Abstract
A sensitive vertical denaturing gradient gel electrophoresis (DGGE) method, using 13 unipolar psoralen-clamped PCR primer pairs, was developed for detecting sequence variants in the 22 tRNA genes and flanking regions (together spanning ~21%) of the human mitochondrial genome. A study was conducted to determine (i) if mitochondrial DNA (mtDNA) polymorphisms and/or mutations were detectable in healthy newborns and (ii) if prepartum 3′-azido-2′,3′-dideoxythymidine (AZT) based HIV-1 prophylaxis was associated with significant increases in mtDNA mutations and changes in the degree of heteroplasmy of sequence variants in uninfected infants born to HIV-1-infected mothers. DGGE analysis of umbilical cord tissue (where vascular endothelium and smooth muscle cells are the major source of mtDNA) showed that mtDNA sequence variants were significantly elevated by threefold in AZT-treated infants compared with unexposed controls (P < 0.001), with 24 changes observed in 19/52 (37%) treated newborns (averaging 0.46 changes/subject) versus only eight changes found in 7/55 (13%) unexposed newborns (averaging 0.15 changes/subject). Six distinct sequence variants occurring in unexposed controls were predominately synonymous and homoplasmic, representing previously reported polymorphisms. Uninfected infants exposed to a combination of AZT and 2′,3′-dideoxy-3′-thiacytidine and “maternal HIV-1” had a significant shift in the spectrum of mutations (P = 0.04) driven by increases in nonsynonymous heteroplasmic sequence variants at polymorphic sites (10 distinct variants) and novel sites (four distinct variants). While the weight of evidence suggests that prepartum AZT-based prophylaxis produces mtDNA mutations, additional research is needed to determine the degree to which fetal responses to maternal HIV-1 infection, in the absence of antiretroviral treatment, contribute to prenatal mtDNA mutagenesis.
Keywords: antiretrovirals, AZT, DGGE, mitochondrial DNA mutation, nucleoside analogs, 3TC, tRNA genes, transplacental exposure
INTRODUCTION
The global spread of HIV-1, with more than 33 million living with the infection in 2007, confirms the need for safe and effective antiretroviral (ARV) drugs for treating viral infection and preventing mother-to-child transmission (MTCT) of the virus [Watts, 2006; Kallings, 2008]. In well-resourced settings, “highly active antiretroviral therapy” (HAART) including one or more nucleoside reverse transcriptase inhibitors (NRTIs) plus a protease inhibitor or a non-nucleoside reverse transcriptase inhibitor is typically used to control viral replication in HIV-1-infected patients. A combination of zidovudine (3′-azido-3′-deoxythymidine, AZT) and lamivudine (2′-deoxy-3′-thiacytidine, 3TC) (referred to as AZT-3TC) is most commonly given to HIV-1-infected pregnant women to reduce MTCT of the virus. Without perinatal ARV drugs, vertical transmission of HIV-1 occurs ~25% of the time [Cooper et al., 2002; Thorne and Newell, 2007]. The advent of HAART in the management of HIV-1 infection during pregnancy has had an enormous impact on pediatric HIV-1 associated morbidity and mortality, with the rate of MTCT of HIV-1 being reduced to <2% [Minkoff et al., 2001; Cooper et al., 2002; Dorenbaum et al., 2002; European Collaborative Study, 2003, 2005; Thorne and Newell, 2007].
The importance of NRTI-based ARV therapy/prophylaxis to public health cannot be underestimated; however, NRTIs are transplacental carcinogens in rodents and have been implicated as potential carcinogens and mitochondrial toxins in humans [Blanche et al., 1999; IARC, 2000; Wutzler and Thust, 2001; Divi et al., 2005, 2007; Venhoff and Walker, 2006; Walker et al., 2007; Wogan, 2007; Foster and Lyall, 2008]. In HIV-1-infected pregnant women, HAART begins at conception or after the first trimester, followed by intravenous administration of AZT during labor and delivery and then treatment of the infant with AZT during the first 6 weeks of life [Cooper et al., 2002]. Prepartum AZT-3TC has been associated with increased frequencies of cytogenetic events and somatic mutations during fetal life and may pose a risk for persistent genetic damage and cancer in children later in life [O'Neill et al., 2001; Escobar et al., 2007; Witt et al., 2007]. The mutagenic effects of NRTIs may result in part from the mechanism by which they prevent viral replication. NRTIs are analogs of natural nucleosides, but lack the 3′-OH of the deoxyribose sugar that serves as the site for the 5′ to 3′-phosphoester bond with the proceeding nucleic acid. They are able to inhibit viral replication by taking the place of an endogenous nucleotide during reverse transcription of viral RNA and, thus, cause premature termination of proviral DNA synthesis [Kakuda, 2000]. This mode of action also allows for the incorporation of these drugs into host cell nuclear DNA (nDNA) and mitochondrial DNA (mtDNA) [Olivero et al., 1997, 1999, 2000, 2002; Meng et al., 2007].
Substantial evidence supports the role of prolonged treatment with AZT and/or other NRTIs in the induction and/or progression of a number of clinical syndromes in HIV-1-infected patients that are believed to arise from underlying mitochondrial toxicities [reviewed in Lewis and Dalakas, 1995; Moyle, 2000; Dagan et al., 2002; Powderly, 2002; Lewis et al., 2003; Kohler and Lewis, 2007], but the implication of findings related to clinical mitochondrial dysfunction in uninfected children receiving perinatal NRTIs is uncertain [Blanche et al., 2006; Spector and Saitoh, 2006; Thorne and Newell, 2007; Foster and Lyall, 2008]. Concerns about mitochondrial toxicity in NRTI-exposed children were first raised by a report of symptoms and laboratory abnormalities suggesting mitochondrial dysfunction in eight uninfected children in the French Perinatal Cohort Study that examined a total of 2,644 infants [Blanche et al., 1999]. Clinical signs included mild metabolic abnormalities, neurological symptoms, and one case of transient cardiomyopathy, with the most serious changes in infants receiving prepartum AZT-3TC. In subsequent reviews of U.S. and European cohorts consisting of thousands of children born to HIV-1-infected mothers, no deaths were attributable to mitochondrial dysfunction and symptoms of mitochondrial toxicity were lacking in NRTI-exposed children [Lindegren et al., 2000; Perinatal Safety Review Working Group, 2000; European Collaborative Study, 2003]. However, Thorne and Newell [2007] noted that no clinical work-up or tests specific for mitochondrial function were included in these studies and that most investigations lacked the sensitivity to identify mild cases of mitochondrial dysfunction. Since 1999, studies specific for mitochondrial toxicity have identified more than two dozen cases of mitochondrial dysfunction in uninfected children exposed perinatally to AZT and other ARVs in the French Perinatal Cohort Study, with additional cases reported in Spain, Italy, and Canada [Barret et al., 2003; Noguera et al., 2004; Tovo et al., 2005; Blanche et al., 2006; Benhammou et al., 2007]. In the U.S., severe mitochondrial dysfunction in an infant receiving prepartum AZT-3TC and nevirapine during the last 6 weeks of pregnancy was confirmed by muscle biopsy [Cooper et al., 2004], and a recent retrospective review of records for 1,037 uninfected children enrolled in PACTG protocols 219/219C identified 20 cases of unexplained signs of mitochondrial dysfunction according to the Enquête Périnatale Française criteria, suggesting a possible link between first exposure to 3TC or AZT-3TC in the third trimester and mitochondrial dysfunction [Brogly et al., 2007].
Mitochondrial toxicity induced by NRTIs may result from several mechanisms, including (i) direct inhibition of mtDNA polymerase γ (pol γ) without NRTI incorporation, (ii) mtDNA chain termination due to incorporation of the NRTI into mtDNA by pol γ, (iii) inhibition of the 3′ to 5′ exonuclease activity of pol γ, (iv) inhibition of nucleoside metabolizing enzymes, and (v) alterations in mitochondrial metabolism affecting oxidative phosphorylation (OXPHOS) enzyme activity [reviewed in Dagan et al., 2002; Lewis, 2003, 2004, 2005; Kohler and Lewis, 2007]. Pol γ is preferentially inhibited by NRTIs over other cellular polymerases, with inhibition of the polymer-ase and exonuclease activities of pol γ leading to mtDNA depletion or disruption of mtDNA structure [Brinkman et al., 1998; Kakuda, 2000]. Depletion of mtDNA results in a decrease in the synthesis of proteins that are essential for OXPHOS, causing a disruption in OXPHOS and subsequent energy loss via a decrease in the production of ATP [Lewis et al., 2003]. This phenomenon is followed by an increase in leakage of electrons from the electron-transport chain that reacts with oxygen, forming reactive oxygen species, which damage proteins, lipids, nDNA [Bialkowska, 2000], and mtDNA. The net effect of these deleterious NRTI-associated changes in mitochondrial function is an increase in genetic instability of mtDNA, predisposing to the induction of persistent, heritable mutations.
Perhaps due to the difficulty of detecting drug-induced mutations amidst abundant wild-type mtDNA, studies documenting the occurrence of mtDNA mutations in NRTI-treated HIV-1-infected patients are relatively few and provide limited evidence of the mutagenic activity of NRTIs in mtDNA. Several groups reported decreased mtDNA levels, oxidative lesions, and deletion mutations associated with NRTI-related lactic acidosis and lipodys-trophy in HIV-1-infected patients [Miro et al., 2000; Zaera et al., 2001; Walker and Venhoff, 2001; Hashiguchi et al., 2004; Maagard et al., 2006]. Initial studies of small-scale mtDNA mutations in peripheral blood mononuclear cells (PBMCs) of HIV-1-infected adults suggest that NRTI-based therapy provides conditions permissive for the development of “random” mutations in vivo over time [Martin et al., 2003], albeit further research is needed to determine if NRTIs induced these mutations or unmask silent variations [McComsey et al., 2002, 2005].
The purpose of the present study was to determine (i) if a sensitive and specific vertical denaturing gradient gel electrophoresis (DGGE) technique using psoralen-clamped PCR primers could be developed for the detection of mtDNA polymorphisms and novel mutations in umbilical cord tissue from healthy infants born to HIV-1-uninfected mothers and (ii) if the frequency, spectrum, and degree of heteroplasmy of sequence variants in uninfected newborns receiving prepartum AZT-based HIV-1 prophylaxis was similar to or distinct from those found in NRTI-naïve uninfected infants born to healthy mothers. The current work focused on the potential occurrence of sequence variants in the 22 mtDNA tRNA genes because they are thought to represent “hotspots” for heritable mutation-based mitochondrial disease in humans [Levinger et al., 2004]. Notably, mutations in the tRNA genes account for nearly two-thirds of these diseases, testifying to the importance of mitochondrial tRNA gene integrity in maintaining normal mitochondrial and cellular function [Kelley et al., 2000; Levinger et al. 2004; McFarland et al., 2004; Ruiz-Pesini et al., 2007]. The critical question is whether multiple drug-induced mtDNA mutations, as opposed to a single pathogenic maternally inherited mutation, are linked to the pathogenesis and clinical consequences of prepartum exposure to NRTIs.
MATERIALS AND METHODS
Patient Population and Sample Collection
Samples were obtained within the context of a larger observational cohort study [Meng et al., 2007] that included infants of uninfected women (n = 68) and infants of HIV-1-infected women receiving ARV therapy during pregnancy (n = 71). The cohort study included infants born at several different hospitals and was designed to evaluate clastogenic and mutagenic events occurring in NRTI-exposed infants [Walker and Poirier, 2007]. However, in response to reports of mitochondrial toxicity in NRTI-exposed infants [Blanche et al., 1999; Barret et al., 2003; Poirier et al., 2003; Shiramizu et al., 2003; Divi et al., 2004; Noguera et al., 2004; Tovo et al., 2005; Benhammou et al., 2007], a study of the potential of AZT-3TC to cause mtDNA mutations during fetal life was performed using fresh umbilical cord collected from infants born at study sites during the day shift. After delivery of the neonate, several inches of the umbilical cord were snap frozen and stored at –70°C for future DNA extraction. Samples from each newborn were coded with patient identification (PID) numbers at the clinical centers, and subsequently analyzed in a “blinded” fashion. The original PID numbers were changed for this report to provide an additional level of patient identity protection.
The protocol for the cohort study was reviewed and approved by the Institutional Review Boards at the participating medical centers. Written informed consent was obtained from each participating woman and from each child's parent/guardian. Questionnaire, medical history, and chart data provided information on maternal HIV-1 status, alcohol consumption, smoking history, and medications and illicit drugs (amphetamines, cannabinoids, cocaine, and opiates) used during the pregnancy. Infant data collected from the records included, in part, birth weight, estimated gestational age at delivery, and HIV-1 status. Infants were defined as uninfected if they had at least two negative HIV PCR tests on separate occasions after the age of 1 month.
Disinfection of Biological Specimens and Isolation of DNA
Before DNA was extracted from sections of umbilical cord, virus from potential contamination of the tissue with maternal blood or from possible HIV-1 infection of the infant was inactivated using DISCIDE-TB (Fisher Scientific, Houston, TX), a quaternary based disinfectant that is effective in eliminating the risk associated with HIV-1 [URL: http://www.nurseslearning.com/courses/fice/fde0008/coursebook.pdf]. Briefly, a thin cross-section of umbilical cord was sliced from the frozen sample, and then placed in a 50% solution of DISCIDE-TB in water for 15 min. The thin sections were then rinsed in distilled water to remove all DISCIDE-TB. Total DNA was isolated from the sections of umbilical cords using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI).
PCR Amplification, Heteroduplex Formation, and Photoinduced Crosslinking
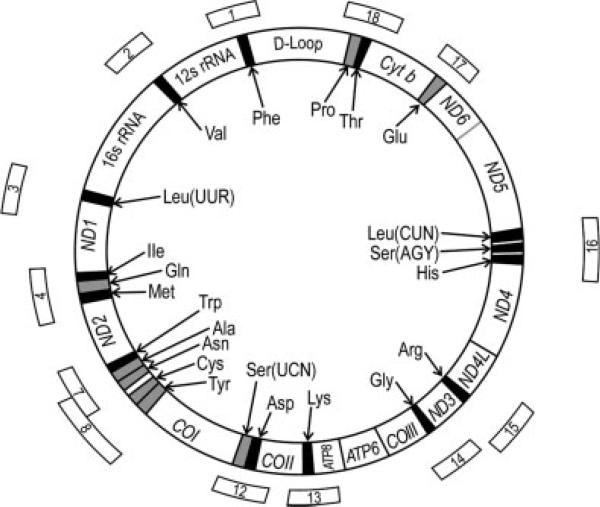
The report of Michikawa et al. [1997] was used as the basis for designing primers and selecting regions for PCR amplification of mtDNA segments encoding the 22 tRNA genes and flanking regions. Thirteen PCR primer pairs were designed using the Primo Melt 3.4 computer program (Chang Biosciences, San Francisco, CA) on the basis of the melting characteristics of psoralen-oligonucleotides conjugated with the DNA region of interest (Fig. 1). To reduce the melting map of each mtDNA segment to a single domain, a psoralen clamp (psoralen C6 phosphoroamidite) was linked to one oligonucleotide of each PCR primer pair, and the chosen primer oligonucleotides were synthesized by Alpha DNA (Montreal, Canada). These 13 unipolar psoralen-clamped PCR primer sets were used to produce PCR products ranging from 134 to 419 bases, and to yield a single melting domain with a uniform melting temperature of 61°C for all segments selected for screening the 22 tRNA genes and flanking regions that together spanned ~3,500 of the 16,569 nucleotides (21%) of the human mitochondrial genome. The primer names, sequences, PCR variables, and amplified products are shown in Table I.
Fig. 1.
Map positions of human mtDNA segments amplified by PCR for sequence variations in the tRNA genes. Thirteen segments of human mtDNA (Table I) were selected to screen all tRNA-coding regions using the Primo Melt 3.4 computer program.
TABLE I.
Oligonucleotide Primers for PCR Amplification and Vertical DGGE Analysis of the 22 Human tRNA Genes and Flanking Regions
| PCR conditions |
PCR product |
|||||
|---|---|---|---|---|---|---|
| Oligonucleotide namea | Primer sequence | mMol MgCl2 | PCR cycles | Sequence position | Length (bases) | tRNA gene(s) amplified |
| H tRNA 1 Sense | TGCTAACCCCATACCCCGAA | 2.0 | 32 | 5′-528 | 193 | tRNAPhe |
| H tRNA 1 Antisense PS | ACTCACTGGAACGGGGATGCTT | 3′-720 | ||||
| H tRNA 2 Sense PS | TTAGTTGAACAGGGCCCTGAAGC | 2.0 | 32 | 5′-1451 | 279 | tRNAVal |
| H tRNA 2 Antisense | AATGGTTTGGCTAAGGTTGTCTGG | 3′-1729 | ||||
| H tRNA 3 Sense | GTGCAGCCGCTATTAAAGGTTCGT | 2.0 | 35 | 5′-3014 | 407 | tRNALeu |
| H tRNA 3 Antisense PS | GTTGGGGCCTTTGCGTAGTTGTAT | 3′-3420 | ||||
| H tRNA 4 Sense | ATTACAATCTCCAGCATTCCCCCT | 2.2 | 30 | 5′-4231 | 329 | tRNAIle, tRNAGln, tRNAMet |
| H tRNA 4 Antisense PS | TCAGGTAAAAAATCAGTGCGAGCTT | 3′-4559 | ||||
| H tRNA 7 Sense | CCCTTACCACGCTACTCCTACC | 1.5 | 32 | 5′-5461 | 254 | tRNATrp, tRNAAla, tRNAAsn |
| H tRNA 7 Antisense PS | TGATTAGGGTGCTTAGCTGTTAACT | 3′-5714 | ||||
| H tRNA 8 Sense | TTAAGCTAAGCCCTTACTAGACCAA | 2.5 | 30 | 5′-5641 | 289 | tRNACys, tRNATyr |
| H tRNA 8 Antisense PS | GAGAATAGTCAACGGTCGGCGAA | 3′-5929 | ||||
| H tRNA 12 Sense | CATTCGAAGAACCCGTATACATAAAA | 2.0 | 32 | 5′-7414 | 315 | tRNASer, tRNAAsp |
| H tRNA 12 Antisense PS | GTTGTGAGTGTTAGGAAAAGGGCA | 3′-7728 | ||||
| H tRNA 13 Sense | CCCATCGTCCTAGAATTAATTCCCC | 2.0 | 30 | 5′-8207 | 277 | tRNALys |
| H tRNA 13 Antisense PS | TTGGTGAGGGAGGTAGGTGGTAGTTT | 3′-8483 | ||||
| H tRNA 14 Sense | CGCCTGATACTGGCATTTTGTAG | 2.0 | 32 | 5′-9920 | 230 | tRNAGly |
| H tRNA 14 Antisense PS | TGTAGCCGTTGAGTTGTGGTAGTC | 3′-10149 | ||||
| H tRNA 15 Sense | AGCCCTAAGTCTGGCCTATGAGTGA | 2.2 | 32 | 5′-10352 | 134 | tRNAArg |
| H tRNA 15 Antisense PS | TGTAAATGAGGGGCATTTGGT | 3′-10485 | ||||
| H tRNA 16 Sense | TATCCCCCATTCTCCTCCTATCCCT | 2.0 | 32 | 5′-12081 | 419 | tRNAHis, tRNASer, tRNALeu |
| H tRNA 16 Antisense PS | TGAATATTGTTGTGGGGAAGAGACTG | 3′-12499 | ||||
| H tRNA 17 Sense | CGACCACACCGCTAACAATCAA | 2.0 | 32 | 5′-14559 | 215 | tRNAGlu |
| H tRNA 17 Antisense PS | GGGGTTAATTTTGCGTATTGGGG | 3′-14773 | ||||
| H tRNA 18 Sense PS | CATCATTGGACAAGTAGCATCCGT | 2.0 | 32 | 5′-15790 | 286 | tRNAThr, tRNAPro |
| H tRNA 18 Antisense | ATGGGTGAGTCAATACTTGGGTGG | 3′-16075 | ||||
The oligonucleotide denoted with PS has a psoralen derivative linked to the 5′ end.
Amplification was carried out in 50-μl PCR reactions containing 1× reaction buffer (without MgCl2), 1.5 to 2.2 mM MgCl2 (see Table I), 0.2 mM of each deoxynucleotide triphosphate, 0.2 μM of each primer, 0.5 U GoTaq Flexi (Promega, Madison, WI), and 100 ng of total DNA. PCR amplification was performed using a Perkin Elmer 9600 thermal cycler as follows: 94°C for 2 min, followed by 30–32 cycles (see Table I) of 94°C for 15 sec, 61°C for 30 sec, and 68°C for 1 min. The PCR reaction was completed by a final extension of 72°C for 5 min. Following amplification, 5 μl of each sample were electrophoresed on 8% polyacrylamide gel to assess PCR efficiency. The remaining PCR product was heat-denatured at 99°C for 10 min, and then allowed to cool at 12°C per min to 37°C and held at 37°C for 30 min. After reannealing, the samples were placed on ice and irradiated for 25 min using a 366-nm UV source (Ultra Violet Products, San Gabriel, CA) to form a psoralen-DNA cross-link at one end of the individual PCR fragments. Samples were then vacuum-dried and resuspended in 5 μl 6× loading dye.
Vertical Denaturing Gradient Gel Electrophoresis
DGGE is based on the fact that the electrophorectic mobility of a DNA molecule in a polyacrylamide gel is considerably reduced as the molecule becomes partially melted (denatured). Mismatched heteroduplexes are always less stable than the corresponding perfectly base-paired homoduplexes and consequently melt at a lower concentration of denaturant. Any mutant/wild-type heteroduplex always will travel a shorter distance relative to wild-type or mutant homoduplexes in a gel containing a gradient of denaturant. Substitution of psoralen-oligonucleotide conjugates for the commonly used GC-tailed oligonucleotides (“GC clamps”) for PCR amplification of mtDNA fragments allows simultaneous analysis of a large number of different amplified DNA fragments over a broad range of denaturing gradient because of the lack of strand separation in the “clamp” region [Lerman et al., 1987; Michikawa et al., 1997].
Vertical DGGE was performed utilizing a DGGE-4001 system (C.B.S. Scientific Company, Solana Beach, CA). The resuspended PCR products were loaded onto a 1-mm thick, 8% polyacrylamide gel with a linear denaturant gradient of 28–54% (100% denaturant is defined as 7 M urea plus 40% v/v formamide). The gels were submerged into the DGGE tank, with 1× TBE buffer continuously circulating between the upper reservoir and the buffer tank. Electrophoresis was conducted at 100 V and a constant temperature of 58°C for 17 hr. Following electrophoresis, the gels were stained for 15 min in ethidium bromide (1 μg/ml), destained for 10 min, visualized with a Spectroline UV transilluminator (Spectronics, Westbury, NY), and photographed using Polaroid 667 black and white film. Bands that contained presumptive resolved and unresolved mutant/wild-type heteroduplexes or mutant homoduplexes were carefully excised from the gel, crushed, and eluted overnight in 200 μl of sterile H2O at 4°C.
To purify the DNA, a second round of PCR was conducted following the same conditions as previously described. The samples were once again heat-denatured at 99°C for 10 min, and then allowed to cool at 12°C per min to 37°C and held at 37°C for 30 min. Following reannealing, the samples were placed on ice and irradiated as above. Samples were then vacuum-dried, resuspended in loading dye, and run on a secondary DGGE gel using the same gradient (28–54%) to confirm the identification of the heteroduplexes or mutant homoduplexes representing presumptive sequence variants. The resulting bands were excised carefully from the gel, crushed, and eluted overnight as described above to serve as template for an additional round of PCR and DNA sequencing. In the few instances where a pair of heteroduplexes occurred along with the corresponding mutant homoduplex, all three bands were excised and processed to confirm that all represented the same sequence variant.
DNA Sequencing and Estimation of Degree of Heteroplasmy or Mutant Fraction
For DNA sequencing, the final PCR products from excised mutant homoduplexes or pairs of heteroduplexes were purified of salts, primers, and unincorporated dNTPs using Millipore Montage PCR centrifugal filter devices (Bedford, MA) per the manufacturer's instructions. The purified samples were then further prepared for DNA sequencing by removal of polymerase according to the instructions set forth by Millipore Micropure-EZ centrifugal filter devices (Bedford. MA). Twenty-nine of 36 sequence variants found in unexposed and NRTI-exposed infants were identified first as mutant homoduplexes on primary DGGE gels (data presented below); for the remaining seven sets of mutant/wild-type heteroduplexes (excised from secondary DGGE gels), strand-biased PCR was performed to identify which strand contained the mutation in the mismatch for each heteroduplex [Walker and Skopek, 1993]. Forward and reverse primers were used to generate PCR products for each set of mutant/wild-type heteroduplexes; the expectation was that DNA sequencing for one heteroduplex would show one strand containing a mutation with the opposite strand being wild-type, while the reverse would occur for the matching heteroduplex. The purified PCR products were sequenced using an Applied Biosystem 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA).
The relative amounts of somatic mutations in a region of interest in nDNA from mutagen-exposed cells/animals, or the relative proportion of mutant DNA to wild-type DNA (mutant fraction or degree of heteroplasmy) for a given sequence variant in mtDNA, can be estimated from the banding patterns and relative densities of mutant/wild-type heteroduplexes, mutant homoduplexes, and/or wild-type homoduplexes appearing on primary DGGE gels of PCR fragments [Cariello et al., 1990]. One option was to use a densitometer for assessing the relative densities of bands and deriving a mutant fraction estimate for each mtDNA sequence variant identified. Since this semiquantitative method requires additional quantitative PCR-based techniques to define the mutant fractions more precisely, we elected to estimate the degree of heteroplasmy/mutant fraction for each mtDNA sequence variant in a qualitative fashion based upon the banding patterns on primary DGGE gels: low mutant fraction (L), only mutant/wild-type heteroduplexes were present; medium mutant fraction (M), either mutant/wild-type heteroduplexes and a mutant homoduplex (with/without a wild-type homoduplex) or resolved mutant and wild-type homoduplexes were present; high mutant fraction (H), only a mutant-homoduplex was present suggesting that the sequence variant approached homoplasmy.
In Vitro Exposure of Human Lymphoblastoid Cells to AZT and/or 3TC, and DGGE-Based Screening for mtDNA Sequence Variants
Human TK6 B-lymphoblastoid cells were cultured as described [Torres et al., 2007], and optimal growth conditions were maintained throughout the experiment [Carter et al., 2007]. Briefly, TK6 cells in log phase growth were exposed to 0 or 100 μM AZT, 3TC or AZT-3TC (equimolar) (n = 4 flasks per group) through 14 rounds of cell replication. During culture in T-flasks, cells were maintained at a density of 400,000 cell/ml by removing excess cells and adding medium containing the appropriate amount of NRTI(s) to maintain the molar concentration of drug(s). Cell numbers and survival were measured each day using Trypan blue exclusion. Periodically through the exposure period and at the end of 14 rounds of cell replication, aliquots of cells were taken from each flask, pelleted, washed, and stored for DNA isolation and vertical DGGE analysis for mtDNA sequence variants (using the techniques described above).
Statistical Analyses
Chi-square and Mann-Whitney tests were used to assess differences in the distribution and numbers of mtDNA sequence variants in the AZT-3TC treatment and unexposed groups of infants. A Chi-square test was also used to evaluate differences in the proportions of homoplasmic versus heteroplasmic sequence variants in treated versus unexposed groups. The chi-square tests a null hypothesis that the relative frequencies of occurrence of observed events follow a specified frequency distribution, while the null hypothesis for the Mann-Whitney test states that there is no difference between the unexposed and treated groups. Multivariate analyses were used to assess associations between maternal/fetal factors or potential confounders and the numbers of sequence variants found in NRTI-exposed versus unexposed infants. Simple linear regression analysis was performed to assess the relationship between the duration of prepartum NRTI exposure and the number of mtDNA sequence variants in the treatment group of newborns. A P-value < 0.05 was considered significant.
RESULTS
Maternal NRTI Therapy and HIV-1Status of Infants
Biological samples were collected as part of an observational cohort study [Meng et al., 2007], in which all HIV-1-infected women were given ARVs to control their viral infection and reduce MTCT. The current study included 55 infants born to uninfected mothers and 53 infants born to HIV-1-infected mothers who received AZT or AZT-3TC prior to labor and delivery (Tables II and III). Of the NRTI-treated infants in whom sequence variants were identified (n = 20/53), three were exposed to AZT only, 12 were exposed to AZT-3TC, and three were exposed to AZT alone followed by AZT-3TC during the later portion of the pregnancy (5.9 months or less) (Table III). Two of the HIV-1-infected mothers received combinations of stavudine (d4T) with AZT or 3TC for different periods of time before receiving AZT-3TC later in pregnancy.
TABLE II.
Maternal and Fetal Demographics
| Uninfected mothers (n = 55) | HIV-1 infected mothers (n = 53) | |
|---|---|---|
| Maternal factors | ||
| Ethnicity/Race | ||
| American Indian | 0 | 1 |
| Black, not Hispanic | 9 | 15 |
| Hispanic/Latino | 23 | 29 |
| White (not Hispanic) | 19 | 5 |
| Not known | 4 | 3 |
| Age at delivery (years; mean ± SD) | 27.2 ± 1.1 | 28.3 ± 0.7 |
| Plasma HIV-1 RNA (log10 copies/ml; range) | Not tested | 50–172,000a |
| CD4 cell count (×106 cells/l; range) | Not tested | 15–850a |
| CD4 cell % (range) | Not tested | 1–48a |
| Fetal factors | ||
| Gender | ||
| Female | 30 | 26 |
| Male | 25 | 27 |
| Birth weight (g; mean ± SD) | 3265.8 ± 74.5 | 3098.6 ± 84.1 |
| Gestational age at delivery (weeks; mean ± SD) | 39.1 ± 0.3 | 38.0 ± 0.2 |
| HIV-1 status | Not tested | Negative for 52/53 infantsb |
Values represent the range across HIV-1 infected mothers.
For all 52 infants included in the analyses, at least two negative HIV-1 PCR tests were obtained on separate occasions after the age of 1 month.
TABLE III.
Unexposed and NRTI-Exposed Infants with Sequence Variants Identified by PCR-Based DGGE and DNA Sequence Analysis of mtDNA from Umbilical Cord Tissuea
| PID numberb | Gender | Ethnicity or racec | NRTI drugs and gestational months of prepartum therapyd | Location (base No.) | Sequence variant | Mutant fractione |
|---|---|---|---|---|---|---|
| Unexposed (n = 7/55, 13%)f | ||||||
| B208 | M | 3 | None | 10115 | T → C | H |
| B253 | F | 3 | None | 9966 | G → A | H |
| B254 | M | 3 | None | 10115 | T → C | H |
| B255 | F | 3 | None | (1) 8271 | –9 base deletiong | H |
| (2) 9950 | T → C | H | ||||
| B310 | M | 2 | None | 15940 | –T deletion | H |
| B345 | M | 2 | None | 8269 | G → A | H |
| B377 | F | 1 | None | 10115 | T → C | L |
| AZT-exposed (n = 20/53, 38%)h | P < 0.001i | P = 0.04i | ||||
| B101 | M | 2 | AZT, 1.6; AZT-3TC, 3.0 | 10115 | T → C | H |
| B202 | F | 3 | AZT-d4T, 5.8; AZT-3TC, 3.2 | (1) 8271 | –9 base deletiong | M |
| (2) 9950 | T → C | H | ||||
| B218j | F | 3 | AZT, 6.0 | (1) 10398 | A → G | H |
| (2) 10454 | T → C | H | ||||
| B225 | F | 3 | AZT, 5.0 | 8271 | –9 base deletiong | M |
| B231k | F | 3 | AZT-3TC, 2.7 | (1) 14392 | C → G | M |
| (2) 14558 | C → T | M | ||||
| B233 | F | 3 | AZT, 3.3; AZT-3TC, 2.0 | 9966 | G → A | H |
| B234k | M | 3 | AZT, 0.7; AZT-3TC, 5.3 | 14392 | C → G | M |
| B243 | F | 3 | AZT, 5.5 | 8271 | –9 base deletiong | H |
| B322 | M | 2 | AZT-3TC, 1.5 | 15942 | T → C | H |
| B324 | M | 2 | AZT-3TC, 2.3 | 9986 | G → A | H |
| B333 | M | 2 | AZT-3TC, 9.0 | 8374 | A → G | H |
| B335 | M | 1 | AZT-3TC, 5.5 | 9966 | G → A | H |
| B356 | M | 2 | AZT-/3TC, 5.0 | (1) 10115 | T → C | M |
| (2) 10398 | A → G | L | ||||
| B371 | M | 3 | AZT-3TC, 4.1 | (1) 10398 | A → G | M |
| (2) 10454 | T → C | M | ||||
| B601 | F | 3 | d4T-3TC, 3.0; AZT-3TC, 5.9 | 8271 | –9 base deletiong | H |
| B603 | M | 3 | AZT-3TC, 5.3 | (1) 14766 | C → T | M |
| (2) 14759 | +19 base insertionl | M | ||||
| B606 | M | 3 | AZT-3TC, 4.8 | 15924 | A → G | M |
| B701 | F | 2 | AZT-3TC, 5.2 | 10398 | A → G | M |
| B702 | F | 2 | AZT-3TC, 5.4 | 10398 | A → G | M |
| B703 | F | 3 | AZT-3TC, 3.7 | 10398 | A → G | M |
Vascular endothelial and smooth muscle cells serve as the major source of DNA in umbilical cord tissue.
In this report, patient identification numbers were replaced with substitute numbers, and only infants with identifiable sequence variants are listed (in sequential order) for the unexposed and treatment groups.
Maternal ethnicity/race: 1 = white, not Hispanic; 2 = black, not Hispanic; 3 = Hispanic (Spanish culture/origin regardless of color).
Duration (in months) of maternal NRTI therapy during the final months of pregnancy; NRTI drug regimens are shown in temporal order ending with the birth of the infant.
The degree of heteroplasmy, or mutant fraction (proportion of mutant DNA to wild-type DNA), for each sequence variant was assessed qualitatively based upon the banding patterns on primary DGGE gels: Low mutant fraction (L), only mutant/wild-type heteroduplexes were present; medium mutant fraction (M), either mutant/wild-type heteroduplexes and a mutant homoduplex (with/without a wild-type homoduplex) or resolved mutant and wild-type homoduplexes were present; high mutant fraction (H), only a mutant-homoduplex was present suggesting that the sequence variant approached homoplasmy.
Infants born to healthy HIV-1-uninfected mothers.
Deletion of 9 bases, accccctct or ccccctcta [Hertzberg et al., 1989; Harihara et al., 1992; Lorenz et al., 1994; Torroni et al., 1995].
Infants born to HIV-1-infected mothers receiving prepartum AZT-based therapy.
Frequencies of sequence variants, as well as variants exhibiting heteroplasmy, were significantly different in AZT-exposed infants compared with unexposed controls; Chi-square test.
Infant # B218 was HIV-1 positive by HIV PCR tests at 1, 3, and 5 months of age; thus, sequence variants detected in this infant are shown here but were excluded from statistical analyses and consideration in Tables IV and V.
A ~70-base insertion found at nucleotide 14392 in NADH dehydrogenase subunit 3 in each of two infants was excluded from statistical analyses of sequence variants because the base sequence of the entire insertion could not be determined with certainty without the development of a new primer set, which was beyond the scope of this study.
Insertion of 19 bases, ccccaatacgcaaaattaa.
In considering the sequence variants found in mtDNA of newborns in this study (data presented below), we wanted to ascertain whether or not maternal health factors (such as age at delivery, viral load, and CD4 cell count) or fetal health factors (such as gestational age at delivery and birth weight) had an impact on the occurrence of mutations at polymorphic sites or novel mutations. A comparison of maternal and fetal demographics in exposed and unexposed infants is shown in Table II. There was no difference between the age at delivery for uninfected women and HIV-1-infected women receiving AZT-based therapies. For HIV-1-infected women, values for plasma HIV-1 RNA, CD4 cell count, and CD4 cell percent are shown in Table II. There were no differences in gender or mean birth weights in unexposed versus NRTI-exposed newborns; however, the mean estimated gestational age of 38.0 ± 0.2 (SD) weeks in infants born to NRTI-treated HIV-1-infected mothers was significantly reduced by 1.1 weeks compared to infants born to uninfected mothers. Infant HIV-1 status was evaluated using HIV PCR tests on two or more separate occasions after 1 month of age and infection was excluded based on the presence of at least two negative HIV PCR tests. With the exception of one infant, all infants born to HIV-1-infected mothers had two or more negative HIV PCR tests between 1 and 6 months of age and were confirmed uninfected (Tables II and III). Infant # B218 received prepartum AZT alone, but was HIV-1 positive by HIV PCR tests at 1, 3, and 5 months of age, and was excluded from statistical analyses for the treatment group (Table III). History of drug use, and drug use during pregnancy, was reported for both HIV-1-infected and uninfected mothers. For maternal drug use, there were no differences between exposed infants and unexposed control infants as well as between exposed infants who had mtDNA sequence variants and exposed infants who did not have any detectable mtDNA sequence changes. The same observation was made for maternal cigarette smoking before and during pregnancy, in which no difference was seen between NRTI-exposed infants and unexposed control infants and also between exposed infants with mtDNA sequence variants versus those without detectable variants. Thus, results of these statistical analyses indicate that neither maternal/fetal factors nor potential confounders accounted for differences in the numbers and types of sequence variants found in NRTI-exposed compared to unexposed infants.
DGGE and Sequence Analysis of mtDNA from Unexposed and NRTI-Exposed Infants
DNA isolates from umbilical cord tissue, where vascular endothelium and smooth muscle cells are the major source of DNA (see Discussion), were examined by vertical DGGE for sequence variants in the 22 tRNA genes and flanking regions. Table III lists the location and type of individual sequence variants and the degree of heteroplasmy for each variant found in unexposed and NRTI-exposed infants. Table III also shows data for the gender, ethnicity/race, and prepartum NRTI treatment and duration for each child with an identified mtDNA change. Heteroduplex or mutant homoduplex bands representing presumptive sequence variants were identified using PCR primer sets 13, 14, and 18 for amplification of DNA from unexposed infants, and primer sets 13, 14, 15, 17, and 18 for amplification of DNA from infants who received prepartum AZT-based prophylaxis. Table IV shows the distribution of each distinct sequence variant found in unexposed and/or NRTI-exposed newborns. Table V summarizes the mtDNA changes identified in unexposed and NRTI-exposed infants, and provides additional information for each distinct mtDNA sequence variant including the amino acid change, whether the variant was novel or occurred at a polymorphic site, any reported evidence of pathogenicity, and the numbers of unexposed and exposed newborns with a particular sequence change.
TABLE IV.
Distribution of Sequence Variants in mtDNA from Umbilical Cord Tissue of Control and HIV-1-Uninfected Infantsa
| Primer set number |
||||||
|---|---|---|---|---|---|---|
| Population | 13 | 14 | 15 | 17 | 18 | Total |
| Infants born to healthy HIV-1-uninfected mothers | 1(a) | 1(d) | 0 | 0 | 1(n) | Eight changes in 7/55 (13%) subjects; |
| 1(b) | 1(e) | Mean = 0.15/subject | ||||
| 3(f) | ||||||
| Uninfected infants born to HIV-1-infected mothers receiving prepartum AZT-based therapy | 4(b) | 1(d) | 6(h) | 2(j) | 1(o) | 24 changes in 19/52 (37%) subjects |
| 1(c) | 2(e) | 2(i) | 1(k) | 1(p) | Mean = 0.46/subject | |
| 1(f) | 1(l) | P <0.001b | ||||
| 2(g) | 1(m) | |||||
Each number below individual primer set numbers represents the number of patients with the same distinct sequence variant; each letter in parentheses indicates a distinct sequence variant (see Table V). Infant # B218 was HIV-1 positive by HIV PCR tests at 1, 3, and 5 months of age; thus, sequence variants detected in this infant are not included in this table.
Chi-Square test.
TABLE V.
Summary of Sequence Variants in mtDNA from Umbilical Cord Tissue of Control and HIV-1-Uninfected Infants Receiving AZT-Based Prepartum Therapy
| Sequence and locationa | Amino acid change | Polymorphic site (PS) or novel (N) | Evidence of pathogenicity | No. of unexposed infants | No. of exposed infants |
|---|---|---|---|---|---|
| (a) G8269A transition in COII | Noncoding | PSb | No | 1 | 0 |
| (b) 9-base deletion at nucleotide 8271 in NC7 | Noncoding | PSc | No | 1 | 4 |
| (c) A8374G transition in ATPase8 | Synonymous | N | Unknownd | 0 | 1 |
| (d) T9950C transition in COIII | Synonymous | PSc | No | 1 | 1 |
| (e) G9966A transition in COIII | Val → Ile | PSc | No | 1 | 2 |
| (f) G9986A transition in COIII | Gly → Asp | PSe | Unknownd | 0 | 1 |
| (g) T10115C transition in ND3 | Synonymous | PSc | No | 3 | 2 |
| (h) A10398G transition in ND3 | Thr → Ala | PSc | Yesf | 0 | 5 |
| (i) T10454C transition in tRNAArg | Noncoding | PSc | Nog | 0 | 1 |
| (j) C14392G transversion in ND6 | His → Asp | N | Unknownd | 0 | 2 |
| (k) C14558T transition in ND6 | Pro → Leu (2) | N | Unknownd | 0 | 1 |
| (l) 19-base insertion at 14759 in Cytb | Miscoding | N | Yesh | 0 | 1 |
| (m) C14766T transition in Cytb | Thr → Ile | PSc | No | 0 | 1 |
| (n) –T single-base deletion at 15940-15944 in tRNAThr | Noncoding | PSc | Nog | 1 | 0 |
| (o) A15924G transition in tRNAThr | Noncoding | PSc | Yesg,i | 0 | 1 |
| (p) T15942C transition in NC10 | Noncoding | PSc | No | 0 | 1 |
Abbreviations for relevant mtDNA function locations: COII, Cytochrome c oxidase subunit 2; NC7, noncoding nucleotides in NC7 region; ATPase8, ATP synthase F0 subunit 8; COIII, Cytochrome c oxidase subunit 3; ND3, NADH dehydrogenase subunit 3; ND6, NADH dehydrogenase subunit 6; Cytb, Cytochrome b; NC10, noncoding nucleotides in NC10 region.
Reports evaluating the potential impact of these mtDNA sequences changes on protein function were not found.
McFarland et al. [2004]; sequence changes in mt tRNA genes may be noncoding but still may have significant impact on tRNA molecule function.
Large insertions/frameshift mutations in coding regions would presumably result in altered protein function.
Among unexposed newborns, eight changes were identified in 7 of 55 (13%) infants (Tables III and IV). The six distinct sequence variants found in unexposed controls included four transitions, a 9-base deletion, and a single-base deletion (Tables IV and V). Each of these sequence changes occurred at a polymorphic site, confirming that the DGGE assay had sufficient sensitivity to detect polymorphisms previously identified in populations of humans [Saxena et al., 2006; Ruiz-Pesini et al., 2007].
The numbers of sequence variants in uninfected infants receiving prepartum NRTIs were significantly elevated compared with unexposed newborns (P < 0.001, Chi-Square test; P = 0.02, Mann-Whitney test), with 19 of 52 (37%) NRTI-exposed infants having one or more different mtDNA changes for a total of 24 sequence variants (Tables III and IV). The number of NRTI-exposed infants with sequence variants was threefold higher than the number of unexposed infants with sequence variants (P < 0.001). In addition, there was a threefold increase in the average number of changes per subject in exposed infants (0.46/subject) compared with controls (0.15/subject). Only one of the unexposed control infants had more than one sequence variant, while five of the NRTI-exposed infants had two mtDNA changes (Table III). Infants receiving prepartum NRTIs generally had an increase in the number of sequence variants at polymorphic sites (10 distinct variants), plus novel mutations (four distinct variants) not found in unexposed controls. Most sequence variants in coding regions of exposed infant mtDNA [i.e., 13/17 (76%) coding region variants] occurred at positions that resulted in an amino acid change (referred to as a nonsynonymous change), while only 1 of 5 (20%) coding region variants in unexposed controls was nonsynonymous (Table V). In contrast, the numbers of synonymous changes in the two groups were similar (four synonymous transitions in each group). The distinct variants in NRTI-exposed infants included 11 transitions (two novel), one novel transversion, a 9-base deletion, and one novel multibase insertion. While the numbers of sequence variants found in NRTI-exposed newborns were significantly greater than those in unexposed infants, there was not a significant correlation between the duration of maternal AZT-based therapies and the numbers of mtDNA changes in their newborn children (R = 0.10, linear regression).
Mutant Fraction and Pathogenicity of Identified mtDNA Sequence Variants
The estimated mutant fraction, or proportion of mutant DNA to wild-type DNA, was assessed qualitatively based on the banding patterns of the DGGE gels, and was considered to reflect the degree of heteroplasmy. The last column of Table III shows these data. Among the sequence variants found in unexposed or AZT-exposed newborns, 30/32 (94%) mtDNA changes were identified first by the occurrence of mutant homoduplexes (with or without the presence of mutant/wild-type heteroduplexes and/or wild-type homoduplexes) on primary DGGE gels. In unex-posed infants, 7/8 (88%) of the sequence variants were identified on primary DGGE gels by the presence of only mutant homoduplexes. These homoduplexes were identifiable because they had a different migration pattern than the wild-type homoduplexes for mtDNA of samples from children with no alterations in corresponding PCR fragments. Therefore, the sequence changes were likely homoplasmic and maternally inherited polymorphisms [Saxena et al., 2006]. In contrast, only 8/19 (42%) of the sequence variants occurring at polymorphic sites in a nearly equal number of AZT-exposed children (n = 52 vs. 55 unexposed controls) appeared to be homoplasmic. Notably, the mutant fractions for the two sequence variants found in increased numbers in exposed infants (9-base deletion at base 8271 in four exposed subjects vs. one unexposed newborn; A10398C transition in five exposed subjects vs. none in unexposed controls) ranged from low to high (see Table III, footnote “e”), suggesting a treatment-related increase in sequence changes at these polymorphic sites. Only one of the novel mutations found in exposed infants appeared to approach homoplasmy (A8374G transition in PID # 333). In contrast, a moderate degree of heteroplasmy was estimated for the other three previously unreported mutations occurring in three different exposed subjects (PID #s 231, 234, and 606).
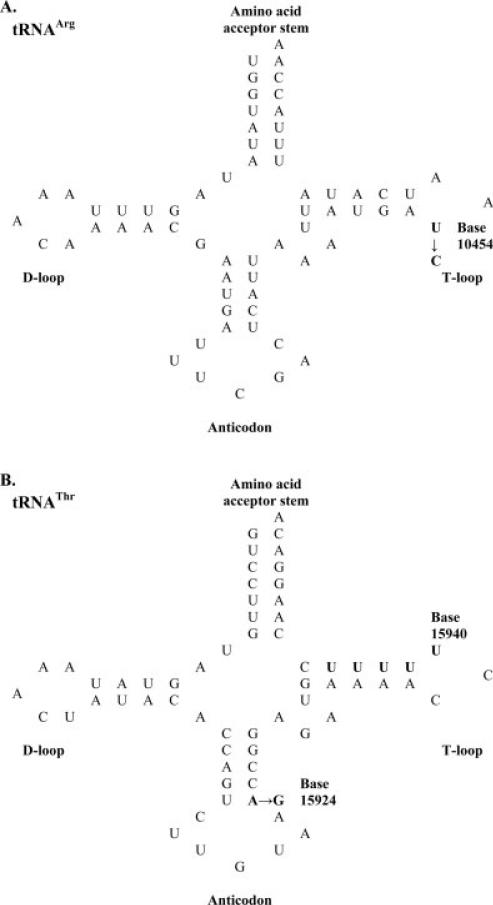
Sequence changes in tRNA genes were of particular interest since they are responsible for more than half of all known mitochondrial pathogenic mutations despite the fact that they comprise only 10% of the human mitochondrial genome [Kondrashov, 2005], while mutations in flanking regions examined have a lower probability of being pathogenic [Ruiz-Pesini et al., 2007]. Using published reports (see Table V and footnotes for column 4) and criteria for estimating pathogenicity of tRNA gene mutations [McFarland et al., 2004; Kondrashov, 2005], the 17 distinct sequence variants identified in the current study were classified as including four with pathogenic characteristics, nine with no evidence of pathogenicity, and four with unknown effects on mitochondrial function (Table V). Of the 11 polymorphisms identified in this study, three occurred in tRNA genes of two infants exposed to AZT-based therapies (T10454C in PID # 371 and A15924G in PID # 606) and in one unexposed infant (base 15940, -T deletion in infant # B310). To estimate the effect of the tRNA gene mutations upon their secondary structure, the location of sequence change(s) were identified in schematic cloverleaf structures for each mutated tRNA molecule [Helm et al., 2000] and individual variants were considered to have pathogenic characteristics if one or more criteria of McFarland et al. [2004] were met (Fig. 2). T10454C transitions found in two exposed infants occurred in the T-loop of tRNAArg, but this sequence change likely has little consequence because of the small size of the loop (three bases) (Fig. 2A). Likewise, the single-base deletion of thymidine occurring at base 15940 in the T-loop of tRNAThr in one unexposed infant is unlikely to be pathogenic based upon the small size of the loop. This deletion is presumed to occur at the first base in a series of five thymidine nucleotides (bases 15940–15944) because deletion of any of the other thymidines would occur in the T-loop stem, causing a shift in bases with multiple mispairings in the T-loop stem and amino acid acceptor stem, and would have pathogenic consequences. The A15924G transition identified in one exposed infant was located in the anticodon stem of tRNAThr (Fig. 2B, criteria 1 and 2) and disrupted Watson-Crick base pairing (Fig. 2B, criterion 3), characteristic of ~73% of pathogenic mutations [McFarland et al., 2004]. Three groups have connected the G variant with respiratory enzyme deficiency, mitochondrial myopathy, and cardiomyopathy [Ozawa et al., 1991; Yoon et al., 1991; Brown et al., 1992; Rupert et al., 2004].
Fig. 2.
Schematic representations of cloverleaf structures for mitochondrial tRNAArg (A) and tRNAThr (B) showing the location of each mutation (shown in bold) in umbilical cord tissue from two infants receiving prepartum AZT-3TC for prophylaxis of HIV-1 (PID #s 371 and 606) and one unexposed infant (PID # 310). D-loop, dihydrouridine loop; T-loop, TCC loop. Criteria used for assessing pathogenicity are those described by McFarland et al. [2004]: (1) the mutation occurs in a stem structure, (2) the mutation occurs in an acceptor or anticodon stem structure, (3) the mutation disrupts Watson-Crick base pairing, (4) the mutation disrupts GC base pairing, (5) the mutation, if in a loop, is in a loop of unusual size or affects tertiary folding. The A15924G transition meets criteria 1–3, while the T10454C transition and single-base deletion at base 15940 occur in small T-loops and fail to meet criterion 5.
Among the remaining mtDNA mutations found in regions flanking tRNA genes, the A10398G polymorphism found in five exposed infants (PID #s 356, 371, 701, 702, 703) was previously identified by Brandon et al. [2006] as a tumor-specific somatic mtDNA mutation in a thyroid tumor, and is thought to play a functional role in neoplastic transformation. The 10398G variant in the ND3 subunit of complex I (Table V) also has been found to be associated with an increased risk for breast cancer in European-American women [Bai et al., 2007], with the A variant causing a significant increased risk of invasive breast cancer in African-American women [Canter et al., 2005]. The two novel multibase insertions found in NADH dehydrogenase subunit 3 and cytochrome b of exposed children (Table III) are miscoding lesions of sufficient size that diminution of protein function would be reasonably expected.
The mtDNA isolate from infant # B218, who received prepartum AZT but was HIV-infected, had two sequence variants at polymorphic sites including one (i.e., A10398G) seen in several AZT-exposed uninfected infants and one (i.e., T10454C) not observed in any other children in the study (Table III). Both base substitutions were homoplasmic, suggesting that these variants were maternally inherited polymorphisms and unrelated to potential HIV-infection at birth.
Analysis of mtDNA Changes in TK6 Cells Exposed In Vitro to AZTand/or 3TC
An experiment was designed to determine, in part, if TK6 cells could be used as a model for distinguishing the nature of mtDNA sequence variants associated with exposure to AZT or 3TC alone versus these NRTIs combined. The DGGE technique described herein, and successfully used to identify mtDNA sequence variants in control and AZT-3TC exposed infants, was not able to detect sequence variants in mtDNA from aliquots of TK6 cells exposed in vitro to 0 or 100 lM AZT, 3TC or AZT-3TC for up to 14 rounds of replication (~10 days). In contrast, exposure of TK6 cells to 10 μM AZT-3TC (equivalent to peak plasma levels in humans) for up to 30 days produced highly significant accumulation and increases in the frequencies of somatic mutations in nuclear genes [Torres et al., 2007]. The effects of treatment of TK6 cells with 100 μM AZT, 3TC, or AZT-3TC, versus vehicle-treated cells, on cell replication and cell survival are described elsewhere [Torres et al., 2007]. In brief, cell replication rates and levels of cell survival in each of the drug exposure groups were significantly decreased compared to vehicle-exposed cells. These combined findings suggest that TK6 cells serve as a good model for investigating NRTI-induced cytotoxicity and nuclear gene mutations, but do not appear to be an appropriate cell culture system for detecting and characterizing mtDNA sequence variants associated with in vitro exposure to NRTIs.
DISCUSSION
The primary reason for conducting this study was to discover whether AZT-based HIV-1 prophylaxis was significantly associated with the induction of mtDNA mutations in uninfected infants born to HIV-1-infected mothers. Answers to this question were obtained in a stepwise fashion. First, we determined that a previously reported horizontal DGGE method [Michikawa et al., 1997] could be adapted to a vertical DGGE procedure for comprehensive, rapid, and sensitive detection of rare to common mutations/polymorphisms occurring in the tRNA genes and flanking regions of the human mitochondrial genome. Second, analysis of umbilical cord tissue using vertical DGGE revealed modest numbers of mainly synonymous polymorphisms in unexposed newborns compared to increased levels of nonsynonymous sequence variants at both polymorphic and novel sites in AZT-exposed infants. Third, estimates of the mutant fractions for each mtDNA change showed a range in the degree of heteroplasmy among sequence variants from AZT-exposed infants versus mostly homoplasmic polymorphisms in unexposed controls. Last, classification of the sequence variants based upon reports in the literature or the nature of the mutations suggested that several mtDNA changes were potentially pathogenic.
Umbilical cord tissue was selected for analysis because it is accessible and isolated mitochondria originate primarily from energy-demanding endothelium and smooth muscle cells of the umbilical vein and two umbilical arteries. The remaining structural elements of the umbilical cord are arranged to support these blood vessels crucial to the developing fetus. Very few mitochondria occur in the enveloping amnion epithelial cell layer and mucous connective tissue called Wharton's jelly, that consists of relatively few stellate cells, collagen fibers, and abundant extracellular matrix of various glycosaminoglycans and cavernous spaces that contribute to the elasticity of the umbilical cord [Takechi et al., 1993; Eyden et al., 1994; Sexton et al., 1996; Sobolewski et al., 1997]. Thus, mtDNA alterations found in umbilical cord tissue may reflect events occurring in vascular cells throughout the body.
DGGE or related techniques, such as denaturing HPLC or temporal temperature gradient gel electrophoresis (TTGE), have significant advantages over other methods used for mtDNA mutation detection in that it permits the simultaneous detection of more than one mutation occurring in an amplified region of DNA, it can detect several types of mutations including any base substitution, frame-shift, or small deletion/insertion, and it can provide limited information about the mutant fraction for a mtDNA sequence variant [Myers et al., 1985; Lerman et al., 1987; Walker and Skopek, 1993; Michikawa et al., 1997]. Other methods, including Southern blotting, long-extension PCR, single-stranded conformation polymorphism (SSCP), PCR-restriction fragment length polymorphism analysis, and solid-phase mini-sequencing, using PCR with allele-specific oligonucleotides, are limited in the scope of mutations that can be identified using a single approach [Kajander et al., 1999; Boles et al., 2003]. The current study used DGGE to screen for sequence variants in ~21% of the mitochondrial genome of umbilical cord tissue from AZT-exposed versus healthy unexposed infants. As discussed below, however, two research groups have previously employed sensitive methods for genome-wide analysis of small-scale mtDNA mutations in PBMCs of NRTI-treated HIV-1-infected adult patients.
McComsey et al. [2002] investigated the molecular mechanisms of NRTI-associated mitochondrial dysfunction by, in part, using TTGE and DNA-sequencing analysis to screen for mtDNA sequence variants in PBMCs from 10 NRTI-treated HIV-1-infected patients, four ARV-naïve HIV-1-infected individuals, and 10 healthy adult controls. More than 20 different base substitutions (including changes at polymorphic sites and novel mutations) were found in NRTI-naïve and NRTI-treated patients; most of these sequence variants denoted benign homoplasmic polymorphisms. The authors suggested that NRTI-induced mutations are tissue-specific or that NRTIs may unmask pre-existing mtDNA variations in HIV-1 disease and promote clinical mitochondrial dysfunction. A follow-up study was conducted to assess the utility of serial blood samples in characterizing the relationships between HIV-1 infection, NRTI therapies, acquisition of mtDNA mutations, and clinical mitochondrial toxicity [McComsey et al., 2005]. The study population included NRTI-treated HIV-1-infected adults (n = 54), ARV-naïve HIV-1-infected patients (n = 33), and healthy adult controls (n = 48) followed for up to 52-months. The report only included mtDNA mutation data for the two HIV-1-infected patients who showed detectable differences in TTGE banding patterns over time in serial blood samples. Both patients (one drug-naïve subject given d4T plus emtricitabine and one drug-experienced patient receiving AZT-3TC) had one or more sequence variants that were heteroplasmic in the first blood specimen but homoplasmic after months of NRTI treatment, suggesting a dynamic process where NRTI therapy appears associated with acquisition, segregation, and expansion of these mtDNA changes. The authors believed that NRTI-treated patients might have additional variants at a low level of heteroplasmy undetectable by their analytical methods.
Martin et al. [2003] used SSCP and DNA-sequencing analysis to investigate the impact of NRTI therapy on the development, frequency, and nature of mtDNA mutations in PBMCs prior to (T1 samples) and after 6–77 months of treatment (T2 samples). Their study population consisted of 16 drug-naïve HIV-1-infected patients who were placed on NRTI treatment and a reference group of 10 drug-naïve, untreated HIV-1-infected subjects [Martin et al., 2003]. Based upon T1 and T2 samples from these 26 patients, five NRTI-treated subjects incurred increased numbers of novel heteroplasmic mutations over time. Four of five patients with mtDNA mutations developed evidence of peripheral fat wasting (lipoathrophy) between blood sample intervals (P = 0.031), suggestive of a pathogenic potential for NRTI-related mtDNA mutations. There also was a trend toward more nonsynonymous base substitutions in T2 samples than T1 samples, with the numbers of synonymous substitutions being similar in the before and after NRTI-treatment samples. The authors concluded that NRTI therapy provides conditions permissive for the emergence over time of “random” mtDNA mutations; however, they could not cite evidence for positive selection of pre-existing non-wild-type sequences.
The finding of seemingly “random” mtDNA mutations in earlier studies of HIV-1-infected adults may be partly a consequence of the variety of NRTI treatment protocols used. In the reports of McComsey et al. [2002, 2005], ARV-experienced HIV-1-infected patients, as a group, received more than a dozen different triple-drug therapies including one or two NRTIs. In the work of Martin et al. [2003], NRTI-treated patients were given one of three different NRTI drug combinations, including d4T-3TC (n = 10), AZT-3TC (n = 3), or d4T-didanosine (n = 3) [with sequence variants detected in 3, 1, and 1 subject(s) in their respective groups]. In these earlier studies, the finding of mutations formed in a “random” fashion would be highly likely in that different individual NRTIs, or pairs of NRTIs, should not cause the same specific mtDNA mutations. Rather, individual NRTIs, or combinations of NRTIs, possess different mutagenic potencies and specificities in human lymphoblastoid cells [Carter et al., 2007]. Therefore, it is not surprising to find no overlap in the sequence variants occurring in AZT-3TC exposed newborns in the current work (Table III) versus those detected in earlier studies of NRTI-treated HIV-1-infected adults. It is also noteworthy that adults have slower rates of cell replication than developing fetuses, possibly leading to the finding of greater numbers of sequence variants at birth. These dissimilarities in sequence variants found in PBMCs versus umbilical cord tissue of NRTI-exposed subjects are unlikely due to tissue-specific differences because preliminary data from our group suggests that AZT-exposed newborns have similar mtDNA mutations in umbilical cord tissue (current study) and cord blood lymphocytes (unpublished data). On the other hand, the finding of two AZT-3TC exposed infants with the same novel mtDNA mutation (Table III, PID #s 231 and 234) is suggestive of targeted chemical mutagenesis at a potential mutational hot spot.
In our study, we cannot establish whether the increased detection of sequence changes at two polymorphic sites in AZT-exposed infants (i.e., 9-base deletion at nucleotide 8271 and A10398G transition, Table V) resulted from the unmasking of silent sequence variants or chemical induction of mutations at predisposed sites in mtDNA. However, given that the 9-base pair deletion at nucleotide 8271 is typically a polymorphism associated with Asian, North American Indian, and Polynesian populations [Hertzberg et al., 1989; Harihara et al., 1992; Lorenz and Smith, 1994], or isolated cases in European or Latin populations [Torroni et al., 1995], the occurrence of this deletion in one Hispanic unexposed infant versus four Hispanic AZT-exposed infants (Table IV) raises the question as to whether these variants are rare preexisting polymorphisms that have been unmasked or are caused by NRTI treatment.
Among the NRTIs used as ARVs, AZT has clearly been shown to be genotoxic in standard in vitro and in vivo assays [IARC, 2000; Walker and Poirier, 2007; Wogan, 2007], and mounting evidence suggests that it also triggers mitochondrial damage and mutation [Poirier et al., 2004; Walker et al., 2004; Kohler and Lewis, 2007]. In infants receiving prepartum AZT-3TC, a significant increase in the frequency of HPRT reporter gene mutations was driven by an increase in transversion mutations [O'Neill et al. 2001]. Likewise, mutations found in the K-ras and p53 cancer genes of lung neoplasms from mice exposed transplacentally to AZT were predominantly transversions [Hong et al., 2007]. In contrast, 11/12 heteroplasmic base substitutions observed in AZT-3TC exposed infants in the current study involved transition mutations, leaving open the question of the mechanism(s) leading to the formation of these mtDNA mutations. Lim and Copeland [2001] have shown that AZT-monophosphate (at clinically relevant levels) inhibits the exonuclease activity of DNA pol γ, contributing to an increase in spontaneous mutations which are preferentially transitions. These authors further found that AZT-related mitochondrial toxicity in vivo may result, in part, from moderately efficient incorporation and very inefficient removal of AZT from mtDNA, yielding an increase in mutations. AZT has also been shown to inhibit TK2, the mitochondrial targeted thymidine kinase, which could cause a decrease or alteration in the dNTP precursor pools needed for mtDNA replication [Lynx and McKee, 2006; Susan-Resiga et al., 2007]. Imbalanced mitochondrial dNTP pools have been shown to alter the fidelity of DNA pol γ [Song et al., 2005]. Other mechanisms underlying the toxicities of AZT have been proposed, but additional work is needed to gain a better understanding of the mutagenic modes of action of AZT and their long-term consequences.
For ethical reasons, the current study did not include a treatment-control group of drug-naïve infants born to untreated HIV-1-infected mothers to assess the mutagenic potential of the fetal environment in the presence of maternal HIV-1 infection. In the United States, women are routinely treated with ARVs upon diagnosis of HIV-1 infection, making it problematic to address the degree to which fetal responses to “HIV-1 exposure,” in the absence of maternal ARV treatment, contributes to perinatal toxicities and long-term health risks in the offspring [Poirier et al., 2003, 2004; Funk et al., 2007]. Consequently, there is limited evidence that fetal responses and/or “HIV-1 exposure” damage mitochondria of fetal cells/tissues, perhaps increasing the risk for mtDNA mutations. In collaboration with the Womens and Infants Transmission Study (WITS), Poirier et al. [2003] found significant mtDNA depletion in leukocytes of uninfected infants born to HIV-1-infected mothers who received no treatment, with this depletion being further increased in infants of mothers receiving AZT during pregnancy. These findings suggest that factors related to maternal HIV-1 infection can affect the integrity of mitochondria and possibly play an undefined role in the perturbations in genetic markers observed in uninfected children. In the future, it is important to test the hypothesis that fetal environment in the face of maternal HIV-1 infection and absence of NRTI treatment has mutagenic potential leading to changes in nDNA and mtDNA that may be preventable in uninfected infants [Funk et al., 2007].
The findings related to clinical mitochondrial dysfunction in uninfected children receiving perinatal NRTIs have been the subject of considerable debate as illustrated by the recent opinion piece [Blanche et al., 2006] and editorial response [Spector and Saitoh, 2006] published in the journal AIDS. Interim reports of cord blood leukocyte mtDNA depletion and umbilical cord mitochondrial morphological damage and depletion indicated that most clinically asymptomatic infants exposed to NRTIs in utero do have molecular evidence of mitochondrial compromise in the absence of clinical manifestations of mitochondrial disease [Benhammou et al., 2007]. In the current study, the discovery of increases in mtDNA sequence variants in umbilical cord tissue of uninfected newborns receiving prepartum AZT-3TC is troublesome because umbilical vessel endothelium and smooth muscle cells are the predominant source of mtDNA, and the same cell types occur throughout the body including mitochondrial-rich tissues such as the heart. Initial reports of Lipshultz et al. [2000, 2002] suggested that perinatal exposure to AZT monotherapy was not associated with cardiac abnormalities in uninfected children. More recent preliminary findings in children receiving multiple ARV drug therapies suggest different risks. The 2- to 3-year follow-up of larger numbers of HIV-1-uninfected ARV-exposed infants suggest that these children have significantly less ventricular mass and wall thickness compared to HIV-1-uninfected children who were not exposed in utero to ARV-based therapy. These differences appear to persist for 2–3 years after birth, mandating long-term cardiac outcome studies [Lipshultz, 2008; Lipshultz et al., 2005, 2006]. As NRTI-exposed children age, long-term studies also are needed to delineate the expression and clinical consequences of multiple induced mtDNA mutations compared to single pathogenic mutations arising from maternal inheritance. Foster and Lyall [2008] argue that the cumulative NRTI-associated toxicities are becoming progressively apparent in the clinic, and, in an era of expanding treatment options, understanding and minimizing toxicities becomes a possibility.
ACKNOWLEDGMENTS
The authors thank Wendy Piper for assistance in preparing the figures and David J. H. Walker for identifying additional enlightening reports. We acknowledge sequencing by DNA Research Services at the University of New Mexico. The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Grant sponsor: National Institute of Environmental Health Sciences; Grant numbers: R01 HD033648, R01 HL072727, 1 F31 HL081928; Grant sponsors: The National Institute of Child Health and Human Development, the National Cancer Institute, the Office of AIDS Research, The National Heart, Lung, and Blood Institute.
Abbreviations
- 3TC
lamivudine or 2′,3′-dideoxy-3′-thiacytidine
- ARV
antiretroviral
- AZT
zidovudine or 3′-azido-2′,3′-dideoxythymidine
- d4T
stavudine
- DGGE
denaturing gradient gel electrophoresis
- EM
electron microscopy
- MTCT
mother-to-child transmission
- mtDNA
mitochondrial DNA
- nDNA
nuclear DNA
- NRTIs
nucleoside reverse transcriptase inhibitors
- OXPHOS
oxidative phosphorylation enzyme activity
- PBMCs
peripheral blood mononuclear cells
- pol γ
DNA polymerase gamma
- SSCP
single-stranded conformation polymorphism
- TTGE
temporal temperature gradient gel electrophoresis
REFERENCES
- Bai R-K, Leal SM, Covarrubias D, Liu A, Wong L-JC. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007;67:4687–4694. doi: 10.1158/0008-5472.CAN-06-3554. [DOI] [PubMed] [Google Scholar]
- Barret B, Tardieu M, Rustin P, Lacroix C, Chabrol B, Desguerre I, Dollfus C, Mayaux MJ, Blanche S. Persistent mitochondrial dysfunction in HIV-1-exposed but uninfected infants: Clinical screening in a large prospective cohort. AIDS. 2003;17:1769–1785. doi: 10.1097/00002030-200308150-00006. [DOI] [PubMed] [Google Scholar]
- Benhammou V, Tardieu M, Warszawski J, Rustin P, Blanche S. Clinical mitochondrial dysfunction in uninfected children born to HIV-infected mothers following perinatal exposure to nucleoside analogues. Environ Mol Mutagen. 2007;48:173–179. doi: 10.1002/em.20279. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lopman DJ, Ostell J, Rapp BA, Wheeler DL. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialkowska A, Bialkowski K, Gerschenson M, Diwan BA, Jones AB, Olivero OA, Poirier MC, Anderson LM, Kasprzak KS, Sipowicz MA. Oxidative DNA damage in fetal tissues after transplacental exposure to 3′-azido-3′-deoxythymidine (AZT). Carcinogenesis. 2000;21:1059–1062. doi: 10.1093/carcin/21.5.1059. [DOI] [PubMed] [Google Scholar]
- Blanche S, Tardieu M, Rustin P, Slama A, Barret B, Firtion G, Ciraru-Vigneron N, Lacroix C, Rouzioux C, Mandelbrot L, Desguerre I, Rotig A, Mayaux MJ, Delfraissy JF. Persistent mitochondrial dysfunction and perinatal exposure to antiretroviral nucleoside analogues. Lancet. 1999;354:1084–1089. doi: 10.1016/S0140-6736(99)07219-0. [DOI] [PubMed] [Google Scholar]
- Blanche S, Tardieu M, Benhammou V, Warszawski J, Rustin P. Mitochondrial dysfunction following perinatal exposure to nucleoside analogues. AIDS. 2006;20:1685–1690. doi: 10.1097/01.aids.0000242814.42344.77. [DOI] [PubMed] [Google Scholar]
- Boles RG, Chaudhari D, Soderkvist J, Podberezin M, Ito M. Quantification of mitochondrial DNA heteroplasmy by temporal temperature gradient gel electrophoresis. Clin Chem. 2003;49:198–200. doi: 10.1373/49.1.198. [DOI] [PubMed] [Google Scholar]
- Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- Brinkman K, ter Hofstede JM, Burger DM, Smeitink JAM, Koopmans PP. Adverse effects of reverse transcriptase inhibitors: Mitochondrial toxicity as common pathway. AIDS. 1998;12:1735–1744. doi: 10.1097/00002030-199814000-00004. [DOI] [PubMed] [Google Scholar]
- Brogly SB, Ylitalo N, Mofenson LM, Oleske J, Van Dyke R, Crain MJ, Abzug MJ, Brady M, Jean Phillipe P, Hughes D, Seage GR., III In utero nucleoside reverse transcriptase inhibitor exposure and signs of possible mitochondrial dysfunction in HIV-uninfected children. AIDS. 2007;21:929–938. doi: 10.1097/QAD.0b013e3280d5a786. [DOI] [PubMed] [Google Scholar]
- Brown MD, Torroni A, Shoffner JM, Wallace DC. Mitochondrial tRNA(Thr) mutations and lethal infantile mitochondrial myopathy. Am J Hum Genet. 1992;51:446–447. [PMC free article] [PubMed] [Google Scholar]
- Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005;65:8028–8033. doi: 10.1158/0008-5472.CAN-05-1428. [DOI] [PubMed] [Google Scholar]
- Cariello NF, Keohavong P, Kat AG, Thilly WG. Molecular analysis of complex human cell populations: Mutational spectra of MNNG and ICR-191. Mutat Res. 1990;231:165–176. doi: 10.1016/0027-5107(90)90023-w. [DOI] [PubMed] [Google Scholar]
- Carter MM, Torres SM, Cook DL, Jr, McCash CL, Yu M, Griegos J, Walker DM, Walker VE. Relative mutagenic potencies of several nucleoside reverse transcriptase inhibitors at the HPRT and TK loci of exposed human TK6 lymphoblastoid cells. Environ Mol Mutagen. 2007;48:239–247. doi: 10.1002/em.20282. [DOI] [PubMed] [Google Scholar]
- Cooper ER, Charurat M, Mofenson L, Hanson IC, Pitt J, Diaz C, Hayani K, Handelsman E, Smeriglio V, Hoff R, Blattner W. Combination antiretroviral strategies for the treatment of pregnant HIV-1-infected women and prevention of perinatal HIV-1 transmission. J Acquir Immune Defic Syndr. 2002;29:484–494. doi: 10.1097/00126334-200204150-00009. [DOI] [PubMed] [Google Scholar]
- Cooper ER, DiMauro S, Sullivan M, Jones-Evans D, Kay L, Moloney C, Regan AM. Biopsy-confirmed mitochondrial dysfunction in an HIV-exposed infant whose mother received combination antiretrovirals during the last 6 weeks of pregnancy.. 15th International AIDS Conference Bangkok; Bankok, Thailand. 11–16 July; 2004. Abstract TuPeB4394. [Google Scholar]
- Dagan T, Sable C, Bray J, Gerschenson M. Mitochondrial dysfunction and antiretroviral nucleoside analog toxicities: What is the evidence? Mitochondrion. 2002;1:397–412. doi: 10.1016/s1567-7249(02)00003-x. [DOI] [PubMed] [Google Scholar]
- Divi RL, Walker VE, Wade NA, Nagashima K, Seilkop SK, Adams ME, Nesel CJ, O'Neill JP, Abrams EJ, Poirier MC. Mitochondrial damage and DNA depletion in cord blood and umbilical cord from infants exposed in utero to Combivir. AIDS. 2004;18:1013–1021. doi: 10.1097/00002030-200404300-00009. [DOI] [PubMed] [Google Scholar]
- Divi RL, Leonard SL, Kuo MM, Walker BL, Orozco CC, St. Claire MC, Nagashima K, Harbaugh SW, Harbaugh JW, Thamire C, Sable CA, Poirier MC. Cardiac mitochondrial compromise in 1-yr-old Erythrocebus patas monkeys perinatally-exposed to nucleoside reverse transcriptase inhibitors. Cardiovasc Toxicol. 2005;5:333–346. doi: 10.1385/ct:5:3:333. [DOI] [PubMed] [Google Scholar]
- Divi RL, Leonard SL, Kuo MM, Nagashima K, Thamire C, St. Claire MC, Wade NA, Walker VE, Poirier MC. Transplacentally exposed human and monkey newborn infants show similar evidence of nucleoside reverse transcriptase inhibitor-induced mitochondrial toxicity. Environ Mol Mutagen. 2007;47:201–209. doi: 10.1002/em.20201. [DOI] [PubMed] [Google Scholar]
- Dorenbaum A, Cunningham CK, Gelber RD, Culnane M, Mofenson L, Britto P, Rekacewicz C, Newell ML, Delfraissy JF, Cunningham-Schrader B, Mirochnick M, Sullivan JL, International PACTG 316 Team Two-dose intrapartum/newborn nevirapine and standard antiretroviral therapy to reduce perinatal HIV transmission: a randomized trial. JAMA. 2002;288:189–198. doi: 10.1001/jama.288.2.189. [DOI] [PubMed] [Google Scholar]
- Escobar P, Olivero OA, Day B, Wade NA, Abrams EJ, Nesel CJ, Ness RB, Day RD, Day BW, Meng Q, O'Neill JP, Walker DM, Poirier MC, Walker VE, Bigbee WL, for the study team Genotoxicity assessed by the Comet and GPA assays following in vitro exposure of human lymphoblastoid cells (H9) or perinatal exposure of mother-child pairs to AZT or AZT-3TC. Environ Mol Mutagen. 2007;48:330–343. doi: 10.1002/em.20285. [DOI] [PubMed] [Google Scholar]
- European Collaborative Study Exposure to antiretroviral therapy in utero or early life: The health of uninfected children born to HIV-infected women. J Acquir Immune Defic Syndr. 2003;32:380–387. doi: 10.1097/00126334-200304010-00006. [DOI] [PubMed] [Google Scholar]
- European Collaborative Study Mother to child transmission of HIV infection in the era of highly active antiretroviral therapy. Clin Infect Dis. 2005;40:458–465. doi: 10.1086/427287. [DOI] [PubMed] [Google Scholar]
- Eyden BP, Ponting J, Davies H, Bartley C, Torgersen E. Defining the myofibroblast: Normal tissues, with special reference to the stromal cells of Wharton's jelly in human umbilical cord. J Submicrosc Cytol Pathol. 1994;26:347–355. [PubMed] [Google Scholar]
- Foster C, Lyall H. HIV and mitochondrial toxicity in children. J Antimicrob Chemother. 2008;61:8–12. doi: 10.1093/jac/dkm411. [DOI] [PubMed] [Google Scholar]
- Funk MJ, Belinson SE, Pimenta JM, Morsheimer M, Gibbons DC. Mitochondrial disorders among infants exposed to HIV and antiretroviral therapy. Drug Safety. 2007;30:845–859. doi: 10.2165/00002018-200730100-00004. [DOI] [PubMed] [Google Scholar]
- Harihara S, Hirai M, Suutou Y, Shimizu K, Omoto K. Frequency of a 9-bp deletion in the mitochondrial DNA among Asian populations. Hum Biol. 1992;64:161–166. [PubMed] [Google Scholar]
- Hashiguchi K, Bohr VA, de Souza-Pinto NC. Oxidative stress and mitochondrial DNA repair: implications for NRTIs induced DNA damage. Mitochondrion. 2004;4:215–222. doi: 10.1016/j.mito.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Helm M, Brule H, Friede D, Giege R, Putz D, Florentz C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–1379. doi: 10.1017/s1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg M, Mickleson KN, Serjeantson SW, Prior JF, Trent RJ. An Asian-specific 9-bp deletion of mitochondrial DNA is frequently found in Polynesians. Am J Hum Genet. 1989;44:504–510. [PMC free article] [PubMed] [Google Scholar]
- Hong H-H, Dunnick J, Herbert R, Devereux TR, Kim Y, Sills RC. Genetic alterations in K-ras and p53 cancer genes in lung neoplasms of Swiss (CD-1) male mice following AZT transplacental exposure. Environ Mol Mutagen. 2007;48:299–306. doi: 10.1002/em.20197. [DOI] [PubMed] [Google Scholar]
- IARC . IARC Monographs on the evaluation of carcinogenic risks to humans, some antiviral and antineoplastic drugs, and pharmaceutical agents. IARC Scientific Publications No. 76; Lyon: 2000. General remarks; and the Monograph on Zidovudine. pp. 35–42.pp. 73–127. [Google Scholar]
- Kajander OA, Kunnas TA, Perola M, Lehtinen SK, Karhunen PJ, Jacobs HT. Long-extension PCR to detect deleted mitochondrial DNA molecules is compromized by technical artefacts. Biochem Biophys Res Commun. 1999;254:507–514. doi: 10.1006/bbrc.1998.9975. [DOI] [PubMed] [Google Scholar]
- Kakuda TM. Pharmacology of nucleoside and nucleotide reverse transcriptase inhibitor-induced mitochondrial toxicity. Clin Ther. 2000;22:685–708. doi: 10.1016/S0149-2918(00)90004-3. [DOI] [PubMed] [Google Scholar]
- Kallings LO. The first postmodern pandemic: 25 years of HIV/AIDS. J Intern Med. 2008;263:218–243. doi: 10.1111/j.1365-2796.2007.01910.x. [DOI] [PubMed] [Google Scholar]
- Kelley SO, Steinberg SV, Schimmer P. Functional defects of pathogenic human mitochondrial tRNAs related to structural fragility. Nat Struct Biol. 2000;7:862–865. doi: 10.1038/79612. [DOI] [PubMed] [Google Scholar]
- Kohler JJ, Lewis W. A brief overview of mechanisms of mitochondrial toxicity from NRTIs. Environ Mol Mutagen. 2007;48:166–172. doi: 10.1002/em.20223. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA. Prediction of pathogenic mutations in mitochondrially encoded human tRNAs. Hum Mol Genet. 2005;14:2415–2419. doi: 10.1093/hmg/ddi243. [DOI] [PubMed] [Google Scholar]
- Lerman LS, Silverstein K. Computational simulation of DNA melting and its application to denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:482–501. doi: 10.1016/0076-6879(87)55032-7. [DOI] [PubMed] [Google Scholar]
- Levinger L, Morl M, Florentz C. Mitochondrial tRNA 3′ end metabolism and human disease. Nucleic Acids Res. 2004;32:5430–5441. doi: 10.1093/nar/gkh884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis W. Mitochondrial dysfunction and nucleoside reverse transcriptase inhibitor therapy: Experimental clarifications and persistent clinical questions. Antiviral Res. 2003;58:189–197. doi: 10.1016/s0166-3542(03)00069-x. [DOI] [PubMed] [Google Scholar]
- Lewis W. Cardiomyopathy, nucleoside reverse transcriptase inhibitors and mitochondria are linked through AIDS and its therapy. Mitochondrion. 2004;4:141–152. doi: 10.1016/j.mito.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Lewis W. Nucleoside reverse transcriptase inhibitors, mitochondrial DNA, AIDS therapy. Antivir Ther. 2005;10(Suppl 2):M13–M27. [PubMed] [Google Scholar]
- Lewis W, Dalakas MC. Mitochondrial toxicity of antiviral drugs. Nat Med. 1995;1:417–422. doi: 10.1038/nm0595-417. [DOI] [PubMed] [Google Scholar]
- Lewis W, Day BJ, Copeland WC. Mitochondrial toxicity of NRTI antiviral drugs: An integrated cellular perspective. Nat Rev Drug Discov. 2003;2:812–822. doi: 10.1038/nrd1201. [DOI] [PubMed] [Google Scholar]
- Lim SE, Copeland WC. Differential incorporation and removal of antiviral deoxynucleotides by human DNA polymerase gamma. J Biol Chem. 2001;276:23616–23623. doi: 10.1074/jbc.M101114200. [DOI] [PubMed] [Google Scholar]
- Lindegren ML, Rhodes P, Gordon L, Fleming P. Drug safety during pregnancy and in infants. Lack of mortality related to mitochondrial dysfunction among perinatally HIV-exposed children in pediatric HIV surveillance. Ann N Y Acad Sci. 2000;918:222–235. [PubMed] [Google Scholar]
- Lipshultz SE. The effect of ART exposure during childhood on cardiovascular structure and function in children born to HIV infected mothers. Toxicologist. 2008;102(1):481. [Google Scholar]
- Lipshultz SE, Easley KA, Orav EJ, Kaplan S, Starc TJ, Bricker JT, Lai WW, Moodie DS, Sopko G, McIntosh K, Colan SD. Absence of cardiac toxicity of zidovudine in infants. Pediatric Pulmonary and Cardiac Complications of Vertically Transmitted HIV Infection Study Group. N Engl J Med. 2000;343:759–766. doi: 10.1056/NEJM200009143431102. [DOI] [PubMed] [Google Scholar]
- Lipshultz SE, Easley KA, Orav EJ, Kaplan S, Starc TJ, Bricker JT, Wyman WL, Moodie DS, Sopko G, Schluchter MD, Colan SD, for the P2C2 HIV Study Group Cardiovascular status of infants and children of women infected with HIV-1 (P2C2 HIV): A cohort study. Lancet. 2002;360:368–373. doi: 10.1016/S0140-6736(02)09607-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshultz SE, Shearer WT, Rich T, Thompson B, Cheng I, Harmon T, Orav EJ, Pignatelli RH, Bezold LI, LaRussa P, Starc TJ, Glickstein J, O'Brian S, McIntosh K, Cooper ER, Coaln SD. Antiretroviral therapy (ART)-associated cardiotoxicity in uninfected but ART-exposed infants born to HIV-infected women: The prospective NHLBI CHAART-1 study. Mutat Res. 2005;577(Suppl 1):e28. [Google Scholar]
- Lipshultz SE, Shearer WT, Thompson B, Rich K, Cheng I, Milton A, Orav EJ, Pignatelli RH, Bezold LI, LaRussa P, Starc TJ, Glickstein J, McIntosh K, Cooper ER, O'Brien S, Colan SD. Antiretroviral therapy (ART)-associated cardiotoxicity in uninfected and ART-exposed infants born to HIV-infected women: The prospective NHLBI CHAART-I Study.. Late-Breaker Abstract # 8, Proceedings of 2006 Pediatric Academic Societies Annual Meeting; San Francisco, CA. 2006. [Google Scholar]
- Lorenz JG, Smith DG. Distribution of the 9-bp mitochondrial DNA region V deletion among North American Indians. Hum Biol. 1994;66:777–788. [PubMed] [Google Scholar]
- Lynx MD, McKee EE. 3′-Azido-3′-deoxythymidine (AZT) is a competitive inhibitor of thymidine phosphorylation in isolated rat heart and liver mitochondria. Biochem Pharmacol. 2006;72:239–243. doi: 10.1016/j.bcp.2006.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maagard A, Holberg-Petersen M, Kollberg G, Oldfors A, Sandvik L, Brun JN. Mitochondrial (mt)DNA changes in tissue may not be reflected by depletion of mtDNA in peripheral blood mononuclear cells in HIV-infected patients. Antivir Ther. 2006;11:601–608. [PubMed] [Google Scholar]
- Martin AM, Hammond E, Nolan D, Pace C, Den Boer M, Taylor L, Moore H, Martinez OP, Christiansen FT, Mallal S. Accumulation of mitochondrial DNA mutations in human immunodeficiency virus-infected patients treated with nucleoside-analogue reverse-transcriptase inhibitors. Am J Hum Genet. 2003;72:549–560. doi: 10.1086/367849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McComsey G, Tan DJ, Lederman M, Wilson E, Wong LJ. Analysis of the mitochondrial DNA genome in the peripheral blood leukocytes of HIV-infected patients with or without lipoatrophy. AIDS. 2002;16:513–518. doi: 10.1097/00002030-200203080-00001. [DOI] [PubMed] [Google Scholar]
- McComsey G, Bai RK, Maa JF, Seekins D, Wong LJ. Extensive investigations of mitochondrial DNA genome in treated HIV-infected subjects: Beyond mitochondrial DNA depletion. J Acquir Immune Defic Syndr. 2005;39:181–188. [PubMed] [Google Scholar]
- McFarland R, Elson JL, Taylor RW, Howell N, Turnbull DM. Assigning pathogenicity to mitochondrial tRNA mutations: When “definitely maybe” is not good enough. Trends Genet. 2004;20:591–596. doi: 10.1016/j.tig.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Meng Q, Olivero OA, Fasco MJ, Bellisario R, Kaminsky LS, Pass KA, Abrams EJ, Wade NA, Poirier MC, Walker VE, protocol team Plasma and cellular markers of 3′-azido-3′-dideoxythymidine (AZT) metabolism as indicators of DNA damage in cord blood mononuclear cells from infants receiving prepartum NRTIs. Environ Mol Mutagen. 2007;48:307–321. doi: 10.1002/em.20298. [DOI] [PubMed] [Google Scholar]
- Michikawa Y, Hofhaus G, Lerman LS, Attardi G. Comprehensive, rapid and sensitive detection of sequence variants of human mitochondrial tRNA genes. Nucleic Acids Res. 1997;25:2455–2463. doi: 10.1093/nar/25.12.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkoff H, Ahdieh L, Watts DF, Greenblatt RM, Schmidt J, Schneider M, Stek A. The relationship of pregnancy to the use of highly active antiretroviral therapy. Am J Obstet Gynecol. 2001;184:1221–1227. doi: 10.1067/mob.2001.113871. [DOI] [PubMed] [Google Scholar]
- Miró Ò , Gómez M, Pedrol E, Cardellac F, Nunes V, Casadamont J. Respiratory chain dysfunction associated with multiple mitochondrial DNA deletions in antiretroviral therapy-related lipodystrophy. AIDS. 2000;14:1855–1857. doi: 10.1097/00002030-200008180-00024. [DOI] [PubMed] [Google Scholar]
- Moyle G. Clinical manifestations and management of antiretroviral nucleoside analog-related mitochondrial toxicity. Clin Ther. 2000;22:911–936. doi: 10.1016/S0149-2918(00)80064-8. discussion 898. [DOI] [PubMed] [Google Scholar]
- Myers RM, Fischer SG, Maniatis T, Lerman LS. Modification of the melting properties of duplex DNA by attachment of a GC-rich DNA sequence as determined by denaturing gradient gel electrophoresis. Nucleic Acids Res. 1985;13:3111–3129. doi: 10.1093/nar/13.9.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguera A, Fortuny C, Munoz-Almagro C, Sanchez E, Vilaseca MA, Artuch R, Pou J, Jimenez R. Hyperlactatemia in human immunodeficiency virus-uninfected infants who are exposed to antiretrovirals. Pediatrics. 2004:114598–114603. doi: 10.1542/peds.2004-0955. [DOI] [PubMed] [Google Scholar]
- Olivero OA, Anderson LM, Diwan BA, Haines DC, Harbaugh SW, Moskal TJ, Jones AB, Rice JM, Riggs CW, Logsdon D, Yuspa SH, Poirier MC. Transplacental effects of 3′-azido-2′,3′-dideoxythymidine (AZT): Tumorigenicity in mice and genotoxicity in mice and monkeys. J Natl Cancer Inst. 1997;89:1602–1608. doi: 10.1093/jnci/89.21.1602. [DOI] [PubMed] [Google Scholar]
- Olivero OA, Shearer GM, Chougnet CA, Kovacs AA, Landay AL, Baker R, Stek AM, Khoury MM, Proia LA, Kessler HA, Sha BE, Tarone RE, Poirier MC. Incorporation of zidovudine into leukocyte DNA from HIV-1-positive adults and pregnant women, and cord blood from infants exposed in utero. AIDS. 1999;13:919–925. doi: 10.1097/00002030-199905280-00007. [DOI] [PubMed] [Google Scholar]
- Olivero OA, Shearer GM, Chougnet CA, Kovacs AA, Baker R, Stek AM, Khoury MM, Poirier MC. Incorporation of zidovudine into cord blood DNA of infants and peripheral blood DNA of their HIV-1-positive mothers. Ann N Y Acad Sci. 2000;918:262–268. doi: 10.1111/j.1749-6632.2000.tb05495.x. [DOI] [PubMed] [Google Scholar]
- Olivero OA, Fernandez JJ, Antiochos BB, Wagner JL, St. Claire ME, Poirier MC. Transplacental genotoxicity of combined antiretroviral nucleoside analogue therapy in Erythrocebus patas monkeys. J Acquir Immune Defic Syndr. 2002;29:323–329. doi: 10.1097/00126334-200204010-00001. [DOI] [PubMed] [Google Scholar]
- O'Neill JP, Gundel RM, Trombley LM, Walker VE. HPRT mutant frequency and mutation spectra in newborns with perinatal exposure to antiviral drugs. Environ Mol Mutagen. 2001;37(Suppl 32):58. [Google Scholar]
- Ozawa T, Ito T, Deguchi H, Kawamura K. Patients with idiopathic cardiomyopathy belong to the same mitochondrial DNA gene family of Parkinson's disease and mitochondrial encephalomyopathy. Biochem Biophys Res Commun. 1991;177:518–525. doi: 10.1016/0006-291x(91)92014-b. [DOI] [PubMed] [Google Scholar]
- Perinatal Safety Review Working Group Nucleoside exposure in children of HIV-infected women receiving antiretroviral drugs: Absence of clear evidence for mitochondrial disease in children who died before 5 years of age in five United States Cohorts. J Acquir Immune Defic Syndr. 2000;25:261–268. doi: 10.1097/00126334-200011010-00009. [DOI] [PubMed] [Google Scholar]
- Poirier MC, Divi RL, Al-Harthi L, Olivero OA, Nguyen V, Walker B, Landay AL, Walker VE, Charurat M, Blattner WA. Longterm mitochondrial toxicity in HIV-uninfected infants born to HIV-infected mothers. J Acquir Immune Defic Syndr. 2003;33:175–183. doi: 10.1097/00126334-200306010-00010. [DOI] [PubMed] [Google Scholar]
- Poirier MC, Olivero OA, Walker DM, Walker VE. Perinatal genotoxicity and carcinogenicity of the antiretroviral nucleoside analog drugs. Toxicol Appl Pharmacol. 2004;199:151–161. doi: 10.1016/j.taap.2003.11.034. [DOI] [PubMed] [Google Scholar]
- Powderly WG. Long-term exposure to lifelong therapies. J Acquir Immune Defic Syndr. 2002;29(Suppl 1):S28–S40. doi: 10.1097/00126334-200202011-00005. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Lott MT, Procaccio V, Poole J, Brandon MC, Mishmar D, Yi C, Kreuziger J, Baldi P, Wallace DC. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35(Database issue):D823–D828. doi: 10.1093/nar/gkl927. URL: http://www.mitomap.org. [DOI] [PMC free article] [PubMed]
- Rupert V, Nolte D, Aschenbrenner T, Pankuweit S, Funck R, Maisch F. Novel point mutations in the mitochondrial DNA detected with dilated cardiomyopathy by screening the whole mitochondrial genome. Biochem Biophys Res Commun. 2004;318:535–543. doi: 10.1016/j.bbrc.2004.04.061. [DOI] [PubMed] [Google Scholar]
- Saxena R, de Bakker PI, Singer K, Mootha V, Burtt N, Hirschhorn JN, Gaudet D, Isomaa B, Daly MJ, Groop L, Ardlie KG, Altshuler D. Comprehensive association testing of common mitochondrial DNA variation in metabolic disease. Am J Hum Genet. 2006;79:54–61. doi: 10.1086/504926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton AJ, Turmaine M, Cai WQ, Burnstock G. A study of the ultrastructure of developing human umbilical cord vessels. J Anat. 1996;188:75–85. [PMC free article] [PubMed] [Google Scholar]
- Shiramizu B, Shikuma KM, Kamemoto L, Gerschenson M, Erdem G, Pinti M, Cossarizza A, Shikuma C. Placenta and cord blood mitochondrial DNA toxicity in HIV-infected women receiving nucleoside reverse transcriptase inhibitors during pregnancy. J Acquir Immune Defic Syndr. 2003;32:370–374. doi: 10.1097/00126334-200304010-00004. [DOI] [PubMed] [Google Scholar]
- Sobolewski K, Bañkowski E, Chyczewski L, Jaworski S. Collagen and glycosaminoglycans of Wharton's jelly. Biol Neonate. 1997;71:11–21. doi: 10.1159/000244392. [DOI] [PubMed] [Google Scholar]
- Song S, Pursell ZT, Copeland WC, Longley MJ, Kukel TA, Mathews CK. DNA precursor asymmetries in mammalian tissue mitochondria and possible contribution to mutagenesis through reduced replication fidelity. Proc Natl Acad Sci USA. 2005;102:4990–4995. doi: 10.1073/pnas.0500253102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector SA, Saitoh A. Mitochondrial dysfunction: Prevention of HIV-1 mother-to-infant transmission outweighs fear. AIDS. 2006;20:1777–1778. doi: 10.1097/01.aids.0000242825.97495.a7. [DOI] [PubMed] [Google Scholar]
- Susan-Resiga D, Bentley AT, Lynx MD, LaClair DD, McKee EE. Zidovudine inhibits thymidine phosphorylation in isolated perfused rat heart. Antimicrob Agents Chemother. 2007;51:1142–1149. doi: 10.1128/AAC.01227-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takechi K, Kuwabara Y, Mizuno M. Ultrastructural and immunohistochemical studies of Wharton's jelly umbilical cord cells. Placenta. 1993;14:235–245. doi: 10.1016/s0143-4004(05)80264-4. [DOI] [PubMed] [Google Scholar]
- Thorne C, Newell ML. Safety of agents used to prevent mother-to-child transmission of HIV: Is there any cause for concern? Drug Safety. 2007;30:203–213. doi: 10.2165/00002018-200730030-00004. [DOI] [PubMed] [Google Scholar]
- Torpey JL. Honors Thesis. Department of Chemistry; Tufts University; Medford, MA: 2007. Developing a DNA microarray for human ancestry and migration. [Google Scholar]
- Torres SM, Walker DM, Carter MM, Cook DL, Jr, McCash CL, Cordova EM, Olivero OA, Poirier MC, Walker VE. Cytoxicity and mutagenicity of zidovudine, Lamivudine, and abacavir following in vitro exposure of human lymphoblastoid cells or in utero exposure of CD-1 mice to single agents or drug combinations. Environ Mol Mutagen. 2007;48:224–238. doi: 10.1002/em.20264. [DOI] [PubMed] [Google Scholar]
- Torroni A, Petrozzi M, Santolamazza P, Sellitto D, Cruciani F, Scozzari R. About the “Asian”-specific 9-bp deletion of mtDNA. Am J Hum Genet. 1995;57:507–508. [PMC free article] [PubMed] [Google Scholar]
- Tovo PA, Chiapello N, Gabiano C, Zeviani M, Spada M. Zidovudine administration during pregnancy and mitochondrial disease in the offspring. Antivir Ther. 2005;10:697–699. [PubMed] [Google Scholar]
- Venhoff N, Walker UA. Mitochondrial disease in the offspring as a result of antiretroviral therapy. Expert Opin Drug Saf. 2006;5:373–381. doi: 10.1517/14740338.5.3.373. [DOI] [PubMed] [Google Scholar]
- Walker DM, Poirier MC, Campen MJ, Cook DL, Jr, Divi RL, Nagashima K, Lund AK, Cossey PY, Hahn FF, Walker VE. Persistence of mitochondrial toxicity in hearts of female B6C3F1 mice exposed in utero to 3′-azido-3′-deoxythymidine. Cardiovasc Toxicol. 2004;4:133–153. doi: 10.1385/ct:4:2:133. [DOI] [PubMed] [Google Scholar]
- Walker DM, Malarkey DE, Seilkop SK, Ruecker FA, Funk KA, Wolfe MJ, Treanor CP, Foley JF, Hahn FF, Hardisty JF, Walker VE. Transplacental carcinogenicity of 3′-azido-3′-deoxythymidine in B6C3F1 mice and F344 rats. Environ Mol Mutagen. 2007;48:283–298. doi: 10.1002/em.20297. [DOI] [PubMed] [Google Scholar]
- Walker UA, Venhoff N. Multiple mitochondrial DNA deletions and lactic acidosis in an HIV-infected patient under antiretroviral therapy. AIDS. 2001;15:1449–1450. doi: 10.1097/00002030-200107270-00020. [DOI] [PubMed] [Google Scholar]
- Walker VE, Poirier MC. Foreword: Special issue on health risks of perinatal exposure to nucleoside reverse transcriptase inhibitors. Environ Mol Mutagen. 2007;48:159–165. doi: 10.1002/em.20296. [DOI] [PubMed] [Google Scholar]
- Walker VE, Skopek TR. A mouse model for the study of in vivo mutational spectra: Sequence specificity of ethylene oxide at the Hprt locus. Mutat Res. 1993;288:151–162. doi: 10.1016/0027-5107(93)90216-3. [DOI] [PubMed] [Google Scholar]
- Watts DH. Treating HIV during pregnancy. An update on safety issues. Drug Safety. 2006;29:467–490. doi: 10.2165/00002018-200629060-00002. [DOI] [PubMed] [Google Scholar]
- Witt KL, Cunningham CK, Patterson KB, Kissling GE, Dertinger SD, Livingston E, Bishop JB. Elevated frequencies of micronucleated erythrocytes in infants exposed to zidovudine in utero and postpartum to prevent mother-to-child transmission of HIV. Environ Mol Mutagen. 2007;48:322–329. doi: 10.1002/em.20266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wogan G. Does perinatal antiretroviral therapy create an iatrogenic cancer risk? Environ Mol Mutagen. 2007;48:210–214. doi: 10.1002/em.20283. [DOI] [PubMed] [Google Scholar]
- Wutzler P, Thust R. Genetic risks of antiviral nucleoside analogues—A survey. Antiviral Res. 2001;49:55–74. doi: 10.1016/s0166-3542(00)00139-x. [DOI] [PubMed] [Google Scholar]
- Yoon KL, Aprille JR, Ernst SG. Mitochondrial tRNA(thr) mutation in fatal infantile respiratory enzyme deficiency. Biochem Res Commun. 1991;176:1112–1115. doi: 10.1016/0006-291x(91)90399-r. [DOI] [PubMed] [Google Scholar]
- Zaera MG, Miró Ò , Pedrol E, Soler A, Picón M, Cardellach F, Casademont J, Nunes V. Mitochondrial involvement in antiretroviral therapy-related lipodystrophy. AIDS. 2001;15:1643–1651. doi: 10.1097/00002030-200109070-00006. [DOI] [PubMed] [Google Scholar]