Abstract
Humans share 96% of our 30,000 genes with Chimpanzees. The 1,200 genes that differ appear at first glance insufficient to describe what makes us human and them apes. However, we are now discovering that the mechanisms that regulate how genes are expressed tell a much richer story than our DNA alone. Sections of our DNA are constantly being turned on or off, marked for easy access, or secluded and hidden away, all in response to on-going cellular activity. In the brain, neurons encode information – in effect memories – at the cellular level. Yet while memories may last a lifetime, neurons are dynamic structures. Every protein in the synapse undergoes some form of turnover, some with half-lives of only hours. How can a memory persist beyond the lifetimes of its constitutive molecular building blocks? Epigenetics – changes in gene expression that do not alter the underlying DNA sequence – may be the answer. In this article, epigenetic mechanisms including DNA methylation and acetylation or methylation of the histone proteins that package DNA are described in the context of animal learning. Through the interaction of these modifications a “histone code” is emerging wherein individual memories leave unique memory traces at the molecular level with distinct time courses. A better understanding of these mechanisms has implications for treatment of memory disorders caused by normal aging or diseases including schizophrenia, Alzheimer’s, depression, and drug addiction.
Keywords: epigenetics, genes, memory, learning, histone, hippocampus, behavior
The human genome contains approximately 30,000 genes responsible for the inner workings of some 100 trillion cells in the human body. These genes manifest themselves in both obvious ways, such as eye and hair color, but also in more subtle ways, such as predispositions to physical and mental conditions including heart disease and schizophrenia. However, the idea that individual genes are responsible for complex cognitive dysfunctions, (e.g. autism, dyslexia), rather than a set of genes with interacting influences (epistasis), is still a topic of research. Certainly there are cases where a defect or deletion in a single gene may give rise to rampant cognitive or health deficits, as is discussed below. However, the vast majority of cognitive phenotypes will probably be multifaceted amalgams of numerous genes working in concert. The interaction of these genes goes far beyond simply knowing if a gene is present or absent in an individual.
Humans share about 96% of our genes with chimpanzees (Pan troglodytes). At first glance, this implies that a mere 1,200 genes are responsible not just for differences in our physical appearance but also for language, critical thinking, and a host of other “uniquely human” measures of interest to psychologists. The 4% of our genome that differs from chimps is likely a large part of what makes us human. However, the other 28,800 genes present in both humans and chimpanzees are not sitting idle. The same genes may actually be performing different roles in cognitive and other processes across species.
The term epigenetics, literally “above genetics,” has been coined to describe mechanisms that alter gene expression without altering the underlying DNA sequence. This article focuses on the ways in which DNA is regulated and how precise control over gene expression exponentially extends the computational power of the genome. Specifically, mechanisms for direct methylation of the DNA double helix are discussed along with the role that DNA methylation plays in normal cellular function and also in terms of disorders that emerge from improper DNA methylation. In addition, two modifications to the histone proteins responsible for packaging and regulating access to DNA are discussed: histone acetylation and histone methylation. Finally, experimental methodologies and paradigms for studying the role of DNA regulation in learning and memory are described along with the application of these findings to models of aging, disease and brain dysfunction. But first, it is necessary to understand why mechanisms for regulating DNA are important and how they work.
The Need for DNA Regulation
Following fertilization, stem cells are totipotent, capable of becoming any cell type; and through repeated divisions become progressively more restricted until they become fully differentiated into specific cell types such as cells lining the stomach, liver cells, or pyramidal neurons in the hippocampus. Each of our 100 trillion cells, regardless of cell type, contains the same DNA, but expresses only a subset of those genes (Reik, 2007).
Inactive or silent genes are, generally speaking, the norm. Liver cells are very good at making proteins to break down toxins in the body. Liver cells do not make brain-specific proteins just as the brain does not specialize in creating liver enzymes even though neurons contain the DNA code for liver enzymes. Inside the cell nucleus, genes that are not used are silenced through the addition of methyl groups to the DNA. Generally, DNA methylation occurs at CpG sites where cytosine and guanine nucleotides occur next to each other, separated by a phosphate. Methylation of CpGs frequently occurs in a gene’s promoter region and interferes with binding of the transcriptional machinery responsible for making an mRNA copy, which will later be converted into a protein by the cell’s ribosomes. By interfering with promoter binding, the downstream DNA sequence cannot be “read” and no protein is made (Li, Beard, & Jaenisch, 1993). The patterns of gene silencing are more or less fixed when a cell divides into its final cell type and this lineage is passed along to daughter cells during cell division. Thus, liver cells divide to become liver cells. Daughter cells do not spontaneously become a different cell type than their parent cells. In a sense, the DNA becomes hard coded by cell type.
The brain is different. In the adult nervous system, the vast majority of neurons are terminally differentiated, meaning they do not continue to divide and do not produce daughter cells. Humans start with a fixed number of brain cells and this number declines with age, except in the Dentate Gyrus region of the hippocampus and possibly in a few other small brain regions where adult neurogenesis does occur; however, this is the exception, rather than the rule (Landgren & Curtis, 2010). Perhaps surprisingly, mechanisms for turning genes on and off by adding or removing methyl groups are present in the brain (Levenson & Sweatt, 2005). Neurons dynamically regulate the pattern of DNA methylation on a gene-by-gene basis in response to neuronal activity, as will be discussed in greater depth below.
Beyond turning off genes extraneous to a particular cell types, DNA must be regulated for logistical reasons. Unraveled, the DNA contained in each cell would stretch nearly 2 meters, yet the DNA must fit inside the cell nucleus, a structure about 6 microns (10−6 meters) in diameter. This is equivalent to stuffing 10,000 miles of spaghetti into a regulation-sized basketball (Peterson & Laniel, 2004). To accomplish this feat, the DNA is compressed about 10,000-fold by wrapping the DNA around histone proteins. This histone-DNA complex is termed chromatin and is responsible for strengthening the DNA, preventing damage, and repairing damage when it occurs (Bhaumik, Smith, & Shilatifard, 2007).
There are four main histone proteins: H2A, H2B, H3, and H4. These proteins are structurally nearly identical both within and between plants and animals. In other words, they are “conserved” across species. This structural conservation suggests functional similarity. Indeed, regardless of species, the histone proteins assemble into an octomer containing two of each histone protein around which 146 base pairs of DNA wrap. These octomers are connected by H1 linker histones that hold the DNA in place (Farkas, 1996). The chromatin complex continues to roll itself up until it forms coils, and eventually individual chromosomes.
The DNA interacts with histone proteins in several ways, but the simplest explanation boils down to charge (García-Pérez, Pinto, & Subirana, 2003). DNA is negatively charged. Histones are positively charged. The DNA is attracted to the histones and winds around the octomer like thread wrapping around a spool, two wraps per spool. As with DNA methylation, the histone proteins may be activated or inactivated, altering the accessibility of DNA (Burgess-Beusse et al., 2002; Horn & Peterson, 2002).
Gaining access to specific genes in a timely manner is no small feat. It has been estimated that only 1.5% of our DNA codes for proteins (Lander et al., 2001). The remainder of the genome consists of non-coding DNA, regulatory sequences, or sequences of unknown function, sometimes termed “junk” DNA. Gene regulation comes down to locating specific genes when they are needed, but also compressing the unused DNA to get it out of the way.
When a cell needs to make new proteins in response to activation—be it a neuron firing or a liver cell making new enzymes to break down a toxin—the relevant DNA must be unwound from the histone proteins and the cellular machinery must then locate the appropriate DNA sequence and make an mRNA copy. In the case of immediate-early genes, such as zif268, this can happen in as little as 15 minutes. To put the enormity of this task into perspective, finding a single gene is the equivalent of finding a specific 2 cm strand of spaghetti amongst the 10,000 miles stuffed into that basketball. To aid in locating genes of interest, a number of histone modifications act as sign-posts and also regulate access to the DNA.
Histone proteins have “tails” that extend past the wrapped DNA. These tails both influence the interaction between the histones and the DNA but also serve as molecular signposts for the transcriptional machinery (Bártová, Krejcí, Harnicarová, Galiová, & Kozubek, 2008; Nicholson et al., 2004). The tails can be modified in a number of predictable ways at highly specific sites by acetylation, methylation, phosphorylation, ubiquitination, and SUMOylation of the histone tails. This article will focus on two of these modifications: histone tail acetylation and methylation in the context of learning and memory.
Molecular Methods for Studying Epigenetic Changes
Epigenetic studies frequently begin with isolation and/or purification of the protein or DNA of interest. Generally, a sample is collected (e.g. blood, saliva, brain tissue) and everything but the DNA and DNA-interacting proteins are systematically removed. DNA and histone proteins are isolated both mechanically (centrifugation, physical grinding) and chemically (“washes” in different combinations of acids, alcohols, and other solutions). Many of these protocols are freely available on the internet.
Once a sample has been purified, the DNA or histone proteins must be visualized. The form of visualization will depend on the question being asked. Is there a question about the presence or absence of a particular DNA sequence? Is there a difference in the quantity of a particular gene or protein in two different samples? Are multiple proteins thought to interact with the same sequence of DNA? Are multiple histone tail modifications present? Most of these questions can be answered by using an antibody against the protein of interest or a labeled (e.g. radioactive) DNA probe. Two frequently employed techniques for visualizing epigenetic modifications are gel electrophoresis and immunohistochemistry.
Gel electrophoresis separates samples based on size. The gel is a cross-linked polymer matrix that allows small proteins or short sections of DNA to pass easily through the gel while larger elements in the sample move more slowly. DNA is negatively charged and proteins are mixed with a negatively charged solution such that when a current is passed through the gel, the sample is pushed through the matrix. The samples can be labeled with a radioactive marker or visualized using a variety of detection techniques, including antibodies that only bind to the specific protein of interest. Gel electrophoresis is an excellent tool for identifying the presence of a DNA sequence or a particular protein or epigenetic modification (e.g. acetylated or methylated histones).
It is also possible to determine which DNA sequences interact with a particular protein by performing a chromatin immunoprecipitation or ChIP. This assay strengthens the bond between DNA and connected proteins using formaldehyde. The combined protein/DNA is then broken into small pieces using ultrasonic noise. An immunoprecipitation is then performed against the protein of interest. In short, the sample is poured through a tube containing antibodies that trap the target protein and the attached DNA while other proteins pass through, unbound, and are discarded. The protein/DNA complex is then washed from the tube and collected. The binding is reversed and the DNA can be run on a gene chip (discussed below) or compared against a smaller group of candidate DNA sequences using a combination of DNA amplification to increase sample size (Polymerase Chain Reaction or PCR) and gel electrophoresis.
Immunohistochemistry is a way to simultaneously visualize where multiple proteins are located inside a tissue sample. Ultra-thin slices are made of the structure of interest (e.g. the hippocampus) on the order of 5–50 microns. The tissue is then fixed and perforated chemically. Antibodies against the target proteins are washed into the tissues, one at a time, with each type of antibody being attached to a different fluorophore – a tag that will fluoresce under a specific wavelength of light. The tissue is then photographed on a confocal microscope to create a 3D image of the tissue, one picture per wavelength of light. The pictures are then overlaid and the different colors from different protein-bound antibodies are compared to determine if the proteins are found in the same part of each cell.
Experiments for Identifying Changes in Gene Regulation
Knowing that DNA is regulated is a far cry from knowing when and why that regulation occurs in response to something like learning. At present, scientists are unable to monitor changes in gene regulation in real-time using non-invasive methodologies. The majority of our knowledge of gene expression comes from one of three sources: 1) cheek swabs or blood samples, 2) post-mortem studies of human tissue, and 3) animal studies.
Cheek swabs and blood samples offer a snapshot of a subject’s DNA. As a forensic tool, these measures allow sample matching and can be used to identify the presence or absence of a particular gene. It is also possible to compare genetic profiles between groups using gene chips. Gene chips, sometimes referred to as DNA microarrays, are glass or silicon grids containing thousands of DNA probes. These probes are specifically designed to detect the presence or absence of a particular gene or a gene mutation and can also detect the degree to which a gene is expressed in a sample. Thus a gene chip could simultaneously test for all known genetic diseases (e.g., Tay-Sachs, Huntington’s, etc.) or determine if a hereditary predisposition to a disease is correlated with an increase in the expression of a particular gene not seen in individuals without a family history of the disease (Manolio, 2010). Similarly, such technology might be applied to identification of viral or bacterial infection for deployment in third world countries on “chips” made of paper the size of a postage stamp. These paper chips are essentially cousins of pregnancy tests. A pregnancy test determines the presence or absence of the hormone chorionic gonadotropin (hCG), secreted by the placenta shortly after fertilization. Similar tests can identify protein or DNA markers in blood or saliva and produce a color-coded read-out capable of being interpreted in the field or photographed using a cell phone and sent to a lab for analysis (Whitesides, 2006).
An important caveat to any sample, be it blood, cheek swab, or tissue biopsy, is that different tissues express different genes, even though every cell contains the same DNA. In the hippocampus, even adjoining areas (e.g., CA1 and CA3) express different genes (Torres-Muñoz, Van Waveren, Keegan, Bookman, & Petito, 2004). To determine which genes are involved in learning we need to look at specific changes in gene expression inside the brain regions involved in learning and memory.
Post-mortem studies of gene expression get us closer to this goal. By either sub-dissecting regions of interest for gene chips or by fixing brain slices and using staining or immunohistochemical techniques to observe tissue-specific changes in DNA expression, normal and abnormal brains can be compared side-by-side. Aberrant DNA methylation and histone modifications resulting in altered gene expression have been identified in a number of conditions including autism, schizophrenia, and depression (Akbarian & Huang, 2009; H. S. Huang & Akbarian, 2007). For example, changes in the number or proper function of methyltransferases including MLL1 and GAD1 may result in improper histone tail methylation (Akbarian & Huang, 2009; H. S. Huang & Akbarian, 2007). These studies highlight the role altered gene expression plays in disease and disorders of the nervous system. However, these studies cannot be conducted in living humans.
At present, animal studies are the most effective means of understanding gene regulation. However, using an animal model limits cognitive assessment of “uniquely human” attributes, such as language. Other cognitive abilities lend themselves well to study in animal models. A number of behavioral paradigms have been developed to tease apart subtle differences in learning and memory. By sacrificing the animal following memory formation and sub-dissecting regions of the brain involved in learning – such as the hippocampus – a snapshot of changes in gene regulation in response to memory-inducing events can be determined. These changes are then compared to a baseline of gene expression in animals not exposed to the memory-paradigm.
Most animal behavioral learning paradigms are variations on classical Pavlovian conditioning (McIlwain, Merriweather, Yuva-Paylor, & Paylor, 2001; Paylor, Tracy, Wehner, & Rudy, 1994; Rudy, Huff, & Matus-Amat, 2004). Contextual fear conditioning is commonly used to measure changes in protein or gene expression related to learning and memory (Fanselow, 2000). In this task, the animal is placed in a novel training environment with an electrifiable floor. The animal is allowed to briefly explore the environment before receiving a series of between one and three footshocks, usually in the 0.5–1.0 mA range. The animal is then re-introduced to the context 24 hours later and freezing behavior is measured as an index of the fear memory. The stronger the association between the context and the footshock, the greater the freezing behavior (Blanchard & Blanchard, 1969). These animals are generally compared to naïve animals or to animals exposed to the context but not given a footshock.
To determine which genes or proteins are altered during contextual fear conditioning, the animal is sacrificed at a time point corresponding to peak changes in gene or protein regulation. Immediate early genes, such as Zif-268, are activated within 15 minutes of memory induction (Bozon et al., 2003). Proteins, such as NF-κB are activated between one and two hours after contextual fear conditioning and trigger additional gene expression at two hours (Freudenthal & Romano, 2000; Saccani, Pantano, & Natoli, 2001).
DNA Methylation and Gene Silencing
As already discussed, t h e b e s t-known epigenetic modification is DNA methylation. Within five days of fertilization, cells in the blastocyst begin to show different patterns of gene expression (Reik, 2007). Cells that are destined to become liver cells shut off genes for neuron functions and future brain cells turn off, or silence, genes responsible for liver enzymes.
Gene silencing is accomplished through direct methylation of the DNA encoding particular genes. Adding a methyl group at specific gene loci blocks the transcriptional machinery from binding the DNA and prevents gene copying (Schübeler et al., 2000). Improper DNA methylation gives rise to a number of developmental disorders, including Angelman and Prader-Willi syndromes (Jaenisch & Bird, 2003; Li et al., 1993; Maeda & Hayashizaki, 2006; Paulsen & Ferguson-Smith, 2001; A. J. Wood & Oakey, 2006).
Patterns of DNA methylation may be initially established through imprinting, the inheritance of on/off DNA states from either paternal or maternal genes. Some genes inherit gene alleles specific to the father (e.g. insulin-like growth factor-2; IGF2) whereas other gene activation/inactivations are inherited from the mother while the father’s DNA is suppressed (e.g., the receptor for IGF2, IGF2r) (Biliya & Lee A. Bulla, 2010). Errors of imprinting can lead to diseases, including Prader-Willi/Angelman syndrome (Buiting, 2010).
A particularly interesting example of DNA methylation gone awry is Rett syndrome. Rett syndrome appears to be a rare example of a single gene giving rise to a complex disease phenotype. Individuals with Rett Syndrome have a mutation in the X-linked MECP2 gene that encodes a protein for methylating DNA (Chahrour & Zoghbi, 2007; Gonzales & LaSalle, 2010). Mutated MECP2 proteins cannot properly bind DNA and add methylation markers to silence genes. Consequently, genes that should normally be turned off are active, resulting in severe mental retardation, including autistic-like behavior. Females with Rett Syndrome develop language normally at first, before regressing and losing language entirely. Constant hand-wringing is another characteristic of the disease. In animal models of Rett where mice have a mutated form of MECP2, the mice exhibit a similar paw-wringing behavior.
DNA Methylation in Learning and Memory
Rett syndrome is interesting, in part, for the characteristic development of linguistic ability followed by total loss of language. However, language cannot be assessed in an animal model. In contrast, in animal models of learning and memory, increased DNA methylation is observed following behavioral fear conditioning. This finding suggests that learning results in the silencing of DNA (Miller & Sweatt, 2007). Turning off genes during learning is counterintuitive. Learning is an active process that should require activation of new genes. In fact, learning does activate a host of genes (zif268, bdnf) (Gupta et al., 2010). However, some genes may be active to suppress new memory formation except in cases where that learning is beneficial to the animal’s survival. Injecting animals with DNA methylation inhibitors prior to contextual fear conditioning, prevents freezing behavior when the animal is later re-exposed to the shock-paired context (Miller & Sweatt, 2007). This finding suggests that DNA methylation is actively modulated in the brain to regulate gene expression. Which genes are being regulated and to what degree is an active area of research.
At present, DNA methylation inhibitors lack the specificity needed to target only those sections of DNA that are improperly methylated. Theoretically, probes could be designed to identify individual DNA sequences that are improperly methylated, however these molecular medicines are far from clinical trials. Thus, while aberrant DNA methylation is suspected to play a large role in schizophrenia, Rett syndrome, autism, and numerous forms of mental retardation, altering DNA methylation through systemic administration of DNA methylation inhibitors could lead to the onset of other disorders, including cancer (Lechner, Boshoff, & Beck, 2010).
Histone Acetylation in Learning and Memory
Much of the work on histone modifications and memory has focused on histone acetylation (Guan et al., 2002; Korzus, Rosenfeld, & Mayford, 2004; Levenson et al., 2004; Levenson & Sweatt, 2005; M. A. Wood, Hawk, & Abel, 2006; Yeh, Lin, & Gean, 2004). Histone tails are acetylated by histone acetyltransferases (HATs) and acetyl groups are removed by histone deacetylases (HDACs) (Hake, Xiao, & Allis, 2004). In the non-acetylated state, histone proteins are positively charged, attracting the negatively charged DNA. When acetyl groups are added by HATs, the positive charge is neutralized and the DNA repels itself, thereby moving away from the histones and opening up for transcriptional machinery (Tanner, Langer, & Denu, 2000). HATs and HDACs are highly specialized proteins that only recognize and bind to very specific locations on histone tails. This specificity is even greater for histone methylation, as will be discussed in greater detail below.
Whereas acetylation of one histone protein, H3, is generally permissive for transcription, acetylation of histone H4 is a transcriptional silencer (Braunstein, Sobel, Allis, Turner, & Broach, 1996). Interestingly, H3 and H4 acetylation are differentially regulated in rodent hippocampus during behavioral fear conditioning. H3 acetylation is increased following the pairing of footshock with a short exposure to a novel context whereas a long pre-exposure (2 hours) to the context prior to footshock results in an increase in H4 acetylation and no change in H3 acetylation (Levenson et al., 2004). Differential regulation of H3 and H4 acetylation suggests that different forms of behavioral conditioning will activate or inactivate different genes responsible for encoding that memory.
In many learning and memory experiments, acetylation is a short-lived modification, peaking after 1–2 hours (Levenson et al., 2004). However, a number of chemical compounds including Trichostatin A, sodium butyrate, and valproic acid act as HDAC-inhibitors, prolonging acetylation (de Ruijter, van Gennip, Caron, Kemp, & van Kuilenburg, 2003; Villar-Garea & Esteller, 2004). These chemicals enhance memory acquisition, extinction, and synaptic plasticity (Bredy & Barad, 2008; Levenson et al., 2004; Levenson & Sweatt, 2005; Vecsey et al., 2007). Interestingly, HDAC inhibitors do not increase global levels of acetylation, but seem to target genes active during the memory paradigm (Vecsey et al., 2007).
Pharmacological HDAC inhibitors show promise in the treatment of memory dysfunction and the different classes of HDAC inhibitors may allow targeting of specific HDAC classes, such as those associated with Alzheimer’s, Huntington’s, and other progressive diseases (Abel & Zukin, 2008; Chuang, Leng, Marinova, Kim, & Chiu, 2009). However, the effects of these drugs are not limited to HDACs. Trichostatin A also targets some Interleukins and Nitric Oxide Synthase while Valproic acid blocks some voltage-gated Calcium channels (Rosenberg, 2007). Consequently, these drugs require further refinement before being used to bolster memory performance.
Histone Methylation in Learning and Memory
Histone methylation involves the addition of a methyl group at specific amino acids along the histone tails. Methylation can be either permissive or repressive depending on the degree of methylation (mono-, di-, or tri-methylation) and the site being methylated (Nakayama, Rice, Strahl, Allis, & Grewal, 2001). For example, di-methylation at lysine 9 of histone H3 (H3K9me2) inhibits gene transcription whereas tri-methylation at lysine 4 (H3K4me3) permits transcription. As with HAT proteins that add acetyl groups, a number of histone methyltransferases have been identified. These methyltransferases only target specific amino acid sites along the histone tails. For example, Suv39 methylates both H3K9 and H3K27 but not H3K4 (Allis et al., 2007; Lachner, O'Sullivan, & Jenuwein, 2003). Histone demethylases remain more mysterious, although several have been identified and are being characterized, including LSD1 which demethylates either mono- or di-methylated H3K4, but will not remove methyl groups from tri-methylated H3K4 (Mosammaparast & Shi, 2010).
As with acetylation, histone methylation is altered following contextual fear conditioning. One hour after contextual fear conditioning, changes in methylation on histone H3 are observed in area CA1 of rat hippocampus. Both H3K4me3 (permissive) and H3K9me2 (repressive) are increased (Gupta et al., 2010). Mice lacking the H3K4-specific methyltransferase, Mll, show a contextual fear memory deficit, suggesting that histone methylation at this site is necessary for memory formation. Indeed a number of memory-related genes, including bdnf and zif268 are downstream of H3K4me3 (Gupta et al., 2010). Identifying genes silenced by an increase in H3K9me2 is more difficult, but it can be speculated that a memory thresholding mechanism may exist. Below a certain threshold, a record of the animal’s experience is not important enough for devoting cellular resources. Thus, mechanisms prevent storage of irrelevant or non-valuable information. As with increases in DNA methylation observed during a memory task, these memory-suppressive histone mechanisms would need to be turned off while other epigenetic mechanisms for storing valuable memories are turned on.
The interplay of multiple histone methylation marks is more clearly seen when comparing different behavioral paradigms at different time points and in different brain regions. As noted above, changes in H3K4 and H3K9 were observed at one hour in CA1 (Gupta et al., 2010). However, no changes in Dentate Gyrus or area CA3 of hippocampus were observed one hour after contextual fear conditioning (Molfese, Sweatt, & Lubin, in revision).
However, 48 hours later, an increase in H3K4me3 was observed in Dentate Gyrus, despite no additional behavioral conditioning. In a separate cohort of animals that underwent extinction, also at 48 hours, H3K4me3 had returned to baseline in all brain regions examined, but an increase in H3K9me2 was seen in areas CA1 and CA3. When compared with animals pre-exposed to the context for two hours, increases in H3K9me2 were observed in both CA3 and Dentate Gyrus at one hour, but not at 48 hours (Molfese et al., in revision).
A brief pairing of context plus shock (7 minutes) thus differs from a two-hour pre-exposure to context in two ways. First, pre-exposure increases H3K9me2 in CA3 and DG compared to an increase in both H3K9me2 and H3K4me3 in area CA1 following brief exposure. Second, histone H3 acetylation is increased by brief exposure whereas histone H4 acetylation is increased by pre-exposure. Indeed, treating mice with the HDAC inhibitor sodium butyrate increased trimethylation at H3K4 and decreased di-methylation at H3K9 (Gupta et al., 2010). This result suggests an interaction between histone acetylation and histone methylation.
To summarize these findings, different gene regulation mechanisms, and presumably different genes, are active at different time points in different parts of the brain, even within the same structure, such as the hippocampus. These genes are actively regulated in response to different learning paradigms, such that each type of learning produces a unique gene-regulation profile. Simply knowing what DNA is present in an individual is insufficient to know how that DNA will be expressed in a particular cell.
The Histone Code
The histone code hypothesis states that the combination of multiple histone modifications (including DNA methylation, histone acetylation, and histone methylation) established either concurrently or sequentially, determines gene expression by regulating gene transcription directly or leading to additional DNA/histone remodeling (Strahl & Allis, 2000). Some of these modifications are transient, such as acetylation. Others, including methylation, may last for days whereas DNA methylation can be permanent but may also be reversible.
From a cellular housekeeping perspective, this view of epigenetic mechanisms has great appeal. Individual neurons receive tens of thousands of synaptic inputs and must continue to function as part of a network even as individual proteins are replaced. Consider an individual synapse where excitability is determined by the ratio of AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) receptors to NMDA (N-Methyl-D-aspartic acid) receptors (Wheal et al., 1998). To make an individual synapse more or less likely to cause the neuron to fire, simply alter the AMPA-to-NMDA ratio. Both AMPA and NMDA receptors have limited life spans, with half-lives between 2 and 34 hours, (Huh & Wenthold, 1999). The AMPA/NMDA ratio will shift as old receptors are removed and as new receptors are delivered to the synapse. New NMDA receptors may be inserted into the synaptic membrane within hours of synaptic activation (Quinlan, Philpot, Huganir, & Bear, 1999).
Synaptic activation sends a signal to the cell nucleus. This signal is integrated with the existing epigenetic state of the cell’s DNA. Genes that are already active may be shut down. Inactive genes may be activated. The synaptic activation may call for an increase in NMDA receptors, but with a different sub-unit composition (and thus a different excitability or half-life) than presently exists at the active synapse. The availability of specific genes encoding the different components of the NMDA receptor is determined by the current state of DNA methylation and by the pattern of histone acetylation or methylation (plus histone phosphorylation, ubiquitination, and SUMOylation, all of which likely interact to regulate gene expression). The combinatorial nature of these epigenetic modifications significantly extends the information potential of the genetic code (Jenuwein & Allis, 2001).
Epigenetics and Disease
In addition to Rett syndrome, discussed above, chronic changes in DNA regulation have been observed in autism, schizophrenia, depression, and addiction (Akbarian & Huang, 2009; J. Huang et al., 2007). Under-methylated genes may be over-active, resulting in too much of a protein being produced, while over-methylation may cause too little of a protein to be made. Deficits in DNA methylation have previously been reported in schizophrenics (J. Huang et al., 2007). Changes in histone methylation have also been observed in postmortem tissue from schizophrenic patients (Akbarian & Huang, 2009). As some histone methylation sites are permissive for gene expression and others are repressive, aberrant methylation could similarly affect the quantity of disease-related protein produced.
A central feature of depression is the need for chronic administration of anti-depressant medications to combat the disorder. The molecular mechanisms underlying depression and antidepressant action are poorly understood. However, mice exposed to chronic social defeat stress exhibit down-regulation of brain-derived neurotrophic factor (BDNF) in the hippocampus (Tsankova et al., 2006). Chronic imipramine (a tricyclic antidepressant) reverses BDNF down-regulation and increases histone acetylation by down-regulating HDAC5, an HDAC that targets specific regions of the histone tails for removal of acetylation (Tsankova et al., 2006).
A number of epigenetic mechanisms are sensitive to drugs of abuse. The gene transcription factor, DeltaFosB, accumulates in striatum after repeated drug use and chronic amphetamine exposure represses the gene c-fos by recruiting HDAC1 (Renthal et al., 2008). Concurrently, there is a reduction in SUV39, a specific methyltransferase, at the c-fos gene promoter. Mice lacking HDAC1 do not exhibit this reduction in c-fos. C-fos is also regulated by cocaine. Within 30 minutes of acute cocaine injection histone H4 is acetylated at the c-fos promoter in striatum. These changes are not observed following chronic cocaine exposure; however, chronic cocaine exposure results in histone H3 acetylation at the gene promoters for bdnf and Cdk5 (Kumar et al., 2005). Other histone modifying enzymes also differentiate acute versus chronic drug exposure, including HDAC5, the loss of which results in hypersensitivity to chronic, but not acute, cocaine and stress (Renthal et al., 2007).
Although we do not think about childhood abuse and neglect as a disease, per se, recent findings suggest that abuse may increase susceptibility to schizophrenia, depression, and bipolar disorders (De Bellis, 2005). The mechanisms underlying this predisposition appear to be epigenetic. Roth and colleagues created a stressful environment for rat mothers who then were more likely to abuse or neglect their pups (Roth, Lubin, Funk, & Sweatt, 2009). Littermate pups were placed with either a nurturing or a stressed mother for 30 minutes a day for one week. Changes in bdnf gene methylation were observed in the prefrontal cortex of abused pups. Surprisingly, these changes persisted 30 and 90 days after a single week of maltreatment. Even more surprising, abused pups grew up to be abusive parents and passed along changes in gene methylation to their offspring (possibly through imprinting mechanisms). A tendency to abuse may therefore have a genetic component. Perpetuating abuse to offspring of abused parents has also been observed in monkeys (Sanchez, 2006). Thus, while our DNA may define aspects of who we are, our environment and our experiences may alter the way in which that DNA is expressed.
Finally, Alzheimer’s disease may also result from mutations in specific proteins, either as a result of mis-expression of those genes or because of genetic mutations that pre-dispose individuals to developing Alzheimer’s (Bertram, Lill, & Tanzi, 2010). Alternatively, DNA damage occurring naturally over time may not be properly repaired, resulting in Alzheimer’s and other diseases (Coppedè & Migliore, 2010). Both gene expression and failure to repair damaged DNA are regulated by epigenetic mechanisms
Real-Time Genetic Imaging
Although the most productive areas of epigenetic research are currently monopolized by post-mortem human or invasive animal work, a promising new technology may well allow us to non-invasively visualize gene regulation in action. MR Spectroscopy (MRS) is a variant of Magnetic Resonance Imaging wherein the scanner is tuned to a specific protein. This technology has been used to identify the stage of cancerous cells without doing a biopsy (Law, 2009).
Such technology may one day allow us to visualize changes in gene expression in real time and could theoretically already track proteins responsible for methylating DNA or altering histone tails by adding or removing acetylation and methylation markers. These observations would speak not only to what parts of the brain are active in a given task, but also to how the neurons in that region are altering their function in response to being activated. Imagine a neuroimaging study to determine which genes are being activated by an epigenetic mechanism as identified by that particular protein’s magnetic signature.
Summary
Epigenetic mechanisms, that is, changes in the regulation and expression of DNA, have the capacity to greatly increase the information contained in the genome by precisely regulating gene expression. Some genes are kept active in response to environmental inputs while other genes are permanently silenced. In studies of learning and memory, systematic changes in DNA methylation, histone acetylation, and histone methylation have been observed. These changes are a form of cellular memory, recording the experiences of the animal. The specificity of these changes suggests an epigenetic code. Were we able to “read” the modifications we might, in theory, be able to reconstruct an animal’s memory by identifying which learning paradigm it experienced.
Epigenetics is not limited to studies of learning and memory, although animal models do lend themselves well to this area of research. A better understanding of epigenetic mechanisms may allow for more targeted drug therapies for schizophrenia, depression, Alzheimer’s disease, and addiction; or might allow novel treatments for developmental disorders including Angelman, Prader-Willi, and Rett syndromes. Although a cheek swab may identify a genetic pre-disposition to some of these diseases and disorders, the real power of the genome lies in regulating the expression of individual genes through epigenetic mechanisms on a tissue-by-tissue basis. But while a drop of blood cannot predict an individual’s ability to memorize or their verbal IQ, it will place them at the scene of the crime with nearly 100% accuracy.
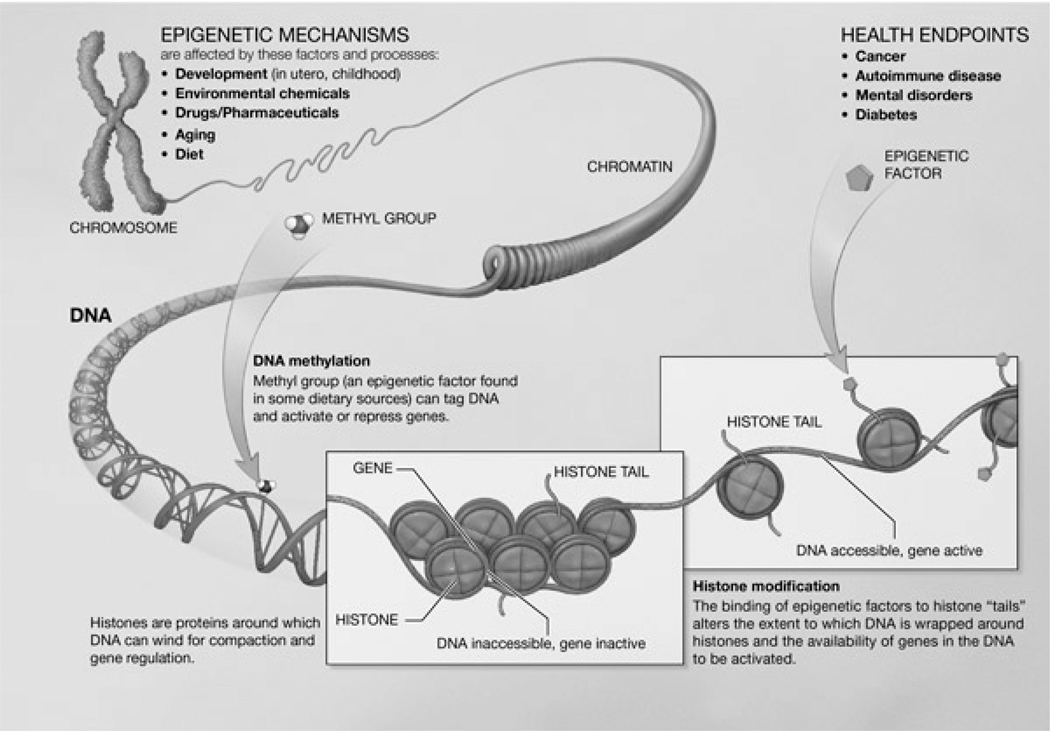
Figure 1.
Epigenetic mechanisms. DNA wraps twice around an octomer of histone proteins. Histone tails extend past the coiled DNA. The DNA-wrapped histones continue to coil, compressing the DNA 10,000-fold. Long sections of this compressed DNA form chromosomes. Image courtesy of NIH (public domain).
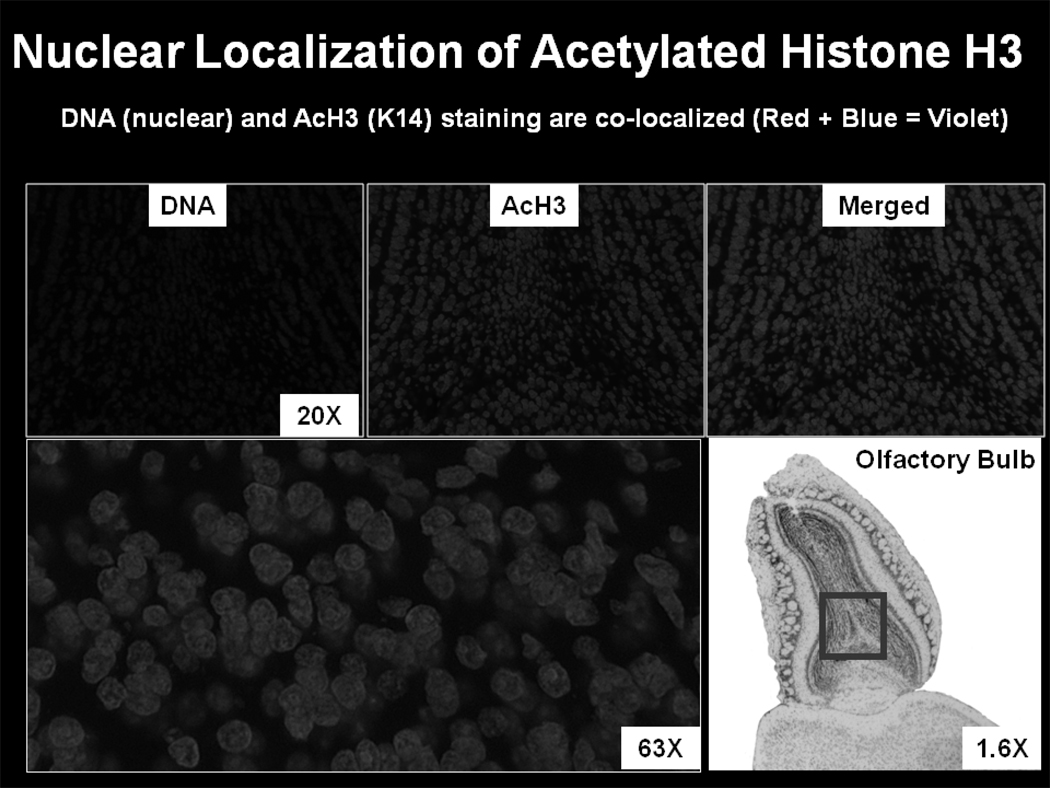
Figure 2.
Immunohistochemistry. DNA co-localizes with acetylated (active) histone H3 in the rodent olfactory bulb. DNA is stained blue while acetylation at lysine 14 on histone H3 is stained in red. Image courtesy of Dr. Caterina M. Hernandez.
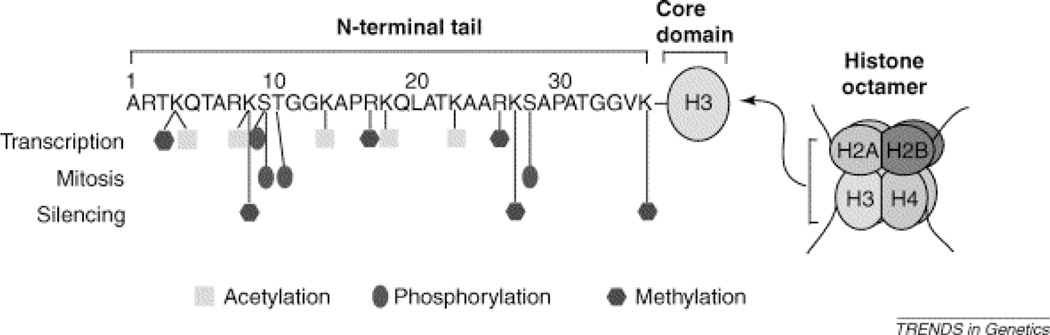
Figure 3.
Histone modifications including acetylation, methylation, and phosphorylation occur at specific amino acids along histone tails. Some histone tail modifications are transcriptionally permissive, including trimethylation of histone H3 at amino acid position 4, a lysine (K). This is denoted H3K4me3 for the histone number, amino acid type and position, and the type and level of modification. (Nowak & Corces, 2004)
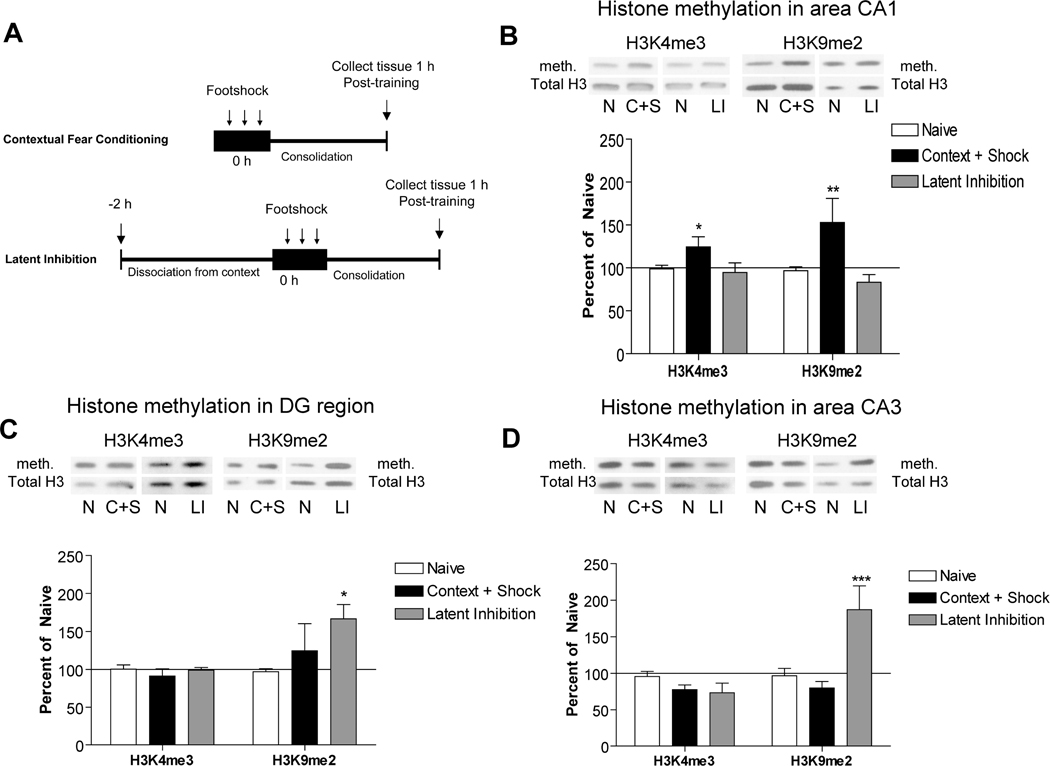
Figure 4. Histone methylation is altered in hippocampal subfields.
A) Experimental behavior paradigms are shown. For contextual fear conditioning, animals were placed in a novel context for 7 minutes wherein they received 3 one-second, 0.5 mA footshocks at two minute intervals. Latent Inhibition animals were pre-exposed to the context for two hours, followed immediately by the contextual fear conditioning protocol. Both groups were immediately returned to their home cages after removal from the context and sacrificed one hour later. B) Histone methylation for H3K4me3 and H3K9me2 were determined using immunoblotting in area CA1 of hippocampus one hour after contextual fear conditioning or latent inhibition. Significant differences between fear conditioned and naïve animals were observed for both histone modifications (*p<0.05 and **p<0.005, respectively). There were no differences between latent inhibition and naïve animals. C) In the Dentate Gyrus, a significant increase in histone H3K9me2 was observed following latent inhibition, as compared to naïve animals (*p<0.05). No other differences were observed. D) As in Dentate Gyrus, a significant difference between naïve and latent inhibition animals was observed for H3K9me2 in area CA3 (***p<0.001). No other differences were observed. Solid line represents normalized Naïve control levels. Error bars are SEM. N, Naïve controls; C+S, Contextual fear conditioning; LI, Latent inhibition; meth, methylation.
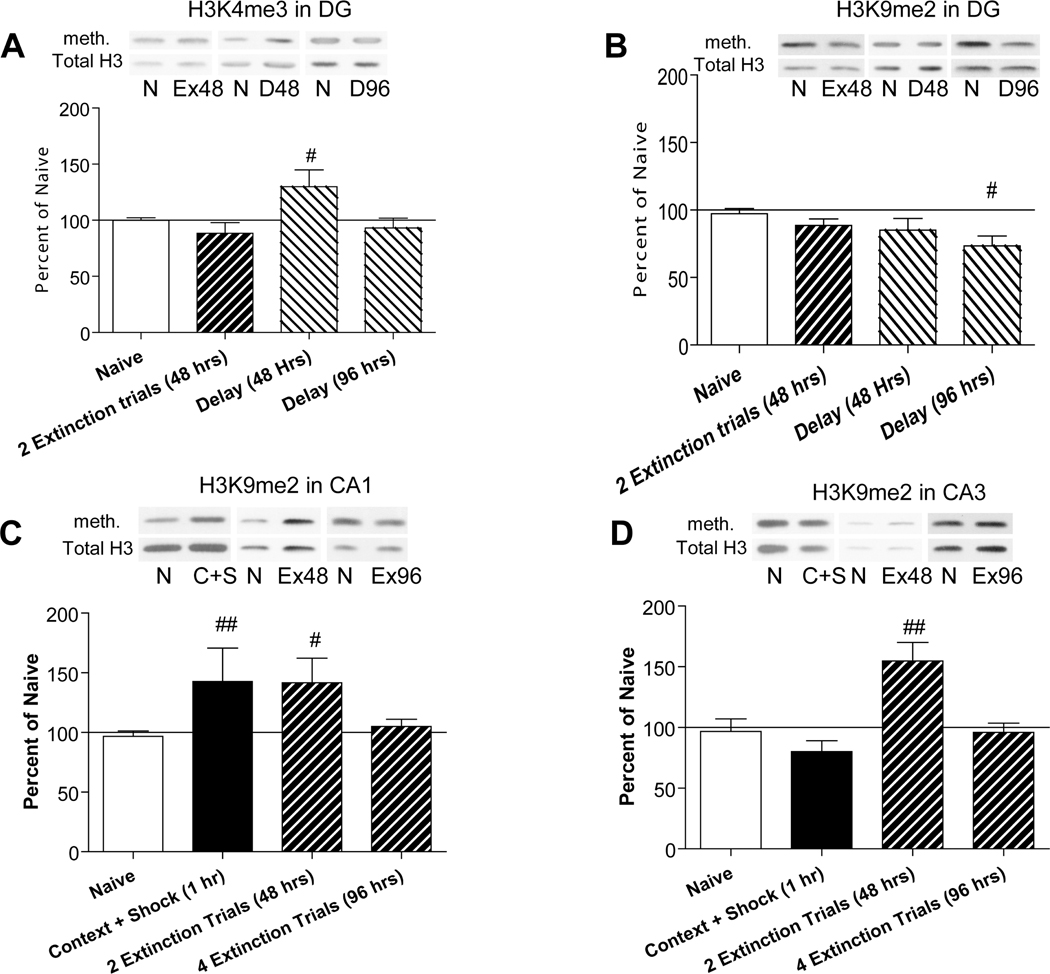
Figure 5. Histone methylation in hippocampal subfields following fear memory extinction.
Extinction animals underwent contextual fear conditioning (0 hours) and were re-exposed to the context for five minutes every 24 hours. The extinction animals were exposed to the context and sacrificed as either a partial (48 hours) or full extinction (96 hours) group. Animals in the Delay group were fear conditioned then handled every 24 hours without re-exposure to the context. At the 48 or 96 hour time points, these animals were sacrificed. Histone methylation was assessed at, 48 and 96 hours for both the delay (D) and extinction (Ex) groups. A) A significant increase in H3K4me3 for delay animals, compared to naïve controls, was observed at 48 h (#p<0.05) in the Dentate Gyrus (DG) region. No changes in H3K4me3 at 48 or 96 h were observed in other brain regions. B) An initial ANOVA of condition×region failed to detect any changes in H3K9me2 at 48 or 96 h. However, a subsequent post-hoc t-test revealed a significant difference between naïve and delay animals at 96 hours (#p<0.05). C) H3K9me2 levels were elevated in area CA1 at one hour (as reported above). Following 2 extinction trials, H3K9me2 levels remained significantly elevated in area CA1 (##p<0.005) but returned to baseline by 96 hours. D) H3K9me2 was also elevated in area CA3 following 2 extinction trials (##p<0.005). This modification was not present either at 1 or 96 hours. No effects of context were observed for H3K9me2 in any region at 48 or 96 hours (data not shown), indicating this is not simply an effect of context re-exposure. Instead, the upregulation of H3K9me2 appears to persist as long as the freezing behavior is present. Once freezing behavior has been extinguished the change in methylation was also been extinguished. Solid line represents normalized Naïve control levels. Error bars are SEM. N, Naïve controls; C+S, Contextual fear conditioning; D, Delay; Ex, Extinction; meth, methylation.
Acknowledgements
The author would like to thank J. David Sweatt and Caterina M. Hernandez for helpful discussions. Funding was provided through NIH (MH82106, MH57014, AG013722, and NS057098) and the Evelyn F. McKnight Brain Research foundation.
References
- Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8(1):57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarian S, Huang H. Epigenetic regulation in human brain-focus on histone lysine methylation. Biol Psychiatry. 2009;65:198–203. doi: 10.1016/j.biopsych.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131(4):633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Bártová E, Krejcí J, Harnicarová A, Galiová G, Kozubek S. Histone modifications and nuclear architecture: a review. J Histochem Cytochem. 2008;56(8):711–721. doi: 10.1369/jhc.2008.951251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68(2):270–281. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14(11):1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- Biliya S, Lee A, Bulla J. Genomic imprinting: the influence of differential methylation in the two sexes. Exp. Biol. Med. 2010;235:139–147. doi: 10.1258/ebm.2009.009251. [DOI] [PubMed] [Google Scholar]
- Blanchard R, Blanchard D. Crouching as an index of fear. J Comp Physiol Psychol. 1969;67:370–375. doi: 10.1037/h0026779. [DOI] [PubMed] [Google Scholar]
- Bozon B, Kelly A, Josselyn SA, Silva AJ, Davis S, Larouche S. MAPK, CREB and zif268 are all required for the consolidation of recognition memory. Philos Trans R Soc Lond, B, Biol Sci. 2003;358(1432):805–814. doi: 10.1098/rstb.2002.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16(8):4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Barad M. The histone deacetylase inhibitor valproic acid enhances acquisition, extinction, and reconsolidation of conditioned fear. Learn Mem. 2008;15(1):39–45. doi: 10.1101/lm.801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buiting K. Prader-Willi syndrome and Angelman syndrome. Am J Med Genet C Semin Med Genetics. 2010;154C(3):365–376. doi: 10.1002/ajmg.c.30273. [DOI] [PubMed] [Google Scholar]
- Burgess-Beusse B, Farrell C, Gaszner M, Litt M, Mutskov V, Recillas-Targa F, et al. The insulation of genes from external enhancers and silencing chromatin. Proc Natl Acad Sci USA. 2002;99 Suppl 4:16433–16437. doi: 10.1073/pnas.162342499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56(8):422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Chuang DM, Leng Y, Marinova Z, Kim HJ, Chiu CT. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 2009;32(11):591–601. doi: 10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppedè F, Migliore L. Evidence linking genetics, environment, and epigenetics to impaired DNA repair in Alzheimer's disease. J Alzheimers Dis. 2010;20(4):953–966. doi: 10.3233/JAD-2010-1415. [DOI] [PubMed] [Google Scholar]
- De Bellis MD. The psychobiology of neglect. Child Maltreatment. 2005;10:150–172. doi: 10.1177/1077559505275116. [DOI] [PubMed] [Google Scholar]
- de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370(Pt 3):737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanselow MS. Contextual fear, gestalt memories, and the hippocampus. Behav Brain Res. 2000;110(1–2):73–81. doi: 10.1016/s0166-4328(99)00186-2. [DOI] [PubMed] [Google Scholar]
- Farkas D. DNA simplified: the hitchhiker's guide to DNA. Washington, D.C: AACC Press; 1996. [Google Scholar]
- Freudenthal R, Romano A. Participation of Rel/NF-kappaB transcription factors in long-term memory in the crab Chasmagnathus. Brain Res. 2000;855(2):274–281. doi: 10.1016/s0006-8993(99)02358-6. [DOI] [PubMed] [Google Scholar]
- García-Pérez M, Pinto M, Subirana JA. Nonsequence-specific arginine interactions in the nucleosome core particle. Biopolymers. 2003;69(4):432–439. doi: 10.1002/bip.10389. [DOI] [PubMed] [Google Scholar]
- Gonzales ML, LaSalle JM. The role of MeCP2 in brain development and neurodevelopmental disorders. Current Psychiatry Reports. 2010;12(2):127–134. doi: 10.1007/s11920-010-0097-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, et al. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111(4):483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, et al. Histone methylation regulates memory formation. J Neurosci. 2010;30(10):3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake SB, Xiao A, Allis CD. Linking the epigenetic 'language' of covalent histone modifications to cancer. Br J Cancer. 2004;90(4):761–769. doi: 10.1038/sj.bjc.6601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, Peterson CL. Molecular biology. Chromatin higher order folding--wrapping up transcription. Science. 2002;297(5588):1824–1827. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS ONE. 2007;2(8):e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449(7158):105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- Huh KH, Wenthold RJ. Turnover analysis of glutamate receptors identifies a rapidly degraded pool of the N-methyl-D-aspartate receptor subunit, NR1, in cultured cerebellar granule cells. J Biol Chem. 1999;274(1):151–157. doi: 10.1074/jbc.274.1.151. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 Suppl:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42(6):961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48(2):303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116(Pt 11):2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Landgren H, Curtis MA. Locating and labeling neural stem cells in the brain. J. Cell Physiology. 2010 doi: 10.1002/jcp.22319. in press. [DOI] [PubMed] [Google Scholar]
- Law M. Advanced imaging techniques in brain tumors. Cancer Imaging. 2009;9(A):S4–S9. doi: 10.1102/1470-7330.2009.9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M, Boshoff C, Beck S. Cancer epigenome. Adv Genet. 2010;70:247–276. doi: 10.1016/B978-0-12-380866-0.60009-5. [DOI] [PubMed] [Google Scholar]
- Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279(39):40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6(2):108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366(6453):362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Maeda N, Hayashizaki Y. Genome-wide survey of imprinted genes. Cytogenet Genome Res. 2006;113(1–4):144–152. doi: 10.1159/000090826. [DOI] [PubMed] [Google Scholar]
- Manolio TA. Genomewide association studies and assessment of the risk of disease. New England Journal of Medicine. 2010;363(2):166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- McIlwain KL, Merriweather MY, Yuva-Paylor LA, Paylor R. The use of behavioral test batteries: effects of training history. Physiol Behav. 2001;73(5):705–717. doi: 10.1016/s0031-9384(01)00528-5. [DOI] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53(6):857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Molfese DL, Sweatt JD, Lubin FD. Histone methylation as an epigenetic marker of memory extinction. (in revision). [Google Scholar]
- Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annual Review of Biochemistry. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292(5514):110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nicholson JM, Wood CM, Reynolds CD, Brown A, Lambert SJ, Chantalat L, et al. Histone structures: targets for modifications by molecular assemblies. Ann N Y Acad Sci. 2004;1030:644–655. doi: 10.1196/annals.1329.075. [DOI] [PubMed] [Google Scholar]
- Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20(4):214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Paulsen M, Ferguson-Smith AC. DNA methylation in genomic imporinting, development, and disease. J. Pathol. 2001;195(1):97–110. doi: 10.1002/path.890. [DOI] [PubMed] [Google Scholar]
- Paylor R, Tracy R, Wehner J, Rudy JW. DBA/2 and C57BL/6 mice differ in contextual fear but not auditory fear conditioning. Behav Neurosci. 1994;108(4):810–817. doi: 10.1037//0735-7044.108.4.810. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14(14):R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Quinlan EM, Philpot BD, Huganir RL, Bear MF. Rapid, experience-dependent expression of synaptic NMDA receptors in visual cortex in vivo. Nat Neurosci. 1999;2(4):352–357. doi: 10.1038/7263. [DOI] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Renthal W, Carle TL, Maze I, Covington HE, Truong HT, Alibhai I, et al. Delta FosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. J Neurosci. 2008;28(29):7344–7349. doi: 10.1523/JNEUROSCI.1043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, Xiao G, Kumar A, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56(3):517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Rosenberg G. The mechanisms of action of valproate in neuropsychiatric disorders: can we see the forest for the trees? Cell Mol Life Sci. 2007;64(16):2090–2103. doi: 10.1007/s00018-007-7079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting Epigenetic Influence of Early-Life Adversity on the BDNF Gene. Biol Psychiatry. 2009 doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy JW, Huff NC, Matus-Amat P. Understanding contextual fear conditioning: insights from a two-process model. Neuroscience and biobehavioral reviews. 2004;28(7):675–685. doi: 10.1016/j.neubiorev.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G. Two waves of nuclear factor kappaB recruitment to target promoters. J Exp Med. 2001;193(12):1351–1359. doi: 10.1084/jem.193.12.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MM. The impact of early adverse care of HPA axis development: Nonhuman primate models. Hormones and Behavior. 2006;50:623–631. doi: 10.1016/j.yhbeh.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Tanner KG, Langer MR, Denu JM. Kinetic mechanism of human histone acetyltransferase P/CAF. Biochemistry. 2000;39(39):11961–11969. doi: 10.1021/bi001272h. [DOI] [PubMed] [Google Scholar]
- Torres-Muñoz JE, Van Waveren C, Keegan MG, Bookman RJ, Petito CK. Gene expression profiles in microdissected neurons from human hippocampal subregions. Brain Res Mol Brain Res. 2004;127(1–2):105–114. doi: 10.1016/j.molbrainres.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9(4):519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27(23):6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar-Garea A, Esteller M. Histone deacetylase inhibitors: understanding a new wave of anticancer agents. Int J Cancer. 2004;112(2):171–178. doi: 10.1002/ijc.20372. [DOI] [PubMed] [Google Scholar]
- Wheal HV, Chen Y, Mitchell J, Schachner M, Maerz W, Wieland H, et al. Molecular mechanisms that underlie structural and functional changes at the postsynaptic membrane during synaptic plasticity. Prog Neurobiol. 1998;55(6):611–640. doi: 10.1016/s0301-0082(98)00026-4. [DOI] [PubMed] [Google Scholar]
- Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Oakey RJ. Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet. 2006;2(11):e147. doi: 10.1371/journal.pgen.0020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Hawk JD, Abel T. Combinatorial chromatin modifications and memory storage: a code for memory? Learn Mem. 2006;13(3):241–244. doi: 10.1101/lm.278206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh SH, Lin CH, Gean PW. Acetylation of nuclear factor-kappaB in rat amygdala improves long-term but not short-term retention of fear memory. Mol Pharmacol. 2004;65(5):1286–1292. doi: 10.1124/mol.65.5.1286. [DOI] [PubMed] [Google Scholar]