Abstract
Asparagine N-linked glycosylation is one of the most important forms of protein post-translational modification in eukaryotes and is one of the first metabolic pathways described at a biochemical level. Here, we report a new annotation of this pathway for the Human species, published after passing a peer-review process in Reactome. The new annotation presented here offers a high level of detail and provides references and descriptions for each reaction, along with integration with GeneOntology and other databases. The open-source approach of Reactome toward annotation encourages feedback from its users, making it easier to keep the annotation of this pathway updated with future knowledge. Reactome's web interface allows easy navigation between steps involved in the pathway to compare it with other pathways and resources in other scientific databases and to export it to BioPax and SBML formats, making it accessible for computational studies. This new entry in Reactome expands and complements the annotations already published in databases for biological pathways and provides a common reference to researchers interested in studying this important pathway in the human species. Finally, we discuss the status of the annotation of this pathway and point out which steps are worth further investigation or need better experimental validation.
Keywords: aspargine N-linked glycosylation, Reactome database
Introduction
Among the metabolic pathways described in the scientific literature, the pathway of asparagine N-linked glycosylation is one of the better characterized and one of the earliest to have been described at a gene level. Most of the reactions involved in the first part of the pathway, the synthesis of the lipid-linked oligosaccharide (LLO), were already described in the 1980s (Hubbard and Ivatt 1981; Huffaker and Robbins 1983; Kornfeld and Kornfeld 1985), and over the years, the book “Essentials of Glycobiology” (Stanley et al. 2009; Varki and Sharon 2009) has become the reference for summarizing the literature on this topic. However, in order to be useful for computational studies, a biological process not only has to be described in prose in the literature but also must be codified in a unique and well-defined electronic format, to which different studies can refer in order to obtain reproducible and comparable results.
In this paper, we describe the asparagine N-linked glycosylation pathway newly annotated in Reactome. Reactome (http://www.reactome.org) is a freely available and open-source database of human biological pathways (Vastrik et al. 2007; Matthews et al. 2009; Croft et al. 2011). Expert scientists, together with in-house curators, enter information into a central repository which is then peer-reviewed to obtain a consensus representation of the process or pathway. These data are extensively cross-linked to major protein and nucleotide sequence databases as well as to Gene Ontology (GO; Ashburner et al. 2000) and PubMed databases. A new website for Reactome was recently released, which will allow users to interact with curated pathways to a greater extent for searching and visualizing them and for analyzing user-supplied data sets.
Asparagine N-glycosylation in other databases
Reactions involved in the asparagine N-linked glycosylation pathway are already annotated in other scientific databases. The finest annotation is provided in KEGG/Pathways (Kanehisa and Goto 2000; Kanehisa et al. 2010), in the entry hsa00510 N-Glycan BioSynthesis (http://www.genome.jp/dbget-bin/show_pathway?hsa00510). The synthesis of the LLO, corresponding to the first steps of N-glycosylation, is also described in the MetaCyc database in the entry “Mannosyl-Chito-Dolichol-Biosynthesis” (Caspi et al. 2010). Details of a subset of the reactions in N-glycosylation are also described in Uniprot/Pathways (http://www.grenoble.prabi.fr/obiwarehouse/unipathway/upa?upid=UPA00378) and in BioCarta (http://www.biocarta.com/genes/index.asp). The GlycoGene database provides fine annotations on the genes involved in asparagine-linked and other forms of glycosylation, along with their tissue distribution, substrates specificities and homologs (Ito et al. 2010). Another resource comes from the CFG-Nature Functional Glycomics Gateway which characterized more than 7500 glycan structures (http://www.functionalglycomics.org/). The efforts on annotating N-glycan and other carbohydrate structures have been recently re-organized in the Glycome-DB database (Ranzinger et al. 2008, 2009). However, in none of these databases, the pathway of asparagine N-glycosylation is completely annotated, from the synthesis of the precursor to the late branching steps.
Structure of a pathway entry in Reactome
In Reactome, pathways are composed by a conjunction of two different entries: “reactions”, which are entries describing the details of a single metabolic reaction or of other kinds of gene-to-gene interaction; and “pathways”, which are placeholders entries that group together different reactions with a similar function or role. For each reaction, the most important details provided are: a reference to GO for the localization of each component of the reaction and the molecular function of the enzyme involved in the reaction; the Uniprot (Jain et al. 2009; Uniprot Consortium 2010) and the Chemical Entities of Biological Interest identification numbers (ChEBI, De Matos et al. 2010) of substrates and products; the references to publications that validate experimentally the reaction in the human species; a descriptive summary explaining the reaction in detail and eventual implications of the genes involved in a reaction with a congenital disease are also described in the summary. Pathway entries are similar to reaction entries, but instead of a reference to a specific experimental evidence they contain references to reviews and books related to all the reactions included. They also are assigned GO Biological Process terms.
Results and discussion
The pathway of N-glycosylation and its subdivision into subpathways
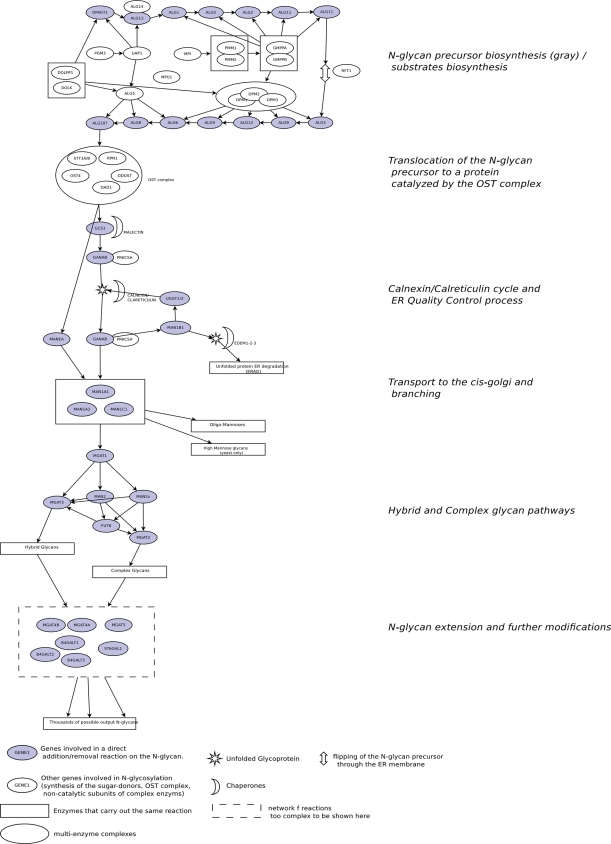
Figure 1 shows a schematic representation of the pathway used during the curation process, and Supplementary data, Table S1 shows the list of genes annotated in the pathway, and their subdivision into subpathways. The classification into subpathways is based on the current literature. For each of the subpathways, we provided at least one review describing it. As Reactome focuses on human pathways, we have provided only human-specific references, even if many steps on this pathway were originally identified and studied in other species, mainly in yeast.
Fig. 1.
A manually drawn representation of the asparagine N-linked glycosylation pathway, used as reference during the curation.
The first subpathway described is “Biosynthesis of the N-glycan precursor” (Hubbard and Ivatt 1981; Huffaker and Robbins 1983; Kornfeld and Kornfeld 1985; Yarema and Bertozzi 2001; Kelleher and Gilmore 2006; Weerapana and Imperiali 2006; Stanley et al. 2009). It corresponds to the first step in asparagine N-glycosylation and consists of the synthesis of a precursor molecule composed of 14 sugar/monosaccharide units and usually referred to as N-glycan precursor or LLO. This process occurs on the endoplasmic reticulum (ER) membrane and produces a glycan with a defined structure, which constitutes a core common to all the N-glycans observed on mature proteins. Along with the entry for the “Biosynthesis of the N-glycan precursor”, we decided to include the main reactions leading to the “Synthesis of substrates in N-glycan biosythesis”, to describe the reactions leading to the synthesis of the small molecules required as sugar donors for the sugar-addition reactions in separate sections. In this case, the grouping of these reactions does not correspond to a single specific metabolic process, but has the simple role of organizing the reactions that lead to the synthesis of these biomolecules.
After being synthesized on the ER membrane, the N-glycan precursor is co-translationally attached to an unfolded protein, while it is still being synthesized by a ribosome attached to the ER membrane and transported to the ER through the Sec61 channel (Kelleher and Gilmore 2006). This step is described in the subpathway “Transfer of N-glycan to the protein”.
The subsequent step in the pathway is “N-glycan trimming in the ER and Calnexin/Calreticulin cycle” where the N-glycan is trimmed of glucose residues and many of the mannose residues and is involved in the process of folding of the protein to which it is attached (Ferna et al. 1998; Parodi 2000; Ruddock and Molinari 2006; Caramelo and Parodi 2008; Roth et al. 2008; Aebi et al. 2010; Freeze et al. 2009; Lederkremer 2009; Shental-Bechor and Levy 2009; Määttänen et al. 2010). In this step, an intermediately trimmed form of the N-glycan is used as a signal for the binding of the unfolded glycoprotein to a chaperone protein (calnexin or calreticulin) with the role of providing an environment where the former can fold properly without engaging in incorrect interactions with other proteins. The presence of at least one N-glycan sugar is important for this interaction in the majority of the proteins in the secretory pathway. If the glycoprotein does not correctly fold after interacting with the calnexin or calreticulin complex, it is transported to a compartment called “Endoplasmic Reticulum Quality Control Compartment (ERQC)” (Avezov et al. 2008; Kamhi-Nesher et al. 2001), where it can be directed toward degradation in the case of major folding problems, or returned back to the calnexin or calreticulin complex.
If the glycoprotein is instead folded properly, it is transported to the Golgi complex where the N-glycan undergoes further modifications (Murshid and Presley 2004; Appenzeller-herzog and Hauri 2006). The mechanism of transport has been described in the subpathway “Transport to the Golgi and subsequent modification”; here, we also added a reference to another mechanism for N-glycosylation-independent ER-to-Golgi transport, through which some glycoproteins can be transported even in the presence of inhibitors of the calnexin/calreticulin-related glucosydases (Wieland et al. 1987; Martínez-Menárguez et al. 1999). N-Glycans observed in human cells on mature proteins are usually classified into three different classes: oligomannoses, complex glycans and hybrid glycans (Stanley et al. 2009). We decided to describe the mannose trimming reactions occurring in the medial-Golgi, where the first choice between synthesis of oligomannoses and hybrid/complex N-glycans is made, as a separated subpathway, “N-glycan trimming and elongation in the cis-Golgi”. After the trimming of the mannoses on the α1,6-branch has been carried out, the pathway of N-glycosylation becomes very complex and becomes a network of reactions in which few enzymes originate thousands of different N-glycan structures, as described in the Glycome-DB database (Ranzinger et al. 2008, 2009). Since most of these reactions have not yet been characterized experimentally, it has not been possible to include the detailed structure of this network of reactions in Reactome. Although some reconstructions on the structure of the network of complex asparagine N-glycosylation are available (Kornfeld and Kornfeld 1985; Murrell et al. 2004; Hossler et al. 2006; Hossler et al 2007; Kim et al. 2009), these refer only to computationally inferred reconstructions of the network of N-glycosylation; therefore, they cannot be included in Reactome. Instead, a reaction has been created for each enzyme known to be involved in the modification of the N-glycan antennae, using a generic “N-glycan” entry for input and output. For example, we described only a single reaction for the addition of α1,6-fucose to the first GlcNAc of the N-glycan by the enzyme FUT8, using an “N-glycoprotein” as input and an “N-Glycoprotein with fucosyl α1,6-GlcNAc” as output, without specifying the exact structures of all the possible inputs and outputs of this modification. Among these reactions, we decided to annotate in separate subpathways the “reactions specific to the hybrid N-glycan synthesis pathway” such as the addition of a bifurcating GlcNAc by MGAT3, and the “reactions specific to the complex N-glycan synthesis pathway”, such as mannose trimming by MAN2 (Crispin et al 2007), and finally, we put all the N-glycan antennae modification events known to be common to both hybrid and complex glycans under a common subpathway, entitled “N-Glycan antennae elongation”.
Advantages of annotating a pathway in Reactome
One of the strongest points of Reactome is its open-source-like approach toward annotation, which allows a good interaction with users who want to submit corrections or feedback. The inclusion of a reference and a description for each reaction facilitates the tracing back of the sources used for each annotation and prevents incorrect interpretations of the data. The names of authors and reviewers of each reaction are shown publicly, so they are available for discussion and clarification. Moreover, the Reactome web interface makes it easy to submit error reports, providing a “Let us know what you think of this article” button on each reaction page.
Thanks to the Reactome web interface, a pathway can also be exported to advanced formats like BioPax (Luciano 2005), SBML (Hucka 2003), Protegè (http://protege.stanford.edu) and Cytoscape (Cline et al. 2007), facilitating the integration with other tools used in computational biology and bioinformatics. Each reaction can be navigated and integrated with other pathways annotated in the same database, making it possible to see which of the entities involved in a given pathway participate to other biological processes.
Points worth further investigation
The process of annotating this pathway in Reactome has been an opportunity to identify and report various errors present in other databases. These changes are describes in Supplementary data, Document S1. Other unclear or missing information have been found in the literature and may deserve further investigation.
First, the gene ALG10 has been found to have experienced a recent duplication in some primates (Homo sapiens, Pan troglodytes, Pan paniscus, Gorilla gorilla, Pongo pygmaeus but not in Macaca mulatta; T Marquès-Bonet, personal communication), so in these species, it is present in two copies, called ALG10A and ALG10B. To date, no literature explains which of these copies is the functional one, or whether they are both functional genes. It would be interesting to study this recent duplication specific to great apes on a gene involved in a process so conserved and functionally constrained such as the synthesis of the N-glycan precursor.
Second, there is no specific literature to demonstrate that the addition of the third, fourth and fifth mannoses are added by the genes identified as ALG2 and ALG11 in humans, during the synthesis of the LLO; further investigation could validate these reactions in humans.
The high quality of the newly annotated pathway will allow researchers to use it with a high accuracy and confidence and will help to disseminate the idea that accurate biological knowledge and automatic use of database information can go together in good scientific setting
Availability
The new entry is available at the address http://www.reactome.org/entitylevelview/PathwayBrowser.html#DB=gk_current_pathway_diagram&FOCUS_SPECIES_ID=48887&FOCUS_PATHWAY_ID=446203&ID=446203, or from the alternative url http://tinyurl.com/nglyco-reactome. A screenshot of the pathway from the Reactome website is shown in Supplementary data, Figure S1.
Materials and methods
For the annotation of the pathway, we followed the Reactome's standard procedure on how to submit a new pathway. For every reaction proposed, we provided a reference showing that the reaction involved the described components and that it occurs in humans. In the cases where it has not been possible to provide the evidence that it occurs in human, we annotated a reference relative to another species and created a reaction in that species which is then inferred for the human reaction. For historical reasons, most of these references are relative to Saccharomyces cerevisiae.
Along with the literature reference(s) for each reaction, the standard information captured by Reactome is as follows: GO terms are used to describe the molecular function of catalysts (GO molecular function), locations of entities and events (GO cellular component) and overall processes occurring (GO biological process). A reaction contains input and output entities which may or may not be catalyzed by a protein. Proteins and chemicals are assigned UniProt IDs and ChEBI IDs (Chemical Entities of Biological Interest), respectively. A summary is included describing the details of the event in question. Outputs of reactions can become inputs for other reactions and in this way a connected pathway can be built up.
The information is provided to a Reactome curator who enters the information into the database using an in-house GUI called the Curator Tool. A second expert is recruited to check the data for biological correctness. Any comments are discussed between author, curator and reviewer until a consensus pathway is reached.
Most of the ChEBI IDs used to identify glycans in the pathway have been explicitly created in collaboration with the ChEBI maintainers. In these cases, we took the entries in the Glycome-DB (Ranzinger et al. 2008, 2009) database as models and requested a ChEBI ID with the same chemical structure. Differently from the convention used in the KEGG database and similar to what has been made for Glycome-DB, we decided to not include the Aglyca part of the glycan (i.e. the amino acid to which it is attached) in the structure, but only to annotate all the possible Aglycas known in the literature, taking the Glycome-DB entries as reference. The list of reactions used during the curation process and details of each of the reactions annotated are provided in Supplementary data, Table S2.
Supplementary data
Supplementary data for this article is available online at http:// glycob.oxfordjournals.org/.
Funding
This research was funded by grants SAF-2007-63171 and BFU2010-19443 (subprogram BMC) awarded by Ministerio de Educación y Ciencia (Spain), the Direcció General de Recerca, Generalitat de Catalunya (Grup de Recerca Consolidat 2009 SGR 1101), a grant from the National Human Genome Research Institute at the National Institutes of Health (P41 HG003751) and from the European Union 6th Framework Programme “ENFIN” (LSHG-CT-2005-518254). G.M.D. is supported by a PhD fellowship from the Programa de becas FPI (BES-2009-017731) del Ministerio de Educación y Ciencia, Spain.
Supplementary Material
Acknowledgements
We would like to thank Gerardo Lederkremer for solving doubts on the ER mannosidase genes, KEGG and UniProt technical support, Renè Ranzinger from Glycome-DB for helping to choose the right structures from the Glycome-DB, Patrick Hossler from Glycovis for helping to annotate the latter part of the pathway, and Tomas Marquès-Bonet for confirming the ALG10 duplication. We would also want to acknowledge two anonymous reviewers for their comments and suggestions that greatly improved the manuscript, Kevin Keys and Brandon Invergo for revising the grammar in the manuscript and Anna Bauer-Mehren for counseling about Reactome. Bioinformatics services were kindly provided by the Genomic Diversity node, Spanish Bioinformatic Institute (http://www.inab.org).
Conflict of interest
None declared.
Abbreviations
ChEBI, chemical Entities of Biological Interest identification numbers; ER, endoplasmic reticulum; GO, Gene Ontology; LLO, lipid-linked oligosaccharide.
References
- Aebi M, Bernasconi R, Clerc S, Molinari M. N-glycan structures: Recognition and processing in the ER. Trends Biochem Sci. 2010;35:74–82. doi: 10.1016/j.tibs.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Appenzeller-herzog C, Hauri H-P. The ER-Golgi intermediate compartment (ERGIC): In search of its identity and function. J Cell Sci. 2006;119:2173–83. doi: 10.1242/jcs.03019. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avezov E, Frenkel Z, Ehrlich M, Herscovics A, Lederkremer GZ. Endoplasmic reticulum (ER) mannosidase I Is compartmentalized and required for N-glycan trimming to Man5–6GlcNAc2 in glycoprotein ER-associated degradation. Mol Biol Cell. 2008;19:216–225. doi: 10.1091/mbc.E07-05-0505. doi:10.1091/mbc.E07-05-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramelo JJ, Parodi AJ. Getting in and out from calnexin/calreticulin cycles. J Biol Chem. 2008;283:10221–10225. doi: 10.1074/jbc.R700048200. doi:10.1074/jbc.R700048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38(Database issue):D473–D479. doi: 10.1093/nar/gkp875. doi:10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2(10):2366–2382. doi: 10.1038/nprot.2007.324. doi:10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispin M, Aricescu AR, Chang VT, Jones EY, Stuart DI, Dwek RA, Davis SJ, Harvey DJ. Disruption of α-mannosidase processing induces non-canonical hybrid-type glycosylation. FEBS Lett. 2007;581(10):1963–1968. doi: 10.1016/j.febslet.2007.04.020. doi:10.1016/j.febslet.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq1018. 39(Database issue):D691–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matos P, Alcántara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical entities of biological interest: An update. Nucleic Acids Res. 2010;38(Database issue):D249–D254. doi: 10.1093/nar/gkp886. doi:10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferna F, Fanchiotti S, Parodi AJ. A misfolded protein conformation is not a sufficient condition for in vivo glucosylation by the UDP-Glc:glycoprotein glucosyltransferase. EMBO J. 1998;17(20):5877–5886. doi: 10.1093/emboj/17.20.5877. doi:10.1093/emboj/17.20.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeze H, Esko J, Parodi A. Glycans in glycoprotein quality control. Cold Spring Harbor Laboratory Press New York (USA); 2009. pp. 1–10. In: Essentials of Glycobiology. [PubMed] [Google Scholar]
- Hossler P, Goh L-T, Lee MM, Hu W-S. GlycoVis: Visualizing glycan distribution in the protein N-glycosylation pathway in mammalian cells. Biotechnol Bioeng. 2006;5:946–960. doi: 10.1002/bit.21062. doi:10.1002/bit.21062. [DOI] [PubMed] [Google Scholar]
- Hossler P, Mulukutla BC, Hu W-S. Systems analysis of N-glycan processing in mammalian cells. PLoS One. 2007;2(1):e713. doi: 10.1371/journal.pone.0000713. doi:10.1371/journal.pone.0000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard CS, Ivatt RJ. Synthesis and processing of asparagine-linked oligosaccharides. Ann Rev Biochem. 1981;50:555–83. doi: 10.1146/annurev.bi.50.070181.003011. [DOI] [PubMed] [Google Scholar]
- Hucka M. The systems biology markup language (SBML): A medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. doi:10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Huffaker TC, Robbins PW. Yeast mutants deficient in protein glycosylation. Proc Natl Acad Sci USA. 1983;80:7466–7470. doi: 10.1073/pnas.80.24.7466. doi:10.1073/pnas.80.24.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Chiba Y, Kameyama A, Sato T, Narimatsu H. In vitro and in vivo enzymatic syntheses and mass spectrometric database for N-glycans and O-glycans. Methods Enzymol. 2010;478:127–149. doi: 10.1016/S0076-6879(10)78005-8. doi:10.1016/S0076-6879(10)78005-8. [DOI] [PubMed] [Google Scholar]
- Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek BE, Martin MJ, McGarvey P, Gasteiger E. Infrastructure for the life sciences: Design and implementation of the UniProt website. BMC Bioinformatics. 2009;10:136. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamhi-Nesher S, Shenkman M, Tolchinsky S, Fromm SV, Ehrlich R, Lederkremer GZ. A novel quality control compartment derived from the endoplasmic reticulum. Molecular biology of the cell. 2001;12(6):1711–23. doi: 10.1091/mbc.12.6.1711. doi:10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38(Database issue):D355–D360. doi: 10.1093/nar/gkp896. doi:10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. doi:10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16(4):47R–62R. doi: 10.1093/glycob/cwj066. doi:10.1093/glycob/cwj066. [DOI] [PubMed] [Google Scholar]
- Kim P-J, Lee D-Y, Jeong H. Centralized modularity of N-linked glycosylation pathways in mammalian cells. PLoS One. 2009;4(10):e7317. doi: 10.1371/journal.pone.0007317. doi:10.1371/journal.pone.0007317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Ann Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. doi:10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Lederkremer GZ. Glycoprotein folding, quality control and ER-associated degradation. Curr Opin Struct Biol. 2009;19(5):515–523. doi: 10.1016/j.sbi.2009.06.004. doi:10.1016/j.sbi.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Luciano JS. PAX of mind for pathway researchers. Drug Discov Today. 2005;10(13):937–942. doi: 10.1016/S1359-6446(05)03501-4. doi:10.1016/S1359-6446(05)03501-4. [DOI] [PubMed] [Google Scholar]
- Määttänen P, Gehring K, Bergeron JJ, Thomas DY. Protein quality control in the ER: The recognition of misfolded proteins. Semin Cell Dev Biol. 2010;21:500–511. doi: 10.1016/j.semcdb.2010.03.006. doi:10.1016/j.semcdb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, Kanapin A, Lewis S, Mahajan S, May B, Schmidt E, Vastrik I, Wu G, Birney E, Stein L, D'Eustachio P. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37(Database issue):D619–22. doi: 10.1093/nar/gkn863. doi:10.1016/S0092-8674(00)80608-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Menárguez JA, Geuze HJ, Slot JW, Klumperman J. Vesicular tubular clusters between the ER and Golgi mediate concentration of soluble secretory proteins by exclusion from COPI-coated vesicles. Cell. 1999;98:81–90. doi: 10.1016/S0092-8674(00)80608-X. doi:10.1016/S0092-8674(00)80608-X. [DOI] [PubMed] [Google Scholar]
- Murrell MP, Yarema KJ, Levchenko A. The systems biology of glycosylation. Chembiochem. 2004;5:1334–1347. doi: 10.1002/cbic.200400143. doi:10.1002/cbic.200400143. [DOI] [PubMed] [Google Scholar]
- Murshid A, Presley JF. ER-to-Golgi transport and cytoskeletal interactions in animal cells. Cell Mol Life Sci. 2004;61:133–45. doi: 10.1007/s00018-003-3352-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi AJ. Protein glucosylation and its role in protein folding. Ann Rev Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. doi:10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- Ranzinger R, Frank M, von der Lieth C-W, Herget S. Glycome-DB.org: A portal for querying across the digital world of carbohydrate sequences. Glycobiology. 2009;19(12):1563–1567. doi: 10.1093/glycob/cwp137. doi:10.1093/glycob/cwp137. [DOI] [PubMed] [Google Scholar]
- Ranzinger R, Herget S, Wetter T, von der Lieth C-W. GlycomeDB—integration of open-access carbohydrate structure databases. BMC Bioinformatics. 2008;9:384. doi: 10.1186/1471-2105-9-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J, Yam GH-F, Fan J, Hirano K, Gaplovska-Kysela K, Le Fourn V, Guhl B, Santimaria R, Torossi T, Ziak M, et al. Protein quality control: The who's who, the where's and therapeutic escapes. Histochem Cell Biol. 2008;129(2):163–177. doi: 10.1007/s00418-007-0366-7. doi:10.1007/s00418-007-0366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddock LW, Molinari M. N-glycan processing in ER quality control. J Cell Sci. 2006;119(Pt 21):4873–4380. doi: 10.1242/jcs.03225. [DOI] [PubMed] [Google Scholar]
- Shental-Bechor D, Levy Y. Folding of glycoproteins: Toward understanding the biophysics of the glycosylation code. Curr Opin Struct Biol. 2009;19(5):524–533. doi: 10.1016/j.sbi.2009.07.002. doi:10.1016/j.sbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Stanley P, Schachter H, Taniguchi N. Cold Spring Harbor Laboratory Press. 2009. N-Glycans. In: Essentials of Glycobiology. New York (USA). [PubMed] [Google Scholar]
- Uniprot Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38(Database issue):D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Sharon N. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press. New York (USA); 2009. Historical background and overview. [PubMed] [Google Scholar]
- Vastrik I, D'Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, et al. Reactome: A knowledge base of biologic pathways and processes. Genome Biol. 2007;8(3):R39. doi: 10.1186/gb-2007-8-3-r39. doi:10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Imperiali B. Asparagine-linked protein glycosylation: From eukaryotic to prokaryotic systems. Glycobiology. 2006;16(6):91R–101R. doi: 10.1093/glycob/cwj099. doi:10.1093/glycob/cwj099. [DOI] [PubMed] [Google Scholar]
- Wieland FT, Gleason ML, Serafini TA, Rothman JE. The rate of bulk flow from the endoplasmic reticulum to the cell surface. Cell. 1987;50(2):289–300. doi: 10.1016/0092-8674(87)90224-8. doi:10.1016/0092-8674(87)90224-8. [DOI] [PubMed] [Google Scholar]
- Yarema KJ, Bertozzi CR. Characterizing glycosylation pathways. Genome Biol. 2001;2(5):REVIEWS0004. doi: 10.1186/gb-2001-2-5-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.