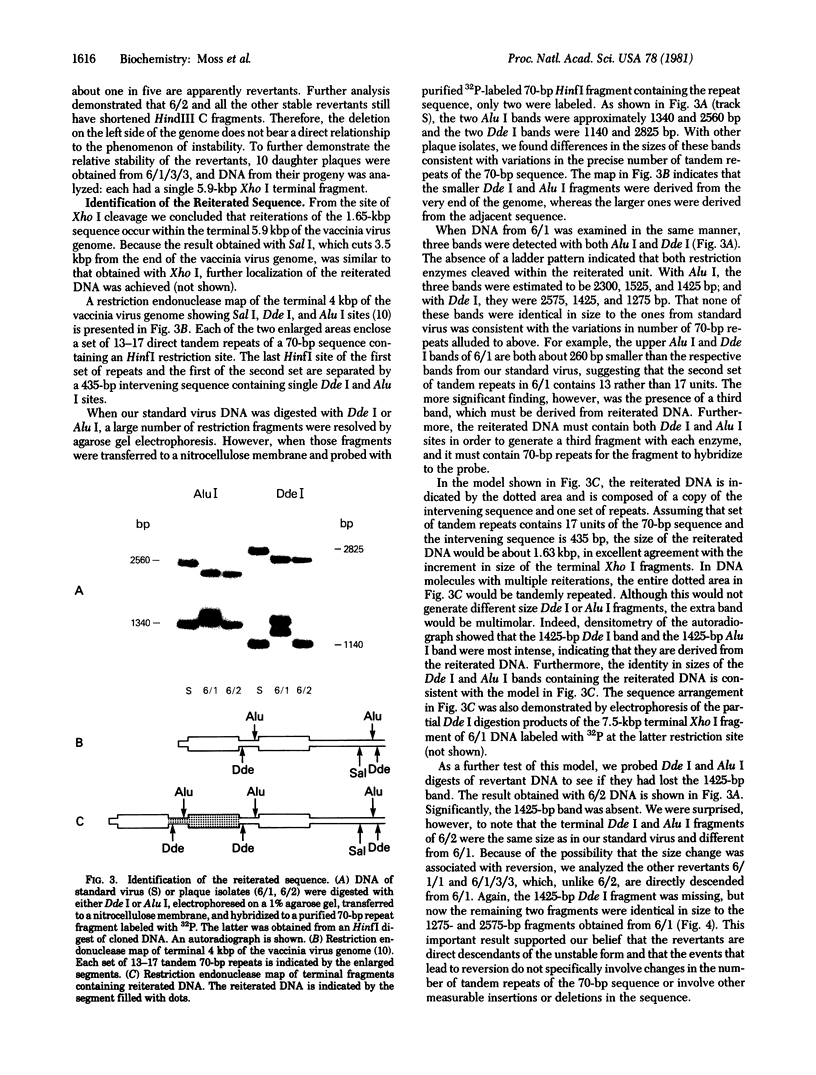
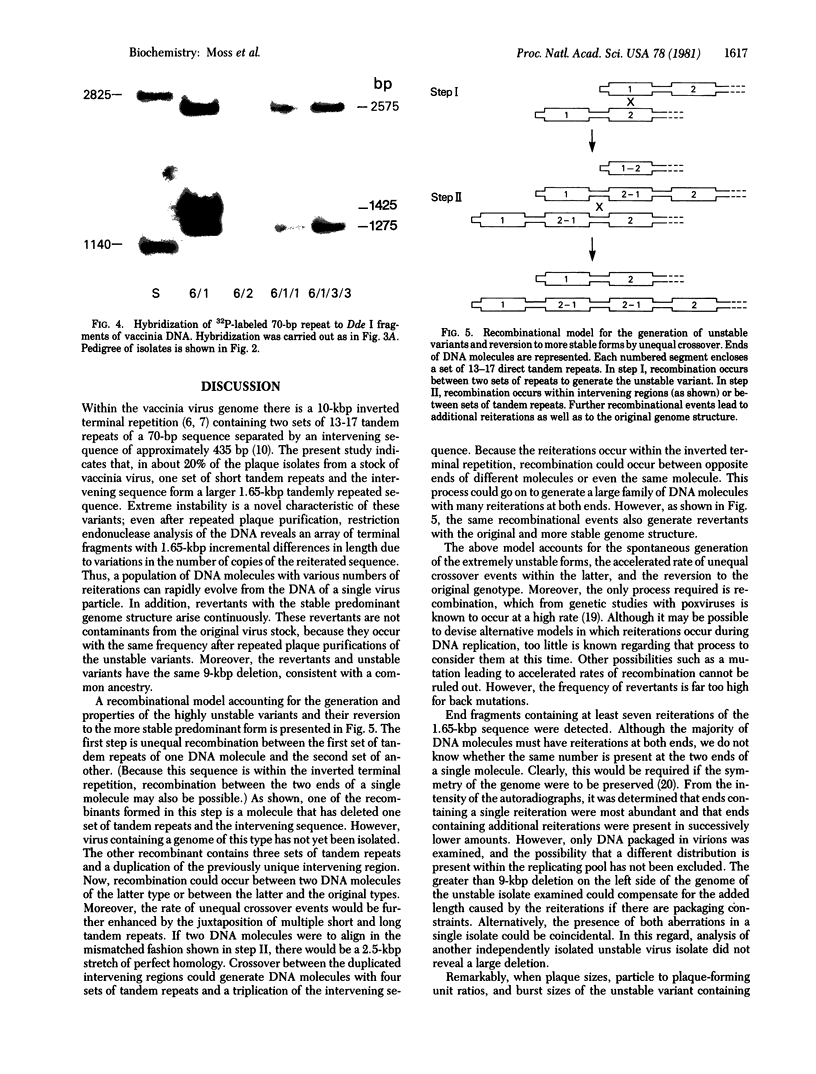
The sequence arrangement within the nontranscribed portion of the inverted terminal repetition of the vaccinia virus genome exists in quasi-stable and unstable forms that are not distinguishable on the basis of viral infectivity. The unstable forms, which composed about 20% of a serially passaged stock of virus, were recognized by terminal heterogeneity on restriction endonuclease analysis. Instead of a single terminal fragment from each end of the genome, an array of eight or more fragments differing in size by 1650-base-pair increments was detected. This feature was not eliminated by repeated plaque purification, indicating that the population of DNA molecules with various numbers of reiterations can rapidly evolve from the DNA of a single virus particle. However, at each successive round of plaque purification, about 20% of the unstable isolates revert back to the more stable form. Stable forms are characterized by the presence of a set of 13-17 tandem 70-base-pair repeats on each side of a 435-base-pair intervening sequence near both ends of the genome. In contrast, the unstable forms possess sets of tandem repeats and intervening sequences that alternate many times in series. The transition between the two genomic forms and the evolution of the unstable form appear to be mediated by recombinational events.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Archard L. C., Mackett M. Restriction endonuclease analysis of red cowpox virus and its white pock variant. J Gen Virol. 1979 Oct;45(1):51–63. doi: 10.1099/0022-1317-45-1-51. [DOI] [PubMed] [Google Scholar]
- Boone R. F., Moss B. Sequence complexity and relative abundance of vaccinia virus mRNA's synthesized in vivo and in vitro. J Virol. 1978 Jun;26(3):554–569. doi: 10.1128/jvi.26.3.554-569.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- DeFilippes F. Restriction enzyme digests of rapidly renaturing fragments of vaccinia virus DNA. J Virol. 1975 Jan;17(1):227–238. doi: 10.1128/jvi.17.1.227-238.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L., Gillespie D. Restriction site periodicities in highly repetitive DNA of primates. J Mol Biol. 1979 Nov 15;134(4):805–834. doi: 10.1016/0022-2836(79)90487-x. [DOI] [PubMed] [Google Scholar]
- Doolittle W. F., Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980 Apr 17;284(5757):601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Fenner F. Conditional lethal mutants of animal viruses. Curr Top Microbiol Immunol. 1969;48:1–28. doi: 10.1007/978-3-642-46163-7_1. [DOI] [PubMed] [Google Scholar]
- Garon C. F., Barbosa E., Moss B. Visualization of an inverted terminal repetition in vaccinia virus DNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4863–4867. doi: 10.1073/pnas.75.10.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geshelin P., Berns K. I. Characterization and localization of the naturally occurring cross-links in vaccinia virus DNA. J Mol Biol. 1974 Oct 5;88(4):785–796. doi: 10.1016/0022-2836(74)90399-4. [DOI] [PubMed] [Google Scholar]
- McCarron R. J., Cabrera C. V., Esteban M., McAllister W. T., Holowczak J. A. Structure of vaccinia DNA: analysis of the viral genome by restriction endonucleases. Virology. 1978 May 1;86(1):88–101. doi: 10.1016/0042-6822(78)90010-7. [DOI] [PubMed] [Google Scholar]
- McFadden G., Dales S. Biogenesis of poxviruses: mirror-image deletions in vaccinia virus DNA. Cell. 1979 Sep;18(1):101–108. doi: 10.1016/0092-8674(79)90358-1. [DOI] [PubMed] [Google Scholar]
- Moyer R. W., Rothe C. T. The white pock mutants of rabbit poxvirus. I. Spontaneous host range mutants contain deletions. Virology. 1980 Apr 15;102(1):119–132. doi: 10.1016/0042-6822(80)90075-6. [DOI] [PubMed] [Google Scholar]
- Orgel L. E., Crick F. H. Selfish DNA: the ultimate parasite. Nature. 1980 Apr 17;284(5757):604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Smith G. P. Evolution of repeated DNA sequences by unequal crossover. Science. 1976 Feb 13;191(4227):528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittek R., Barbosa E., Cooper J. A., Garon C. F., Chan H., Moss B. Inverted terminal repetition in vaccinia virus DNA encodes early mRNAs. Nature. 1980 May 1;285(5759):21–25. doi: 10.1038/285021a0. [DOI] [PubMed] [Google Scholar]
- Wittek R., Cooper J. A., Barbosa E., Moss B. Expression of the vaccinia virus genome: analysis and mapping of mRNAs encoded within the inverted terminal repetition. Cell. 1980 Sep;21(2):487–493. doi: 10.1016/0092-8674(80)90485-7. [DOI] [PubMed] [Google Scholar]
- Wittek R., Menna A., Müller H. K., Schümperli D., Boseley P. G., Wyler R. Inverted terminal repeats in rabbit poxvirus and vaccinia virus DNA. J Virol. 1978 Oct;28(1):171–181. doi: 10.1128/jvi.28.1.171-181.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittek R., Moss B. Tandem repeats within the inverted terminal repetition of vaccinia virus DNA. Cell. 1980 Aug;21(1):277–284. doi: 10.1016/0092-8674(80)90135-x. [DOI] [PubMed] [Google Scholar]
- Wittek R., Müller H. K., Wyler R. Length heterogeneity in the DNA of vaccinia virus is eliminated on cloning the virus. FEBS Lett. 1978 Jun 1;90(1):41–46. doi: 10.1016/0014-5793(78)80293-2. [DOI] [PubMed] [Google Scholar]