Abstract
Clear and unambiguous associations have been established between therapeutic exposures and specific complications. However, considerable inter-individual variability is observed in the risk of developing an outcome for a given therapeutic exposure. Genetic predisposition and especially its interaction with therapeutic exposures can potentially exacerbate the toxic effect of treatment on normal tissues and organ systems, and can possibly explain the inter-individual variability. This article provides a brief overview of the current knowledge regarding the role of genomic variation in the development of therapy-related complications. Relatively common outcomes with strong associations with therapeutic exposures, including cardiomyopathy, obesity, osteonecrosis, ototoxicity, and subsequent malignancies are discussed here. In order to develop a deeper understanding of the molecular underpinnings of therapy-related complications, comprehensive and near-complete collection of clinically-annotated samples is critical. Methodological issues such as study design, definition of the endpoints or phenotypes, identification of appropriate and adequately sized study population together with a reliable plan for collecting and maintaining high quality DNA, and selection of an appropriate approach or platform for genotyping are also discussed. Understanding the etiopathogenetic pathways that lead to the morbidity is critical to developing targeted prevention and intervention strategies, optimizing risk-based health care of cancer survivors, thus minimizing chronic morbidities and improving quality of life.
The 5-year overall survival rate for all invasive cancer types has increased from 50% in 1975–1977 to 67% in 1999–2005 and exceeds 80% for many of the common cancer types, including breast, prostate, testicular, thyroid, bladder, endometrial cancer, melanoma, and Hodgkin lymphoma (HL) (1). As a result of this improvement in survival, the number of cancer survivors has quadrupled in the last 4 decades and now exceeds 12 million, of which 8 million have survived 5 or more years. This population is growing at a rate of almost 2% per year (2).
The increasing number of long-term survivors is attended by a growing awareness that many will develop health conditions as a direct or an indirect consequence of their cancer therapy (3–6). While some of these conditions occur during therapy and persist well after the therapy has been completed (such as steroid-induced osteonecrosis, or alkylator/radiation-induced hypogonadism), many outcomes are not evident until 10–20 years later, such as subsequent malignant neoplasms (SMNs) and anthracycline-related late onset congestive heart failure (CHF).
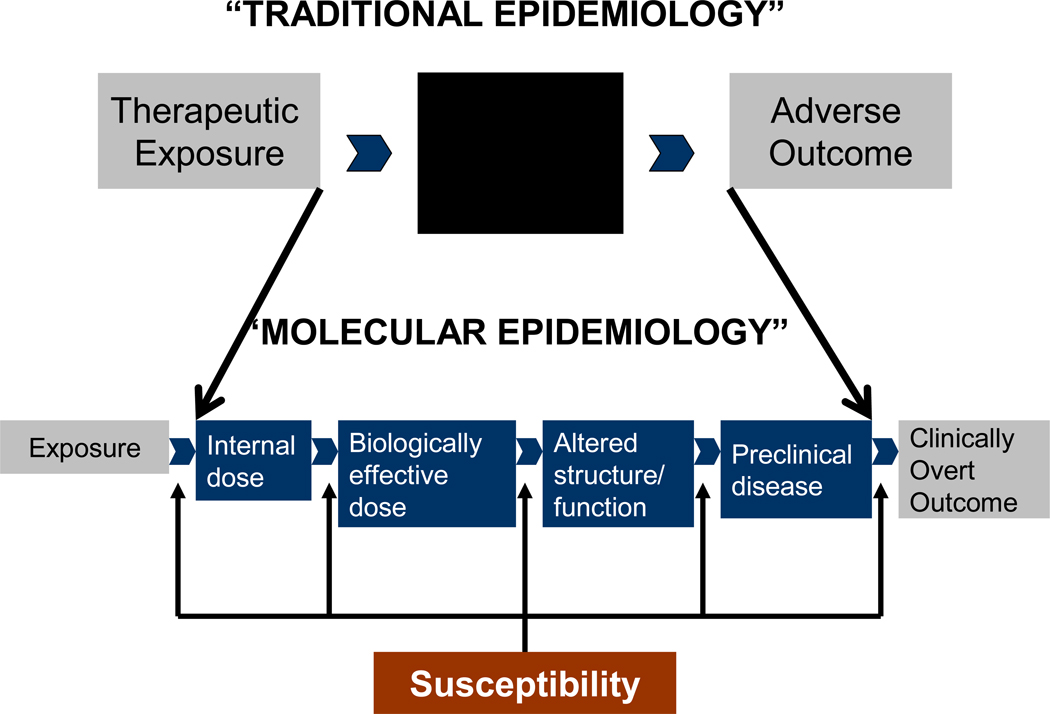
Individuals exposed to radiation and chemotherapy are vulnerable to long-lasting organ toxicity; the very young because their organs are developing and the elderly because of organ senescence. In addition, genetic predisposition and its interaction with therapeutic exposures can potentially exacerbate the toxic effect of treatment on normal tissues and organ systems. Thus, it becomes imperative to understand the individual variability in: i) the internal dose of the therapeutic agent; ii) the biologically effective dose; iii) the alterations in structure or function of the tissue or organ; and iv) the consequent development of preclinical disease, (Figure 1) in order to understand the pathogenesis of therapy-related complications, and also develop a better idea of the individual susceptibility.
Figure 1.
Proposed mechanisms to understand etiology and pathogenesis of therapy-related adverse outcomes
The sections below describe a brief overview of the current knowledge regarding the role of genomic variation in the development of therapy-related complications. The focus is on outcomes that are relatively common, have clearly established associations with therapeutic exposures, and are associated with significant long-term morbidity and hence carry the potential to negatively impact the quality of life of the survivors. These outcomes include cardiomyopathy, obesity, osteonecrosis, ototoxicity, and SMNs. An attempt has been made to present scientifically robust studies that have utilized a variety of methodologies. Thus, the studies presented here included: i) GWAS studies utilizing large populations with successful validation of the findings in independent cohorts or extension of the findings with some functional data; ii) candidate gene studies utilizing large carefully and comprehensively curated sets of genes that were biologically plausible; and iii) single gene studies, where there was ample pre-clinical (in vitro and/or in vivo) data that provided a compelling rationale for examination of the association in a single gene setting. The fact that there are not enough scientifically or methodologically robust studies highlights the issue that there are methodological challenges to conducting such studies, and these are detailed here. These challenges notwithstanding, understanding the etiopathogenetic pathways that lead to long-term morbidity is critical to developing targeted prevention and intervention strategies, optimizing risk-based health care of cancer survivors, and improving quality of life.
Methodological Issues
In order to develop a deeper understanding of the molecular underpinnings of therapy-related complications, careful attention needs to be devoted to developing the appropriate study design with an appropriate and adequately sized study population and a precise definition of endpoints or phenotypes and a reliable plan for collecting high-quality DNA. It is critical to have a comprehensive and near-complete collection of clinically-annotated samples. Ideally, this should be in the form of blood, collected to allow subsequent extraction of DNA and RNA, as well as to establish lymphoblastoid cell lines. The study design, study question, and the available sample size, should help selection of an appropriate approach or platform for genotyping.
If the intent is to analyze a single endpoint, a case-control study design is most efficient; however a cohort design is more appropriate for the study of complex or multiple phenotypes. Furthermore, consideration should be given to issues related to survival bias when designing prevalent case-control studies, especially where the endpoint is associated with high early lethality. Study design must include rigorous power estimations to determine the number of subjects necessary to meet statistical objectives. Another efficient and cost-effective methodology is the use of a nested case-control study design, especially when the samples have been banked on the entire cohort and a comprehensive longitudinal follow-up of the cohort has resulted in a near-complete ascertainment of the outcome of interest. Finally, use of appropriate comparison groups is critical. Previous studies have either used healthy individuals or used individuals with histologically identical de novo cancer as comparison groups (e.g., de novo AML as a reference group for therapy-related myelodysplasia/ acute myeloid leukemia [t-MDS/AML]). This strategy could be problematic because of the possibility of shared genotoxic insults (e.g., benzene), driving the association towards null. The ideal comparison group should consist of individuals identical to the cases with respect to primary cancer but who do not develop the outcome of interest. It is also important to ensure that the controls have been followed for at least as long as the cases from the time of diagnosis and preferably for a longer duration. This is done to ensure that the “controls” have had ample opportunity to develop the outcome of interest. Ideally, the outcome of interest should be clinically validated in order to avoid misclassification. It is also important that the controls be subjected to similar validation as the cases to confirm the absence of disease.
There are two approaches to the study of genetic variation in disease: (1) candidate gene studies based on the selection of a limited number of biologically relevant genes and pathways; and (2) genome-wide association studies (GWAS) using DNA arrays capable of detecting a million or more SNPs. The candidate gene approach is guided by a specific hypothesis whereas a genome-wide approach is necessary for comprehensive discovery analysis. Both strategies have their own strengths and limitations. A GWAS approach offers the ability to study complex pathways, allowing for an assessment of the action/ interaction of many genes; importantly, it allows for new genes to be identified. It has gained significant favor, because of the fact that several studies that have utilized a candidate gene approach have failed replication. However, a GWAS approach requires a large sample size, in order to account for false discovery, and is accordingly an expensive endeavor. In addition, there is the need for a replication cohort so that the genes identified in the discovery set can be validated in the test set. The GWAS approach does not have an a priori hypothesis, and is considered to be hypothesis-generating and more suited for complex disorders where a clear etiologic lead is not established. However, this is not true for outcomes observed in survivors, where for the most part there is a clearly established etiologic association between the exposure and outcome (e.g., radiation and SMNs; anthracyclines and CHF). In such a case, an argument could be made for a comprehensively selected (and biologically plausible) list of genes identified along the path of the action of therapeutic exposure on the target organ. The need for a large sample size due to issues related to multiple testing is less of an issue with a candidate gene approach if limited to a few genetic variants, and the cost is accordingly less than that of a GWAS approach. Finally, while gene-gene interactions would require prohibitively large samples in a GWAS setting, they are logistically feasible when conducting a candidate gene study.
Anthracycline-related Cardiomyopathy
Anthracyclines are among the most widely-used chemotherapeutic agents (7). However, cardiomyopathy is a dose-limiting complication. The incidence of congestive heart failure (CHF) is less than 10% in patients exposed to cumulative dose of anthracycline of less than 500 mg/m2, and approaches 36% for doses exceeding 600mg/m2 (8). The risk of anthracycline-related cardiomyopathy is higher among those exposed at a younger age (<5 years), among girls, among those with pre-existing heart disease, and among those who received chest irradiation (8, 9). Extended follow-up of childhood cancer survivors has made it clear that lower cumulative doses of anthracyclines may place children at risk for cardiac compromise; exposure to 250 mg/m2 increased the relative hazard of CHF by 5-fold when compared to children with cancer who were not exposed to anthracyclines (10). CHF is associated with a poor prognosis; 5-year overall survival rates are reported to be less than 50% (11). Utilization of non-invasive cardiac imaging has identified a growing population of survivors with asymptomatic left ventricular dysfunction who may be at risk for late CHF (12, 13).
Pathogenesis of anthracycline-related cardiomyopathy
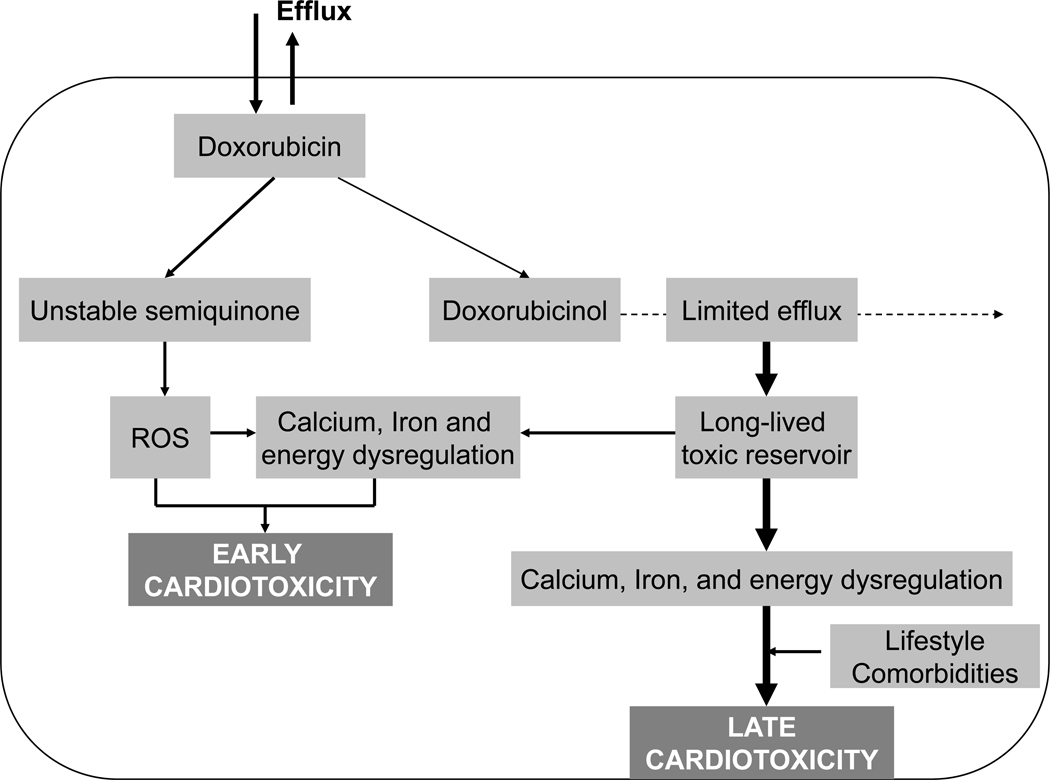
Anthracyclines cause direct myocardial injury due to free radical formation. Myocardial injury results in thinning of the left ventricular wall; increased myocardial stress; and decreased contractility (14). A one- or two-electron reductive activation of anthracyclines leads to cardiotoxicity. A one-electron reduction of the quinone moiety of doxorubicin results in formation of a semiquinone free radical, which in turn regenerates its parent quinone by reducing molecular oxygen to superoxide anion and hydrogen peroxide – members of the family of reactive oxygen species (ROS). The ROS then cause oxidative stress and energy depletion in cardiomyocytes. A two-electron reduction of the side chain carbonyl moiety converts anthracyclines to secondary alcohol metabolites that dysregulate calcium and iron homeostasis. Oxidative stress and ion dysregulation eventually combine to induce cardiomyopathy (Figure 2) (15).
Figure 2.
Proposed mechanisms of anthracycline-induced cardiotoxicity
Studies using transgenic and knockout mice demonstrated altered sensitivity to anthracyclines, suggesting a role for genetic susceptibility in the development of anthracycline-induced CHF in humans. Thus, transgenic overexpression of the multiple drug resistance gene (MDR1) protects the heart from doxorubicin-induced toxicity (16), consistent with the transport activity of MDR1 for anthracyclines. Furthermore, a deficiency (17) and overexpression (18) of the doxorubicin-metabolizing enzyme carbonyl reductase in mice protects and accentuates (respectively) anthracycline-induced CHF. However, a limitation of the clinical application of studies using genetically engineered mouse strains is the strong penetrance of the introduced mutations, which do not necessarily mimic the more subtle genetic variability in the humans.
Nonetheless, it is increasingly evident that the conventionally described clinical and therapeutic risk factors may not fully explain the wide inter-individual variability in susceptibility to anthracycline-related cardiomyopathy. While cardiotoxicity has been reported at cumulative exposure of <250–300 mg/m2 in some patients (19, 20), doses exceeding 1000 mg/m2 have been tolerated well by others (21).
Investigators have begun examining the role of genetic susceptibility in the development of therapy-related CHF (Table 1). Using a candidate gene approach, studies have identified genetic polymorphisms involved in the metabolism of anthracyclines, the myocardial response to the drug, as well as others thought to play a role in susceptibility to de novo disease, which could place survivors at increased risk for therapy-related CHF (22). A recent report from the Childhood Cancer Survivor Study (CCSS) (23) demonstrated a trend towards an association between a polymorphism in carboxyl reductase 3, CBR3 V244M and risk of CHF (OR=8.16, P=0.056 for G/G vs. A/A; OR=5.44, P=0.09 for G/A vs. A/A), suggesting that functional CBR3 V244M polymorphism may impact the risk of anthracycline-related CHF by modulating the intracardiac formation of cardiotoxic anthracycline alcohol metabolites. Wojnowski et al (24) selected single nucleotide polymorphisms (SNPs) from 82 genes with biological relevance to anthracycline-related cardiotoxicity. CHF was associated with a variant of the NAD(P)H oxidase subunit NCF4 (rs1883112; OR=2.5, 95% CI, 1–3.9). In agreement with these findings, mice deficient in NAD(P)H oxidase activity were resistant to chronic doxorubicin treatment. Thus, this report suggests that genetic variants in free radical metabolism may modulate the individual risk to develop anthracycline-related cardiotoxicity. However, these studies utilized small samples (30 and 54 cases respectively), and the findings need to be validated in larger independent replication cohorts.
Table 1.
Role of Genetic Susceptibility in the Development of treatment-related adverse Events
| STUDY | GWAS VS. CANDIDATE GENE |
STUDY DESIGN |
REPLICATION STUDY |
SAMPLE SIZE | RESULTS |
|---|---|---|---|---|---|
| Anthracycline-related cardiomyopathy | |||||
| Wojnowski et al, 2005 (24) | Candidate gene (82 genes with conceivable relevance to anthracycline-related cardiomyopathy) | Prospective cohort study | No Instead, follow-up with mice studies |
1697 patients (54 with chronic anthracycline-related cardiomyopathy) | Anthracycline-related cardiomyopathy was associated with a variant of the NAD(P)H oxidase subunit NCF4 (rs1883112) (OR=2.5, 95%CI, 1.3–5.0). Mice deficient in NAD(P)H oxidase activity were resistant to chronic doxorubicin exposure, unlike the wild-type mice. |
| Blanco et al, 2008 (23) | Candidate gene (CBR3, NQO1) | Nested case-control study | No Instead, follow-up with functional studies |
30 cases with CHF; 115 matched cancer survivors with no CHF | A trend toward an association between the CBR V244M polymorphism and risk of CHF (OR=8.16, p=0.056 for G/G vs. A/A; OR=5.44, p=0.09 for GA vs. AA). Recombinant CBR3 V244 (G allele) synthesized 2.6-fold more cardiotoxic doxorubicinol per unit of time than CBR3 M244 (A Allele; CBR V244, p=0.01) |
| Osteonecrosis | |||||
| Kawedia et al, 2011 (52) | GWAS | Prospective cohort study | No | 364 patients (69 with symptomatic osteonecrosis) | Polymorphisms of ACP1 (e.g., rs12714403: OR=5.6, 95%CI, 2.7–11.3) were associated with symptomatic osteonecrosis; ACP1 regulates lipid levels and osteoblast differentiation. |
| Relling, et al, 2004 (32) | Candidate gene (16 polymorphisms in genes likely to play a role in pharmacogenetics/ pharmacokinetics of antileukemic therapy | Prospective cohort study | No | 64 (25 patients with osteonecrosis) | Vitamin D receptor Fokl start site CC genotype (OR=4.5, p=0.045) and thymidylate synthase low activity 2/2 enhancer repeat genotype (OR=7.4, p=0.049) |
| French et al, 2008 (47) | Candidate gene (TYMS, MTHFR, ABCB1, BLGAP, ACP5, LRP5, ESR1, PAI-1, VDR, PTH, PTHR – 12 polymorphisms – chosen based on putative mechanisms underlying osteonecrosis risk | Prospective cohort study | No | 361 patients (51 with osteonecrosis) | PAI-1 polymorphism (rs6092) was associated with risk of osteonecrosis (OR=2.89, p=0.002). PAI-1 polymorphisms and PAI-1 serum levels have been associated with thrombosis. |
| Obesity | |||||
| Ross et al, 2004 (84) | Candidate gene Polymorphism in leptin receptor (LEPR) gene, Gln223Arg | Retrospective cohort study | No | 600 patients (278 with BMI ≥25 Kg/m2) | Female patients with BMI > 25 Kg/m2 were more likely Srg homozygous than those with BMI less than 25 Kg/m2 (24% vs. 12%, p=0.007). This difference was not observed in males. Among females treated with >20 Gy cranial irradiation, Arg/Arg individuals had six times higher odds of having BMI >25 Kg/m2 (95%CI, 2.1–22) than those with Gln allele (p=0.04 for interaction). Thus, LEPR polymorphism may influence obesity in female survivors of childhood ALL, particularly those exposed to cranial irradiation. |
| Ototoxicity | |||||
| Ross et al, 2009 (70) | Candidate gene 220 drug metabolism genes (1,949 polymorphisms) | Case-control | Yes | Discovery set: 33 cases; 20 controls Replication set: 73 cases; 36 controls Controls treated with cisplatinum but with no ototoxicity |
Genetic variants in TPMT (rs12201199) – OR=17.0, 95%CI, 2.3–125.9 and COMT (rs9332377) – OR=5.5, 95%CI, 1.9–15.9 were associated with cisplatinum-induced hearing loss in children |
| Oldenburg et al, 2007 (67) | Candidate gene Known functional polymorphisms in GSTT1 and GSTM1 and codon 105 A/G (Ile/Val) in GSTP1 | Prospective cohort study | No | 173 patients 89 with hearing impairment | Risk of inferior audiometric result was higher in testicular cancer survivors with 105Ile/105Ile-GSTP1 or 105Val/105Ile-GSTP1 compared with 105Val-GSTP1 (OR=4.21, 95%CI, 1.99–8.88). GSTM1 positivity increased risk of hearing loss. Two combined genotypes were associated with hearing ability. Presence of pattern 1 (GSTT1 positive, GSTM1 positive, and 105Ile/105Ile GSTP1) was associated with hearing impairment (OR=2.76, 95%CI, 1.35–5.64). Presence of pattern 2 (GSTT1 positive, GSTM1 positive, and 105Val/105Val-GSTP1) resulted in better hearing ability (OR=5.35, 95%CI, 2.25–12.76) |
| Riedemann et al, 2008 (69) | Candidate gene Polymorphisms in megalin gene (rs2075252 and rs4668123) Megalin is a member of the low-density lipoprotein receptor family, and is highly expressed in the marginal cells of the stria vascularis of the inner ear – resulting in high accumulation of platinum-DNA adducts. | Case control | No | 25 cases; 25 controls (cancer patients exposed to cisplatin with no hearing loss) | An association was found between the A allele of rs2075252 and hearing impairment (OR=3.45, 95%CI, 1.11–11.2), indicating that SNPs at the megalin gene may impact the individual susceptibility against cisplatin-induced toxicity. |
| Therapy-related Leukemia | |||||
| Knight et al., 2009 (174) | GWAS | Case-control (healthy controls) | Yes | Discovery set: 80 cases; 150 controls Replication set: 70 cases; 95 controls | Among patients with acquired abnormalities of chromosomes 5 or 7; 3 SNPs (rs1394384 [OR=0.29, 95% CI, 0.15–0.56], rs1381392 [OR=2.08, 95%CI, 1.29–3.35], and rs1199098 [OR=0.46, 95% CI, 0.27–0.79]) were associated with t-MDS/AML; rs1394384 is intronic to ACCN1, a gene encoding an amiloride-sensitive cation channel that is a member of the degenerin/epithelial sodium channel; rs1199098 is in LD with IPMK, which encodes a multikinase that positively regulates the prosurvival AKT kinase and may modulate Wnt/beta-catenin signaling; rs1381392 is not near any any known genes, miRNAs, or regulatory elements, although it lies in a region recurrently deleted in lung cancer. |
| Ellis et al, 2008 (173) | Candidate gene (2 common functional p53-pathway variants, the MDM2 SNP309 and the tP53 codon 72 | Case-control (healthy controls) | yes | Discovery set: 80 cases Replication set: 91 cases | Neither polymorphism alone influenced the risk of t-MDS/AML; however an interactive effect was detected such that MDM2 TT TP53 Arg/Arg double homozygotes, and individuals carrying both a MDM2 G allele and a TP53 Pro allele, were at increased risk of t-MDS/AML (OR=2.04, 95%CI, 1.20–3.48, Pinteraction=0.009) |
| Allan et al, 2001 (141) | Candidate gene approach Polymorphisms in GSTM1, GSTT1, GSTP1 | Case control | No | 89 cases; 420 patients with de novo AML; 1022 healthy controls | Individuals with at least one GSTP1 codon 105 Val allele were significantly over-represented in t-AML cases compared with de novo AML cases (OR=1.81, 95%CI, 1.11–2.94). Also, relative to do novo AML, the GSTP1 codon 105 allele occurred more often among t-AML patients with prior exposure to chemotherapy (OR=2.66, 95%CI, 1.39–5.09), particularly among those with prior exposure to known GSTP1 substrates (OR=4.34, 95%CI, 1.43–13.20) and not among t-AML patients with exposure to radiation alone. |
| Worrillow et al, 2003 (134) | Candidate gene hMSH2 –6exon 13 polymorphism Evaluation of MSI | Case control | No Verification performed by direct sequencing |
91 cases; 420 patients with de novo AML, 837 healthy controls | The variant (C) hMSH2 allele was significantly overrepresented in t-AML cases that had previously been treated with O6-guanine alkylating agents, including cyclophosphamide and procarbazine, compared with controls (OR=4.02, 95%CI 1.40–11.37); 38% of the patients were MSI positive; hMSH2 -6 exon 13 (C) allele confers a nondisabling DNA mismatch repair defect and predisposes to development of t-AML via induction of DNA mismatch repair mutations and high grade MSI. |
| Worrillow et al, 2008 (137) | Candidate gene Polymorphism of MLH1 (position -93, rs1800734) | Case control | No | 133 cases 420 patients with de novo AML, 242 patients with primary HL, 1177 healthy controls | Carrier frequency of MLH1 -93 variant was higher in patients who developed t-AML or secondary breast cancer after alkylating agents exposure for HL, compared to patients without alkylating agent exposure. The MLH1 -93 variant allele was also over-represented in t-AML cases when compared to de novo AML cases and was associated with increased risk of t-AML (OR=5.31, 95%CI, 1.40–20.15) among patients exposed to alkylating agents |
| Seedhouse et al, 2002 (166) | Candidate gene Polymorphisms in XRCC1, XRCC3, XPD, NQO1 | Case control | No | 34 cases; 134 patients with de novo AML; 178 healthy controls | Presence of at least one XRCC1 399Gln allele indicated a protective effect for the allele in controls compared with patients with t-AML (OR=0.44, 95%CI, 0.20–0.93) |
| Jawad et al, 2006 (154) | Candidate gene C/T-3’ untranslated region (UTR) polymorphism in HLX; Polymorphism in RAD51 (135G/C-5’ UTR) | Case control | No | 42 cases; 166 patients with de novo AML; 189 healthy controls | Presence of the variant HLX1 allele significantly increased the risk of t-AML (OR=3.36, 95%CI, 1.65–6.84). Polymorphism in RAD51 (135G/C-5’ UTR) also increased the risk of t-AML. Combined analysis revealed, a synergistic 9.5-fold increase (95%CI, 2.22–40.64) in risk for t-AML |
| Subsequent Solid Malignancies | |||||
| Best et al, 2011 (175) | GWAS | Case-control (controls: cancer survivors with no SMNs) | Yes | Discovery set: 100 cases; 89 controls Replication set: 62 cases; 71 controls | Two variants at chromosome 6q21 (rs4946728 [OR=11.4, 95%CI, 3.23–40.25]; rs1040411 [OR=6.57, 95%CI, 3.19–13.52]) were associated with SMNs in childhood Hodgkin lymphoma (HL) survivors, but not in adult-onset HL survivors. The variants comprise a risk locus associated with decreased basal expression of PRDM1 and impaired induction of PRDM1 protein after radiation exposure. PRDM1 encodes a zinc finger transcriptional repressor involved in cellular processes such as proliferation, differentiation, and apoptosis. |
| Mertens et al, 2004 (136) | Candidate gene Polymorphisms in GSTM1, GSTT1, XRCC1 | Cohort study | No | 650 patients with HL (178 with subsequent malignancy) | Individuals lacking GSTM1 were at increased risk of any subsequent malignancy (OR=1.5, 95%CI, 1.0–2.3). A non-significant increased risk for thyroid cancer was observed in individuals lacking either GSTM1 (OR, 2.9, 95%CI, 0.8–10.9) or GSTT1 (OR, 3.7, 95%CI, 0.6–23.5). Individuals with the genotype of the Arginine/glutamine polymorphism at codon 399 in the XRCC1 gene (R399)r showed a nonsignificant increased risk of breast cancer (OR, 1.4, 95%Ci, 0.7–2.7). |
Osteonecrosis
Osteonecrosis is the death of bone that results in collapse of the bone architecture, leading to progressive joint damage, pain, limited range of motion, articular collapse, arthritis, and loss of function (25, 26). Weight-bearing joints are commonly affected, and the destruction is often severe enough to require total joint replacement surgery for functional rehabilitation and symptomatic relief. Osteonecrosis accounts for 10% of the 500,000 total joint replacement procedures performed annually in the US (25).
Osteonecrosis is a well-recognized complication of corticosteroid use (25, 27–29). Corticosteroids are integral to the management of acute lymphoblastic leukemia (ALL), NHL, and Hodgkin lymphoma (HL). Mattano et al described the incidence of symptomatic osteonecrosis and associated risk factors in children with ALL treated with intensive chemotherapy including multiple, prolonged courses of corticosteroids (28). The cumulative incidence of osteonecrosis was 9.3%±0.9% at 3 years. The incidence was higher among older children (≥10 years: 14.2%±1.3% vs. <10 years: 0.9%±0.4%, p<0.001). Among 10 to 20 year-olds, the incidence was higher for females (17.4%±2.1% vs. 11.7%±1.6%, p=0.03). White children had the highest incidence of osteonecrosis (16.7%±1.4%) and black children the lowest (3.3%±2.3%). However, although the incidence of osteonecrosis was higher for patients randomized to receive two 21-day dexamethasone courses versus one course (23.2%±4.8% vs. 16.4%±4.3%), the difference did not reach statistical significance (p=0.27). Mattano et al extended these findings to patients treated on HR-ALL study COG AALL0232 (30). Induction rapid early responders (RER) receive single Delayed Intensification (DI) while slow responders received double DI; all patients receive monthly 5-day dexamethasone pulses during maintenance. To limit the risk of ON in adolescents, children ≥13 years of age received discontinuous dexamethasone during single or double DI, and those <13 years received continuous dexamethasone. The incidence of ON was 10.4%, and was higher for those age ≥10 vs. <10 (15.2 vs. 2.6%, p<0.0001, HR=6.38). Among all patients, the incidence of ON incidence was higher in the dexamethasone regimen (11.6 vs. 8.7%, p=0.014, HR 1.64). Among patients ≥13y, the incidence of ON was higher in the dexamethasone regimen (18.9 vs. 9.9%, p=0.02, RHR 1.97). There was no difference between regimens for children less than 10 years of age. These findings are informing the utilization of dexamethasone in the future trials.
The contribution of dexamethasone versus prednisone during induction to the development of osteonecrosis has been explored systematically by Teuffel et al. (31). This review included all randomized controlled trials comparing dexamethasone with prednisone during induction in children with ALL. These studies included CALGB7111, CCG-1922, MRC ALL-97/99, TCCSG L95-14, EORTC 58951, COG AALL0232, ALL-BFM 2000, and AIEOP-BFM ALL 2000. The authors concluded that there was no statistically significant difference in the risk of osteonecrosis between the patients treated with dexamethasone or prednisone.
Increased susceptibility to osteonecrosis between 10 and 20 years of age (32–35) is possibly related to puberty-related hormonal and physiologic changes combined with the maturing phase of the adolescent bone.
Certain non-glucocorticoids have also been linked with osteonecrosis, including methotrexate and asparaginase (36, 37), bleomycin/ vinblastine (38), melphalan/ doxorubicin/ cyclophosphamide/ 5-fluorouracil (39), cyclophosphamide/ doxorubicin (40), and daunomycin/ cytarabine/ all-transretinoic acid (ATRA) (41). However, these observations are in the form of case reports; a systematic effort is needed to explore the role of non-corticosteroid exposures in the development of osteonecrosis.
Pathogenesis of osteonecrosis
Vascular compromise to the bone underlies the development of osteonecrosis. This could be due to vascular occlusion resulting in ischemia, to altered fat metabolism and fat emboli, intravascular coagulation, increased intracortical pressure, inhibition of angiogenesis, mechanical stress, and finally because of direct death of the osteocytes. Glucocorticoids may induce osteonecrosis because of their propensity to cause hyperlipidemia, hypercoagulation, and hypofibrinolysis (42, 43) resulting in intravascular thrombotic occlusion and extravascular lipid deposition. Glucocorticoids also increase the size of intraosseous lipocytes, leading to increased intracortical pressure, vascular compromise, marrow ischemia and apoptosis followed by necrosis (44, 45). Glucocorticoids also cause direct injury to the osteocytes (46).
Previous studies have identified polymorphisms in genes putatively related to the development of osteonecrosis, including SERPINE 1 (47), VDR (32), and CYP3A4 (48), but with conflicting results (47, 49) (Table 1). Kawedia et al prospectively screened children with ALL and identified older age (>10 years), more intensive treatment, lower albumin and elevated cholesterol to be associated with symptomatic osteonecrosis; severe osteonecrosis was linked to poor dexamethasone clearance. After adjusting for clinical features, polymorphisms in ACP1 were associated with osteonecrosis risk, as well as with lower albumin and higher cholesterol. Thus, polymorphisms of ACP1 (e.g., rs12714403: OR=5.6, 95%CI, 2.7–11.3) were associated with symptomatic osteonecrosis. ACP1 is associated with serum cholesterol and triglyceride levels (50), and regulates osteoblast differentiation via Src kinase (51). These findings suggest that ACP1 might act via multiple mechanisms to affect bone homeostasis and dexamethasone-induced osteonecrosis (52). The strengths of this study include the use of clinically validated internal controls (i.e., ALL patients screened with MRI to confirm absence of osteonecrosis); inclusion of clinical risk factors, and markers of bone metabolism as well as markers involved in the proposed pathogenetic pathway for development of osteonecrosis. Although there appears to be biological plausibility to this association, the findings need to be validated in an independent replication cohort.
Ototoxicity
Several potentially ototoxic agents including platinum-based chemotherapy, aminoglycoside antibiotics, loop diuretics, and radiation therapy are commonly used in management of patients with cancer, and can result in sensorineural hearing loss (53). Platinum-containing chemotherapy, particularly cisplatin and myeloablative doses of carboplatin, are well-established risk factors for therapy-related hearing loss. Cisplatin causes serious, permanent, bilateral hearing loss in 10–25% of adults and in 41–61% of children (54–60) receiving the drug. Risk of ototoxicity increases with cumulative exposure to platinum agents. Radiation-related hearing loss is infrequent (<3%) when cochlear radiation dose is less than 35 Gy (61). Mild to moderate hearing loss occurs in as many as 37% of children exposed to doses exceeding 60 Gy (61). Similar to platinum-related ototoxicity, radiation-induced injury is caused by dose-dependent injury to the cochlea, and specifically affects high frequency hearing. Concomitant use of platinum agents with radiation may have synergistic sensorineural ototoxicity (62). Use of proton beam radiation has made it possible to limit cochlear radiation doses to nearly zero (63). Risk of platinum-related hearing loss is modified by treatment involving multiple ototoxic agents; and young age (less than 4 years) at exposure to ototoxic agents (53).
Pathogenesis of ototoxicity
Platinum-related ototoxicity results from destruction of cochlear hair cells of the organ of Corti, probably as a result of oxidative stress (64). Because of the tonotypical arrangement (in order of pitch) of these specialized hair cells, the initial hearing loss is generally in frequency ranges that exceed 2000 Hz. As cumulative exposure to platinum chemotherapy increase, so does the progression of injury toward the cochlear apex where frequencies between 500 and 2000 Hz are affected (65). High dose radiation may also result in tympanosclerosis, otosclerosis, and Eustachian tube dysfunction (66).
Interindividual variations in ototoxicity among patients receiving similar cumulative cisplatin exposure, suggests the role for single nucleotide polymorphisms in genes encoding drug metabolizing enzymes (results of extant literature summarized in Table 1). Oldenburg et al examined the association between functional polymorphisms in cisplatinum-detoxifying enzymes – especially glutathione S-transferases (GSTs) in cisplatin-treated testicular cancer survivors (67). Known functional polymorphisms in GSTT1 and GSTM1 and codon 105 A/G (Ile/Val) in GSTP1 were analyzed. The risk of an inferior audiometric test result was four times higher in testicular cancer survivors with 105Ile/105Ile-GSTP1 or 105Val/105Ile-GSTP1 compared with 105Val/105Val-GSTP1. Presence of 105Val-GSTP1 offered protection against cisplatin-induced hearing impairment. Other candidate gene studies have also reported associations of cisplatin ototoxicity with genetic variants in genes encoding glutathione S-transferases and megalin (68, 69). Ross et al reported association analyses for 220 candidate drug-metabolism genes in genetic susceptibility to cisplatin-induced hearing loss in children (70). A total of 1,949 SNPs were genotyped in an initial cohort of 54 children with a replication cohort of 112 children. Genetic variants in TPMT (rs1220119) and COMT (rs9332377) were identified to be associated with cisplatin-induced hearing loss in children. TPMT and COMT are methyltransferases and are dependent on the S-adenosylmethionine (SAM) methyl donor substrate in the methionine pathway. Reduced TPMT and COMT activity result in increased levels of SAM. Administration of SAM and cisplatin together has been shown to increase cisplatin toxicity substantially, while the administration of SAM alone was not ototoxic, and cisplatin alone resulted in a moderate increase the ototoxicity (71). These results suggest that cisplatin-induced ototoxicity could be related to increased levels of SAM because of reduced TPMT or COMT activity. Although the samples sizes were small, the investigators were successful in confirming the findings in an independent replication set. An additional strength of the study was the utilization of an appropriate comparison group i.e., cancer patients exposed to cisplatin but with no hearing loss.
Obesity
Several studies suggest a role for therapeutic exposures in the development of obesity in cancer survivors (72–74). Using a cross-sectional approach, Oeffinger et al (73) identified cranial radiotherapy in doses of 20 Gy or more as the primary risk factors for the increased prevalence of obesity noted among ALL survivors, with the highest risk observed in survivors exposed to radiation at age 4 years or younger. Recently, they extended their findings to describe the rate of increase in body mass index (BMI) after final height attainment in childhood ALL survivors, over a mean interval of 7.8 years of observation (75). ALL survivors treated with cranial radiation had a significantly greater increase in BMI (women, 0.41 units/yr; men, 0.29 units/year) compared with sibling comparison group (women, 0.25 units/ year; men, 0.23 units/ year). Patients exposed to chemotherapy alone did not differ from the siblings with respect to the change in BMI. Younger age at cranial radiation significantly modified the rate of increase.
Pathogenesis of obesity
Leptin insensitivity possibly plays a role in the development of cranial radiation-related obesity. Brennan et al reported significantly higher levels of leptin in 32 survivors of childhood ALL who had been treated with high doses of cranial radiation compared to age- and BMI-matched controls (76). Leptin is an adipocyte-derived hormone that binds to the biologically active receptor in the hypothalamus (77). It has been proposed that radiation-induced damage to the pituitary-hypothalamus axis disrupts leptin signal, resulting in obesity (76). However, the individual variability is possibly related to genetic susceptibility.
Polygenic variants presumably account for most of the genetic variation relevant to body weight regulation in the general population. Obesity results via the interaction of several polygenic variants and their interaction in turn with environmental factors. Two gene variants with small but replicable effects on body weight have been identified so far (melanocortin-4 receptor genes [MC4R]); (78–80) and fat mass and obesity associated genes (FTO) (81, 82). Overall, the mechanism of action of most obesity genes is not well understood; adipose tissue and skeletal muscle are sites for storage, release, and metabolism of fatty acids, and therefore may be involved. The observed genetic variation in obesity explains only a minor fraction (2–4%) of the total genetic variation explained to be present in the general population (83).
Given that the magnitude of risk for obesity is much higher among cancer survivors than in the general population, it is possible that there is a role for gene-environment (therapy) interactions that influence obesity in the survivors. Ross et al evaluated the potential association between polymorphism Gln223Arg, in the leptin receptor (LEPR) gene and risk of obesity in ALL survivors enrolled in the CCSS (84), Table 1. Overweight female ALL survivors were significantly more likely to possess a homozygous Arg genotype compared to female survivors with BMI<25 KG/m2 (24% vs. 12%, p=0.007). In contrast no significant difference was observed for male survivors (25% vs. 21%, p=0.37). Further, for females, there was a statistically significant (p=0.04) interaction with higher levels of radiation treatment; survivors treated with >20 Gy who were homozygous for the Arg allele were over six times more likely to be overweight/ obese compared to those who possessed at least one Gln allele. The risk was considerably lower for females treated with <20 Gy radiation or with chemotherapy alone. This study supported the role for LEPR polymorphism as a potential mechanism for obesity in female survivors of childhood ALL exposed to cranial radiation. However, there is a paucity of data in the literature examining the role of genetic susceptibility in the development of obesity in cancer survivors. There is a need to examine the role of genes implicated in obesity in the general population in the cancer survivor population, with careful attention to exploration of gene-environment interactions.
Subsequent Malignant Neoplasms
The cumulative incidence of SMNs exceeds 20% at 30 years after diagnosis of childhood cancer, representing a 4–6-fold increased risk for cancer survivors, compared to the general population (85, 86). SMNs are a leading cause of non-relapse late mortality (87, 88). Unique associations with specific therapeutic exposures have resulted in the classification of SMNs into two distinct groups: chemotherapy-related myelodysplasia and acute myeloid leukemia (t-MDS/AML) and radiation-related solid SMNs. Characteristics of t-MDS/AML include a short latency (<3 years from primary cancer diagnosis) and association with alkylating agents and/or topoisomerase II inhibitors. Solid SMNs have a strong and well-defined association with radiation, and are characterized by a latency that exceeds 10 years (85, 89–91).
t-MDS/AML has been reported after conventional treatment of HL, NHL, ALL, sarcomas, and breast, ovarian and testicular cancers (89, 92–97), and after autologous hematopoietic cell transplantation (HCT) for HL or NHL, where it is the major cause of non-relapse mortality (98–103). The cumulative incidence of t-MDS/AML ranges from 2% at 15 years after conventional therapy (89) to 8.6% at 6 years after autologous HCT (98).
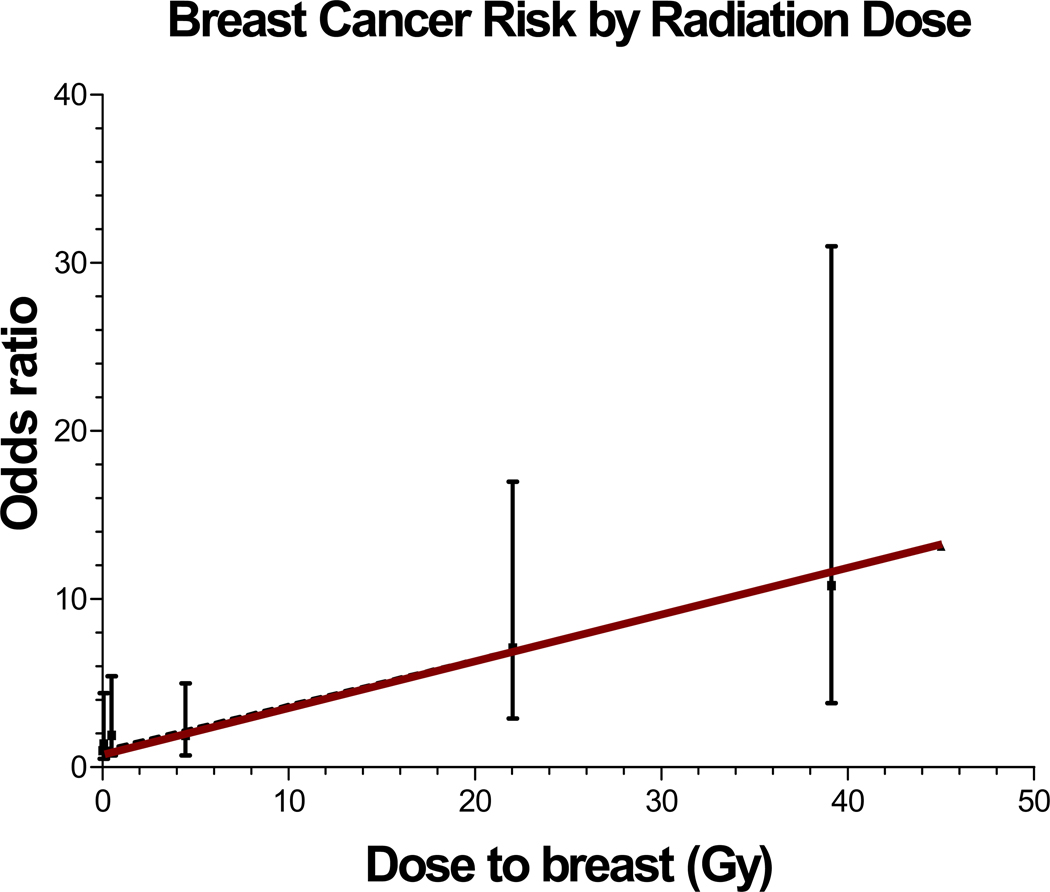
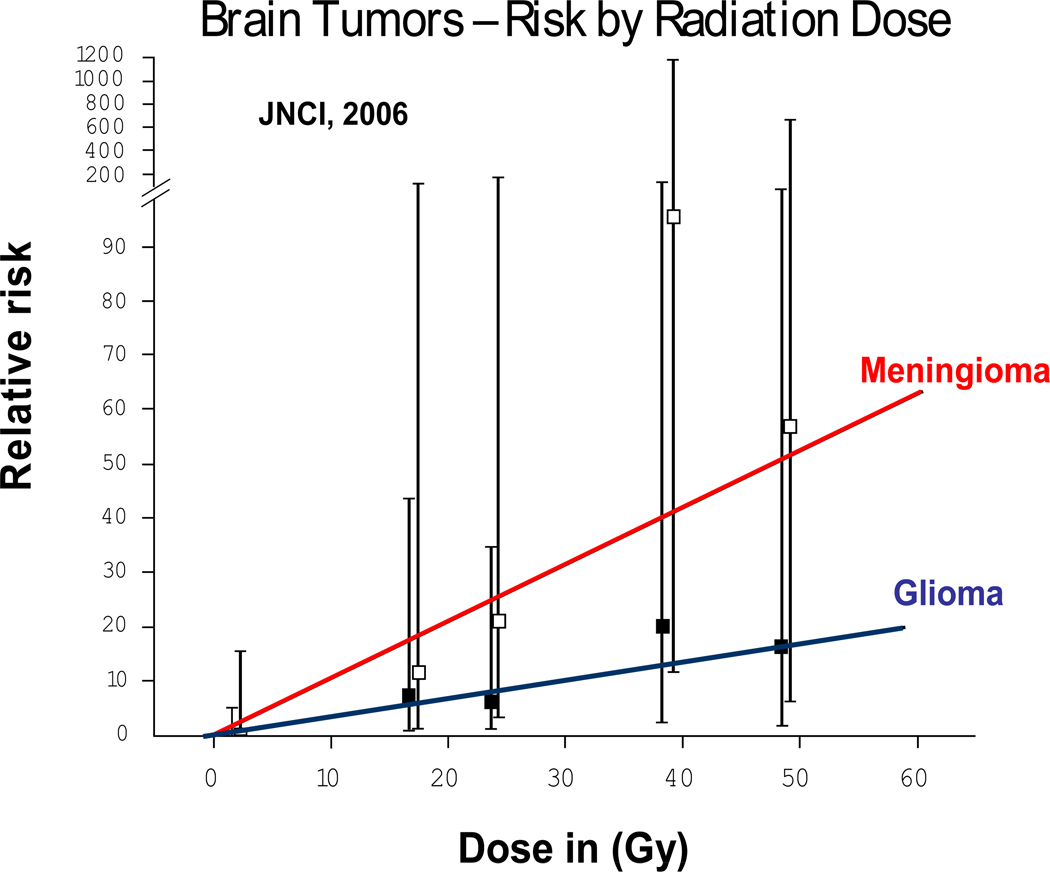
Radiation induces solid SMNs within the radiation field (89, 90, 93, 104, 105). The latency for radiation-related solid SMNs usually exceeds 10 years (89, 90, 93, 105). The risk is highest when radiation exposure occurs at a younger age (89, 91, 105–112), and increases with increasing doses of radiation (Figure 3) and with increasing time since radiation (90, 93). Eighty percent of the entire burden of subsequent malignancies is accounted for by radiation-related solid SMNs. Some of the well-established radiation-related solid SMNs include breast cancer, thyroid cancer, brain tumors, sarcomas and basal cell carcinomas (BCCs) (85, 89–91, 93, 110, 113). Breast cancer is the most common solid SMN after HL, largely due to high-dose chest radiation for HL (standardized incidence ratio [SIR]=25 to 55) (89, 93, 113). For female HL patients treated with chest radiation at less than 16 years of age, the cumulative incidence of breast cancer approaches 20% by age 45 years (93). Thyroid cancer develops after neck radiation for HL or ALL (89, 91, 93, 94, 105). Brain tumors develop after cranial radiation for histologically distinct brain tumors (90), or for management of ALL or NHL (91, 94, 105). Sarcomas develop within the radiation field after a latency period of ~10 years. Over 90% of BCCs develop within radiation field (89–91, 94, 95, 105).
Figure 3.
Radiation-related risk of subsequent malignant neoplasms
Pathogenesis of subsequent malignant neoplasms
t-MDS/AML is a clonal disorder characterized by distinct chromosomal changes (114–116). Two types are recognized by the WHO classification: alkylating agent-related type, and topoisomerase II inhibitor-related type (117). Alkylating agent-related t-MDS/AML: Alkylating agents associated with t-MDS/AML include cyclophosphamide, ifosfamide, mechlorethamine, melphalan, busulfan, nitrosureas, chlorambucil, dacarbazine (118), and platinum compounds (96). Mutagenicity is related to the ability of alkylating agents to form crosslinks and/or transfer alkyl groups to form DNA monoadducts. Alkylation results in inaccurate base pairing during replication and single- and double-strand breaks in the double helix as the alkylated bases are repaired. The risk of alkylating agent-related t-MDS/AML is dose-dependent, with a latency of 3 to 5 years after exposure; it is associated with abnormalities involving chromosomes 5 (−5/del[5q]) and 7 (−7/del[7q]), and a high frequency of multidrug-resistance phenotype (119).
The magnitude of association between specific chemotherapeutic agents and radiation and SMNs are moderate to large (OR: 3.1 to 15.9), with a clear dose-response relationship adding further biological credibility to that association. The risk of second breast cancer after chest radiation increases in a linear fashion with radiation dose (p for trend <0.001) (113, 120), as does the risk for second brain tumors (90), (Figure 3) and second sarcomas (121). Literature clearly supports the role of chemotherapy and radiation in the development of SMNs (118), but interindividual variability suggests a role for genetic variation in susceptibility to genotoxic exposures. The risk of SMNs could potentially be modified by mutations in high-penetrance genes that lead to serious genetic diseases e.g., Li-Fraumeni syndrome (122), and Fanconi anemia (123–126). However, the attributable risk is expected to be very small because of their extremely low prevalence. The interindividual variability in risk of therapy-related SMNs is more likely related to common polymorphisms in low-penetrance genes that regulate the availability of active drug metabolite, or those responsible for DNA repair. Genetic variation contributes 20% to 95% of the variability in cytotoxic drug disposition (127). Polymorphisms in genes involved in drug metabolism and transport are relevant in determining disease-free survival and drug toxicity (128). Variation in DNA repair plays a role in susceptibility to de novo cancer (129–133), and likely modifies SMN risk after exposure to DNA-damaging agents, such as radiation and chemotherapy. Gene-environment interactions may magnify subtle functional differences resulting from genetic variations (134–137). Results from studies examining genetic susceptibility in the development of SMNs are summarized in Table 1.
Drug Metabolism
Metabolism of genotoxic agents occurs in two phases. Phase I involves activation of substrates into highly reactive electrophilic intermediates that can damage DNA – a reaction principally performed by the cytochrome p450 (CYP) family of enzymes. Phase II enzymes (conjugation) function to inactivate genotoxic substrates. The phase II proteins comprise the glutathione S-transferase (GST), NAD(P)H:quinone oxidoreductase-1 (NQO1), and others. The balance between the two sets of enzymes is critical to the cellular response to xenobiotics; e.g., high activity of phase I enzyme and low activity of a phase II enzyme can result in DNA damage from the excess of harmful substrates. Given that these enzymes activate/ detoxify chemotherapeutic agents – their role in the development of SMNs applies mainly to t-MDS/AML. The xenobiotic substrates of CYP proteins include cyclophosphamide, ifosfamide, thiotepa, doxorubicin, and dacarbazine (138). The CYPs transfer singlet oxygen onto their substrates creating highly reactive intermediates which, unless detoxified by phase II enzymes, have a strong ability to damage DNA (139). The expression of these enzymes is highly variable among individuals because of several functionally relevant genetic polymorphisms. GSTs detoxify reactive electrophiles via conjugation to reduced glutathione, preventing damage to DNA. Polymorphisms exist in cytosolic subfamilies: µ [M], π [P), θ [T], and others. GSTs detoxify doxorubicin, lomustine, busulfan, chlorambucil, cisplatin, cyclophosphamide, melphalan, etc. (140). Quinone oxidoreductase NQO1 uses the cofactors NADH and NADPH to catalyze the electron reduction of its substrates, produces less reactive hydroquinones, and therefore prevents generation of reactive oxygen species and free radicals which may subsequently lead to oxidative damage of cellular components. Allan et al utilized a candidate gene approach to examine associations between polymorphisms in the glutathione S-transferase genes (GSTM1, GSTT1 and GSTP1) and t-MDS/AML (141). A case-control study design was used (80 cases; 420 patients with de novo AML; 1022 healthy controls). Individuals with at least one GSTP1 codon 105 Val allele were significantly over-represented in t-AML cases compared with de novo AML cases (OR=1.81, 95%CI, 1.11–2.94). Also, relative to de novo AML, the GSTP1 codon 105 allele occurred more often among t-AML patients with prior exposure to chemotherapy (OR=2.66, 95%CI, 1.39–5.09), particularly among those with prior exposure to known GSTP1 substrates (OR=4.34, 95%CI, 1.43–13.20) and not among t-AML patients with exposure to radiation alone. While the findings were biologically plausible, they were not replicated in this study. Furthermore, the comparison groups consisting of healthy controls and patients with de novo AML could possibly compromise the findings.
Drug transport
P-glycoprotein (encoded by MDR1) traps hydrophobic drugs in the plasma membrane of cells and effluxes them using an ATP-dependent process; many chemotherapeutic drugs are substrates of this protein. A number of polymorphisms exist in the MDR1 gene, some proposed to be functional and evaluated as risk factors for t-MDS/AML (142).
DNA repair
DNA repair mechanisms protect somatic cells from mutations in tumor suppressor genes and oncogenes that can lead to cancer initiation and progression. An individual’s DNA repair capacity appears to be genetically determined (143). A number of DNA repair genes contain polymorphic variants, resulting in large inter-individual variations in DNA repair capacity (143). Even subtle differences in an individual’s DNA repair capacity may be important in the presence of large external influences such as chemotherapy or radiotherapy. Individuals with altered DNA repair mechanisms are likely susceptible to the development of genetic instability that drives the process of carcinogenesis as it relates to both chemotherapy-related t-MDS/AML as well as radiation-related solid SMNs.
Mismatch repair (MMR) functions to correct mismatched DNA base pairs that arise as a result of misincorporation errors that have avoided polymerase proofreading during DNA replication (144). Defects in the MMR pathway result in genetic instability or a mutator phenotype, manifested by an elevated rate of spontaneous mutations characterized as multiple replication errors in simple repetitive DNA sequences (microsatellites) – functionally identified as microsatellite instability (MSI). Approximately 50% of t-MDS/AML patients have MSI, associated with methylation of the MMR family member MLH1 (145, 146), low expression of MSH2 (147), or polymorphisms in MSH2 (134, 148–150). The appearance of MMR-deficient, drug-resistant clones during genotoxic treatment for a primary cancer could be a vital factor in SMN susceptibility, particularly because the mutator phenotype (inherent of MMR-deficient cells) would be expected to accelerate the accumulation of further mutations and eventually SMN initiation. In addition, loss of MMR may result in deregulation of homologous recombination repair and consequent chromosomal instability (151).
Double-Strand Breaks (DSBs) in DNA may lead to loss of genetic material, resulting in chromosomal aberrations. High levels of DSBs arise following ionizing radiation and chemotherapy exposures. Cellular pathways available to repair DSBs include homologous recombination (HR), non-homologous end-joining (NHEJ), and single-strand annealing (152). HR uses the second, intact copy of the chromosome as a template to copy the information lost at the DSB site on the damaged chromosome – a high-fidelity process. RAD51 is one of the central proteins in the HR pathway, functioning to bind to DNA and promote ATP-dependent homologous pairing and strand transfer reactions (153, 154). RAD51-G-135C polymorphism is significantly over-represented in patients with t-MDS/AML compared with controls (C allele: OR=2.7) (155). XRCC3 also functions in the HR DSB repair pathway by directly interacting with, and stabilizing RAD51 (156, 157). XRCC3 is a paralog of RAD51, also essential for genetic stability (158, 159). A polymorphism at codon 241 in the XRCC3 gene results in a Thr→Met amino acid substitution (160). The variant XRCC3-241Met allele has been associated with a higher level of DNA adducts compared with cells with the wild type allele, implying aberrant repair (161) and has also been associated with increased levels of chromosome deletions in lymphocytes after exposure to radiation (162). Although XRCC3-Thr241Met was not associated with t-MDS/AML (OR=1.4, 95%CI, 0.7–2.9), a synergistic effect resulting in an 8-fold increased risk of t-MDS/AML (OR=8.1, 95% CI, 2.2–29.7) was observed in the presence of XRCC3-241Met and RAD51-135C allele in patients with t-MDS/AML compared with controls (155). NHEJ pathway joins broken DNA ends containing very little homology. This process is not always precise and can result in small regions of non-template nucleotides around the site of the DNA break, potentially relevant in MLL-translocation associated with t-MDS/AML. Many of the translocation junctions have been cloned and sequenced and found to contain regions of microhomology consistent with the operation of the NHEJ pathway and an impairment of this pathway has been hypothesized to modulate t-MDS/AML risk (163).
Base Excision Repair (BER) pathway corrects individually damaged bases occurring as a result of ionizing radiation and exogenous xenobiotic exposure. The XRCC1 protein plays a central role in the BER pathway and also in the repair of single strand breaks, by acting as a scaffold and recruiting other DNA repair proteins (164, 165). The protein also has a BRCA1 C-terminus (BRCT) domain – a characteristic of proteins involved in DNA damage recognition and response. The presence of variant XRCC1-399Gln has been shown to be protective for t-MDS/AML (166), and BCC (167).
Nucleotide Excision Repair (NER) removes structurally unrelated bulky damage induced by radiation and chemotherapy. The NER pathway is linked to transcription, and components of the pathway comprise the basal transcription factor IIH complex (TFIIH), which is required for transcription initiation by RNA polymerase II. One of the genes involved in the NER pathway (ERCC2) is a member of the TFIIH complex. The polymorphic Gln variant (ERCC2 Lys751Gln) is associated with t-MDS/AML (135).
Association of variants in XPD, XRCC1, XRCC2, XRCC3, ERCC1 and APE1 with several primary cancers has been reported (130), usually with the risk increased 4- to 5-fold (129). Furthermore, individuals with a reduced capacity in two different pathways exhibit an even higher risk (168). Adaptive response to low-dose radiation in human lymphocytes displays heterogeneity with respect to chromosomal aberrations (169). Chromosomal assays, micronucleus tests (163), hprt gene mutation and comet assays for DNA damage demonstrate significant inter-individual variation (170, 171). Over 80 DNA repair genes have been screened and demonstrate evidence of extensive polymorphic variation (172).
Ellis et al utilized a case-control study design (171 cases) and examined the association between patients with t-MDS/AML and 2 common functional p53-pathway variants – the MDM2 SNP309 and the TP53 codon 72 polymorphism (173). Neither polymorphism demonstrated a significant association. However, an interactive effect was detected such that MDM2 TT TP53 Arg/Arg double homozygotes, and individuals carrying both a MDM2 G allele and a TP53 Pro allele were at increased risk of chemotherapy-related t-MDS/AML. The strengths of the study included the utilization of a discover set (n=80 case) and replication set (n=91 cases). However, the investigators utilized healthy controls as a comparison group – thus precluding the ability to assess whether the case control differences reflected differences in susceptibility to primary disease or t-MDS/AML.
Knight et al utilized a case-control study design to conduct a GWAS in patients who had developed therapy-related leukemia (cases) and healthy controls (174). The discovery set included 80 cases and 150 controls. The relevant findings were replicated in an independent set of 70 cases and 95 controls. The investigators identified 3 SNPs (rs1394384 [OR=0.29, 95% CI, 0.15–0.56], rs1381392 [OR=2.08, 95%CI, 1.29–3.35], and rs1199098 [OR=0.46, 95% CI, 0.27–0.79]) to be associated with t-MDS/AML with chromosome 5/7 abnormalities. rs1394384 is intronic to ACCN1, a gene encoding an amiloride-sensitive cation channel that is a member of the degenerin/ epithelial sodium channel; rs1199098 is in LD with IPMK, which encodes a multikinase that positively regulates the prosurvival AKT kinase and may modulate Wnt/beta-catenin signaling; rs1381392 is not near any known genes, miRNAs, or regulatory elements, although it lies in a region recurrently deleted in lung cancer. Although the investigators were able to confirm findings in an independent replication cohort, the utilization of a non-cancer healthy control group raises concerns about the case-control differences being generated by the genetics of the primary cancer vs. t-MDS/AML.
Best et al performed a GWAS to identify variants associated with radiation-related solid malignancies in survivors of Hodgkin lymphoma (175). They identified two variants at chromosome 6q21 associated with second malignancies. The variants comprise a risk locus associated with decreased basal expression of PRDM1 and impaired induction of the PRDM1 protein after radiation exposure. These data suggest new gene-exposure interaction that may implicate PRDM1 in the etiology of radiation therapy-induced second malignancies.
Summary and Future Directions
Several studies have examined single gene polymorphisms in small heterogeneous samples, contributing to the largely inconclusive results. Functional redundancy often results in the availability of more than one gene product to detoxify the same substrate or repair the same damage type. Hence, a variant in one gene may have minimal consequences, whereas the combination of variants in two or more genes could have more serious consequences resulting in the emergence of a malignant phenotype. Furthermore, previous studies have generally failed to systematically examine gene-therapy interactions, because of the absence of detailed therapeutic exposure data collected by the previous studies and the small sample sizes. A systematic assessment of the role of drug-metabolizing enzymes, DNA repair genes and drug transport in the development of SMNs is currently under way in a Children’s Oncology Group-wide study, funded by the National Cancer Institute. This study utilizes a case-control study design to assess the role of genetic susceptibility in the development of key treatment-related adverse outcomes, including SMNs.
The current report has focused on the current state of the knowledge related to the pathogenesis of adverse events that have clearly-defined associations with therapeutic exposures, and that result in significant long-term morbidity. There are several other adverse outcomes with clearly defined-associations with therapeutic exposures that were not included in this report, largely because of paucity of published literature with respect to the mechanistic relationship between therapy and outcome. These outcomes include endocrine dysfunction, pulmonary compromise, stroke, peripheral neuropathy, etc.
This review is not meant to serve as a comprehensive overview of all published studies; rather it provides examples of some of the more scientifically and methodologically robust studies and demonstrates that genetic variation could possibly interact with therapeutic exposure and increase the risk of adverse outcomes. However, there remains a critical need to replicate these findings in large independent cohorts before these findings can be incorporated into the clinical management of the patients.
The discovery of functional genetic variants associated with key outcomes will have significant implications for future research aimed at improving risk assessment and conducting mechanistic research. Identification of new and informative genetic markers have utility for developing objective pre-therapy risk assessment and patient counseling, and serving as rational tools for clinical management and treatment planning. The ultimate goal is to identify those at highest risk, such that targeted prevention and intervention strategies can be instituted.
Acknowledgments
Supported in part by NCI grant U10 CA98543 (P Adamson); U24 55727 (LL Robison); R01 CA 139633 (S Bhatia); and The Leukemia and Lymphoma Society: 6093-08 (S Bhatia)
REFERENCES
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.OCS. National Cancer Institute. 2011. Office of Cancer Survivorship. [Google Scholar]
- 3.Hewitt M, Greenfield S, Stovall E, editors. From Cancer Patient to Cancer Survivor: Lost in Transition. Washington, D.C.: Committee on Cancer Survivorship: Improving Care and Quality of Life, National Cancer Policy Board, Institute of Medicine and National Research Council, National Academies Press; 2006. [Google Scholar]
- 4.Bhatia S, Robison LL. Cancer survivorship research: opportunities and future needs for expanding the research base. Cancer Epidemiol Biomarkers Prev. 2008;17:1551–1557. doi: 10.1158/1055-9965.EPI-08-0490. [DOI] [PubMed] [Google Scholar]
- 5.Ganz PA. Why and how to study the fate of cancer survivors: observations from the clinic and the research laboratory. Eur J Cancer. 2003;39:2136–2141. doi: 10.1016/s0959-8049(03)00489-1. [DOI] [PubMed] [Google Scholar]
- 6.Oeffinger KC, Robison LL. Childhood cancer survivors, late effects, and a new model for understanding survivorship. JAMA. 2007;297:2762–2764. doi: 10.1001/jama.297.24.2762. [DOI] [PubMed] [Google Scholar]
- 7.Lipshultz SE, Alvarez JA, Scully RE. Anthracycline associated cardiotoxicity in survivors of childhood cancer. Heart. 2008;94:525–533. doi: 10.1136/hrt.2007.136093. [DOI] [PubMed] [Google Scholar]
- 8.Kremer LC, van Dalen EC, Offringa M, Voute PA. Frequency and risk factors of anthracycline-induced clinical heart failure in children: a systematic review. Ann Oncol. 2002;13:503–512. doi: 10.1093/annonc/mdf118. [DOI] [PubMed] [Google Scholar]
- 9.Giantris A, Abdurrahman L, Hinkle A, Asselin B, Lipshultz SE. Anthracycline-induced cardiotoxicity in children and young adults. Crit Rev Oncol Hematol. 1998;27:53–68. doi: 10.1016/s1040-8428(97)10007-5. [DOI] [PubMed] [Google Scholar]
- 10.Mulrooney DA, Yeazel MW, Kawashima T, Mertens AC, Mitby P, Stovall M, et al. Cardiac outcomes in a cohort of adult survivors of childhood and adolescent cancer: retrospective analysis of the Childhood Cancer Survivor Study cohort. BMJ. 2009;339:b4606. doi: 10.1136/bmj.b4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felker GM, Thompson RE, Hare JM, Hruban RH, Clemetson DE, Howard DL, et al. Underlying causes and long-term survival in patients with initially unexplained cardiomyopathy. N Engl J Med. 2000;342:1077–1084. doi: 10.1056/NEJM200004133421502. [DOI] [PubMed] [Google Scholar]
- 12.Hudson MM, Rai SN, Nunez C, Merchant TE, Marina NM, Zalamea N, et al. Noninvasive evaluation of late anthracycline cardiac toxicity in childhood cancer survivors. J Clin Oncol. 2007;25:3635–3643. doi: 10.1200/JCO.2006.09.7451. [DOI] [PubMed] [Google Scholar]
- 13.Shankar SM, Marina N, Hudson MM, Hodgson DC, Adams MJ, Landier W, et al. Monitoring for cardiovascular disease in survivors of childhood cancer: report from the Cardiovascular Disease Task Force of the Children's Oncology Group. Pediatrics. 2008;121:e387–e396. doi: 10.1542/peds.2007-0575. [DOI] [PubMed] [Google Scholar]
- 14.Adams MJ, Lipshultz SE. Pathophysiology of anthracycline- and radiation-associated cardiomyopathies: implications for screening and prevention. Pediatr Blood Cancer. 2005;44:600–606. doi: 10.1002/pbc.20352. [DOI] [PubMed] [Google Scholar]
- 15.Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev. 2004;56:185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- 16.Dell'Acqua G, Polishchuck R, Fallon JT, Gordon JW. Cardiac resistance to adriamycin in transgenic mice expressing a rat alpha-cardiac myosin heavy chain/human multiple drug resistance 1 fusion gene. Hum Gene Ther. 1999;10:1269–1279. doi: 10.1089/10430349950017950. [DOI] [PubMed] [Google Scholar]
- 17.Olson LE, Bedja D, Alvey SJ, Cardounel AJ, Gabrielson KL, Reeves RH. Protection from doxorubicin-induced cardiac toxicity in mice with a null allele of carbonyl reductase 1. Cancer Res. 2003;63:6602–6606. [PubMed] [Google Scholar]
- 18.Forrest GL, Gonzalez B, Tseng W, Li X, Mann J. Human carbonyl reductase overexpression in the heart advances the development of doxorubicin-induced cardiotoxicity in transgenic mice. Cancer Res. 2000;60:5158–5164. [PubMed] [Google Scholar]
- 19.van Dalen EC, van der Pal HJ, Kok WE, Caron HN, Kremer LC. Clinical heart failure in a cohort of children treated with anthracyclines: a long-term follow-up study. Eur J Cancer. 2006;42:3191–3198. doi: 10.1016/j.ejca.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Paulides M, Kremers A, Stohr W, Bielack S, Jurgens H, Treuner J, et al. Prospective longitudinal evaluation of doxorubicin-induced cardiomyopathy in sarcoma patients: a report of the late effects surveillance system (LESS) Pediatr Blood Cancer. 2006;46:489–495. doi: 10.1002/pbc.20492. [DOI] [PubMed] [Google Scholar]
- 21.Henderson IC, Allegra JC, Woodcock T, Wolff S, Bryan S, Cartwright K, et al. Randomized clinical trial comparing mitoxantrone with doxorubicin in previously treated patients with metastatic breast cancer. J Clin Oncol. 1989;7:560–571. doi: 10.1200/JCO.1989.7.5.560. [DOI] [PubMed] [Google Scholar]
- 22.Deng S, Wojnowski L. Genotyping the risk of anthracycline-induced cardiotoxicity. Cardiovasc Toxicol. 2007;7:129–134. doi: 10.1007/s12012-007-0024-2. [DOI] [PubMed] [Google Scholar]
- 23.Blanco JG, Leisenring WM, Gonzalez-Covarrubias VM, Kawashima TI, Davies SM, Relling MV, et al. Genetic polymorphisms in the carbonyl reductase 3 gene CBR3 and the NAD(P)H:quinone oxidoreductase 1 gene NQO1 in patients who developed anthracycline-related congestive heart failure after childhood cancer. Cancer. 2008;112:2789–2795. doi: 10.1002/cncr.23534. [DOI] [PubMed] [Google Scholar]
- 24.Wojnowski L, Kulle B, Schirmer M, Schluter G, Schmidt A, Rosenberger A, et al. NAD(P)H oxidase and multidrug resistance protein genetic polymorphisms are associated with doxorubicin-induced cardiotoxicity. Circulation. 2005;112:3754–3762. doi: 10.1161/CIRCULATIONAHA.105.576850. [DOI] [PubMed] [Google Scholar]
- 25.Mankin HJ. Nontraumatic necrosis of bone (osteonecrosis) N Engl J Med. 1992;326:1473–1479. doi: 10.1056/NEJM199205283262206. [DOI] [PubMed] [Google Scholar]
- 26.Ohzono K, Saito M, Takaoka K, Ono K, Saito S, Nishina T, et al. Natural history of nontraumatic avascular necrosis of the femoral head. J Bone Joint Surg Br. 1991;73:68–72. doi: 10.1302/0301-620X.73B1.1991778. [DOI] [PubMed] [Google Scholar]
- 27.Heimann WG, Freiberger RH. Avascular necrosis of the femoral and humeral heads after high-dosage corticosteroid therapy. N Engl J Med. 1960;263:672–675. doi: 10.1056/NEJM196010062631404. [DOI] [PubMed] [Google Scholar]
- 28.Mattano LA, Jr., Sather HN, Trigg ME, Nachman JB. Osteonecrosis as a complication of treating acute lymphoblastic leukemia in children: a report from the Children's Cancer Group. J Clin Oncol. 2000;18:3262–3272. doi: 10.1200/JCO.2000.18.18.3262. [DOI] [PubMed] [Google Scholar]
- 29.Kadan-Lottick NS, Dinu I, Wasilewski-Masker K, Kaste S, Meacham LR, Mahajan A, et al. Osteonecrosis in adult survivors of childhood cancer: a report from the childhood cancer survivor study. J Clin Oncol. 2008;26:3038–3045. doi: 10.1200/JCO.2007.14.9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattano LA, Jr., Nachman JB, Devidas M, Winick N, Raetz E, Carroll WL, et al. Increased Incidence of Osteonecrosis (ON) with a Dexamethasone (DEX) Induction for High Risk Acute Lymphoblastic Leukemia (HR-ALL): A Report from the Children's Oncology Group (COG) [Abstract] Blood. 2008 Nov;16:333–334. [Google Scholar]
- 31.Teuffel O, Kuster SP, Hunger SP, Conter V, Hitzler J, Ethier MC, et al. Dexamethasone versus prednisone for induction therapy in childhood acute lymphoblastic leukemia: a systematic review and meta-analysis. Leukemia. 2011;25:1232–1238. doi: 10.1038/leu.2011.84. [DOI] [PubMed] [Google Scholar]
- 32.Relling MV, Yang W, Das S, Cook EH, Rosner GL, Neel M, et al. Pharmacogenetic risk factors for osteonecrosis of the hip among children with leukemia. J Clin Oncol. 2004;22:3930–3936. doi: 10.1200/JCO.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 33.Patel B, Richards SM, Rowe JM, Goldstone AH, Fielding AK. High incidence of avascular necrosis in adolescents with acute lymphoblastic leukaemia: a UKALL XII analysis. Leukemia. 2008;22:308–312. doi: 10.1038/sj.leu.2405032. [DOI] [PubMed] [Google Scholar]
- 34.Ribeiro RC, Fletcher BD, Kennedy W, Harrison PL, Neel MD, Kaste SC, et al. Magnetic resonance imaging detection of avascular necrosis of the bone in children receiving intensive prednisone therapy for acute lymphoblastic leukemia or non-Hodgkin lymphoma. Leukemia. 2001;15:891–897. doi: 10.1038/sj.leu.2402139. [DOI] [PubMed] [Google Scholar]
- 35.Arico M, Boccalatte MF, Silvestri D, Barisone E, Messina C, Chiesa R, et al. Osteonecrosis: An emerging complication of intensive chemotherapy for childhood acute lymphoblastic leukemia. Haematologica. 2003;88:747–753. [PubMed] [Google Scholar]
- 36.Hanada T, Horigome Y, Inudoh M, Takita H. Osteonecrosis of vertebrae in a child with acute lymphocytic leukaemia during L-asparaginase therapy. Eur J Pediatr. 1989;149:162–163. doi: 10.1007/BF01958270. [DOI] [PubMed] [Google Scholar]
- 37.Sala A, Mattano LA, Jr., Barr RD. Osteonecrosis in children and adolescents with cancer - an adverse effect of systemic therapy. Eur J Cancer. 2007;43:683–689. doi: 10.1016/j.ejca.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Harper PG, Trask C, Souhami RL. Avascular necrosis of bone caused by combination chemotherapy without corticosteroids. Br Med J (Clin Res Ed) 1984;288:267–268. doi: 10.1136/bmj.288.6413.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marymont JV, Kaufman EE. Osteonecrosis of bone associated with combination chemotherapy without corticosteroids. Clin Orthop Relat Res. 1986:150–153. [PubMed] [Google Scholar]
- 40.Ishii E, Yoshida N, Miyazaki S. Avascular necrosis of bone in neuroblastoma treated with combination chemotherapy. Eur J Pediatr. 1984;143:152–153. doi: 10.1007/BF00445806. [DOI] [PubMed] [Google Scholar]
- 41.Abhyankar D, Nair R, Menon H, Kapoor B, Advani S. Avascular necrosis of head of femur in a patient with acute promyelocytic leukemia. Leuk Lymphoma. 2000;37:635–637. doi: 10.3109/10428190009058519. [DOI] [PubMed] [Google Scholar]
- 42.Smith RW, Margulis RR, Brennan MJ, Monto RW. The influence of ACTH and cortisone on certain factors of blood coagulation. Science. 1950;112:295–297. doi: 10.1126/science.112.2907.295. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto T, Hirano K, Tsutsui H, Sugioka Y, Sueishi K. Corticosteroid enhances the experimental induction of osteonecrosis in rabbits with Shwartzman reaction. Clin Orthop Relat Res. 1995:235–243. [PubMed] [Google Scholar]
- 44.Glueck CJ, Freiberg RA, Fontaine RN, Tracy T, Wang P. Hypofibrinolysis, thrombophilia, osteonecrosis. Clin Orthop Relat Res. 2001:19–33. doi: 10.1097/00003086-200105000-00004. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto T, Irisa T, Sugioka Y, Sueishi K. Effects of pulse methylprednisolone on bone and marrow tissues: corticosteroid-induced osteonecrosis in rabbits. Arthritis Rheum. 1997;40:2055–2064. doi: 10.1002/art.1780401119. [DOI] [PubMed] [Google Scholar]
- 46.Yun SI, Yoon HY, Jeong SY, Chung YS. Glucocorticoid induces apoptosis of osteoblast cells through the activation of glycogen synthase kinase 3beta. J Bone Miner Metab. 2009;27:140–148. doi: 10.1007/s00774-008-0019-5. [DOI] [PubMed] [Google Scholar]
- 47.French D, Hamilton LH, Mattano LA, Jr, Sather HN, Devidas M, Nachman JB, et al. A PAI-1 (SERPINE1) polymorphism predicts osteonecrosis in children with acute lymphoblastic leukemia: a report from the Children's Oncology Group. Blood. 2008;111:4496–4499. doi: 10.1182/blood-2007-11-123885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asano T, Takahashi KA, Fujioka M, Inoue S, Satomi Y, Nishino H, et al. Genetic analysis of steroid-induced osteonecrosis of the femoral head. J Orthop Sci. 2003;8:329–333. doi: 10.1007/s10776-003-0646-7. [DOI] [PubMed] [Google Scholar]
- 49.Hadjigeorgiou G, Dardiotis E, Dardioti M, Karantanas A, Dimitroulias A, Malizos K. Genetic association studies in osteonecrosis of the femoral head: mini review of the literature. Skeletal Radiol. 2008;37:1–7. doi: 10.1007/s00256-007-0395-2. [DOI] [PubMed] [Google Scholar]
- 50.Bottini N, MacMurray J, Peters W, Rostamkhani M, Comings DE. Association of the acid phosphatase (ACP1) gene with triglyceride levels in obese women. Mol Genet Metab. 2002;77:226–229. doi: 10.1016/s1096-7192(02)00120-8. [DOI] [PubMed] [Google Scholar]
- 51.Zambuzzi WF, Granjeiro JM, Parikh K, Yuvaraj S, Peppelenbosch MP, Ferreira CV. Modulation of Src activity by low molecular weight protein tyrosine phosphatase during osteoblast differentiation. Cell Physiol Biochem. 2008;22:497–506. doi: 10.1159/000185506. [DOI] [PubMed] [Google Scholar]
- 52.Kawedia JD, Kaste SC, Pei D, Panetta JC, Cai X, Cheng C, et al. Pharmacokinetic, pharmacodynamic, and pharmacogenetic determinants of osteonecrosis in children with acute lymphoblastic leukemia. Blood. 2011;117:2340–2347. doi: 10.1182/blood-2010-10-311969. quiz 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schell MJ, McHaney VA, Green AA, Kun LE, Hayes FA, Horowitz M, et al. Hearing loss in children and young adults receiving cisplatin with or without prior cranial irradiation. J Clin Oncol. 1989;7:754–760. doi: 10.1200/JCO.1989.7.6.754. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Womer RB, Silber JH. Predicting cisplatin ototoxicity in children: the influence of age and the cumulative dose. Eur J Cancer. 2004;40:2445–2451. doi: 10.1016/j.ejca.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 55.Coradini PP, Cigana L, Selistre SG, Rosito LS, Brunetto AL. Ototoxicity from cisplatin therapy in childhood cancer. J Pediatr Hematol Oncol. 2007;29:355–360. doi: 10.1097/MPH.0b013e318059c220. [DOI] [PubMed] [Google Scholar]
- 56.Knight KR, Kraemer DF, Neuwelt EA. Ototoxicity in children receiving platinum chemotherapy: underestimating a commonly occurring toxicity that may influence academic and social development. J Clin Oncol. 2005;23:8588–8596. doi: 10.1200/JCO.2004.00.5355. [DOI] [PubMed] [Google Scholar]
- 57.Kushner BH, Budnick A, Kramer K, Modak S, Cheung NK. Ototoxicity from high-dose se of platinum compounds in patients with neuroblastoma. Cancer. 2006;107:417–422. doi: 10.1002/cncr.22004. [DOI] [PubMed] [Google Scholar]
- 58.Blakley BW, Gupta AK, Myers SF, Schwan S. Risk factors for ototoxicity due to cisplatin. Arch Otolaryngol Head Neck Surg. 1994;120:541–546. doi: 10.1001/archotol.1994.01880290051009. [DOI] [PubMed] [Google Scholar]
- 59.Bokemeyer C, Berger CC, Kuczyk MA, Schmoll HJ. Evaluation of long-term toxicity after hemotherapy for testicular cancer. J Clin Oncol. 1996;14:2923–2932. doi: 10.1200/JCO.1996.14.11.2923. [DOI] [PubMed] [Google Scholar]
- 60.Fossa SD. Long-term sequelae after cancer therapy--survivorship after treatment for testicular cancer. Acta Oncol. 2004;43:134–141. doi: 10.1080/02841860310023174. [DOI] [PubMed] [Google Scholar]
- 61.Hua C, Bass JK, Khan R, Kun LE, Merchant TE. Hearing loss after radiotherapy for pediatric brain tumors: effect of cochlear dose. Int J Radiat Oncol Biol Phys. 2008;72:892–899. doi: 10.1016/j.ijrobp.2008.01.050. [DOI] [PubMed] [Google Scholar]
- 62.Low WK, Toh ST, Wee J, Fook-Chong SM, Wang DY. Sensorineural hearing loss after radiotherapy and chemoradiotherapy: a single, blinded, randomized study. J Clin Oncol. 2006;24:1904–1909. doi: 10.1200/JCO.2005.05.0096. [DOI] [PubMed] [Google Scholar]
- 63.Merchant TE, Hua CH, Shukla H, Ying X, Nill S, Oelfke U. Proton versus photon radiotherapy for common pediatric brain tumors: comparison of models of dose characteristics and their relationship to cognitive function. Pediatr Blood Cancer. 2008;51:110–117. doi: 10.1002/pbc.21530. [DOI] [PubMed] [Google Scholar]
- 64.van Ruijven MW, de Groot JC, Smoorenburg GF. Time sequence of degeneration pattern in the guinea pig cochlea during cisplatin administration. A quantitative histological study. Hear Res. 2004;197:44–54. doi: 10.1016/j.heares.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 65.Roland JT, Cohen NL. Vestibular and auditory toxicity. In: Cummings CW, Fredrickson JM, Harker LA, et al., editors. Otolaryngology Head and Neck Surgery. ed 3rd. St. Louis, MO: Mosby; 1998. pp. 3186–3197. [Google Scholar]
- 66.Paulino AC, Simon JH, Zhen W, Wen BC. Long-term effects in children treated with radiotherapy for head and neck rhabdomyosarcoma. Int J Radiat Oncol Biol Phys. 2000;48:1489–1495. doi: 10.1016/s0360-3016(00)00799-9. [DOI] [PubMed] [Google Scholar]
- 67.Oldenburg J, Kraggerud SM, Cvancarova M, Lothe RA, Fossa SD. Cisplatin-induced long-term hearing impairment is associated with specific glutathione s-transferase genotypes in testicular cancer survivors. J Clin Oncol. 2007;25:708–714. doi: 10.1200/JCO.2006.08.9599. [DOI] [PubMed] [Google Scholar]
- 68.Peters U, Preisler-Adams S, Hebeisen A, Hahn M, Seifert E, Lanvers C, et al. Glutathione S-transferase genetic polymorphisms and individual sensitivity to the ototoxic effect of cisplatin. Anticancer Drugs. 2000;11:639–643. doi: 10.1097/00001813-200009000-00007. [DOI] [PubMed] [Google Scholar]
- 69.Riedemann L, Lanvers C, Deuster D, Peters U, Boos J, Jurgens H, et al. Megalin genetic polymorphisms and individual sensitivity to the ototoxic effect of cisplatin. Pharmacogenomics J. 2008;8:23–28. doi: 10.1038/sj.tpj.6500455. [DOI] [PubMed] [Google Scholar]
- 70.Ross CJ, Katzov-Eckert H, Dube MP, Brooks B, Rassekh SR, Barhdadi A, et al. Genetic variants in TPMT and COMT are associated with hearing loss in children receiving cisplatin chemotherapy. Nat Genet. 2009;41:1345–1349. doi: 10.1038/ng.478. [DOI] [PubMed] [Google Scholar]
- 71.Ochoa B, Bobadilla N, Arrellin G, Herrera LA. S-Adenosyl-L-methionine increases serum BUN and creatinine in cisplatin-treated mice. Arch Med Res. 2009;40:54–58. doi: 10.1016/j.arcmed.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 72.Dalton VK, Rue M, Silverman LB, Gelber RD, Asselin BL, Barr RD, et al. Height and weight in children treated for acute lymphoblastic leukemia: relationship to CNS treatment. J Clin Oncol. 2003;21:2953–2960. doi: 10.1200/JCO.2003.03.068. [DOI] [PubMed] [Google Scholar]
- 73.Oeffinger KC, Mertens AC, Sklar CA, Yasui Y, Fears T, Stovall M, et al. Obesity in adult survivors of childhood acute lymphoblastic leukemia: a report from the Childhood Cancer Survivor Study. J Clin Oncol. 2003;21:1359–1365. doi: 10.1200/JCO.2003.06.131. [DOI] [PubMed] [Google Scholar]
- 74.Razzouk BI, Rose SR, Hongeng S, Wallace D, Smeltzer MP, Zacher M, et al. Obesity in survivors of childhood acute lymphoblastic leukemia and lymphoma. J Clin Oncol. 2007;25:1183–1189. doi: 10.1200/JCO.2006.07.8709. [DOI] [PubMed] [Google Scholar]
- 75.Garmey EG, Liu Q, Sklar CA, Meacham LR, Mertens AC, Stovall MA, et al. Longitudinal changes in obesity and body mass index among adult survivors of childhood acute lymphoblastic leukemia: a report from the Childhood Cancer Survivor Study. J Clin Oncol. 2008;26:4639–4645. doi: 10.1200/JCO.2008.16.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brennan BM, Rahim A, Blum WF, Adams JA, Eden OB, Shalet SM. Hyperleptinaemia in young adults following cranial irradiation in childhood: growth hormone deficiency or leptin insensitivity? Clin Endocrinol (Oxf) 1999;50:163–169. doi: 10.1046/j.1365-2265.1999.00622.x. [DOI] [PubMed] [Google Scholar]
- 77.Strosberg AD, Issad T. The involvement of leptin in humans revealed by mutations in leptin and leptin receptor genes. Trends Pharmacol Sci. 1999;20:227–230. doi: 10.1016/s0165-6147(99)01313-9. [DOI] [PubMed] [Google Scholar]
- 78.Young EH, Wareham NJ, Farooqi S, Hinney A, Hebebrand J, Scherag A, et al. The V103I polymorphism of the MC4R gene and obesity: population based studies and meta-analysis of 29 563 individuals. Int J Obes (Lond) 2007;31:1437–1441. doi: 10.1038/sj.ijo.0803609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Geller F, Reichwald K, Dempfle A, Illig T, Vollmert C, Herpertz S, et al. Melanocortin-4 receptor gene variant I103 is negatively associated with obesity. Am J Hum Genet. 2004;74:572–581. doi: 10.1086/382490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heid IM, Vollmert C, Hinney A, Doring A, Geller F, Lowel H, et al. Association of the 103I MC4R allele with decreased body mass in 7937 participants of two population based surveys. J Med Genet. 2005;42:e21. doi: 10.1136/jmg.2004.027011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 83.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ross JA, Oeffinger KC, Davies SM, Mertens AC, Langer EK, Kiffmeyer WR, et al. Genetic variation in the leptin receptor gene and obesity in survivors of childhood acute lymphoblastic leukemia: a report from the Childhood Cancer Survivor Study. J Clin Oncol. 2004;22:3558–3562. doi: 10.1200/JCO.2004.11.152. [DOI] [PubMed] [Google Scholar]
- 85.Friedman DL, Whitton J, Leisenring W, Mertens AC, Hammond S, Stovall M, et al. Subsequent neoplasms in 5-year survivors of childhood cancer: the Childhood Cancer Survivor Study. J Natl Cancer Inst. 2010;102:1083–1095. doi: 10.1093/jnci/djq238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reulen RC, Frobisher C, Winter DL, Kelly J, Lancashire ER, Stiller CA, et al. Long-term risks of subsequent primary neoplasms among survivors of childhood cancer. JAMA. 2011;305:2311–2319. doi: 10.1001/jama.2011.747. [DOI] [PubMed] [Google Scholar]
- 87.Bhatia S, Robison LL, Francisco L, Carter A, Liu Y, Grant M, et al. Late mortality in survivors of autologous hematopoietic-cell transplantation: report from the Bone Marrow Transplant Survivor Study. Blood. 2005;105:4215–4222. doi: 10.1182/blood-2005-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mertens AC, Liu Q, Neglia JP, Wasilewski K, Leisenring W, Armstrong GT, et al. Cause-specific late mortality among 5-year survivors of childhood cancer: the Childhood Cancer Survivor Study. J Natl Cancer Inst. 2008;100:1368–1379. doi: 10.1093/jnci/djn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bhatia S, Robison LL, Oberlin O, Greenberg M, Bunin G, Fossati-Bellani F, et al. Breast cancer and other second neoplasms after childhood Hodgkin's disease. N Engl J Med. 1996;334:745–751. doi: 10.1056/NEJM199603213341201. [DOI] [PubMed] [Google Scholar]
- 90.Neglia JP, Robison LL, Stovall M, Liu Y, Packer RJ, Hammond S, et al. New primary neoplasms of the central nervous system in survivors of childhood cancer: a report from the Childhood Cancer Survivor Study. J Natl Cancer Inst. 2006;98:1528–1537. doi: 10.1093/jnci/djj411. [DOI] [PubMed] [Google Scholar]
- 91.Neglia JP, Meadows AT, Robison LL, Kim TH, Newton WA, Ruymann FB, et al. Second neoplasms after acute lymphoblastic leukemia in childhood. N Engl J Med. 1991;325:1330–1336. doi: 10.1056/NEJM199111073251902. [DOI] [PubMed] [Google Scholar]
- 92.Bhatia S, Krailo MD, Chen Z, Burden L, Askin FB, Dickman PS, et al. Therapy-related myelodysplasia and acute myeloid leukemia after Ewing sarcoma and primitive neuroectodermal tumor of bone: A report from the Children's Oncology Group. Blood. 2007;109:46–51. doi: 10.1182/blood-2006-01-023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bhatia S, Yasui Y, Robison LL, Birch JM, Bogue MK, Diller L, et al. High Risk of Subsequent Neoplasms Continues With Extended Follow-Up of Childhood Hodgkin's Disease: Report From the Late Effects Study Group. J Clin Oncol. 2003;21:4386–4394. doi: 10.1200/JCO.2003.11.059. [DOI] [PubMed] [Google Scholar]
- 94.Bhatia S, Sklar C. Second cancers in survivors of childhood cancer. Nat Rev Cancer. 2002;2:124–132. doi: 10.1038/nrc722. [DOI] [PubMed] [Google Scholar]
- 95.Bhatia S, Sather HN, Pabustan OB, Trigg ME, Gaynon PS, Robison LL. Low incidence of second neoplasms among children diagnosed with acute lymphoblastic leukemia after 1983. Blood. 2002;99:4257–4264. doi: 10.1182/blood.v99.12.4257. [DOI] [PubMed] [Google Scholar]
- 96.Travis LB, Andersson M, Gospodarowicz M, van Leeuwen FE, Bergfeldt K, Lynch CF, et al. Treatment-associated leukemia following testicular cancer. J Natl Cancer Inst. 2000;92:1165–1171. doi: 10.1093/jnci/92.14.1165. [DOI] [PubMed] [Google Scholar]
- 97.Travis LB, Holowaty EJ, Bergfeldt K, Lynch CF, Kohler BA, Wiklund T, et al. Risk of leukemia after platinum-based chemotherapy for ovarian cancer. N Engl J Med. 1999;340:351–357. doi: 10.1056/NEJM199902043400504. [DOI] [PubMed] [Google Scholar]
- 98.Krishnan A, Bhatia S, Slovak ML, Arber DA, Niland JC, Nademanee A, et al. Predictors of therapy-related leukemia and myelodysplasia following autologous transplantation for lymphoma: an assessment of risk factors. Blood. 2000;95:1588–1593. [PubMed] [Google Scholar]
- 99.Howe R, Micallef IN, Inwards DJ, Ansell SM, Dewald GW, Dispenzieri A, et al. Secondary myelodysplastic syndrome and acute myelogenous leukemia are significant complications following autologous stem cell transplantation for lymphoma. Bone Marrow Transplant. 2003;32:317–324. doi: 10.1038/sj.bmt.1704124. [DOI] [PubMed] [Google Scholar]
- 100.Laughlin MJ, McGaughey DS, Crews JR, Chao NJ, Rizzieri D, Ross M, et al. Secondary myelodysplasia and acute leukemia in breast cancer patients after autologous bone marrow transplant. J Clin Oncol. 1998;16:1008–1012. doi: 10.1200/JCO.1998.16.3.1008. [DOI] [PubMed] [Google Scholar]
- 101.Stone RM. Myelodysplastic syndrome after autologous transplantation for lymphoma: the price of progress. Blood. 1994;83:3437–3440. [PubMed] [Google Scholar]
- 102.Govindarajan R, Jagannath S, Flick JT, Vesole DH, Sawyer J, Barlogie B, et al. Preceding standard therapy is the likely cause of MDS after autotransplants for multiple myeloma. Br J Haematol. 1996;95:349–353. doi: 10.1046/j.1365-2141.1996.d01-1891.x. [DOI] [PubMed] [Google Scholar]
- 103.Bhatia S, Ramsay NKC, Steinbuch M, Dusenbury KE, Shapiro RS, Weisdorf DJ, et al. Malignant neoplasms following bone marrow transplantation. Blood. 1996;87:3633–3639. [PubMed] [Google Scholar]
- 104.Sigurdson AJ, Ronckers CM, Mertens AC, Stovall M, Smith SA, Liu Y, et al. Primary thyroid cancer after a first tumour in childhood (the Childhood Cancer Survivor Study): a nested case-control study. Lancet. 2005;365:2014–2023. doi: 10.1016/S0140-6736(05)66695-0. [DOI] [PubMed] [Google Scholar]
- 105.Neglia JP, Friedman DL, Yasui Y, Mertens AC, Hammond S, Stovall M, et al. Second malignant neoplasms in five-year survivors of childhood cancer: childhood cancer survivor study. J Natl Cancer Inst. 2001;93:618–629. doi: 10.1093/jnci/93.8.618. [DOI] [PubMed] [Google Scholar]
- 106.Boice JD, Jr, Land C, Preston DL. Ionizing radiation. In: Schottenfield D, Fraumeni JF Jr, editors. Cancer epidemiology and prevention. New York Oxford University Press; 1996. pp. 319–354. [Google Scholar]
- 107.Preston DL, Kusumi S, Tomonaga M, Izumi S, Ron E, Kuramoto A, et al. Cancer incidence in atomic bomb survivors. Part III. Leukemia, lymphoma and multiple myeloma, 1950–1987. Radiat Res. 1994;137:S68–S97. [PubMed] [Google Scholar]
- 108.Thompson DE, Mabuchi K, Ron E, Soda M, Tokunaga M, Ochikubo S, et al. Cancer incidence in atomic bomb survivors. Part II: Solid tumors, 1958–1987. Radiat Res. 1994;137:S17–S67. [PubMed] [Google Scholar]
- 109.Ron E, Modan B, Preston D, Alfandary E, Stovall M, Boice JD., Jr Radiation-induced skin carcinomas of the head and neck. Radiat Res. 1991;125:318–325. [PubMed] [Google Scholar]
- 110.Wong FL, Boice JD, Jr, Abramson DH, Tarone RE, Kleinerman RA, Stovall M, et al. Cancer incidence after retinoblastoma. Radiation dose and sarcoma risk. JAMA. 1997;278:1262–1267. doi: 10.1001/jama.278.15.1262. [DOI] [PubMed] [Google Scholar]
- 111.Strong LC, Stine M, Norsted TL. Cancer in survivors of childhood soft tissue sarcoma and their relatives. J Natl Cancer Inst. 1987;79:1213–1220. [PubMed] [Google Scholar]
- 112.Land CE, Saku T, Hayashi Y, Takahara O, Matsuura H, Tokuoka S, et al. Incidence of salivary gland tumors among atomic bomb survivors, 1950–1987. Evaluation of radiation-related risk. Radiat Res. 1996;146:28–36. [PubMed] [Google Scholar]
- 113.Kenney LB, Yasui Y, Inskip PD, Hammond S, Neglia JP, Mertens AC, et al. Breast cancer after childhood cancer: a report from the Childhood Cancer Survivor Study. Ann Intern Med. 2004;141:590–597. doi: 10.7326/0003-4819-141-8-200410190-00006. [DOI] [PubMed] [Google Scholar]
- 114.Schoch C, Kern W, Schnittger S, Hiddemann W, Haferlach T. Karyotype is an independent prognostic parameter in therapy-related acute myeloid leukemia (t-AML): an analysis of 93 patients with t-AML in comparison to 1091 patients with de novo AML. Leukemia. 2004;18:120–125. doi: 10.1038/sj.leu.2403187. [DOI] [PubMed] [Google Scholar]
- 115.Smith SM, Le Beau MM, Huo D, Karrison T, Sobecks RM, Anastasi J, et al. Clinical-cytogenetic associations in 306 patients with therapy-related myelodysplasia and myeloid leukemia: the University of Chicago series. Blood. 2003;102:43–52. doi: 10.1182/blood-2002-11-3343. [DOI] [PubMed] [Google Scholar]
- 116.Pedersen-Bjergaard J, Christiansen DH, Desta F, Andersen MK. Alternative genetic pathways and cooperating genetic abnormalities in the pathogenesis of therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2006;20:1943–1949. doi: 10.1038/sj.leu.2404381. [DOI] [PubMed] [Google Scholar]
- 117.Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100:2292–2302. doi: 10.1182/blood-2002-04-1199. [DOI] [PubMed] [Google Scholar]
- 118.van Leeuwen FE, Travis LB. Second Cancers. In: DeVita HS, Rosenberg SA, editors. Cancer: principles and practice of oncology 7th edition. Philadelphia: Lippincott Williams and Wilkins; 2005. pp. 2575–2602. [Google Scholar]
- 119.Michels SD, McKenna RW, Arthur DC, Brunning RD. Therapy-related acute myeloid leukemia and myelodysplastic syndrome: a clinical and morphologic study of 65 cases. Blood. 1985;65:1364–1372. [PubMed] [Google Scholar]
- 120.Travis LB, Hill D, Dores GM, Gospodarowicz M, van Leeuwen FE, Holowaty E, et al. Cumulative absolute breast cancer risk for young women treated for Hodgkin lymphoma. J Natl Cancer Inst. 2005;97:1428–1437. doi: 10.1093/jnci/dji290. [DOI] [PubMed] [Google Scholar]
- 121.Henderson TO, Whitton J, Stovall M, Mertens AC, Mitby P, Friedman D, et al. Secondary sarcomas in childhood cancer survivors: a report from the Childhood Cancer Survivor Study. J Natl Cancer Inst. 2007;99:300–308. doi: 10.1093/jnci/djk052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Limacher JM, Frebourg T, Natarajan-Ame S, Bergerat JP. Two metachronous tumors in the radiotherapy fields of a patient with Li-Fraumeni syndrome. Int J Cancer. 2001;96:238–242. doi: 10.1002/ijc.1021. [DOI] [PubMed] [Google Scholar]
- 123.Alter BP. Cancer in Fanconi anemia, 1927–2001. Cancer. 2003;97:425–440. doi: 10.1002/cncr.11046. [DOI] [PubMed] [Google Scholar]
- 124.Tischkowitz M, Dokal I. Fanconi anaemia and leukaemia - clinical and molecular aspects. Br J Haematol. 2004;126:176–191. doi: 10.1111/j.1365-2141.2004.05023.x. [DOI] [PubMed] [Google Scholar]
- 125.Rosenberg PS, Alter BP, Ebell W. Cancer risks in Fanconi anemia: findings from the German Fanconi Anemia Registry. Haematologica. 2008;93:511–517. doi: 10.3324/haematol.12234. [DOI] [PubMed] [Google Scholar]
- 126.Kennedy RD, D'Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 127.Kalow W, Ozdemir V, Tang BK, Tothfalusi L, Endrenyi L. The science of pharmacological variability: an essay. Clin Pharmacol Ther. 1999;66:445–447. doi: 10.1016/S0009-9236(99)70006-8. [DOI] [PubMed] [Google Scholar]
- 128.Evans WE, McLeod HL. Pharmacogenomics--drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 129.Berwick M, Vineis P. Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J Natl Cancer Inst. 2000;92:874–897. doi: 10.1093/jnci/92.11.874. [DOI] [PubMed] [Google Scholar]
- 130.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–1530. [PubMed] [Google Scholar]
- 131.Bhatti P, Doody MM, Alexander BH, Yuenger J, Simon SL, Weinstock RM, et al. Breast cancer risk polymorphisms and interaction with ionizing radiation among U.S. radiologic technologists. Cancer Epidemiol Biomarkers Prev. 2008;17:2007–2011. doi: 10.1158/1055-9965.EPI-08-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bhatti P, Struewing JP, Alexander BH, Hauptmann M, Bowen L, Mateus-Pereira LH, et al. Polymorphisms in DNA repair genes, ionizing radiation exposure and risk of breast cancer in U.S. Radiologic technologists. Int J Cancer. 2008;122:177–182. doi: 10.1002/ijc.23066. [DOI] [PubMed] [Google Scholar]
- 133.Rajaraman P, Bhatti P, Doody MM, Simon SL, Weinstock RM, Linet MS, et al. Nucleotide excision repair polymorphisms may modify ionizing radiation-related breast cancer risk in US radiologic technologists. Int J Cancer. 2008;123:2713–2716. doi: 10.1002/ijc.23779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Worrillow LJ, Travis LB, Smith AG, Rollinson S, Smith AJ, Wild CP, et al. An intron splice acceptor polymorphism in hMSH2 and risk of leukemia after treatment with chemotherapeutic alkylating agents. Clin Cancer Res. 2003;9:3012–3020. [PubMed] [Google Scholar]
- 135.Allan JM, Smith AG, Wheatley K, Hills RK, Travis LB, Hill DA, et al. Genetic variation in XPD predicts treatment outcome and risk of acute myeloid leukemia following chemotherapy. Blood. 2004;104:3872–3877. doi: 10.1182/blood-2004-06-2161. [DOI] [PubMed] [Google Scholar]
- 136.Mertens AC, Mitby PA, Radloff G, Jones IM, Perentesis J, Kiffmeyer WR, et al. XRCC1 and glutathione-S-transferase gene polymorphisms and susceptibility to radiotherapy-related malignancies in survivors of Hodgkin disease. Cancer. 2004;101:1463–1472. doi: 10.1002/cncr.20520. [DOI] [PubMed] [Google Scholar]
- 137.Worrillow LJ, Smith AG, Scott K, Andersson M, Ashcroft AJ, Dores GM, et al. Polymorphic MLH1 and risk of cancer after methylating chemotherapy for Hodgkin lymphoma. J Med Genet. 2008;45:142–146. doi: 10.1136/jmg.2007.053850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.McFadyen MC, Melvin WT, Murray GI. Cytochrome P450 enzymes: novel options for cancer therapeutics. Mol Cancer Ther. 2004;3:363–371. [PubMed] [Google Scholar]
- 139.Park JY, Shigenaga MK, Ames BN. Induction of cytochrome P4501A1 by 2,3,7,8-tetrachlorodibenzo-p-dioxin or indolo(3,2-b)carbazole is associated with oxidative DNA damage. Proc Natl Acad Sci U S A. 1996;93:2322–2327. doi: 10.1073/pnas.93.6.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 141.Allan JM, Wild CP, Rollinson S, Willett EV, Moorman AV, Dovey GJ, et al. Polymorphism in glutathione S-transferase P1 is associated with susceptibility to chemotherapy-induced leukemia. Proc Natl Acad Sci U S A. 2001;98:11592–11597. doi: 10.1073/pnas.191211198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rund D, Krichevsky S, Bar-Cohen S, Goldschmidt N, Kedmi M, Malik E, et al. Therapy-related leukemia: clinical characteristics and analysis of new molecular risk factors in 96 adult patients. Leukemia. 2005;19:1919–1928. doi: 10.1038/sj.leu.2403947. [DOI] [PubMed] [Google Scholar]
- 143.Collins A, Harrington V. Repair of oxidative DNA damage: assessing its contribution to cancer prevention. Mutagenesis. 2002;17:489–493. doi: 10.1093/mutage/17.6.489. [DOI] [PubMed] [Google Scholar]
- 144.Karran P, Offman J, Bignami M. Human mismatch repair, drug-induced DNA damage, and secondary cancer. Biochimie. 2003;85:1149–1160. doi: 10.1016/j.biochi.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 145.Casorelli I, Offman J, Mele L, Pagano L, Sica S, D'Errico M, et al. Drug treatment in the development of mismatch repair defective acute leukemia and myelodysplastic syndrome. DNA Repair (Amst) 2003;2:547–559. doi: 10.1016/s1568-7864(03)00020-x. [DOI] [PubMed] [Google Scholar]
- 146.Seedhouse CH, Das-Gupta EP, Russell NH. Methylation of the hMLH1 promoter and its association with microsatellite instability in acute myeloid leukemia. Leukemia. 2003;17:83–88. doi: 10.1038/sj.leu.2402747. [DOI] [PubMed] [Google Scholar]
- 147.Zhu YM, Das-Gupta EP, Russell NH. Microsatellite instability and p53 mutations are associated with abnormal expression of the MSH2 gene in adult acute leukemia. Blood. 1999;94:733–740. [PubMed] [Google Scholar]
- 148.Horiike S, Misawa S, Kaneko H, Sasai Y, Kobayashi M, Fujii H, et al. Distinct genetic involvement of the TP53 gene in therapy-related leukemia and myelodysplasia with chromosomal losses of Nos 5 and/or 7 and its possible relationship to replication error phenotype. Leukemia. 1999;13:1235–1242. doi: 10.1038/sj.leu.2401466. [DOI] [PubMed] [Google Scholar]
- 149.Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 150.Brentnall TA, Rubin CE, Crispin DA, Stevens A, Batchelor RH, Haggitt RC, et al. A germline substitution in the human MSH2 gene is associated with high-grade dysplasia and cancer in ulcerative colitis. Gastroenterology. 1995;109:151–155. doi: 10.1016/0016-5085(95)90280-5. [DOI] [PubMed] [Google Scholar]
- 151.Worrillow LJ, Allan JM. Deregulation of homologous recombination DNA repair in alkylating agent-treated stem cell clones: a possible role in the aetiology of chemotherapy-induced leukaemia. Oncogene. 2006;25:1709–1720. doi: 10.1038/sj.onc.1209208. [DOI] [PubMed] [Google Scholar]
- 152.O'Driscoll M, Jeggo PA. The role of double-strand break repair - insights from human genetics. Nat Rev Genet. 2006;7:45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- 153.Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 154.Jawad M, Seedhouse CH, Russell N, Plumb M. Polymorphisms in human homeobox HLX1 and DNA repair RAD51 genes increase the risk of therapy-related acute myeloid leukemia. Blood. 2006;108:3916–3918. doi: 10.1182/blood-2006-05-022921. [DOI] [PubMed] [Google Scholar]
- 155.Seedhouse C, Faulkner R, Ashraf N, Das-Gupta E, Russell N. Polymorphisms in genes involved in homologous recombination repair interact to increase the risk of developing acute myeloid leukemia. Clin Cancer Res. 2004;10:2675–2680. doi: 10.1158/1078-0432.ccr-03-0372. [DOI] [PubMed] [Google Scholar]
- 156.Bishop DK, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum RR, et al. Xrcc3 is required for assembly of Rad51 complexes in vivo. J Biol Chem. 1998;273:21482–21488. doi: 10.1074/jbc.273.34.21482. [DOI] [PubMed] [Google Scholar]
- 157.Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, et al. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- 158.Tebbs RS, Zhao Y, Tucker JD, Scheerer JB, Siciliano MJ, Hwang M, et al. Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc Natl Acad Sci U S A. 1995;92:6354–6358. doi: 10.1073/pnas.92.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604–608. [PubMed] [Google Scholar]
- 161.Matullo G, Palli D, Peluso M, Guarrera S, Carturan S, Celentano E, et al. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22:1437–1445. doi: 10.1093/carcin/22.9.1437. [DOI] [PubMed] [Google Scholar]
- 162.Au WW, Salama SA, Sierra-Torres CH. Functional characterization of polymorphisms in DNA repair genes using cytogenetic challenge assays. Environ Health Perspect. 2003;111:1843–1850. doi: 10.1289/ehp.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Coiteux V, Onclercq-Delic R, Fenaux P, Amor-Gueret M. Predisposition to therapy-related acute leukemia with balanced chromosomal translocations does not result from a major constitutive defect in DNA double-strand break end joining. Leuk Res. 2007;31:353–358. doi: 10.1016/j.leukres.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 164.Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol Cell Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 166.Seedhouse C, Bainton R, Lewis M, Harding A, Russell N, Das-Gupta E. The genotype distribution of the XRCC1 gene indicates a role for base excision repair in the development of therapy-related acute myeloblastic leukemia. Blood. 2002;100:3761–3766. doi: 10.1182/blood-2002-04-1152. [DOI] [PubMed] [Google Scholar]
- 167.Nelson HH, Kelsey KT, Mott LA, Karagas MR. The XRCC1 Arg399Gln polymorphism, sunburn, and non-melanoma skin cancer: evidence of gene-environment interaction. Cancer Res. 2002;62:152–155. [PubMed] [Google Scholar]
- 168.Wu X, Gu J, Amos CI, Jiang H, Hong WK, Spitz MR. A parallel study of in vitro sensitivity to benzo[a]pyrene diol epoxide and bleomycin in lung carcinoma cases and controls. Cancer. 1998;83:1118–1127. [PubMed] [Google Scholar]
- 169.Vijayalaxmi, Leal BZ, Deahl TS, Meltz ML. Variability in adaptive response to low dose radiation in human blood lymphocytes: consistent results from chromosome aberrations and micronuclei. Mutat Res. 1995;348:45–50. doi: 10.1016/0165-7992(95)90020-9. [DOI] [PubMed] [Google Scholar]
- 170.Godderis L, Aka P, Mateuca R, Kirsch-Volders M, Lison D, Veulemans H. Dose-dependent influence of genetic polymorphisms on DNA damage induced by styrene oxide, ethylene oxide and gamma-radiation. Toxicology. 2006;219:220–229. doi: 10.1016/j.tox.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 171.Marcon F, Andreoli C, Rossi S, Verdina A, Galati R, Crebelli R. Assessment of individual sensitivity to ionizing radiation and DNA repair efficiency in a healthy population. Mutat Res. 2003;541:1–8. doi: 10.1016/s1383-5718(03)00171-2. [DOI] [PubMed] [Google Scholar]
- 172.Collins FS, Brooks LD, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8:1229–1231. doi: 10.1101/gr.8.12.1229. [DOI] [PubMed] [Google Scholar]
- 173.Ellis NA, Huo D, Yildiz O, Worrillow LJ, Banerjee M, Le Beau MM, et al. MDM2 SNP309 and TP53 Arg72Pro interact to alter therapy-related acute myeloid leukemia susceptibility. Blood. 2008;112:741–749. doi: 10.1182/blood-2007-11-126508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Knight JA, Skol AD, Shinde A, Hastings D, Walgren RA, Shao J, et al. Genome-wide association study to identify novel loci associated with therapy-related myeloid leukemia susceptibility. Blood. 2009;113:5575–5582. doi: 10.1182/blood-2008-10-183244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Best T, Li D, Skol AD, Kirchhoff T, Jackson SA, Yasui Y, et al. Variants at 6q21 implicate PRDM1 in the etiology of therapy-induced second malignancies after Hodgkin's lymphoma. Nat Med. 2011;17:941–943. doi: 10.1038/nm.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]