Abstract
Current interest in amyloid fibrils stems from their involvement in neurodegenerative and other diseases and from their role as an alternative structural state for many peptides and proteins. Solid state NMR methods have the unique capability of providing detailed structural constraints for amyloid fibrils, sufficient for the development of full molecular models. In this article, recent progress in the application of solid state NMR to fibrils associated with Alzheimer’s disease, prion fibrils, and related systems is reviewed, along with relevant developments in solid state NMR techniques and technology.
Keywords: Alzheimer’s disease, protein structure, prion, nuclear magnetic resonance
I. Introduction
Amyloid fibrils are self-assembled, filamentous protein or peptide aggregates with cross-β molecular structures, meaning that they contain ribbon-like β-sheets in which the β-strands run approximately perpendicular to the long fibril axis and the inter-strand hydrogen bonds run approximately parallel the long fibril axis (1). Figure 1 shows transmission electron microscope (TEM) images of typical amyloid fibrils, including fibrils discussed in detail below, as well as cartoon representations of various possible cross-β structures.
Figure 1.
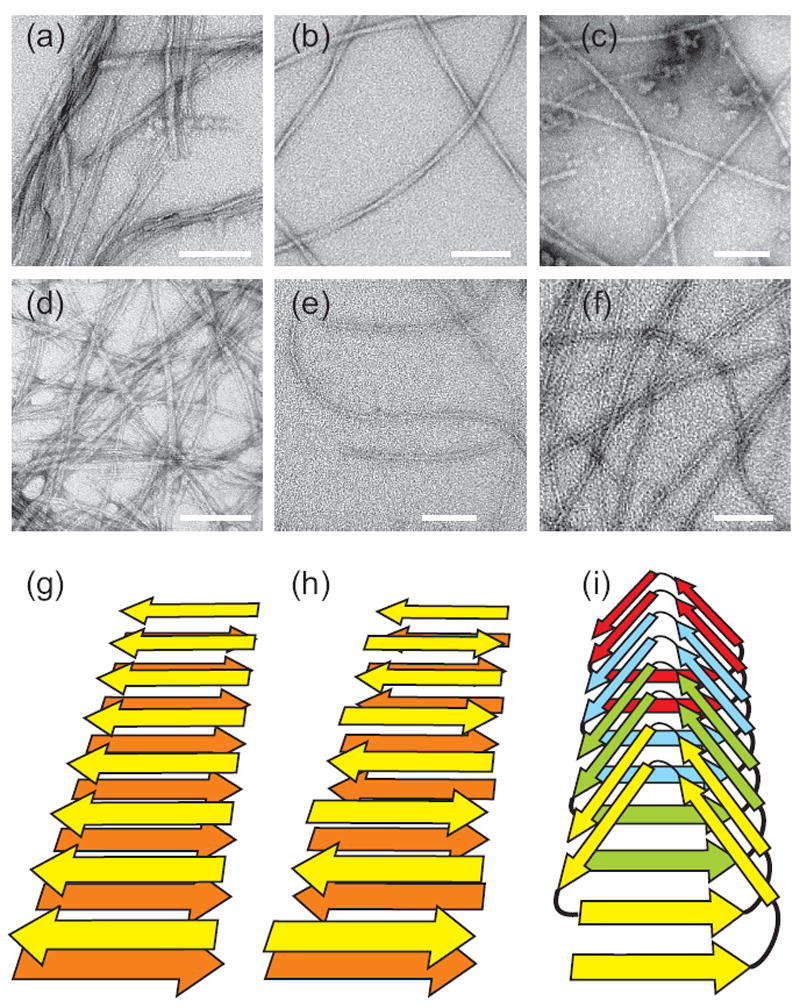
Negatively-stained TEM images of amyloid fibrils. (a) “Striated ribbon” Aβ1-40 fibrils. (b) “Twisted pair” Aβ1-40 fibrils. (c) Brain-seeded Aβ1-40 fibrils. (d) Amylin fibrils. (e) Sup35NM fibrils. (f) HET-s218-289 fibrils. White scale bars are 100 nm. (g,h,i) Schematic representations of cross-β structures formed by parallel β-sheets, antiparallel β-sheets, and a β-helix, respectively.
Current interest in the structural, physical, chemical, and biological properties of amyloid fibrils has two independent sources: (i) Amyloid fibrils form in the affected tissue of a class of diseases called “amyloid diseases” (2). These include several that are major public health problems, including Alzheimer’s disease (AD) and related neurodegenerative diseases, type 2 diabetes, Parkinson’s disease, dialysis-related amyloidosis, familial systemic amyloidosis, and Huntington’s disease. In each case, a different protein or peptide forms the fibrils, and the fibrils and/or their precursors are cytotoxic. Prion proteins that cause transmissible spongiform encephalopathies (TSEs) also form amyloid fibrils or related aggregates in affected brain tissue (3). One goal of amyloid research is therefore to contribute to our understanding of the etiology of amyloid diseases and to their prevention and cure; (ii) Apart from their involvement in disease, polypeptide chains have a generic propensity to form amyloid fibrils in aqueous solution (4). In other words, the amyloid fibril is frequently a stable structural state for a polypeptide that competes with its monomeric folded state (when a folded state exists), or with other non-aggregated states, almost irrespective of the amino acid sequence or amino acid composition. Another goal of amyloid fibril research is therefore to elucidate the physical chemical factors that stabilize amyloid structures and affect fibril assembly mechanisms.
Any understanding of the properties and effects of amyloid fibrils depends on knowledge of their molecular structures. Since amyloid fibrils are inherently noncrystalline and insoluble and are assembled from high-molecular-weight units, standard experimental approaches to structure determination provide limited information. Due to the ineffectiveness of other approaches and for a variety of technical reasons, solid state NMR spectroscopy has become the “method of choice” for amyloid structure determination. Technical reasons include the feasibility of preparing isotopically labeled fibril samples in the 1-10 mg quantities typically required for solid state NMR and the fact that fibril samples have high protein or peptide concentrations (25-100%, depending on hydration level), so that small sample volumes and concomitantly high radio-frequency (rf) field strengths and high magic-angle spinning (MAS) frequencies can be employed. Amyloid fibrils also typically exhibit favorable nuclear spin relaxation properties and can have relatively homogeneous structures, resulting in high-quality solid state NMR data with sharp resonance lines. A variety of sophisticated solid state NMR techniques can therefore be applied, leading to experimental constraints on the identities of and backbone conformations in β-strand and non-β-strand segments, the supramolecular organization of β-sheets (e.g., parallel vs. antiparallel structures, and the registry of inter-strand hydrogen bonds), the conformations and inter-residue contacts of amino acid sidechains, and the overall arrangement and symmetry of cross-β structural units. When sufficient constraints are obtained, full molecular structural models can be developed from the solid state NMR data, especially when supplemented by information from electron microscopy. Most of these constraints are not available from other experimental methods.
Earlier reviews describe solid state NMR studies of amyloid fibrils up to 2005, as well as the basic principles of relevant solid state NMR methods (5, 6). Since 2005, various research groups have obtained results for fibrils formed by the β-amyloid peptide (7-10) and tau protein (11) associated with Alzheimer’s disease (AD), the amylin peptide (or islet amyloid polypeptide, IAPP) associated with type 2 diabetes (12-15), insulin (16), the α-synuclein protein associated with Parkinson’s disease (17-21), the mammalian PrP protein associated with TSEs (22-26), fungal prion proteins (27-36), the β2-microglobulin protein associated with dialysis-related amyloidosis (37, 38), and several other peptides (39-42). A selected set of recent results from solid state NMR is discussed below. This review concludes with a brief discussion of recent advances in solid state NMR methodology that are likely to have an impact on future studies of amyloid fibrils.
II. β-amyloid fibrils and prefibrillar aggregates
A. Fibrils with two-fold and three-fold symmetry
Fibrils of the β-amyloid peptide develop in the brains of AD patients, forming both localized and diffuse deposits (called plaques) in the brain tissue and brain vasculature. The full-length peptide is primarily 40 or 42 residues in length (Aβ1-40 and Aβ1-42). Early solid state NMR investigations focused on the β-sheet structures in fibrils formed by fragments of the β-amyloid peptide (43-48). Experiments on Aβ16-22 fibrils showed that two-dimensional (2D) spectroscopy could be used to resolve and assign 13C NMR resonances in samples that contain multiple uniformly 13C-labeled residues, and that β-strand segments could be identified by their characteristic 13C chemical shift patterns (46). When 2D 13C-13C NMR spectra of full-length Aβ1-40 fibrils with multiple uniformly-labeled residues were obtained (49), it became apparent that these spectra contained more than one set of chemical shifts, implying a coexistence of structurally inequivalent Aβ1-40 molecules within the NMR samples. Structural equivalence could have been a consequence of layering of β-sheets within individual fibrils, as suggested by early structural models (44, 47), which would make the NMR signals from peptides in the inner layers different from those of peptides in the outer layers. However, Petkova et al. subsequently found that variations in chemical shifts were directly correlated with variations in the appearance of the fibrils in TEM images (i.e., fibril morphology), and that both the 2D 13C-13C NMR spectrum and the fibril morphology were “self-propagating” in seeded fibril growth (50). This work proved that the molecular structure in amyloid fibrils is generally not determined uniquely by the amino acid sequence of the fibril-forming polypeptide. Similar polymorphism has now been documented by solid state NMR for α-synuclein fibrils (51), amylin fibrils (13, 14), and other amyloid fibrils (40, 52).
What are the structural differences between fibrils with different morphologies? The predominant morphology in Aβ1-40 fibril samples can be affected by subtle changes in growth conditions, specifically the presence or absence of gentle agitation of the peptide solution during fibril formation (50). Agitation leads to fibrils that appear as straight rods, approximately 6 nm in width, which associate laterally to form “striated ribbons” (see Figure 1a). The individual rods can be considered to be the “protofilaments”, i.e., the basic structural units that combine in various numbers to form ribbons of various widths. Quiescent growth leads to fibrils with an apparent modulation in width and which are less prone to lateral association. Although the modulation period in a quiescently grown sample can be heterogeneous, Paravastu et al. found that samples with a uniform appearance and a specific modulation period (120 ± 20 nm) could be prepared by repetitive seeded growth (9), meaning that many generations of fibrils were created, with generation k+1 being grown from seeds (i.e., short fragments prepared by sonication) of generation k. In negatively stained TEM images, the Aβ1-40 fibrils prepared by Paravastu et al. appear to be “twisted pairs” of protofilaments (see Figure 1b), but in fact they are single protofilaments (i.e., they can not be separated into subunits).
Full structural models for “striated ribbon” and “twisted pair” protofilaments are shown in Figure 2. The two Aβ1-40 protofilaments share several common features, including: (i) Residues 10-22 and 30-40 form β-strands that are separated by a bend or loop in residues 23-29. Residues 1-9 are disordered. Thus, the molecular conformation resembles a “U” in both types of fibrils; (ii) The β-strand segments form in-register, parallel β-sheets with purely intermolecular hydrogen bonds within the β-sheets. In other words, the backbone amide and carbonyl groups of residue k in one Aβ1-40 molecule participate in hydrogen bonds with carbonyl and amide groups of residues k-1 and k+1 of a neighboring molecule. Each molecule participates in two β-sheet layers, through its two β-strand segments. This double-layered sheet is considered to be a single cross-β unit; (iii) The contacts between β-sheets within a cross-β unit include contacts between the F19 sidechains of the N-terminal β-strands and the L34 sidechains of the C-terminal β-strands. These contacts are “staggered”, meaning that F19 of one molecule interacts with L34 of neighboring molecules, not with L34 of the same molecule (53).
Figure 2.
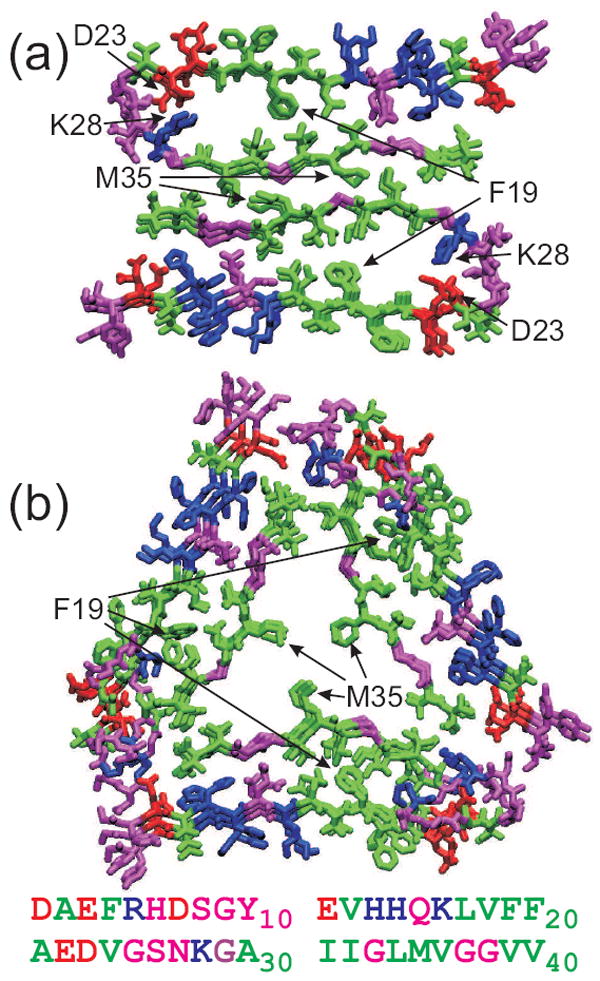
Molecular structural models for “striated ribbon” (a) and “twisted pair” (b) Aβ1-40 fibrils, developed primarily from solid state NMR data with additional constraints from electron microscopy. These models include residues 9-40, with hydrophobic, negatively charged, postively charged (including His), and polar (including Gly) residues colored green, red, blue, and magenta, respectively. Both models are viewed down the long axis of the fibril, with three repeats shown. The full Aβ1-40 sequence is shown below.
The most striking difference between “striated ribbon” and “twisted pair” protofilaments is the overall symmetry with which cross-β units are arranged. The “striated ribbon” protofilament contains two cross-β units, related by an approximate two-fold rotational symmetry axis that coincides with the long axis of the protofilament. The “twisted pair” protofilament contains three cross-β units, related by an approximate three-fold rotational symmetry axis. The conformation of residues 23-29 is also significantly different in “striated ribbon” and “twisted pair” protofilaments. In particular, “striated ribbon” fibrils contain D23-K28 salt bridge interactions, believed to be important in fibril nucleation (54), that are absent in “twisted pair” protofilaments. The details of sidechain-sidechain contacts between cross-β units (i.e., in the central core of the protofilament) are also apparently different, creating a “steric zipper” interface (55) in “striated ribbon” fibrils, while leaving channels that may accomodate internal water molecules in “twisted pair” fibrils.
Compelling evidence for the existence of distinct Aβ1-40 fibril structures comprised of either two or three cross-β units comes from experimental measurements of the mass-per-length (MPL) values of individual fibrils with electron microscopy (9, 50, 56). Figure 3 shows MPL data for “striated ribbon” and “twisted pair” fibrils obtained with a simple and accurate dark-field TEM technique introduced by Chen et al. (57) The symmetries discussed above and shown in Figure 2 are consistent with the observation of a single set of 13C NMR chemical shifts for most sites in solid state NMR spectra of morphologically homogeneous samples.
Figure 3.
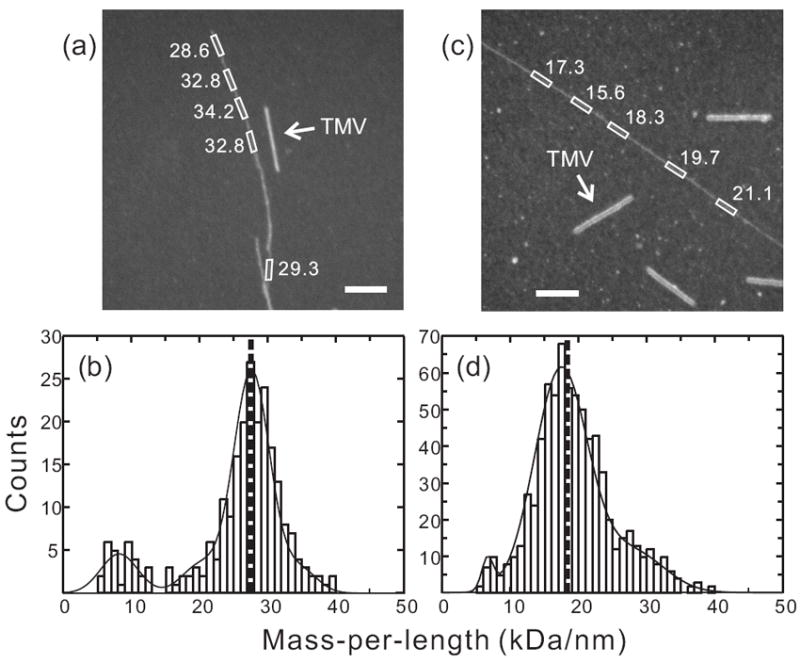
MPL data for Aβ1-40 fibrils. (a,c) Dark-field TEM images of unstained Aβ1-40 fibrils with “twisted pair” and “striated ribbon” morphologies, respectively. Arrows indicate tobacco mosaic virus (TMV) rods that serve as intensity calibration standards. Rectangles enclose fibril segments with the indicated MPL values in kDa/nm. White scale bars are 200 nm. (b,d) MPL histograms obtained from multiple dark-field images. Heavy dashed lines indicate the predicted MPL values for the 3-fold symmetric and 2-fold symmetric structural models in Figure 2, which are comprised of three and two cross-β units, respectively. Solid lines are multiple-Gaussian fits. Minor populations comprised of a single cross-β unit appear at MPL ≈ 9 kDa/nm, possibly arising from disruption of the fibril structures during TEM sample preparation or measurement.
Structural models in Figure 2 were developed from sets of experimental constraints that are described in the relevant publications (9, 49, 50, 53). Constraints include nearly complete sets of 13C chemical shifts (from which backbone torsion angles can be estimated), quantitative measurements of intermolecular 13C-13C and 15N-13C dipole-dipole couplings and intramolecular 15N-15N dipole-dipole couplings, and semi-quantitative measurements of intermolecular 13C-13C proximities arising from sidechain-sidechain contacts. The requirements that the fibril structure be of the cross-β type and possess translational symmetry are also very powerful constraints. Together, these constraints are sufficient to define the principal structural features discussed above.
B. Fibrils from human brain tissue
Since different fibril structures may exert different biological effects (50), it would be interesting to apply solid state NMR methods to fibrils that develop in vivo. Direct structural measurements on fibrils in human tissue or on fibrils extracted from human tissue are not possible, both because the quantity of fibrils is too small and because the fibrils are not isotopically labeled. However, experiments on both amyloid and prion fibrils have shown that fibrils with a given structure can be amplified by seeded growth protocols. Typically, the seeds represent less than 10% of the total peptide or protein in the solution, producing amplification factors of 10 or more in each round of seeded growth. Milligram-scale fibril samples with isotopic labeling suitable for solid state NMR can therefore be prepared from microgram-scale fibril seeds, extracted from the relevant tissue.
The first experiments of this type were performed by Paravastu et al. (8), using highly purified amyloid from frontal lobe brain tissue of deceased AD patients. Three rounds of seeded growth produced ~5 mg samples of labeled Aβ1-40 fibrils. 2D 13C-13C NMR spectra and other measurements showed that the predominant structure in these samples was different from both of the synthetic fibril structures discussed above and shown in Figure 2. More recently, we have modified the amyloid extraction and seeding protocols so that ~1 mg fibril samples can be generated from partially purified amyloid in a single round of seeding, starting with ~1 g of brain tissue. Figure 4 shows 2D 13C-13C and 15N-13C NMR spectra of such a sample, seeded with tissue from the occipital lobe of a deceased AD patient. The observation of a single set of sharp crosspeaks in these spectra indicates the presence of a single molecular structure. Again, the 13C and 15N chemical shifts indicate that this structure is significantly different from synthetic fibril structures.
Figure 4.
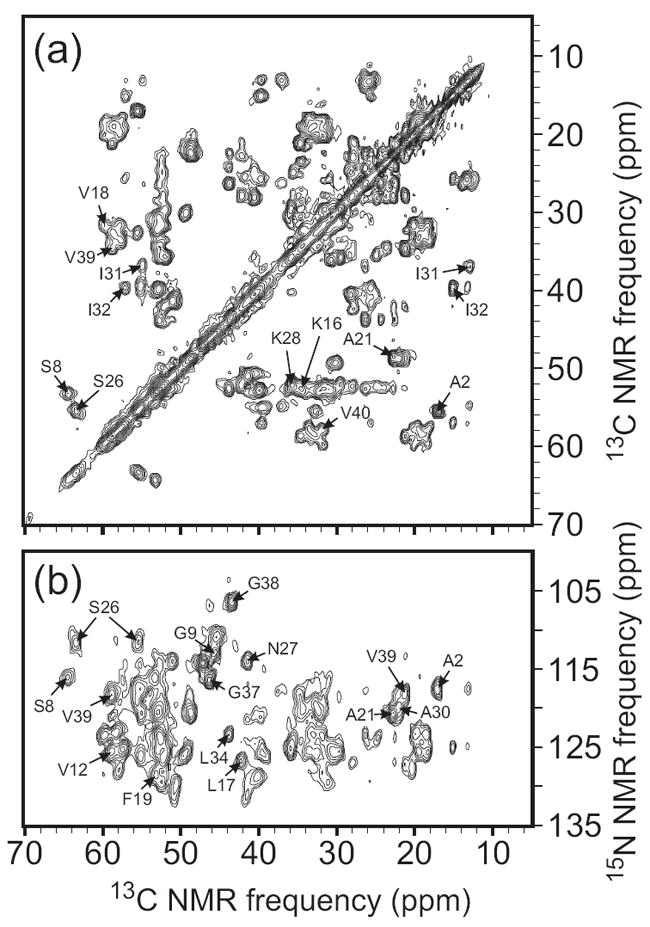
2D solid state NMR spectra of uniformly 15N,13C-labeled Aβ1-40 fibrils prepared by seeding with amyloid extracted from occipital lobe tissue of a deceased Alzheimer’s disease patient. Assignments of selected crosspeaks to specific residues are indicated. (a) 2D 13C-13C NMR spectrum, obtained in a 14.1 T magnetic field with 13.6 kHz MAS, using a 2.94 ms finite-pulse radio-frequency-driven recoupling (fpRFDR) sequence (117) for spin polarization transfer in the mixing period between the two spectral dimensions. (b) 2D 15N-13C spectrum, obtained with frequency-selective 15N-13C cross-polarization followed by fpRFDR in the mixing period.
A prevalent hypotheses in the AD research community is that nonfibrillar β-amyloid aggregates or oligomers, rather than mature amyloid fibrils, may be the primary neurotoxic agents (58, 59). Early support for this hypothesis came from the reported absence of a strong correlation between total amyloid deposition and cognitive impairment in AD patients, although recent data indicate otherwise (60). It is commonly reported that elderly people inevitably develop amyloid plaques (although the quantity of amyloid may vary greatly), while only about 30% of octagenerians develop AD, suggesting that amyloid fibrils alone might not destroy neurons. An alternative hypothesis is that AD results from the neurotoxic action (either direct or indirect) of specific β-amyloid fibril structures, while other structures are relatively benign (8, 50). This hypothesis can be now tested by solid state NMR measurements on fibrils prepared by seeded growth from brain tissue of patients with a variety of medical conditions.
C. Fibrils with anomalous antiparallel β-sheet structures
Evidence for in-register parallel β-sheets in Aβ1-40 fibrils comes from measurements of intermolecular 13C-13C dipole-dipole couplings as in Figure 5a (9, 44, 45, 50). Using similar measurements, in-register parallel β-sheets have been identified in Aβ1-42 fibrils (61), amylin fibrils (14), yeast prion fibrils (32-36), and PrP peptide fibrils (23, 25). An in-register parallel β-sheet naturally aligns hydrophobic residues with themselves, potentially maximizing favorable hydrophobic interactions for any amino acid sequence. An in-register parallel β-sheet also aligns Gln and Asn residues with themselves, maximizing favorable “polar zipper” interactions (62). Antiparallel β-sheets have also been found in amyloid fibrils, but until recently only in fibrils formed by short peptides that contain a single hydrophobic segment in a single β-strand (15, 46, 48, 63, 64).
Figure 5.
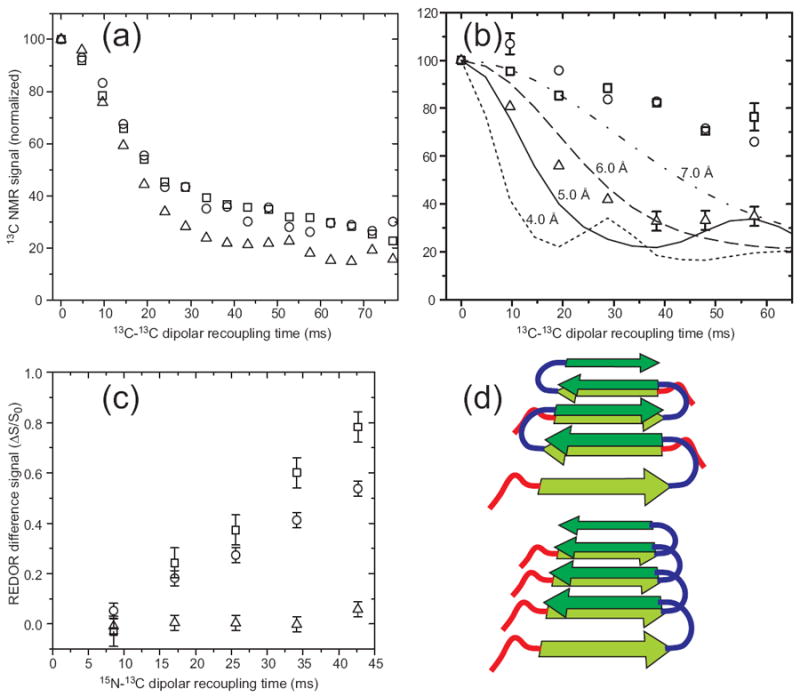
Experimental determination of β-sheet organization in wild-type and D23N mutant Aβ1-40 fibrils with 13C and 15N labels at specific sites. (a) Measurements of intermolecular 13C-13C dipole-dipole couplings in wild-type fibrils using the PITHIRDS-CT technique (98). Squares, circles, and triangles are data for A30 methyl distances in “striated ribbon” fibrils, A30 methyl distances in “twisted pair” fibrils, and M35 methyl distances in “twisted pair” fibrils. Decay of 13C NMR signals on the 30 ms time scale results from ~5 Å intermolecular 13C-13C distances, indicating an in-register parallel β-sheet structure. (b) Measurements of intermolecular couplings among A21 methyl carbons in D23N-Aβ1-40 fibrils containing parallel (triangles) and antiparallel (circles and squares) β-sheet structures. (c) Measurements of intermolecular 15N-13C dipole-dipole couplings between the backbone amide nitrogen of L17 and the methyl carbon of A21 in D23N-Aβ1-40 fibrils containing parallel (triangles) and antiparallel (circles and squares) β-sheet structures, using the REDOR technique (106). Build-up of the normalized REDOR difference signal ΔS/S0 on the 50 ms time scale indicates ~5 Å L17-A21 distances, consistent with an antiparallel β-sheet structure with 17+k ↔ 21-k hydrogen bond registry. (d) Schematic representation of cross-β units in which the two hydrophobic segments of Aβ1-40 (dark and light green arrows) form antiparallel (top) and parallel (bottom) β-sheets. Favorable hydrophobic interactions can occur in both structures.
Surprisingly, certain fibrils formed in vitro by a disease-associated mutant of Aβ1-40, in which Asp at residue 23 is replaced by Asn (D23N-Aβ1-40), have an antiparallel β-sheet structure (7). In vivo, D23N-Aβ1-40 causes neurodegeneration by cerebral amyloid angiopathy (CAA) (65). The presence of antiparallel β-sheets is indicated by a variety of solid state NMR measurements, including the data in Figures 5b and 5c. Schematic structural models in Figure 5d illustrate how apparently similar hydrophobic interactions can be achieved with either in-register parallel or antiparallel β-sheet structures, provided that the peptide adopts the “U” conformation discussed above. The observation of antiparallel β-sheets in D23N-Aβ1-40 fibrils suggests that CAA may be caused specifically by fibrils with these anomalous structures. Other disease-associated Aβ1-40 mutations in residues 21-23 may have similar structural effects (66). Seeded fibril growth using tissue from CAA patients, as discussed above, may permit this possibility to be tested experimentally.
D. Nonfibrillar aggregates
Aggregated species with various morphologies are typically observed during the incubation of fibril-forming polypeptide solutions before the appearance of mature amyloid fibrils. Some of these species (referred to as “amyloid-derived diffusible ligands” (67), “globulomers” (68), or simply oligomers) appear as roughly spherical objects in TEM or atomic force microscope images, with diameters in the 5-50 nm range, while others (called “protofibrils” (69)) appear as worm-like objects, with diameters similar to those of fibrils, but shorter and with greater curvature. In cell cultures, nonfibrillar aggregates can exhibit higher levels of toxicity on a per-molecule basis than mature fibrils (70). Such species have also been shown to occur in human tissue (71, 72). Thus, the hypothesis that nonfibrillar aggregates are the primary causative agents in AD and other amyloid diseases has attracted considerable attention.
The first solid state NMR studies of nonfibrillar aggregates were performed by Ishii and coworkers, who focused on spherical Aβ1-40 oligomers with demonstrated toxicity and a likely on-pathway role in fibril formation (70, 73). 2D 13C-13C NMR spectra of these oligomers, which have diameters in the 15-35 nm range and were prepared specifically at 4° C and pH 7.4, are nearly identical to spectra of Aβ1-40 fibrils, indicating very similar molecular conformations. Measurements of intermolecular 13C-13C dipole-dipole couplings indicate that the β-strands form parallel β-sheets. Sharpe and coworkers have reported related studies of spherical oligomers formed at pH 4.6 by residues 106-126 of the human prion protein (PrP106-126) (74). Again, the solid state NMR data indicate a structure that is very similar to the structure of PrP106-126 fibrils, which form at pH 8.0 (23). In particular, both the oligomers and the fibrils contain in-register parallel β-sheets formed by residues 115-124 (sequence AAAAGAVVGG), with antiparallel stacking of the parallel β-sheets.
The similarity of molecular conformation and supramolecular structure in spherical oligomers and amyloid fibrils is quite surprising, given their qualitatively different morphologies. Future measurements are likely to shed more light on this issue and to provide structural constraints on other aggregated species such as protofibrils.
III. Prion fibrils
A. Mammalian PrP
The mammalian prion protein PrP causes infectious neurodegenerative diseases (TSEs) by structural conversion and aggregation (75). PrP is a 231-residue protein with a ~100-residue N-terminal segment that is disordered in the monomeric state and a ~100-residue C-terminal globular domain that has primarily helical secondary structure (76). The TSE-causing state has not been shown definitively to be a cross-β, amyloid-like state (77), but PrP does form amyloid fibrils (78-80). Moreover, the properties of molecular-level polymorphism and structural self-replication in seeded growth discussed above for amyloid fibrils would account for the existence of self-propagating “strains” or “variants” of TSEs (3, 75). Hydrogen/deuterium exchange and electron paramagnetic resonance data indicate that residues 164-220 (approximately) of PrP form the core of recombinant PrP fibrils, and that the core contains in-register parallel β-sheets (78, 79).
Jaroniec and coworkers (22, 24) have reported studies of fibrils formed by a truncation mutant of PrP (residues 23-144, or PrP23-144) that produces familial CAA. Signals in 2D solid state NMR spectra of uniformly 15N,13C-labeled samples are exceptionally well resolved at 0° C and above, both because the NMR lines are intrinsically narrow (about 1.0 ppm linewidth for 15N lines at 11.7 T; about 0.6 ppm linewidth for 13C lines) and because only 28 residues are sufficiently immobilized in PrP23-144 fibrils to contribute to the solid state NMR spectra. From multidimensional spectra, the solid state NMR signals were assigned to residues 112-141 (24). Quantitative measurements of motionally-averaged heteronuclear dipole-dipole couplings indicate that these residues have rapid fluctuations of backbone Cα-Hα bond directions on the order of ±10°, while the N-terminal segment gives rise to NMR signals that are detectable with solution NMR techniques and have chemical shifts similar to random-coil values, indicating that the N-terminal segment is dynamically disordered (22). A structural model for the rigid core of PrP23-144 fibrils has not yet been reported.
B. Yeast prions
As originally demonstrated by Wickner and coworkers (81, 82), several heritable phenotypes of yeast arise from intracellular aggregation of specific proteins into amyloid states. Such proteins constitute yeast prions. Yeast prions identified to date have Gln- and Asn-rich segments, which are the amyloid-forming segments (or “prion domains”), and which have been shown to be sufficient for prion infectivity and propagation in several cases (83, 84). In a series of solid state NMR studies, amyloid fibrils formed by the prion domains of the Sup35, Ure2p, and Rnq1 prion proteins have been shown to possess in-register parallel β-sheet structures (32-36). These studies depend on measurements of intermolecular 13C-13C dipole-dipole couplings, as in Figure 5a, but performed on samples that are biosynthetically 13C-labeled at backbone carbonyl or sidechain methyl sites of all residues of a specific type (e.g., all Leu or all Tyr residues). The data reflect intermolecular 13C-13C distances as long as the labeled residues do not occur consecutively in the amino acid sequence. The in-register parallel β-sheet structure of yeast prions is consistent with the primacy of polar zippers among Gln and Asn sidechain amide groups as stabilizing interactions for the amyloid fibrils (62). Interestingly, scrambling the sequence of a yeast prion domain does not prevent prion propagation or amyloid formation (85), and preserves the in-register parallel β-sheet structure (33). This is understandable, as scrambling the sequence does not change its capacity to form polar zippers.
Solid state NMR data for yeast prions discussed above are incompatible with alternative structural proposals, including a β-helical model for Sup35 prions (86) and a model for Ure2p prions in which the globular C-terminal domain (rather than the Gln- and Asn-rich N-terminal domain) is the major structural element in the fibril core (31). The observation of strong, sharp solid state NMR signals from the globular C-terminal domain in full-length Ure2p fibrils by Loquet et al. has been presented as support for the latter model (31), but this observation is also consistent with a fibril structure in which the N-terminal domains form the cross-β core while the C-terminal domains (which are largely immobilized both by tethering to the core and by dimerization) form a shell around the core. Baxa et al. have shown biochemically that the C-terminal domains of a variety of chimeric Ure2p protein fibrils retain their globular structures (87).
C. HET-s and functional amyloid
Several cases have been identified in which amyloid fibrils appear to have specific biological functions (88-90). One such case is that of the HET-s protein, whose aggregation into an amyloid state has been shown to be essential for heterokaryon incompatibility in the filamentous fungus Podospora anserina. Briefly, fungal cells that carry the het-s gene, which encodes HET-s, are incapable of fusing viably with cells that carry the het-S gene, which encodes the homologous protein HET-S, but only if HET-s is in its aggregated state. HET-s is therefore considered to be a fungal prion protein, since its structural state encodes a specific phenotype. Unlike yeast and mammalian prions, whose aggregated states are apparently accidental and deleterious to their hosts, HET-s appears to have evolved to control a biological process that protects het-s cells from viral infections (88).
Meier and coworkers have performed solid state NMR studies on the HET-s prion domain (residues 218-289, or HET-s218-289), which is not Gln- or Asn-rich (27-30). HET-s218-289 fibrils prepared in vitro are not polymorphic, consistent with an evolved function and the absence of distinct HET-s prion strains or variants. Initial measurements revealed that the 13C and 15N NMR lines from hydrated HET-s218-289 fibrils are remarkably sharp (linewidths of 0.5 ppm or less), allowing nearly complete resonance assignments to be obtained with standard 2D and 3D solid state NMR techniques from uniformly 15N,13C-labeled samples (30, 91). Only about 60% of the HET-s218-289 sequence contributes strong signals to solid state NMR spectra. These signals arise from two segments (residues 226-248 and 262-282) with homologous sequences, suggesting a cross-β structure in which the two segments alternate within a parallel β-sheet, which then contains both intramolecular and intermolecular hydrogen bonds (unlike the in-register parallel β-sheets discussed above). MPL data that indicate a monomer repeat distance of 9.6 Å along the fibril length support this arrangement (57, 92), and conclusive evidence was obtained from measurements of nonsequential, interstrand contacts between HN and Hα sites using the 2D NHHC solid state NMR technique (28). Additional measurements of nonsequential contacts involving specific sidechains and estimates of backbone torsion angles from 13C chemical shifts provided sufficient constraints to develop the model in Figures 6a and 6b. This model closely resembles the β-helix motifs that have been observed in crystal structures of certain monomeric proteins (93), such as the pertactin structure in Figures 6c and 6d. The HET-s218-289 structure therefore supports earlier suggestions that β-helices might be important structural motifs within amyloid fibrils (94, 95).
Figure 6.
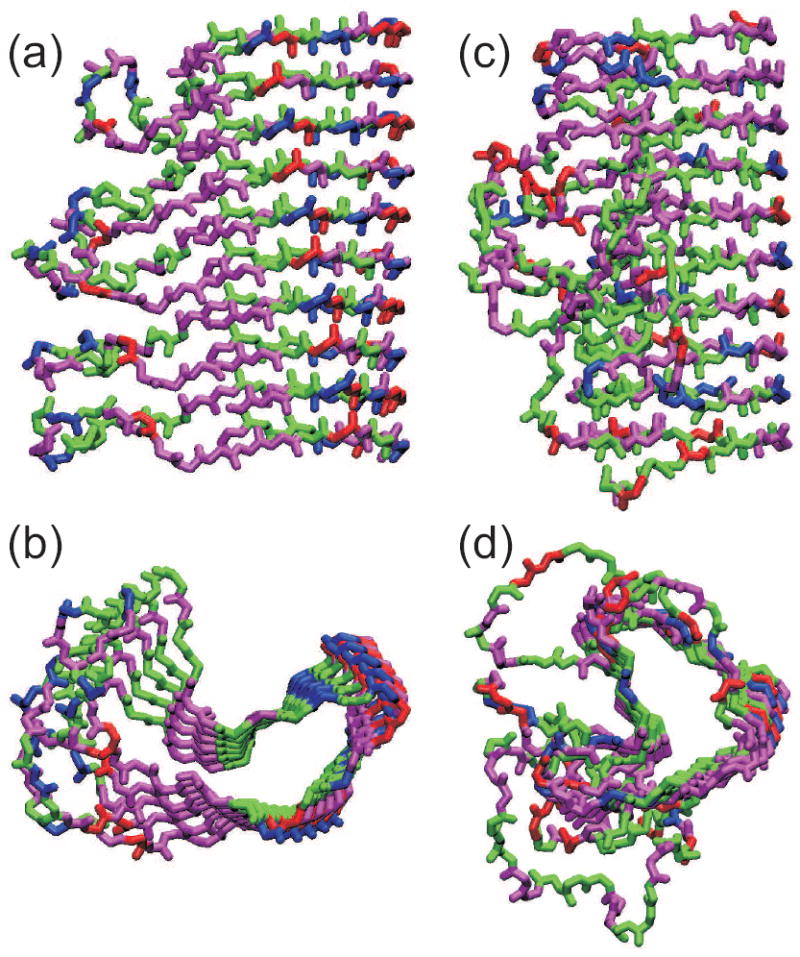
Comparison of the HET-s218-289 prion fibril structure (a,b) with that of the β-helical protein Bordetella pertussis pertactin (c,d), based on Protein Data Bank files 2RNM and 1DAB, respectively. Structures are viewed perpendicular (a,c) and parallel (b,d) to the long axis of the fibril or β-helix. For HET-s218-289, five repeats of residues 226-278 are shown, with each repeat forming two “rungs” of the β-helix. For pertactin, residues 1-285 is shown. Hydrophobic, negatively charged, postively charged (including His), and polar (including Gly) residues are colored green, red, blue, and magenta, respectively.
Solid state NMR data from HET-s218-289 fibrils resemble data from microcrystalline proteins, in that the NMR lines are extraordinarily sharp and the nuclear spin T2 and T1ρ relaxation times are long (leading to strong and well-resolved signals in multidimensional spectra). Data from PrP23-114 fibrils are similar (24). In our experience, the quality of solid state NMR data from amyloid fibrils formed by different polypeptides can be significantly different. Variations in the extent of structural heterogeneity, including heterogeneity in sidechain conformations, sidechain packing, and lateral association (i.e., “bundling” of fibrils), may contribute to variations in linewidths. The time scale and completeness of dynamical averaging of structural heterogeneities probably plays an important role, with sharp solid state NMR lines and long relaxation times occurring only when hetereogeneities are averaged out on sub-microsecond time scales and when the time-averaged structure is homogeneous. For both HET-s218-289 and PrP23-114, solid state NMR linewidths have been shown to increase significantly at low temperatures (24, 96), presumably because molecular motions that average out structural inhomogeneities become too slow.
Curli fibrils of Eschirichia coli, which are formed by the CsgA and CsgB proteins (89), have an adhesive function. The CsgA sequence contain five pseudo-repeat segments, each about 22 residues in length. Solid state NMR measurements on uniformly 15N,13C-labeled CsgA fibrils indicate that nearly the entire sequence is immobilized. Measurements of 13C-13C dipole-dipole couplings in selectively 13C-labeled samples, as discussed above, show only weak couplings, arguing against a simple in-register parallel β-sheet structure. MPL data indicate a monomer repeat distance greater than 9 Å (97). Together, these data suggest a β-helical structure for CsgA fibrils, perhaps with the five pseudo-repeats forming successive rungs of the β-helix.
IV. Advances in solid state NMR methodology
A. New approaches to structural constraints
Under MAS, internuclear distances are measured with dipolar recoupling techniques, i.e., rf pulse sequences synchronized with sample rotation in such a way that the desired dipoledipole couplings are no longer averaged to zero (5). In the case of 13C-13C distances, quantitative distance measurements are possible with established methods when 13C labels are introduced at specific sites (98), as in Figures 5a and 5b. When samples are uniformly 13C-labeled, strong couplings between directly-bonded 13C pairs effectively attenuate the weaker couplings between more distant pairs because of the quantum mechanical non-commutivity of different pairwise couplings, preventing quantitative distance measurements by standard recoupling techniques. This phenomenon is commonly known as “dipolar truncation”. Several approaches to overcoming dipolar truncation have been developed. One approach is to detect long-range couplings by the presence of crosspeaks in 2D 13C-13C spectra obtained with simple “spin diffusion” or “rf-assisted diffusion” techniques (99, 100), i.e., techniques in which 1H-13C couplings are not removed during the 13C-13C polarization exchange period, making the 13C-13C exchange process essentially incoherent. While these approaches allow proximities of 13C pairs to be detected at distances of at least 6 Å (53), they do not provide quantitative distance information. Variants in which 13C-13C exchange proceeds through second-order, three-spin interactions have the advantage of suppressing directly-bonded crosspeaks and enhancing longer-range crosspeaks (101), although dipolar truncation is not eliminated.
A second approach is to devise “frequency selective” recoupling techniques, which take advantage of chemical shift differences to selectively recouple only 13C pairs with particular chemical shifts, thereby avoiding quantum mechanical interferences. Although early versions of frequency selective recoupling required relatively low MAS frequencies and large chemical shift differences (102), the more recent SEASHORE (shift evolution assisted homonuclear recoupling) methods are in principle applicable with high-speed MAS and arbitrary chemical shift differences (10, 103). As shown by Hu and Tycko, a zero-quantum SEASHORE technique can be used to measure couplings among backbone carbonyl 13C sites (103). When combined with measurements of couplings among backbone amide 15N sites by non-selective recoupling techniques (9, 98), these data can provide quantitative constraints on backbone conformations in uniformly 15N,13C-labeled peptides and proteins that may improve the precision and accuracy of structural models such as those in Figures 2 and 6.
We have recently proposed a third approach, called stochastic dipolar recoupling, in which the rf pulse sequence intentionally varies from scan to scan in such a way that the recoupled 13C-13C interactions fluctuate randomly and in a nearly uncorrelated manner. Fluctuations are induced either by random excursions in the rf carrier frequency, in the case of double-quantum stochastic recoupling (104), or random variations in delays between recoupling blocks, in the case of zero-quantum stochastic recoupling (91). In effect, stochastic recoupling mimics the random fluctuations of dipole-dipole interactions that occur in solution NMR due to molecular rotational diffusion. Nuclear spin polarization transfers then become incoherent and can be described by simple kinetic equations, as in the nuclear Overhauser effects observed in solution NMR, rather than by coherent quantum mechanical dynamics (104, 105). In principle, this allows internuclear distances to be extracted from polarization transfer rates in a quantitative manner and without adjustable parameters.
In the case of 15N-13C dipolar recoupling methods, the issue of dipolar truncation does not arise in the same way, since the heteronuclear dipole-dipole couplings created by dipolar recoupling techniques such as REDOR (106) commute with one another. Nonetheless, similar effects occur when couplings are measured from non-selective 15N-13C polarization transfer data. Therefore, frequency-selective 15N-13C recoupling techniques have been developed, and have been shown to furnish quantitative distance constraints in uniformly 15N,13C-labeled proteins (107).
B. Paramagnetic effects
Since the magnetic moments associated with electron spins are ~103 larger than those associated with nuclear spins, dipolar hyperfine couplings between paramagnetic centers and nuclear spins can provide information about significantly longer distances than can be probed by couplings between nuclear spins. Recent work has demonstrated the feasibility of estimating electron-nucleus distances in proteins in the 10-20 Å range from effects on nuclear spin T1 or T2 relaxation times from hyperfine couplings to nitroxide spin labels or paramagnetic metals (108, 109) or from pseudo-contact shifts from hyperfine couplings to paramagnetic metal ions (110).
The general reduction in 1H T1 values that results from paramagnetic doping permits more rapid signal averaging, increasing the effective sensitivity of solid state NMR measurements. As shown by Ishii and coworkers (111), when high-speed MAS is employed, 1H decoupling powers can also be greatly reduced, permitting the use of recycle delays less than 100 ms without excessive sample heating. Data acquisition times can be reduced by factors of 10-100, or sample quantities can be reduced by factors of 3-10.
C. Low-temperature measurements and dynamic nuclear polarization
The sensitivity of solid state NMR measurements can be enhanced by reducing sample temperatures to increase thermal equilibrium nuclear spin polarizations according to the Curie law. We have recently developed an ultra-low-temperature MAS system that allows standard biomolecular solid state NMR techniques to be applied at 25 K, providing sensitivity enhancement factors greater than ten relative to room-temperature measurements (112). Still greater sensitivity enhancements can be achieved by dynamic nuclear polarization (DNP), a phenomenon in which partial saturation of electron spin resonance transitions leads to anomalously large nuclear spin polarizations. Griffin and coworkers have pioneered the development of high-field DNP for biomolecular solid state NMR, using nitroxide dopants and high-power gyrotron microwave sources to generate sensitivity enhancements in the 100-200 range at sample temperatures near 100 K (113, 114). Similar NMR signal amplitudes can be obtained with low microwave powers at lower temperatures (115, 116).
Debelouchina et al. have demonstrated the applicability of DNP to amyloid fibrils, in studies of partially-labeled GNNQQNY peptide fibrils (41). Ultra-low-temperature MAS and DNP technology is likely to have a major impact on future studies, especially of amyloid-forming systems that can be selectively labeled (where the increased MAS NMR linewidths that generally accompany low temperatures are not a serious limitation). A particularly promising application is to studies of oligomers and other intermediate states in amyloid formation, where DNP enhancements should allow solid state NMR measurements at all stages of structural evolution to be performed in frozen solutions of amyloid-forming polypeptides at the 0.01-1.0 mM concentrations that are typically employed in vitro.
Summary Points.
Modern solid state NMR data provide unique insights into the molecular structures of amyloid fibrils that are of real biological and biomedical significance. Full structural models can be developed with assistance from electron microscopy.
Solid state NMR data show that amyloid fibrils are frequently polymorphic, with structural variations that can include variations in symmetry and in the nature of β-sheet structures.
Through seeded growth, structural studies of fibrils that develop in human tissue are now possible. Current data indicate that β-amyloid fibrils in Alzheimer’s disease brain are structurally distinct from fibrils prepared in vitro.
In-register parallel β-sheet structures are most common in amyloid and prion fibrils, but antiparallel and β-helical structures also occur.
Ongoing developments in solid state NMR techniques and technology promise to improve both sensitivity and structural information content substantially.
Future Issues.
In Alzheimer’s disease and other amyloid diseases, does disease severity or progression correlate with molecular structural variations in the associated fibrils?
How do molecular structures of oligomeric and protofibrillar intermediates relate to the structures of mature fibrils?
Are functional amyloids in general structurally distinct from disease-associated amyloids?
How will new solid state NMR methods expand the range of amyloid-forming systems that can be structurally characterized?
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, a component of the National Institutes of Health. Spectra in Figure 4 were obtained by Dr. Jun-Xia Lu, using uniformly labeled Aβ1-40 provided by Dr. Anders Oloffson and brain tissue provided by Dr. Stephen C. Meredith.
Glossary
- amyloid fibril
A filamentous protein or peptide aggregate with a cross-β structure
- cross-β
A ribbon-like β-sheet structure in which the β-strands run perpendicular to and the backbone hydrogen bonds run parallel to the long axis
- prion
A protein with a self-propagating transmissible structural state (usually an amyloid state) that produces a specific phenotype (usually a disease)
- dipolar recoupling
Solid state NMR pulse sequence applied in synchrony with MAS to restore nuclear magnetic dipole-dipole couplings
Acronyms
- NMR
nuclear magnetic resonance
- TEM
transmission electron microscopy
- MAS
magic-angle spinning
- DNP
dynamic nuclear polarization
- Aβ1-40
40-residue β-amyloid peptide associated with Alzheimer’s disease
- AD
Alzheimer’s disease
- TSE
transmissible spongiform encephalopathy
- CAA
cerebral amyloid angiopathy
Literature Cited
- 1.Sunde M, Blake CCF. From the globular to the fibrous state: Protein structure and structural conversion in amyloid formation. Q Rev Biophys. 1998;31:1–39. doi: 10.1017/s0033583598003400. [DOI] [PubMed] [Google Scholar]
- 2.Sacchettini JC, Kelly JW. Therapeutic strategies for human amyloid diseases. Nat Rev Drug Discov. 2002;1:267–75. doi: 10.1038/nrd769. [DOI] [PubMed] [Google Scholar]
- 3.Collinge J. Molecular neurology of prion disease. J Neurol Neurosurg Psychiatry. 2005;76:906–19. doi: 10.1136/jnnp.2004.048660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiti F, Dobson CM. Amyloid formation by globular proteins under native conditions. Nat Chem Biol. 2009;5:15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 5.Tycko R. Molecular structure of amyloid fibrils: Insights from solid state NMR. Q Rev Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. A comprehensive review of solid state NMR studies of amyloid fibrils and relevant methodology up to 2005. [DOI] [PubMed] [Google Scholar]
- 6.Tycko R. Characterization of amyloid structures at the molecular level by solid state nuclear magnetic resonance spectroscopy. In: Kheterpal I, Wetzel R, editors. Methods in Enzymology. Vol. 413. San Diego: Academic Press; 2006. pp. 103–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tycko R, Sciarretta KL, Orgel J, Meredith SC. Evidence for novel β-sheet structures in Iowa mutant β-amyloid fibrils. Biochemistry. 2009;48:6072–84. doi: 10.1021/bi9002666. First evidence for antiparallel β-sheets in fibrils formed by a full-length amyloid polypeptide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paravastu AK, Qahwash I, Leapman RD, Meredith SC, Tycko R. Seeded growth of β-amyloid fibrils from Alzheimer’s brain-derived fibrils produces a distinct fibril structure. Proc Natl Acad Sci U S A. 2009;106:7443–8. doi: 10.1073/pnas.0812033106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer’s β-amyloid fibrils. Proc Natl Acad Sci U S A. 2008;105:18349–54. doi: 10.1073/pnas.0806270105. Reports a structural model for “twisted pair” Aβ1-40 fibrils with three-fold symmetry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paravastu AK, Tycko R. Frequency-selective homonuclear dipolar recoupling in solid state NMR. J Chem Phys. 2006;124:194303. doi: 10.1063/1.2192516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andronesi OC, von Bergen M, Biernat J, Seidel K, Griesinger C, et al. Characterization of Alzheimer’s-like paired helical filaments from the core domain of tau protein using solid state NMR spectroscopy. J Am Chem Soc. 2008;130:5922–8. doi: 10.1021/ja7100517. [DOI] [PubMed] [Google Scholar]
- 12.Smith PES, Brender JR, Ramamoorthy A. Induction of negative curvature as a mechanism of cell toxicity by amyloidogenic peptides: The case of islet amyloid polypeptide. J Am Chem Soc. 2009;131:4470–8. doi: 10.1021/ja809002a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madine J, Jack E, Stockley PG, Radford SE, Serpell LC, Middleton DA. Structural insights into the polymorphism of amyloid-like fibrils formed by region 20-29 of amylin revealed by solid state NMR and x-ray fiber diffraction. J Am Chem Soc. 2008;130:14990–5001. doi: 10.1021/ja802483d. [DOI] [PubMed] [Google Scholar]
- 14.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid state NMR. Biochemistry. 2007;46:13505–22. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen JT, Bjerring M, Jeppesen MD, Pedersen RO, Pedersen JM, et al. Unique identification of supramolecular structures in amyloid fibrils by solid state NMR spectroscopy. Angew Chem Int Edit. 2009;48:2118–21. doi: 10.1002/anie.200804198. [DOI] [PubMed] [Google Scholar]
- 16.Yang YW, Petkova A, Huang K, Xu B, Hua QX, et al. An Achilles’ heel in an amyloidogenic protein and its repair: Insulin fibrillation and therapeutic design. J Biol Chem. 2010;285:10806–21. doi: 10.1074/jbc.M109.067850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heise H, Celej MS, Becker S, Riede D, Pelah A, et al. Solid state NMR reveals structural differences between fibrils of wild-type and disease-related A53T mutant α-synuclein. J Mol Biol. 2008;380:444–50. doi: 10.1016/j.jmb.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 18.Vilar M, Chou HT, Luhrs T, Maji SK, Riek-Loher D, et al. The fold of α-synuclein fibrils. Proc Natl Acad Sci U S A. 2008;105:8637–42. doi: 10.1073/pnas.0712179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madine J, Doig AJ, Middleton DA. Design of an N-methylated peptide inhibitor of α-synuclein aggregation guided by solid state NMR. J Am Chem Soc. 2008;130:7873–81. doi: 10.1021/ja075356q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kloepper KD, Hartman KL, Ladror DT, Rienstra CM. Solid state NMR spectroscopy reveals that water is nonessential to the core structure of α-synuclein fibrils. J Phys Chem B. 2007;111:13353–6. doi: 10.1021/jp077036z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kloepper KD, Zhou DH, Li Y, Winter KA, George JM, Rienstra CM. Temperature-dependent sensitivity enhancement of solid state NMR spectra of α-synuclein fibrils. J Biomol NMR. 2007;39:197–211. doi: 10.1007/s10858-007-9189-z. [DOI] [PubMed] [Google Scholar]
- 22.Helmus JJ, Surewicz K, Surewicz WK, Jaroniec CP. Conformational flexibility of Y145Stop human prion protein amyloid fibrils probed by solid state nuclear magnetic resonance spectroscopy. J Am Chem Soc. 2010;132:2393–403. doi: 10.1021/ja909827v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh P, Simonetti K, Sharpe S. Core structure of amyloid fibrils formed by residues 106-126 of the human prion protein. Structure. 2009;17:417–26. doi: 10.1016/j.str.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 24.Helmus JJ, Surewicz K, Nadaud PS, Surewicz WK, Jaroniec CP. Molecular conformation and dynamics of the Y145Stop variant of human prion protein. Proc Natl Acad Sci U S A. 2008;105:6284–9. doi: 10.1073/pnas.0711716105. Reports remarkably sharp solid state NMR lines in a non-functional amyloid, suggesting that such fibrils can be very highly ordered at the molecular level. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin NS, Chao JCH, Cheng HM, Chou FC, Chang CF, et al. Molecular structure of amyloid fibrils formed by residues 127 to 147 of the human prion protein. Chem -Eur J. 2010;16:5492–9. doi: 10.1002/chem.200903290. [DOI] [PubMed] [Google Scholar]
- 26.Lim KH, Nguyen TN, Damo SM, Mazur T, Ball HL, et al. Solid state NMR structural studies of the fibril form of a mutant mouse prion peptide PrP89-143. Solid State Nucl Magn Reson. 2006;29:183–90. doi: 10.1016/j.ssnmr.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 27.Wasmer C, Schutz A, Loquet A, Buhtz C, Greenwald J, et al. The molecular organization of the fungal prion HET-s in its amyloid form. J Mol Biol. 2009;394:119–27. doi: 10.1016/j.jmb.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 28.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218-289) prion form a β solenoid with a triangular hydrophobic core. Science. 2008;319:1523–6. doi: 10.1126/science.1151839. Reports β-helical structural model for HET-s prion domain fibrils, based on a wealth of solid state NMR data. [DOI] [PubMed] [Google Scholar]
- 29.Siemer AB, Arnold AA, Ritter C, Westfeld T, Ernst M, et al. Observation of highly flexible residues in amyloid fibrils of the HET-s prion. J Am Chem Soc. 2006;128:13224–8. doi: 10.1021/ja063639x. [DOI] [PubMed] [Google Scholar]
- 30.Siemer AB, Ritter C, Steinmetz MO, Ernst M, Riek R, Meier BH. 13C, 15N resonance assignment of parts of the HET-s prion protein in its amyloid form. J Biomol NMR. 2006;34:75–87. doi: 10.1007/s10858-005-5582-7. [DOI] [PubMed] [Google Scholar]
- 31.Loquet A, Bousset L, Gardiennet C, Sourigues Y, Wasmer C, et al. Prion fibrils of Ure2p assembled under physiological conditions contain highly ordered, natively folded modules. J Mol Biol. 2009;394:108–18. doi: 10.1016/j.jmb.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 32.Shewmaker F, Kryndushkin D, Chen B, Tycko R, Wickner RB. Two prion variants of Sup35p have in-register parallel β-sheet structures, independent of hydration. Biochemistry. 2009;48:5074–82. doi: 10.1021/bi900345q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shewmaker F, Ross ED, Tycko R, Wickner RB. Amyloids of shuffled prion domains that form prions have a parallel in-register β-sheet structure. Biochemistry. 2008;47:4000–7. doi: 10.1021/bi7024589. [DOI] [PubMed] [Google Scholar]
- 34.Wickner RB, Dyda F, Tycko R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register β-sheet structure. Proc Natl Acad Sci U S A. 2008;105:2403–8. doi: 10.1073/pnas.0712032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baxa U, Wickner RB, Steven AC, Anderson DE, Marekov LN, et al. Characterization of β-sheet structure in Ure2p1-89 yeast prion fibrils by solid state nuclear magnetic resonance. Biochemistry. 2007;46:13149–62. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- 36.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc Natl Acad Sci U S A. 2006;103:19754–9. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbet-Massin E, Ricagno S, Lewandowski JR, Giorgetti S, Bellotti V, et al. Fibrillar vs crystalline full-length β2-microglobulin studied by high-resolution solid state NMR spectroscopy. J Am Chem Soc. 2010;132:5556–7. doi: 10.1021/ja1002839. [DOI] [PubMed] [Google Scholar]
- 38.Iwata K, Fujiwara T, Matsuki Y, Akutsu H, Takahashi S, et al. 3D structure of amyloid protofilaments of β2-microglobulin fragment probed by solid state NMR. Proc Natl Acad Sci U S A. 2006;103:18119–24. doi: 10.1073/pnas.0607180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nanga RPR, Brender JR, Vivekanandan S, Popovych N, Ramamoorthy A. NMR structure in a membrane environment reveals putative amyloidogenic regions of the SEVI precursor peptide Pap248-286. J Am Chem Soc. 2009;131:17972–9. doi: 10.1021/ja908170s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verel R, Tomka IT, Bertozzi C, Cadalbert R, Kammerer RA, et al. Polymorphism in an amyloid-like fibril-forming model peptide. Angew Chem -Int Edit. 2008;47:5842–5. doi: 10.1002/anie.200800021. [DOI] [PubMed] [Google Scholar]
- 41.Debelouchina GT, Bayro MJ, van der Wel PCA, Caporini MA, Barnes AB, et al. Dynamic nuclear polarization-enhanced solid state NMR spectroscopy of GNNQQNY nanocrystals and amyloid fibrils. Phys Chem Chem Phys. 2010;12:5911–9. doi: 10.1039/c003661g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Wel PCA, Lewandowski JR, Griffin RG. Solid state NMR study of amyloid nanocrystals and fibrils formed by the peptide GNNQQNY from yeast prion protein Sup35p. J Am Chem Soc. 2007;129:5117–30. doi: 10.1021/ja068633m. [DOI] [PubMed] [Google Scholar]
- 43.Lansbury PT, Costa PR, Griffiths JM, Simon EJ, Auger M, et al. Structural model for the β-amyloid fibril based on interstrand alignment of an antiparallel-sheet comprising a C-terminal peptide. Nat Struct Biol. 1995;2:990–8. doi: 10.1038/nsb1195-990. The first amyloid structural model developed from solid state NMR constraints. A 9-residue β-amyloid peptide forms antiparallel β-sheets. [DOI] [PubMed] [Google Scholar]
- 44.Benzinger TLS, Gregory DM, Burkoth TS, Miller-Auer H, Lynn DG, et al. Propagating structure of Alzheimer’s β-amyloid10-35 is parallel β-sheet with residues in exact register. Proc Natl Acad Sci U S A. 1998;95:13407–12. doi: 10.1073/pnas.95.23.13407. The first experimental evidence for in-register parallel β-sheets in amyloid fibrils, based on 13C-13C dipolar recoupling data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gregory DM, Benzinger TLS, Burkoth TS, Miller-Auer H, Lynn DG, et al. Dipolar recoupling NMR of biomolecular self-assemblies: Determining inter- and intrastrand distances in fibrilized Alzheimer’s β-amyloid peptide. Solid State Nucl Magn Reson. 1998;13:149–66. doi: 10.1016/s0926-2040(98)00086-1. [DOI] [PubMed] [Google Scholar]
- 46.Balbach JJ, Ishii Y, Antzutkin ON, Leapman RD, Rizzo NW, et al. Amyloid fibril formation by Aβ16-22, a seven-residue fragment of the Alzheimer’s β-amyloid peptide, and structural characterization by solid state NMR. Biochemistry. 2000;39:13748–59. doi: 10.1021/bi0011330. [DOI] [PubMed] [Google Scholar]
- 47.Burkoth TS, Benzinger TLS, Urban V, Morgan DM, Gregory DM, et al. Structure of the β-amyloid10-35 fibril. J Am Chem Soc. 2000;122:7883–9. [Google Scholar]
- 48.Petkova AT, Buntkowsky G, Dyda F, Leapman RD, Yau WM, Tycko R. Solid state NMR reveals a pH-dependent antiparallel β-sheet registry in fibrils formed by a β-amyloid peptide. J Mol Biol. 2004;335:247–60. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 49.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, et al. A structural model for Alzheimer’s β-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A. 2002;99:16742–7. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petkova AT, Leapman RD, Guo ZH, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer’s β-amyloid fibrils. Science. 2005;307:262–5. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 51.Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length α-synuclein fibrils studied by solid state NMR. 2005;102:15871–6. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paravastu AK, Petkova AT, Tycko R. Polymorphic fibril formation by residues 10-40 of the Alzheimer’s β-amyloid peptide. Biophys J. 2006;90:4618–29. doi: 10.1529/biophysj.105.076927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s β-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. Reports a structural model for “striated ribbon” Aβ1-40 fibrils with two-fold symmetry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sciarretta KL, Gordon DJ, Petkova AT, Tycko R, Meredith SC. Aβ40-lactam(D23/K28) models a conformation highly favorable for nucleation of amyloid. Biochemistry. 2005;44:6003–14. doi: 10.1021/bi0474867. [DOI] [PubMed] [Google Scholar]
- 55.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–7. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 56.Goldsbury C, Frey P, Olivieri V, Aebi U, Muller SA. Multiple assembly pathways underlie amyloid-β fibril polymorphisms. J Mol Biol. 2005;352:282–98. doi: 10.1016/j.jmb.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 57.Chen B, Thurber KR, Shewmaker F, Wickner RB, Tycko R. Measurement of amyloid fibril mass-per-length by tilted-beam transmission electron microscopy. Proc Natl Acad Sci U S A. 2009;106:14339–44. doi: 10.1073/pnas.0907821106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, et al. A specific amyloid-β protein assembly in the brain impairs memory. Nature. 2006;440:352–7. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 59.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: Separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–98. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 60.Sabbagh MN, Cooper K, DeLange J, Stoehr JD, Thind K, et al. Functional, global and cognitive decline correlates to accumulation of Alzheimer’s pathology in MCI and AD. Curr Alzheimer Res. 2010;7:280–6. doi: 10.2174/156720510791162340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Antzutkin ON, Leapman RD, Balbach JJ, Tycko R. Supramolecular structural constraints on Alzheimer’s β-amyloid fibrils from electron microscopy and solid state nuclear magnetic resonance. Biochemistry. 2002;41:15436–50. doi: 10.1021/bi0204185. [DOI] [PubMed] [Google Scholar]
- 62.Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: Their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci U S A. 1994;91:5355–8. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kammerer RA, Kostrewa D, Zurdo J, Detken A, Garcia-Echeverria C, et al. Exploring amyloid formation by a de novo design. Proc Natl Acad Sci U S A. 2004;101:4435–40. doi: 10.1073/pnas.0306786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bu ZM, Shi Y, Callaway DJE, Tycko R. Molecular alignment within β-sheets in Aβ14-23 fibrils: Solid state NMR experiments and theoretical predictions. Biophys J. 2007;92:594–602. doi: 10.1529/biophysj.106.091017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grabowski TJ, Cho HS, Vonsattel JPG, Rebeck GW, Greenberg SM. Novel amyloid precursor protein mutation in an Iowa family with dementia and severe cerebral amyloid angiopathy. Ann Neurol. 2001;49:697–705. doi: 10.1002/ana.1009. [DOI] [PubMed] [Google Scholar]
- 66.Rostagno A, Holton JL, Lashley T, Revesz T, Ghiso J. Cerebral amyloidosis: Amyloid subunits, mutants and phenotypes. Cell Mol Life Sci. 2010;67:581–600. doi: 10.1007/s00018-009-0182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Catalano SM, Dodson EC, Henze DA, Joyce JG, Krafft GA, Kinney GG. The role of amyloid-β derived diffusible ligands (ADDLs) in Alzheimer’s disease. Curr Top Med Chem. 2006;6:597–608. doi: 10.2174/156802606776743066. [DOI] [PubMed] [Google Scholar]
- 68.Yu LP, Edalji R, Harlan JE, Holzman TF, Lopez AP, et al. Structural characterization of a soluble amyloid β-peptide oligomer. Biochemistry. 2009;48:1870–7. doi: 10.1021/bi802046n. [DOI] [PubMed] [Google Scholar]
- 69.Kheterpal I, Chen M, Cook KD, Wetzel R. Structural differences in Aβ amyloid protofibrils and fibrils mapped by hydrogen exchange-mass spectrometry with on-line proteolytic fragmentation. J Mol Biol. 2006;361:785–95. doi: 10.1016/j.jmb.2006.06.066. [DOI] [PubMed] [Google Scholar]
- 70.Chimon S, Shaibat MA, Jones CR, Calero DC, Aizezi B, Ishii Y. Evidence of fibrillike β-sheet structures in a neurotoxic amyloid intermediate of Alzheimer’s β-amyloid. Nat Struct Mol Biol. 2007;14:1157–64. doi: 10.1038/nsmb1345. [DOI] [PubMed] [Google Scholar]
- 71.Tomic JL, Pensalfini A, Head E, Glabe CG. Soluble fibrillar oligomer levels are elevated in Alzheimer’s disease brain and correlate with cognitive dysfunction. Neurobiol Dis. 2009;35:352–8. doi: 10.1016/j.nbd.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kokubo H, Kayed R, Glabe CG, Yamaguchi H. Soluble Aβ oligomers ultrastructurally localize to cell processes and might be related to synaptic dysfunction in Alzheimer’s disease brain. Brain Res. 2005;1031:222–8. doi: 10.1016/j.brainres.2004.10.041. [DOI] [PubMed] [Google Scholar]
- 73.Chimon S, Ishii Y. Capturing intermediate structures of Alzheimer’s β-amyloid, Aβ1-40, by solid state NMR spectroscopy. J Am Chem Soc. 2005;127:13472–3. doi: 10.1021/ja054039l. [DOI] [PubMed] [Google Scholar]
- 74.Walsh P, Neudecker P, Sharpe S. Structural properties and dynamic behavior of nonfibrillar oligomers formed by PrP106-126. J Am Chem Soc. 2010;132:7684–95. doi: 10.1021/ja100431q. [DOI] [PubMed] [Google Scholar]
- 75.Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–83. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garcia FL, Zahn R, Riek R, Wuthrich K. NMR structure of the bovine prion protein. Proc Natl Acad Sci U S A. 2000;97:8334–9. doi: 10.1073/pnas.97.15.8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, et al. The most infectious prion protein particles. Nature. 2005;437:257–61. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cobb NJ, Sonnichsen FD, McHaourab H, Surewicz WK. Molecular architecture of human prion protein amyloid: A parallel, in-register β-structure. Proc Natl Acad Sci U S A. 2007;104:18946–51. doi: 10.1073/pnas.0706522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu XJ, Wintrode PL, Surewicz WK. β-sheet core of human prion protein amyloid fibrils as determined by hydrogen/deuterium exchange. Proc Natl Acad Sci U S A. 2007;104:1510–5. doi: 10.1073/pnas.0608447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Makarava N, Baskakov IV. The same primary structure of the prion protein yields two distinct self-propagating states. J Biol Chem. 2008;283:15988–96. doi: 10.1074/jbc.M800562200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wickner RB. [URE3] as an altered Ure2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–9. doi: 10.1126/science.7909170. First identification of a non-mammalian prion protein. [DOI] [PubMed] [Google Scholar]
- 82.Edskes HK, Gray VT, Wickner RB. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci U S A. 1999;96:1498–503. doi: 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brachmann A, Baxa U, Wickner RB. Prion generation in vitro: Amyloid of Ure2p is infectious. Embo J. 2005;24:3082–92. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–8. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 85.Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci U S A. 2005;102:12825–30. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–72. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baxa U, Speransky V, Steven AC, Wickner RB. Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein. Proc Natl Acad Sci U S A. 2002;99:5253–60. doi: 10.1073/pnas.082097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saupe SJ. A short history of small s, a prion of the fungus Podospora anserina. Prion. 2007;1:110–5. doi: 10.4161/pri.1.2.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang X, Hammer ND, Chapman MR. The molecular basis of functional bacterial amyloid polymerization and nucleation. J Biol Chem. 2008;283:21530–9. doi: 10.1074/jbc.M800466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:100–7. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tycko R, Hu KN. A Monte Carlo/simulated annealing algorithm for sequential resonance assignment in solid state NMR of uniformly labeled proteins with magic-angle spinning. J Magn Reson. 2010;205:304–14. doi: 10.1016/j.jmr.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sen A, Baxa U, Simon MN, Wall JS, Sabate R, et al. Mass analysis by scanning transmission electron microscopy and electron diffraction validate predictions of stacked β-solenoid model of HET-s prion fibrils. J Biol Chem. 2007;282:5545–50. doi: 10.1074/jbc.M611464200. [DOI] [PubMed] [Google Scholar]
- 93.Jenkins J, Pickersgill R. The architecture of parallel β-helices and related folds. Prog Biophys Mol Biol. 2001;77:111–75. doi: 10.1016/s0079-6107(01)00013-x. [DOI] [PubMed] [Google Scholar]
- 94.Govaerts C, Wille H, Prusiner SB, Cohen FE. Evidence for assembly of prions with left-handed β3-helices into trimers. Proc Natl Acad Sci U S A. 2004;101:8342–7. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lazo ND, Downing DT. Fibril formation by amyloid-β proteins may involve β-helical protofibrils. J Pept Res. 1999;53:633–40. doi: 10.1034/j.1399-3011.1999.00057.x. [DOI] [PubMed] [Google Scholar]
- 96.Siemer AB, Ritter C, Ernst M, Riek R, Meier BH. High-resolution solid state NMR spectroscopy of the prion protein HET-s in its amyloid conformation. Angew Chem -Int Edit. 2005;44:2441–4. doi: 10.1002/anie.200462952. [DOI] [PubMed] [Google Scholar]
- 97.Shewmaker F, McGlinchey RP, Thurber KR, McPhie P, Dyda F, et al. The functional curli amyloid is not based on in-register parallel β-sheet structure. J Biol Chem. 2009;284:25065–76. doi: 10.1074/jbc.M109.007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tycko R. Symmetry-based constant-time homonuclear dipolar recoupling in solid state NMR. J Chem Phys. 2007;126:064506. doi: 10.1063/1.2437194. [DOI] [PubMed] [Google Scholar]
- 99.Morcombe CR, Gaponenko V, Byrd RA, Zilm KW. Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J Am Chem Soc. 2004;126:7196–7. doi: 10.1021/ja047919t. [DOI] [PubMed] [Google Scholar]
- 100.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-driven 13C-13C recoupling without 13C rf irradiation in nuclear magnetic resonance of rotating solids. J Chem Phys. 2003;118:2325–41. [Google Scholar]
- 101.De Paepe G, Lewandowski JR, Loquet A, Bockmann A, Griffin RG. Proton assisted recoupling and protein structure determination. J Chem Phys. 2008;129:245101. doi: 10.1063/1.3036928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nomura K, Takegoshi K, Terao T, Uchida K, Kainosho M. Three-dimensional structure determination of a uniformly labeled molecule by frequency-selective dipolar recoupling under magic-angle spinning. J Biomol NMR. 2000;17:111–23. doi: 10.1023/a:1008398906753. [DOI] [PubMed] [Google Scholar]
- 103.Hu KN, Tycko R. Zero-quantum frequency-selective recoupling of homonuclear dipole-dipole interactions in solid state nuclear magnetic resonance. J Chem Phys. 2009;131:045101. doi: 10.1063/1.3176874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tycko R. Stochastic dipolar recoupling in nuclear magnetic resonance of solids. Phys Rev Lett. 2007;99:187601. doi: 10.1103/PhysRevLett.99.187601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tycko R. Theory of stochastic dipolar recoupling in solid state nuclear magnetic resonance. J Phys Chem B. 2008;112:6114–21. doi: 10.1021/jp076808o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gullion T, Schaefer J. Rotational-echo double-resonance NMR. J Magn Reson. 1989;81:196–200. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 107.Helmus JJ, Nadaud PS, Hofer N, Jaroniec CP. Determination of methyl 13C-15N dipolar couplings in peptides and proteins by three-dimensional and four-dimensional magic-angle spinning solid state NMR spectroscopy. J Chem Phys. 2008;128:052314. doi: 10.1063/1.2817638. [DOI] [PubMed] [Google Scholar]
- 108.Nadaud PS, Helmus JJ, Kall SL, Jaroniec CP. Paramagnetic ions enable tuning of nuclear relaxation rates and provide long-range structural restraints in solid state NMR of proteins. J Am Chem Soc. 2009;131:8108–20. doi: 10.1021/ja900224z. [DOI] [PubMed] [Google Scholar]
- 109.Nadaud PS, Helmus JJ, Hofer N, Jaroniec CP. Long-range structural restraints in spin-labeled proteins probed by solid state nuclear magnetic resonance spectroscopy. J Am Chem Soc. 2007;129:7502–+. doi: 10.1021/ja072349t. [DOI] [PubMed] [Google Scholar]
- 110.Balayssac S, Bertini I, Bhaumik A, Lelli M, Luchinat C. Paramagnetic shifts in solid state NMR of proteins to elicit structural information. Proc Natl Acad Sci U S A. 2008;105:17284–9. doi: 10.1073/pnas.0708460105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wickramasinghe NP, Parthasarathy S, Jones CR, Bhardwaj C, Long F, et al. Nanomole-scale protein solid state NMR by breaking intrinsic 1H T1 boundaries. Nat Methods. 2009;6:215–8. doi: 10.1038/nmeth.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thurber KR, Tycko R. Biomolecular solid state NMR with magic-angle spinning at 25 K. J Magn Reson. 2008;195:179–86. doi: 10.1016/j.jmr.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hu KN, Song C, Yu HH, Swager TM, Griffin RG. High-frequency dynamic nuclear polarization using biradicals: A multifrequency EPR lineshape analysis. J Chem Phys. 2008;128:052302. doi: 10.1063/1.2816783. [DOI] [PubMed] [Google Scholar]
- 114.Maly T, Debelouchina GT, Bajaj VS, Hu KN, Joo CG, et al. Dynamic nuclear polarization at high magnetic fields. J Chem Phys. 2008;128:052211. doi: 10.1063/1.2833582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Thurber KR, Tycko R. Prospects for sub-micron solid state nuclear magnetic resonance imaging with low-temperature dynamic nuclear polarization. Phys Chem Chem Phys. 2010;12:5779–85. doi: 10.1039/c0cp00157k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thurber KR, Yau WM, Tycko R. Low-temperature dynamic nuclear polarization at 9.4 T with a 30 mW microwave source. J Magn Reson. 2010;204:303–13. doi: 10.1016/j.jmr.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ishii Y. 13C-13C dipolar recoupling under very fast magic angle spinning in solid state nuclear magnetic resonance: Applications to distance measurements, spectral assignments, and high-throughput secondary-structure determination. J Chem Phys. 2001;114:8473–83. [Google Scholar]


