Abstract
BACKGROUND
The Fifth Evian Annual Reproduction (EVAR) Workshop Meeting discussed knowledge regarding contemporary genetics in female reproduction.
METHODS
Specialist reproductive medicine clinicians and geneticists delivered presentations based on published literature and current research. The content of this report is based on the expert presentations and subsequent group discussions that took place during this Workshop.
RESULTS
Numerous ovarian genes with a role in infertility have been identified. Future challenges for genetic screening of patients, such as those with polycystic ovary syndrome, primary ovarian insufficiency or endometriosis, include the identification of high-throughput strategies and how to apply these findings to infertile patients. The identification of high-quality embryos in IVF using objective technologies remains a high priority in order to facilitate single-embryo transfer. Gene expression profiling of cumulus cells surrounding the oocyte, and proteomic and metabolomic approaches in embryo culture media may significantly improve non-invasive embryo quality assessment.
CONCLUSIONS
The way forward in advancing the knowledge of genes involved in reproduction was considered to be through genome-wide association studies involving large numbers of patients. Establishing international collaboration is required to enable the application of such technologies in sufficient numbers of patients.
Keywords: genes and female reproductive pathologies, embryo assessment, endometriosis, metabolomics, transcriptomics
Introduction
The study of genetics is a rapidly expanding field in all areas of medicine, including fertility. This review focuses on the genetic aspects of a number of topics of importance in fertility medicine: ovarian function, primary ovarian insufficiency (POI) and poor response to ovarian stimulation, polycystic ovary syndrome (PCOS) and endometriosis. In addition, new technologies for the assessment of oocyte and embryo quality are reviewed, including gene expression profiling of cumulus cells, proteomics and metabolomics.
The number of genes known to be expressed in the ovary and involved in reproductive function has increased dramatically in recent years. Increasing our understanding of the genes involved in ovarian ageing and poor response to ovarian stimulation may lead to improved clinical diagnosis and early detection, along with the development of new agents or technologies for use in fertility treatment. Although familial clustering of PCOS cases has been described, the mode of inheritance remains uncertain. Candidate genes for PCOS are discussed, although few studies have satisfied the rigorous criteria of population selection and size, repeatability and correction for multiple comparisons. Genetics also appear to have a role in complex reproductive diseases, such as endometriosis. A number of candidate gene families that may be differentially regulated in endometriosis have been identified. New developments in this field include the identification of a candidate population of endometrial stem cells, the study of which may provide new insights into this condition.
Improving our knowledge of the genetics of the ovary and the genetic causes of infertility should lead to future improvements in fertility treatment. For instance, it is now recognized that the oocyte is not passive in the ovarian follicle but is a fundamental regulator of somatic cell differentiation and function. The cumulus–oocyte complex plays a central role in the regulation of folliculogenesis and is important for the maturation, reprogramming and fertilization of oocytes. Consequently, cumulus cell gene expression profiling is being explored as a new method for the assessment of oocyte competence and embryo developmental competence, which shows promise for the near future.
Currently, IVF represents the only hope for many couples with fertility problems. Embryo assessment has an enormous impact on the success of IVF and, with moves towards single-embryo transfer (SET), assessment of embryo quality is of the utmost importance. Despite its evident importance, until recently, assessment of embryo quality has depended on subjective morphological assessment (Assou et al., 2011). Any objective method of embryo assessment that could determine its potential for implantation, as well as its health status, will increase success rates.
With the recent expansion of the current knowledge and developments in this area, a thorough review of contemporary genetics and ‘omics’ technologies in human reproduction is timely.
Methods
Prior to the Fifth Evian Annual Reproduction (EVAR) Workshop Meeting, held on 16–17 April 2010, expert speakers prepared presentations based on published literature and current research. Presenters were asked to include comprehensive information using systematic literature search criteria. Combinations of the following keywords were used for PubMed searches: genes and fertility-associated pathologies, primary ovarian insufficiency, premature ovarian failure, genomics, proteomics, metabolomics, endometriosis, polycystic ovary syndrome (and PCOS), floating granulosa cell gene expression, cumulus cell gene expression and protein expression profiling.
Following each presentation, a group discussion to reach joint conclusions on the topics covered was facilitated by the chairmen: Professors Fauser and Diedrich. The content of this report is based on the expert presentations and subsequent group discussions that took place during the workshop meeting.
The discussions relating to each topic were complemented as necessary with key publications that were known to the authors and electronic literature searches. Articles of any type and published in English were permitted and were unlimited by the date of publication.
Genetics of the ovary
Key genes involved in ovarian function
Our knowledge of the number of genes known to be involved in ovarian function has increased dramatically in recent years. Approaches to elucidate the role of specific genes involved in human reproduction include the use of PCR, real-time PCR, fluorescence in situ hybridization (FISH), single-nucleotide polymorphisms (SNPs), array SNPs, comparative genomic hybridization (CGH), array CGH (aCGH), genome-wide linkage analysis, candidate gene-association studies, genome-wide association studies (GWAS) and transgenic animal models. PCR and FISH have been available for some time, but the more recent development of microarray technologies has increased substantially the volume of information available on the genetics of reproduction. Indeed, the online Ovarian Kaleidoscope Database now contains information on more than 3100 ovarian-expressed genes and is a useful resource of knowledge on ovarian genetics (http://ovary.stanford.edu).
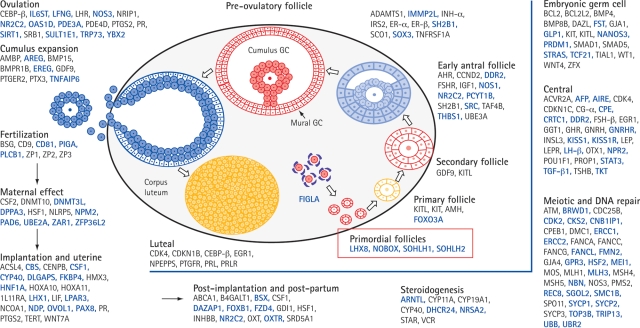
Figure 1 shows a model of recognized genes involved in the hypothalamo–pituitary–ovarian axis (Matzuk and Lamb, 2008). Gene mutations shown to affect gonadotrophin synthesis or actions and to cause infertility in humans have been summarized in a recent review and are shown in Table I (Edson et al., 2009). Pre-antral follicular growth appears to be independent of gonadotrophins, but further functional characterization of thecal cells (through antral, pre-ovulatory, ovulation and corpus luteum phases) requires gonadotrophin stimulation (Edson et al., 2009).
Figure 1.
Model of ovarian genes recognized in 2008. Reproduced with permission from Macmillan Publishers Ltd (Matzuk and Lamb, 2008).
Table I.
Gene mutations affecting gonadotrophin synthesis or actions and causing infertility.
| Gene | Phenotype | OMIM gene [OMIM infertility] |
|---|---|---|
| Bone morphogenetic protein 15 (BMP15) | Hypergonadotrophic ovarian failure (POF4) | 300247 [300510] |
| Bone morphogenetic protein receptor 1B (BMPR1B) | Ovarian dysfunction, hypergonadotrophic hypogonadism and acromesomelic chondrodysplasia | 603248 |
| Chromobox homolog 2, Drosophila polycomb class (CBX2; M33) | Autosomal 46,XY, male-to-female sex reversal (phenotypically perfect females) | 602770 (67) |
| Chromodomain helicase DNA-binding protein 7 (CHD7) | CHARGE syndrome and Kallmann syndrome (KAL5) | 608892 [612370] |
| Diaphanous homolog 2 (DIAPH2) | Hypergonadotrophic, premature ovarian failure (POF2A) | 300108 [300511] |
| Fibroblast growth factor 8 (FGF8) | Normosmic hypogonadotrophic hypogonadism and Kallmann syndrome (KAL6) | 600483 [612702] |
| Fibroblast growth factor receptor 1 (FGFR1) | Kallmann syndrome (KAL2) | 136350 [147950] |
| FSH receptor (FSHR) | Hypergonadotrophic hypogonadism and ovarian hyperstimulation syndrome | 136435 |
| FSHβ (FSHB) | Deficiency of follicle-stimulating hormone, primary amenorrhoea and infertility | 136530 [229070] |
| Forkhead box L2 (FOXL2) | Isolated premature ovarian failure (POF3) associated with BPES type I; FOXL2 402C → G mutations associated with human granulosa cell tumours | 605597 [608996] |
| Fragile X mental retardation 1 (FMR1) | Premature ovarian failure (POF1) associated with premutations | 309550 [311360] |
| GnRH receptor (GNRHR) | Hypogonadotrophic hypogonadism | 138850 |
| GnRH (GNRH1) | Normosmic hypogonadotrophic hypogonadism | 152760 (769, 770) |
| Kallmann syndrome 1 (KAL1) | Hypogonadotrophic hypogonadism and insomnia, X-linked Kallmann syndrome (KAL1) | 308700 |
| KISS1 receptor (KISS1R; GPR54) | Hypogonadotrophic hypogonadism | 604161 |
| LHβ (LHB) | LHB G102S mutations associated with infertility | 152780 |
| LH/choriogonadotrophin receptor (LHCGR) | Hypergonadotrophic hypogonadism (luteinizing hormone resistance) | 152790 |
| Nuclear receptor subfamily 0, group B, member 1 (NROB1; DAX1) | X-linked congenital adrenal hypoplasia with hypogonadotrophic hypogonadism; dosage-sensitive male-to-female sex reversal | 300473 [300200; 300018] |
| Nuclear receptor subfamily 5, group A, member 1 (NR5A1; SF1) | 46,XY male-to-female sex reversal and streak gonads and congenital lipoid adrenal hyperplasia; 46,XX gonadal dysgenesis and 46,XX primary ovarian insufficiency | 184757 (771) |
| Premature ovarian failure 1B (POF1B) | Hypergonadotrophic, primary amenorrhea (POF2B) | 300603 [300604] |
| Prokineticin (PROK2) | Normosmic hypogonadotrophic hypogonadism and Kallmann syndrome (KAL4) | 607002 [610628] |
| Prokineticin receptor 2 (PROKR2) | Kallmann syndrome (KAL3) | 607123 [244200] |
| R-spondin family, member 1 (RSPO1) | 46,XX, female-to-male sex reversal (individuals contain testes) | 609595 |
| Sex-determining region Y (SRY) | Mutations lead to 46,XY females; translocations lead to 46,XX males | 480000 |
| SRY-related HMB-box gene 9 (SOX9) | Autosomal 46,XY male-to-female sex reversal (campomelic dysplasia) | 608160 |
| Tachykinin 3 (TAC3) | Normosmic hypogonadotrophic hypogonadism | 162330 |
| Tachykinin receptor 3 (TACR3) | Normosmic hypogonadotrophic hypogonadism | 162332 |
Reproduced with permission from the Endocrine Society (Edson et al., 2009). Because of space limitations, most cases associated with female-to-male sex reversal due to steroidogenesis defects, syndromes and chromosomal abnormalities are excluded from the table. The primary reference (in parentheses) is included for work not yet described in OMIM.
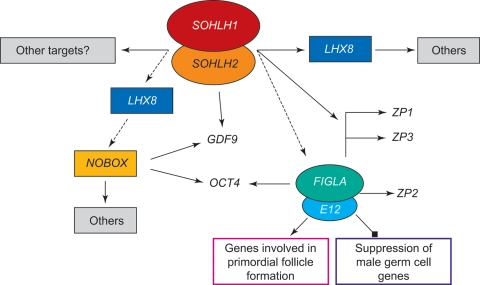
An important basic helix–loop–helix (HLH) transcription factor that is essential for fertility, factor in the germline alpha (FIGLA), was identified in 1997 (Liang et al., 1997). FIGLA is involved in formation of the primordial follicle and knockout mice are infertile. In FIGLA knockout mice ovaries, OCT4, a protein required for primordial germ cell survival (Kehler et al., 2004), is absent (Joshi et al., 2007). FIGLA is also involved in coordinating expression of the zona pellucida (ZP) genes. Analysis of newborn ovaries has identified many oocyte-specific genes that are down-regulated in FIGLA knockout mice (Joshi et al., 2007). Thus, FIGLA acts as a central regulator of oocyte-specific genes that play roles in folliculogenesis, fertilization and early development.
Regulators of the development of primordial to primary follicles include the following genes: newborn ovary homeobox (NOBOX), a homeobox-encoding gene (Suzumori et al., 2002; Rajkovic et al., 2004), spermatogenesis- and oogenesis-specific basic HLH1 (SOHLH1) and 2 (SOHLH2) transcription factors (Pangas et al., 2006; Hao et al., 2008; Toyoda et al., 2009) and Lim homeobox protein 8 (LHX8) (Choi et al., 2008). All of these are expressed in the late embryonic and newborn ovaries, and knockout models lack primordial follicles and are sterile.
SOHLH1 disruption has been shown to down-regulate the expression of FIGLA and NOBOX genes and their targets: ZP1, ZP2, ZP3 and OCT4, growth differentiation factor 9 (GDF9) and LHX8 (Pangas et al., 2006) and up-regulate the expression of stimulated by retinoic acid gene 8 (STRA8), which is directly or indirectly repressed by NOBOX and LHX8 (Choi et al., 2008). SOHLH1 and SOHLH2 appear to be master–master regulators of other master transcription factors. The presence of two master–master regulators (SOHLH1 and SOHLH2) supports the concept that it is important to have more than one transcription factor regulating key genes (Toyoda et al., 2009). A summary of these findings is shown in Fig. 2.
Figure 2.
Working model of the regulation of oocyte gene expression. As described in the text, a complex of SOHLH1/2 is believed to be a master–master regulator of other master transcription factors (boxed or circled). Key downstream transcriptional targets (GDF9, OCT4 and ZP1–3) are also shown.
The transforming growth factor β superfamily, which includes GDF9 and bone morphogenetic protein 15 (BMP15), is the largest family of growth factor ligands in mammals. They function in multiple developmental and physiological processes (Chang et al., 2002). Oocyte-expressed GDF9 is essential for granulosa cell proliferation and differentiation at the one- to two-layer follicle transition (Dong et al., 1996). A key role of GDF9 at this transition is to suppress inhibin-α and a secondary role is to stimulate the formation of a steroidogenically functional theca (Elvin et al., 1999a, b). The GDF9 promoter contains three NOBOX-binding elements (Choi and Rajkovic, 2006).
Novel oocyte-enriched gene, G-protein receptor 149
G-protein receptor 149 (GPR149) is a recently identified ‘antifertility’ gene, expressed in ovaries and oocytes, which is of particular interest. It was identified from searches of online databases for genes with expression patterns similar to GDF9. These genes were grouped by gene ontology into G protein-coupled receptors, three of which were identified as being of potential importance: GPR3, GPR149 and GPR175 (Mehlmann et al., 2004; Aki et al., 2008; Edson et al., 2010).
Unique features of the GPR149 receptor include a very long carboxy-terminal tail of 360 amino acids. Phylogenetic analysis of GPR149 suggests that it is widely conserved in vertebrates (Edson et al., 2010). GPR149-deficient mice have been generated and found to be viable and fertile (Edson et al., 2010). GPR149-null females have increased fertility (large litter sizes, high litter frequency and prolonged time in oestrus). Increased fertility was secondary to ovulation, which was likely due to reduced follicular atresia and a corresponding increase in the number of antral follicles. Importantly, the number of antral follicles was not depleted in mature GPR149-null female mice, which suggests that the ovaries of GPR149-null females do not undergo premature ageing (Edson et al., 2010).
Expression of the follicle-stimulating hormone receptor gene (FSHR) and GDF9 is increased in GPR149-deficient mice. Elevated levels of FSHR increase cyclin D2 levels in granulosa cells and stimulate cell proliferation (Edson et al., 2010). If GPR149 is found to have a similar function in humans, GPR149 antagonists may represent potential new fertility treatments.
Cumulus expansion
Cumulus cells are specialized cells that surround and nourish the oocyte. They have important roles in promoting cytoplasmic maturation of oocytes, which is necessary for pronuclear formation and subsequent developmental capability (Vanderhyden and Armstrong, 1989). Cumulus expansion involves production of a hyaluronic acid-rich extracellular matrix that surrounds the oocyte. In the peri-ovulatory period, this expansion has several important roles including protection of the oocyte, aiding growth of the cumulus–oocyte complex, permitting oocyte capture by oviductrial fimbria and enhancement of the fertilization ability of sperm.
The genes involved in the process of cumulus expansion include GDF9 and BMP15 with involvement of hyaluronan synthase 2 (HAS2), tumour necrosis factor-induced protein-6 (TNFIP6), pentraxin 3 (PTX3), prostaglandin-endoperoxide synthase 2 (PTGS2) and gremlin 1 (GREM1) (Elvin et al., 1999b, 2000; Varani et al., 2002; Pangas et al., 2004). The expression of these genes produces factors involved in the production of granulosa cells, promotion of glycolysis, stimulation of sterol biosynthesis and amino acid transport with bidirectional communication between the oocyte and cumulus cells (Dong et al., 1996; Elvin et al., 1999a, 2000; Yan et al., 2001; Varani et al., 2002; Pangas et al., 2004). When the cumulus cells are removed (e.g. during ICSI procedures), supplying the oocyte with these substances may have benefit. Therefore, cumulus cells could potentially be used in IVF programmes in co-culture with embryos to support development to the blastocyst stage.
Cumulus cell gene expression profiling has been used recently as a method of assessing oocyte quality and embryo development and is discussed later in this review.
POI and poor response to ovarian stimulation
POI, which was known previously as premature ovarian failure, and follicular atresia may share several common mechanisms, including abnormal apoptosis, an abnormal number of primordial follicles and/or abnormal follicular maturation. Ovarian failure, whether natural or induced as a consequence of pathological insults, is driven by depletion of ovarian follicles through apoptosis (Tilly, 2001; De Vos et al., 2010).
The mean age at which the menopause occurs is 51 years and, with a range between 40 and 60 years, is relatively wide-ranging (Broekmans et al., 2009). Hence, exhaustion of the primordial follicle pool in some women takes 50% more time compared with others. Evidence suggests that all women are born with a similar number of follicles (Gosden and Faddy, 1994) and this number steadily dwindles throughout life as a result of atresia and recruitment towards ovulation (Gosden and Faddy, 1998). The rate of depletion of the follicular pool determines the age at which the menopause occurs and is thought to be controlled by multiple genes. Although only 1–2% of women undergo premature menopause, defined as before 40 years of age, studies in this population may provide important information regarding the genes involved in ovarian ageing and may help to predict better the age of menopause for individual patients (Torgerson et al., 1997; Goswami and Conway, 2005). Both infertility and poor response to ovarian hyperstimulation have been associated with subsequent early menopause in several studies (Farhi et al., 1997; Nikolaou et al., 2002; de Boer et al., 2003; Lawson et al., 2003; Broekmans et al., 2009). Poor response to ovarian hyperstimulation is generally taken to be the retrieval of fewer than five oocytes and is associated with lower than average pregnancy rates (van der Gaast et al., 2006; Verberg et al., 2009).
The mean age of menopause is remarkably similar throughout the world and does not appear to have changed over time (Torgerson et al., 1997). There appears to be a strong link between mothers' and daughters' menopausal ages (Torgerson et al., 1997; van Asselt et al., 2004; Murabito et al., 2005) and twin studies suggest that the age of natural menopause is genetically linked (Snieder et al., 1998; Treloar et al., 1998). Other factors, including smoking and alcohol consumption, body mass index and parity, appear to play a minor role.
Aside from loss of fertility, POI underlies the emergence of an array of health problems in ageing women, including increased risks of cardiovascular disease and osteoporosis (Hartge, 2009; De Vos et al., 2010). In women who undergo bilateral oophorectomy before the natural menopause, with subsequent early reduction in oestrogen levels, evidence suggests an increased risk of cognitive impairment and neurological disorders such as Parkinson's disease (Rocca et al., 2007, 2008).
Known causes of POI
In 70–90% of cases, the cause of POI in humans is unknown. Known causes include bilateral oophorectomy, chemotherapy, radiotherapy and Turner syndrome (TS; 45 X). Other known causes of POI include autoimmune disease [including autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy due to rare mutations in the AIRE (autoimmune regulator) gene (Ahonen et al., 1990)], pseudohypoparathyroidism, congenital disorders of glycosylation, galactosaemia, infections (e.g. herpes zoster and cytomegalovirus), syndromes [such as Fragile X (FMR1 gene) (Murray et al., 1999) and blepharophimosis-ptosis-epicanthus inversus (BPES)] and isolated gene defects (Uda et al., 2004). Known isolated gene defects involved in POI are listed in Table II (Miller and Chatten, 1967; Ahonen et al., 1990; Leslie et al., 1992; Aittomaki et al., 1995; Bione et al., 1998; Chun et al., 1999; Murray et al., 1999; Takahashi et al., 1999; Prueitt et al., 2000, 2002; Crisponi et al., 2001; Di Pasquale et al., 2004; Dixit et al., 2004, 2005, 2006; Fogli et al., 2004; Luoma et al., 2004; Sato et al., 2004; Tung et al., 2006; Watkins et al., 2006; Laissue et al., 2007; Qin et al., 2007; Kovanci et al., 2008; Mandon-Pepin et al., 2008; Vinci et al., 2008; Zhao et al., 2008; Lourenco et al., 2009; van Dooren et al., 2009; Chrzanowska et al., 2010; De Vos et al., 2010).
Table II.
Human genes associated with ovarian insufficiency/failure.
| Acronym | Name |
|---|---|
| AIRE (Ahonen et al., 1990) | Autoimmune regulator |
| ATM (Miller and Chatten, 1967) | Ataxia telangiectasia mutated |
| BHLHB9 (De Vos et al., 2010) | Basic helix–loop–helix domain-containing, class B, 9 |
| BMP15 (Di Pasquale et al., 2004) | Bone morphogenetic protein |
| DACH2 (Prueitt et al., 2002) | Drosophila dachsund |
| DIAPH2 (Bione et al., 1998) | Homologue Drosophila diaph |
| FMR1 (Murray et al., 1999) | Fragile X mental retardation 1 |
| FMR2 (Murray et al., 1999) | Fragile X mental retardation 2 |
| XIST (Sato et al., 2004) | X inactivation transcript |
| XPNPEP2 (Prueitt et al., 2000) | Propyl aminopeptidase |
| DAZL (Tung et al., 2006) | Deleted azoospermia |
| DCM1 (Mandon-Pepin et al., 2008) | Disrupted meiotic cDNA 1 |
| EIF2B (Fogli et al., 2004) | Eukaryotic translation initiation |
| ESR1 (Tung et al., 2006) | Oestrogen receptor |
| FIGLA (Zhao et al., 2008) | Murine factor germline alpha |
| FOXL2 (Crisponi et al., 2001) | Forkhead transcription factor |
| FOXO1A (Watkins et al., 2006) | Forkhead box 01A |
| FOXO3a (Vinci et al., 2008) | Forkhead box O3a |
| FSHR (Aittomaki et al., 1995) | FSH receptor |
| GALT (Leslie et al., 1992) | Galactose phosphate transferase |
| GDF9 (Dixit et al., 2005) | Growth differentiation factor 9 |
| GPR3 (Kovanci et al., 2008) | G protein-coupled receptor |
| INHA (Dixit et al., 2004) | Inhibin A |
| LHB (Takahashi et al., 1999) | LH beta |
| MSH5 (Mandon-Pepin et al., 2008) | MutS homolog 5 |
| NOBOX (Qin et al., 2007) | Murine newborn ovary box |
| NOGGIN (Laissue et al., 2007) | Binds and inactivates members of the transforming growth factor-β superfamily signalling proteins |
| NR5A1 (SF-1) (Lourenco et al., 2009) | Nuclear receptor subfamily 5 A1 |
| NSB1 (Chrzanowska et al., 2010) | Nijmegen breakage syndrome 1 |
| PGRMC1 (van Dooren et al., 2009) | Progesterone receptor membrane component-1 |
| POLG (Luoma et al., 2004) | Mitochondrial DNA polymerase gamma mutations |
| TGFBR3 (Dixit et al., 2006) | Tumour growth factor receptor |
| WT1 (Chun et al., 1999) | Wilms Tumor 1 |
POI has been linked with several X chromosome-linked defects. Most women with TS have primary amenorrhoea, but some are fertile. Factors with a positive predictive value for fertility include mosaicism, normal FSH and anti-Müllerian hormone levels, and spontaneous menarche and puberty (Borgstrom et al., 2009). BMP15 has been described as the first gene on the X chromosome with a role in ovarian function (Layman, 2006). As discussed earlier, together with GDF9, BMP15 plays an important role in oocyte–cumulus cell signalling (Layman, 2006; Gilchrist et al., 2008) and it has been suggested that GDF9 and BMP15 mutations are involved in POI (Laissue et al., 2006). A naturally occurring BMP15 mutation in ewes causes increased ovulation rates, and twin and triplet births in heterozygotes, but primary ovarian failure in homozygotes (Galloway et al., 2000). In humans, a natural heterozygous mutation in the propeptide region of the BMP15 gene has been associated with hypergonadotrophic ovarian failure due to ovarian dysgenesis (Di Pasquale et al., 2004). The mutation appeared to be associated with reduced granulosa cell growth. This suggests that BMP15 is required for human folliculogenesis. However, further studies are needed to elucidate the role of BMP15 in humans.
Fragile X syndrome is caused by mutations of the FMR1 gene in the distal part of the long-arm of the X chromosome. This occurs relatively frequently and is the most common cause of mental retardation, behavioural difficulties, autism and learning difficulties in boys. The syndrome is caused by an abnormal number of CGG repeats in the 5′-untranslated region of the gene. Typically, the number of repeats is fewer than 45, while abnormal genes may have more than 200, which causes FMR1 gene inactivation. Women who have between 55 and 200 repeats (1 in 590 women) carry a premutation that causes altered ovarian function and POI (Wittenberger et al., 2007). Some studies have suggested, however, that there is no linear correlation between the number of repeats and POI, with the highest risk between 80 and 100 repeats (Sullivan et al., 2005; Ennis et al., 2006). Up to 28% of Fragile X premutation carriers (45–200 repeats) develop POI (Welt et al., 2004). Such women appear to have normal menstrual cycles, with slightly raised FSH levels (Welt et al., 2004).
FOXL2 is involved in the development of granulosa cells and, in addition to female infertility, FOXL2 mutations have been shown to cause eyelid/forehead dysmorphology (BPES) (Uda et al., 2004). Furthermore, studies have suggested that in a small proportion of patients with BPES, the genetic defect does not reside within the coding region of the FOXL2 gene (De Baere et al., 2001) and incidences of POI with FOXL2 mutations in patients without BPES have also been reported (De Baere et al., 2001; Harris et al., 2002).
Mutations in the NR5A1 [FSH or steroidal factor 1 (SF-1)] gene, a key transcriptional regulator of genes involved in the hypothalamic–pituitary–steroidogenic axis, have also been associated with impairment of ovarian development and function (Lourenco et al., 2009). Such mutations are associated with POI and also with disorders of sex development in men. NR5A1 is expressed in multiple cell types in the foetal, post-natal, prepubertal and mature ovary, in the adult adrenal cortex and in the Sertoli and Leydig cells of the testis (Lavorgna et al., 1991; Luo et al., 1994; Jeyasuria et al., 2004). The protein regulates the transcription of key genes involved in sexual development and reproduction, including STAR (encoding steroidogenic acute regulatory protein), CYP17A1 (encoding 17-alphahydroxylase), CYP11A1 (encoding cytochrome P-450 cholesterol side-chain cleavage), LHB (encoding the beta subunit of LH), AMH (encoding anti-Müllerian hormone), CYP19A1 (encoding aromatase) and INHA (encoding inhibin-α subunit) (Shen et al., 1994; Keri and Nilson, 1996; Sugawara et al., 1997; Hanley et al., 2001; Gurates et al., 2003; Weck and Mayo, 2006; Mendelson and Kamat, 2007).
Women with FSHR-inactivating mutations develop multiple follicles in the early primordial, primary, secondary and pre-antral stages, but no antral or pre-ovulatory follicles (as their development is FSH-dependent) and such mutations are rare causes of POI (Aittomaki et al., 1995; Doherty et al., 2002). LH receptor mutations have also been associated with infertility. In a study of three sisters with an LH receptor mutation (two homozygous, one heterozygous), both homozygous women had normal menarche and regular menstrual cycles initially, but then experienced early oligomenorrhoea. The homozygous sisters underwent fertility treatment, which was unsuccessful in both cases. The heterozygous sister, however, had five natural pregnancies with two live births (Bruysters et al., 2008).
Other candidate genes
Three types of study have been used to investigate the genetics of the natural menopause in humans; genome-wide linkage analysis, candidate gene-association studies and GWAS. Although GWAS have been used in many disease areas, until recently they had not been performed in reproductive medicine. Such studies need large numbers of patients and are expensive to perform, but can provide major breakthroughs in our understanding of disease aetiology.
Candidate genes for premature menopause include those involved in steroidogenesis, estradiol signalling and the cardiovascular system. A recent systematic review summarizes candidate genes and SNPs in association with the natural age at menopause and identified two genomic regions (19q13.42 and 20p12.3), containing two promising candidate genes [BR serine threonine kinase 1 gene (BRKS1) and maintenance complex component (MCM)] (Voorhuis et al., 2010).
Genome-wide linkage analysis studies have suggested the involvement of B-cell lymphoma 2 (BCL2) and gonadotrophin-releasing hormone 1 (GnRH1) as genes with suggestive linkage with the natural age of menopause (van Asselt et al., 2004; Murabito et al., 2005). The first genome-wide linkage search in familial POI identified a region on chromosome 5q14.1–q15 that may harbour a novel POI susceptibility gene (Oldenburg et al., 2008), and a recent GWAS identified ADAMTS19 (5q31) as a possible candidate gene for POI (Knauff et al., 2009).
Other strategies to discover and study new candidate genes include the use of animal models. Inactivation of the proapoptotic BAX gene in mice has been shown to prolong ovarian function into advanced age (Perez et al., 2007) with minimization of many age-related health problems and, importantly, no increase in tumour incidence. Further studies are needed, however, to assess the importance of the BAX gene on ovarian function in humans.
aCGH analysis is able to detect submicroscopic chromosomal rearrangements with a high genomic resolution. This technique was used in a large cohort of Caucasian women with POI and identified eight copy number variations in eight different genes associated with POI. Five of these genes [DNAH5 (dynein axonemal heavy chain), NAIP (neuronal apoptosis inhibitory protein), DUSP22 (dual specificity phosphatases), AKT1 (akt murine thymoma viral oncogene homolog 1) and NUPR1 (nuclear protein 1)] are involved in reproduction; two are involved in reproductive diseases (DNAH5 and NAIP), two in reproductive endocrinology (DUSP22 and NUPR1) and one in folliculogenesis (AKT1). Thus, these genes represent potential candidate genes in POI (Aboura et al., 2009), but the results need to be confirmed in other studies.
Recently, four SNPs on chromosomes 19, 20, 6 and 5 have been associated with normal variation in menopausal age. They were identified through two GWAS and had a significant impact on the odds of having an early menopause in an independent cohort of women from the Breakthrough Generations Study. Thus, these genetic variants, which are associated with normal variation in menopausal age, are also significant risk factors for early menopause (Murray et al., 2011).
Investigating POI
In the clinical setting, causative genes of POI are identified in very few cases. However, some syndromic causes of POI have physical manifestations that can provide clues to its aetiology; e.g. eyelid abnormalities of patients with BPES and short stature in TS (Uda et al., 2004; Borgstrom et al., 2009). A family history of learning difficulties in boys suggests the presence of Fragile X syndrome and the presence of autoimmune disorders may suggest an immunological aetiology (Ahonen et al., 1990; Murray et al., 1999). When investigating POI, after obtaining informed consent, it is important to perform karyotyping and to screen for FMR1 and NR5A1 mutations as both have serious familial implications.
Polycystic ovary syndrome
Genetic factors in the aetiology of PCOS
PCOS is a complex, heterogeneous disorder of uncertain aetiology. Studies in animal models of PCOS, notably in the Rhesus monkey, have given rise to the hypothesis that PCOS has its origin in foetal life (Abbott et al., 2002). In human females, exposure to excess androgen, at any stage from foetal development of the ovary to the onset of puberty, leads to many of the characteristic features of PCOS, including abnormalities of LH secretion and insulin resistance (Abbott et al., 2002; Xita and Tsatsoulis, 2006). In human post-natal life, the natural history of PCOS can be further modified by factors affecting insulin secretion and/or action, most importantly, nutrition (Ciampelli et al., 1999). There is strong evidence that genetic factors play an important part in the aetiology of PCOS (Franks and McCarthy, 2004; Franks et al., 2006; Urbanek, 2007). The familial clustering of cases (Cooper et al., 1968; Ferriman and Purdie, 1979; Givens, 1988; Hague et al., 1988; Lunde et al., 1989; Carey et al., 1993), greater concordance in monozygotic compared with heterozygotic twins (Vink et al., 2006) and heritability of endocrine and metabolic features of PCOS (Legro et al., 1998, 2002; Franks et al., 2008) all provide support for the involvement of genetic factors.
Genetic studies in PCOS
In the search for genes involved in the aetiology of PCOS, the focus has been on genes implicated in folliculogenesis, such as follistatin, genes involved in the androgen biosynthetic pathway (such as CYP11A, CYP17 and CYP19) and those affecting insulin secretion or action [such as transcription factor 7-like 2 (TCF7L2), potassium inwardly rectifying channel, subfamily J, member 11 (KCNJ11), insulin receptor and peroxisome proliferator-activated receptor-γ (PPAR-γ)] (Franks and McCarthy, 2004; Gaasenbeek et al., 2004; Escobar-Morreale et al., 2005; Christopoulos et al., 2008). However, despite the evaluation of more than 70 genes, the candidate gene approach has, to date, proved disappointing. Due to limited sample sizes, and genetic and phenotypic heterogeneity, findings have often been ambiguous or not reproducible. Even the few studies that have been sufficiently well powered to either identify or exclude candidate susceptibility loci have produced limited positive findings.
Two notable exceptions, for which there is convincing evidence for a role in PCOS, are the fibrillin-3 gene (FBN3) and the fat-mass and obesity-associated (FTO) gene loci. Evidence for linkage disequilibrium was observed at D19S884, a dinucleotide repeat marker on chromosome 19p13.2 (Urbanek et al., 1999, 2005; Tucci et al., 2001). Genotyping of the region around D19S884 subsequently identified FBN3 as a likely candidate for a PCOS susceptibility gene.
The association of the FTO locus (on chromosome 16) with type 2 diabetes and obesity (Frayling et al., 2007) led researchers to explore a role for this gene in the aetiology of PCOS. In a case–control study, an association was found between PCOS status and FTO genotype (Barber et al., 2008). FTO variants that were known to predispose to obesity were also implicated in altered susceptibility to PCOS, confirming the mechanistic link between these conditions. FTO variants have also been shown to strongly influence the glucose intolerance component of the metabolic syndrome in patients with PCOS (Attaoua et al., 2008). Based on the results from various studies of FTO in PCOS, the consensus is that the association is predominantly with obesity in women with PCOS, rather than PCOS per se (Barber et al., 2008; Tan et al., 2010; Wehr et al., 2010).
Blastocysts derived from women with PCOS have markedly different transcriptome profiles than blastocysts from normal controls (Katz-Jaffe et al., 2010); proteomic profiling identified 12 biomarkers that displayed significantly decreased expression in PCOS. It was postulated that these differences may, in part, explain the reduction in reproductive capacity of women with PCOS.
Recently, the role of microRNAs (miRNAs) has been studied in the pathogenesis of endometriosis and associated reproductive conditions [for review, see Ohlsson Teague et al. (2010)]. miRNAs are short (about 21 nucleotides), single-stranded RNAs that regulate gene expression at the post-transcriptional level (non-coding). Differences in miRNA profiles in transferable blastocysts from women with PCOS compared with those from donor fertile controls have been shown (McCallie et al., 2010).
Although the search for candidate genes is bound to continue, it seems increasingly likely that GWAS (of the kind that has paid dividends in the search for genes in type 2 diabetes) is the approach that is most likely to uncover the genes contributing to PCOS. Although this would require substantial resources and very large case–control populations, the limitations of earlier studies may be overcome and new studies promise to be more informative. Indeed, results have been published recently of a GWAS in Han Chinese women with PCOS. A number of loci of interest were identified, including one close to the gene encoding the LH receptor (Chen et al., 2011).
Genetics of disorders of the female reproductive system
Endometriosis
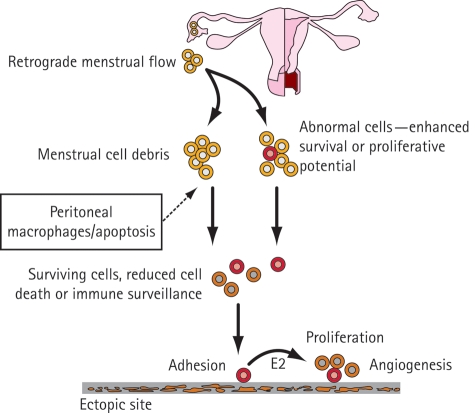
Endometriosis is an oestrogen-dependent condition that is characterized by the presence of endometrium-like tissue at ectopic sites such as the pelvic peritoneum and ovaries (Fig. 3). Pelvic pain and infertility are the most common features of endometriosis. There are several proposed hypotheses to explain the pathogenesis of endometriosis (Sasson and Taylor, 2008); the most widely accepted theory is that of retrograde menstruation. The mechanisms involved in the development of an endometriotic lesion in the pelvic peritoneum may include attachment, invasion into the mesothelium, and survival and proliferation of ectopic endometrial cells.
Figure 3.
A theoretical model of the development of endometriosis.
The basic underlying cause of endometriosis is likely to be multifactorial and involves interplay between several factors. The pathogenesis of endometriosis may, for example, involve retrograde menstruation in the context of an abnormal immune response and a genetic predisposition to developing endometriotic lesions; this may possibly occur after exposure to an unidentified environmental factor (Sasson and Taylor, 2008).
The study of endometriosis is complicated by a number of factors: these include the presence of different cell types within endometriotic lesions and the involvement of different pathogenic mechanisms in the formation of distinct types of endometriotic lesion (such as peritoneal, ovarian and rectovaginal) (Nisolle and Donnez, 1997). In addition, it may also be difficult to distinguish between cause and effect: differences in eutopic endometrium between patients with and without endometriosis, or in eutopic versus ectopic endometrium, may be the cause of this pathological condition or the result of another causative factor.
Although endometriosis can be treated surgically, recurrence of endometriotic lesions can occur. GnRH agonists and long-cycle oral contraceptives can also be used to manage the condition by suppressing ovulation and inducing a pseudo-menopausal state. Although many potential new agents, such as aromatase inhibitors, progesterone antagonists and immunomodulatory drugs, have been investigated, there is still a lack of suitable alternative pharmacological treatments (Guo, 2008; Huang, 2008; Vercellini et al., 2009). A better understanding of the pathogenesis of endometriosis may help to identify new pharmacological targets and facilitate the development of new treatments.
Genetics of endometriosis
It has been proposed that endometriosis results from a series of multiple hits within target genes, in a mechanism similar to the development of cancer (Bischoff and Simpson, 2004). The initial mutation may be either somatic or heritable.
Endometriosis has long been recognized as having a familial association; data are consistent with a complex genetic basis (Malinak et al., 1980; Simpson et al., 1980). Gene expression microarray studies have identified a number of candidate gene families that may be differentially regulated in endometriosis (Kao et al., 2003; Wu et al., 2006; Burney et al., 2007; Hull et al., 2008; Ohlsson Teague et al., 2009). A search for genetic polymorphisms associated with susceptibility to endometriosis has focused mainly on genes involved in inflammation, steroid hormone regulation, metabolism, biosynthesis, detoxification, vascular function and tissue remodelling (Tempfer et al., 2009). However, some polymorphisms have been investigated in only a single or a limited number of studies and, in some cases, conflicting results have been obtained. The majority of polymorphisms studied have, therefore, been found not to be associated with endometriosis. An evaluation of SNPs in 22 miRNAs that were differentially expressed in paired ectopic and eutopic endometria in women with endometriosis, however, identified two SNPs that were significantly associated with endometriosis-related infertility and disease severity: Wolf–Hirschhorn syndrome candidate gene 1 (WHSC1) alleles and solute carrier family 22, member 23 (SLC22A23) haplotypes (Zhao et al., 2011).
Two studies have shown differential expression of miRNAs in microarray analysis of eutopic and ectopic endometrial tissues (Pan et al., 2007; Ohlsson Teague et al., 2009). Although eight miRNAs were differentially expressed in both studies, the direction of dysregulation was not in agreement for any of these (Ohlsson Teague et al., 2010). A recent study using next-generation sequencing reported 22 dysregulated miRNAs (10 up-regulated miRNAs and 12 down-regulated miRNAs) in endometriomas compared with eutopic endometrium; miR-29c was implicated as a key up-regulated miRNA in endometriosis (Hawkins et al., 2011b). It is likely that specific miRNAs have a function in the pathophysiology of endometriosis, and the differences in results could be due to many factors, including the menstrual cycle phase at the time of biopsy. The putative roles of miRNAs in endometriosis and other reproductive diseases are described extensively in a recent review (Hawkins et al., 2011a).
A recent GWAS has identified a strong association signal for endometriosis at locus 7p152 (Painter et al., 2011). This locus is located in an intergenic region upstream of two plausible candidate genes, NFE2L3 and HOXA10.
Epigenetic factors
Single molecular alterations in endometriosis may result in differential regulation of hormone metabolism (Attar and Bulun, 2006). The alterations may arise through epigenetic mechanisms such as DNA methylation. An example is transcriptional activation of SF-1 (NR5A1) in endometriotic stromal cells by hypomethylation of DNA (Xue et al., 2007). The presence of SF-1 in endometriosis and its absence in the endometrium are determined primarily by the methylation of its promoter. In endometriotic lesions, the presence of SF-1 may contribute to increased oestrogen production (Bulun et al., 2009).
Stem cells: a new perspective
Stem cells located in the basal layer of the endometrium are thought to be responsible for cyclical tissue regeneration. Studies have been performed to isolate and characterize this cell population (Kato et al., 2007, 2010; Cervelló et al., 2010; Gargett and Masuda, 2010; Masuda et al., 2010). Such studies have identified a population of candidate endometrial stem cells in human endometrium; the cells demonstrate multipotency and are different from other endometrial cell types. These cells have been termed side population cells (SP cells) as they exhibit a side population phenotype upon staining with Hoechst dye and flow cytometric analysis. SP cells have been demonstrated to proliferate and differentiate into various endometrial cell types in vitro and have unique angiogenic and migratory properties (Masuda et al., 2010). Functional proof-of-concept studies have shown that human endometrium can be reconstructed in immunodeficient mice upon injection with human endometrial SP cells (Cervelló et al., 2010).
It has been suggested that the retrograde menstruation of SP cells and subsequent implantation onto the surface of ectopic sites are responsible for the establishment of endometriotic lesions (Sasson and Taylor, 2008; Masuda et al., 2010). It has also been hypothesized that an initial genetic mutation may cause aberrant behaviour of a subpopulation of endometrial stem cells; clonal expansion of such cells may then occur and result in development of endometriosis (Gargett et al., 2009). It is also conceivable that peritoneal cells could undergo de-differentiation back to endometrial cells, which take on an altered activity (Gargett et al., 2009).
Studies of endometrial stem cells have provided a new focus for endometriosis research. Research on this subpopulation of endometrial cells could be of particular value; the findings may further our understanding of the pathogenesis of endometriosis and provide new therapeutic approaches.
Predictors of oocyte quality and successful embryo development
Currently, many couples with fertility problems resort to IVF or ICSI when other treatments fail. Problems related to multiple pregnancies and poor embryo implantation are among the factors that may limit the success of IVF. The improvement of in vitro techniques, with sequential or co-culture systems, and subsequent reduction in the number of embryos transferred per cycle have ameliorated problems related to multiple pregnancy. However, implantation rates in IVF have not improved in the last decade (Nygren et al., 2006; Andersen et al., 2008). Any objective procedure that could assess the oocyte competence and potential for embryo implantation and development should increase success rates in assisted reproductive technology (ART).
In the study of oocyte competence, in recent years, the cumulus cell complex that surrounds and connects with the oocyte has received renewed attention. In a mouse model, this complex has been shown to play a central role in ovulation, and the capacity of the oocytes to support cumulus gene expression has been shown to be linked to the oocyte's developmental competence (Russell and Robker, 2007). Cumulus gene expression profiling therefore represents a possible means of identifying reliable biomarkers for oocyte quality and competence. It may also be useful to identify biomarkers for embryo development prediction.
Cumulus gene expression profiling
Many studies suggest that cumulus cells: (i) coordinate follicular development with oocyte maturation, (ii) provide energy substrates for oocyte meiotic resumption, (iii) regulate oocyte transcription, (iv) promote nuclear and oocyte molecular maturation, (v) stimulate amino acid transport and sterol biosynthesis, (vi) promote glycolysis and (vii) provide protection for the oocyte (Sugawara et al., 1997; Elvin et al., 1999b, 2000; Varani et al., 2002; Sutton et al., 2003; Pangas et al., 2004; Su et al., 2004; Eppig et al., 2005; Diaz et al., 2007; Gilchrist et al., 2008). The vital supporting role of cumulus cells during in vivo and in vitro maturation has led many groups recently to focus research interest on cumulus cells. Isolated cumulus cells are relatively homogeneous with almost no contamination with other cells, whereas isolated granulosa cells, despite meticulous efforts, generally contain theca and blood cells: this is due to the methods employed for obtaining these cells. In addition, with the advent of the functional genomics and proteomics era, it has become possible to identify the transcriptome and proteome of cumulus cells using high-throughput technologies, such as microarray (Assou et al., 2006) and high-resolution two-dimensional protein electrophoresis (Hamamah et al., 2006). The current concept is that cumulus cells may constitute a reliable model for understanding oocyte quality and ovarian hyperstimulation protocol efficiency and may indirectly predict oocyte aneuploidy, embryo development and pregnancy outcomes (McKenzie et al., 2004; Zhang et al., 2005; Feuerstein et al., 2007; van Montfoort et al., 2008).
Cumulus cells as biomarkers to assess oocyte quality, embryo competence and pregnancy outcome
Cumulus cells are typically discarded during classical IVF and ICSI. These cells are easily accessible and plentiful, which makes them an ideal material to use for the assessment of oocyte quality and embryo development potential. Thus, analysis of gene expression in cumulus cells may provide an indirect indication of the microenvironment in which the oocyte matures, and will help embryologists to better assess embryo quality. Currently, the most common ways of evaluating embryo quality non-invasively are by examining parameters such as morphology and cleavage rate. However, these approaches are subjective and lack precision. Several groups have used microarray technologies, reverse transcriptase PCR (RT–PCR) and quantitative RT–PCR analyses to study the association between cumulus cell gene expression profiles and oocyte competence, embryo quality and pregnancy outcome (Table III) (McKenzie et al., 2004; Cillo et al., 2007; Feuerstein et al., 2007; Assou et al., 2008, 2010; van Montfoort et al., 2008; Anderson et al., 2009; Hamel et al., 2010; Wathlet et al., 2011). In a new and indirect approach for predicting embryo quality and pregnancy outcome, gene expression signatures have been identified recently by Assou et al. (2008, 2010) using transcriptomic data of cumulus cells. This non-invasive approach is based on the level of expression of potential biomarkers in cumulus cells (indicators of successful pregnancy) to assess the potential and quality of the embryo.
Table III.
Association of cumulus cell gene expression with embryo quality and pregnancy outcomes.
| Cumulus cell origin | Approaches | Biomarkers | Outcome | Reference |
|---|---|---|---|---|
| Individual oocytes | Microarray (50 chips) | Including PCK1, BCL2L11, NFIB and others | Predict embryo and pregnancy outcomes | Assou et al. (2008) |
| Individual oocytes | Microarray (16 chips) | CCND2, CXCR4, GPX3, HSPB1, DVL3, DHCR7, CTNND1, TRIM28 | Negatively associated with oocyte competence | van Montfoort et al. (2008) |
| Individual oocytes | RT–PCR | HAS2, GREM1 | Positively associated with oocyte developmental competence | Cillo et al. (2007) |
| Individual oocytes | RT–PCR | STAR, AREG, CX43, PTGS2, SCD1, SCD5 | Negatively associated with oocyte competence | Feuerstein et al. (2007) |
| Individual oocytes | qRT–PCR | HAS2, PTGS2, GREM1 | Positively associated with oocyte competence and embryo development | McKenzie et al. (2004) |
| Individual oocytes | qRT–PCR | GREM1, BDNF | Positive and negative predictors of embryo quality, respectively | Anderson et al., (2009) |
| Mural granulosa cells and individual oocytes | qRT–PCR | PGK1, RGS2, RGS3, CDC42 | Associated with pregnancy | Hamel et al. (2010) |
| Individual oocytes | qRT–PCR | SDC4, PTGS2, VCAN, activated leucocyte cell adhesion molecule, GREM1, TRPM7, ITPKA | Predict embryo development and pregnancy | Wathlet et al. (2011) |
Modified with permission from Assou et al. (2010). AREG, amphiregulin; BCL2L11, BCL-like protein 11; BDNF, brain-derived neurotrophic factor; CCND2, cyclin D2; CDC42, cell division cycle 42; CTNND1, catenin delta 1; CX43, connexin 43; CXCR4, chemokines receptor 4; DHCR7, 7-dehydrocholesterol reductase; DVL3, dishevelled dsh homolog 3; GPX3, glutathione peroxidase; GREM1, gremlin 1; HAS2, hyaluronic acid synthase 2; HSPB1, heatshock 27 kDa protein 1; ITPKA, inositol 1,4,5-trisphosphate 3-kinase A; NFIB, nuclear factor 1B; PCK1, phosphoenolpyruvate carboxykinase 1; PGK1, phosphoglycerate kinase 1; PTGS2, prostaglandin-endoperoxide synthase 2; qRT–PCR, quantitative real-time PCR; RGS2, regulator of G-protein signalling 2; RGS3, regulator of G-protein signalling 3; RT–PCR, real-time PCR; SCD1, stearoyl-co-enzyme A desaturase 1; SCD5, stearoyl-co-enzyme A desaturase; SDC4. syndecan 4; STAR, steroidogenic acute regulatory protein; TRIM28, tripartite motif-containing 28; TRPM7, transient receptor potential cation channel, subfamily M, member 7; VCAN, versican.
Cumulus cell gene expression analysis for optimization of ovarian hyperstimulation protocols
As cumulus cells respond to stimulation protocols with a distinct gene expression profile and are sensitive to changes in environmental conditions, it may be possible to identify the genes that are expressed in the follicular environment. The individual characterization of gene expression may provide important insights for the evaluation of the impact of ovarian hyperstimulation protocols used during IVF.
Using fertilized and non-fertilized oocytes resulting from specific ovarian hyperstimulation protocols, potential differences in the protein expression profiles of cumulus cells have been investigated (Hamamah et al., 2006). In cumulus cells from either fertilized or non-fertilized oocytes, the greatest degree of similarity (more than 85%) was in the protein expression profiles. The analysis of protein expression profiles of cumulus cells from follicles obtained with the same ovarian hyperstimulation protocol showed a strong correlation between protein expression profiles whether or not embryos had been produced successfully. More than 80% of proteins were expressed similarly between cumulus cells from fertilized versus unfertilized oocytes from a cycle in a single patient that used a long GnRH protocol with human menopausal gonadotrophin (HP-hMG). In contrast, when cumulus cell protein expression profiles were analysed in oocytes with the same outcome from the same patient, but originating from different ovarian hyperstimulation protocols, a significant difference was seen in the protein expression profiles. Comparing protein expression profiles in cumulus cells from fertilized oocytes from the same patient, but after GnRH agonist long protocols with HP-hMG compared with those with recombinant FSH, only 55% of the proteins showed similar mobility and expression levels. Comparison of two groups of patients indicated that dissimilarities in protein pattern between patients become very high, even when comparing the same stimulation protocol and oocyte fertilization outcome. These data from protein expression profiling of human cumulus cells suggest that there may be a correlation between the synthesis of specific cumulus cell proteins and the maturity and fecundity of the oocyte. The study reveals three important new findings with respect to human cumulus cell biology: (i) human cumulus cells have robust metabolic activity, (ii) the overall protein expression profiles are highly similar between cumulus cells from oocytes obtained using the same ovarian hyperstimulation protocol and (iii) there are significant differences in protein expression between cumulus cells from oocytes obtained under two different ovarian hyperstimulation protocols, even when the outcomes are the same (Hamamah et al., 2006).
Cumulus cell gene expression analysis in PCOS and aneuploidy identification
Cumulus cells play a major role in the control of oocyte metabolism and, therefore, it is likely that malfunction of these cells might play a role in PCOS. Recently, the gene expression profile of cumulus cells isolated from patients with PCOS has been studied (Kenigsberg et al., 2009). The different gene expression patterns of cumulus cells from lean and obese women with PCOS support the idea that in the two types of patient, the condition may have different pathophysiologies. Furthermore, in recent years, several groups have initiated research aimed at identifying non-invasive biomarkers of chromosome imbalance. Some investigators have detected characteristic transcriptional changes in the cumulus cells attached to aneuploid oocytes (Wells et al., 2008). This suggests that genes involved in the meiotic process in human oocytes are regulated by genes expressed in cumulus cells.
‘Omics’ technologies for embryo assessment
Accurate selection of the most appropriate embryo for SET has been a major aim for reproductive specialists in recent years. To date, the only universally used method to score the potential development and implantation capability of the human embryo is morphological assessment. Therefore, there is a need for new adjunctive technologies for determining the best embryo for transfer and improvement of implantation rates. Successful embryo implantation requires endometrial receptivity, the development of a viable embryo and adequate bi-directional communication between the blastocyst and endometrium. The advent of the era of high-throughput ‘omic’ methodologies (genomics, transcriptomics, proteomics and metabolomics) has facilitated the study of such processes.
Some of these technologies may provide non-invasive methods for embryo evaluation in the future.
Genomics
Preimplantation genetic screening (PGS) can be used during early stages of embryo development to assess aneuploidy. In some countries, PGS is used to maximize the chances of a successful pregnancy in certain patient subpopulations, such as those of advanced maternal age, or women with a history of recurrent implantation failure or miscarriage.
However, the use of PGS is associated with a number of potential drawbacks. Standard PGS methods utilize a Day-3 biopsy, in which one or two blastomeres are removed from the embryo, followed by FISH to detect chromosomal abnormalities. Therefore, the loss of embryos and a reduction in embryo quality are possible. Furthermore, there is a high rate of mosaicism in Day-3 embryos (which might lead to an incorrect diagnosis), and the survival of embryos after blastomere removal and subsequent cryopreservation has been found to be severely reduced (Joris et al., 1999; Platteau et al., 2006).
Several comprehensive chromosome analysis methods are now available, including aCGH, quantitative PCR and SNP microarrays (Johnson et al., 2010). aCGH evaluates aneuploidy of each pair of chromosomes and results can be available within 24 h. Combined with single blastomere biopsy on Day-3 embryos, aCGH has been shown to be robust, with only 2.9% of embryos with no results, and associated with low error rates (1.9%) (Gutiérrez-Mateo et al., 2011). Unlike SNP arrays, aCGH does not require prior testing of parental DNA; therefore, advanced planning and careful scheduling are unnecessary. A short CGH method has also been shown to detect more chromosomal aneuplodies than FISH and has been used successfully to achieve pregnancy (Rius et al., 2011).
Transcriptomics
Over the last decade, microarray studies have characterized the gene expression profile of the human endometrium in different physiological states, such as during decidualization, the window of implantation, endometriosis, the normal menstrual and stimulated cycles, and endometrial cancer (Horcajadas et al., 2007; Haouzi et al., 2009a, b). However, gene expression profiling studies of human embryos are limited, due to legal and ethical issues. Data on early mammalian development have been provided mainly by murine studies (Tanaka et al., 2000; Hamatani et al., 2004; Wang et al., 2004; Jeong et al., 2006).
Gene expression profile studies in cumulus and granulosa cells have been described earlier and are summarized in Table III. Such studies have identified candidate biomarkers for oocyte quality and competence (Cillo et al., 2007; Hamel et al., 2008), early embryo development (McKenzie et al., 2004; van Montfoort et al., 2008; Anderson et al., 2009) and embryo quality and pregnancy outcome (Assou et al., 2008; Hamel et al., 2010). Furthermore, in mice, blastocyst gene expression profiles correlate with outcome, including successful implantation and pregnancy loss (Parks et al., 2011). miRNA expression in human blastocysts is different in transferable blastocysts from infertile patients (male factor infertility and those from women with PCOS) compared with those from donor fertile controls, and it has been suggested that an association of aberrant miRNA profiles may exist with human infertility (McCallie et al., 2010).
Transcriptomic studies may provide valuable information that could lead to the identification of biomarkers. For a comprehensive review, refer to Assou et al. (2011).
Proteomics
Although not as developed as the genomics/transcriptomics methodologies, proteomic analysis of mammalian embryos is also emerging as a powerful assessment tool (Katz-Jaffe et al., 2005; Shankar et al., 2005). Analysis of the proteome of individual human embryos has the potential to provide novel biomarkers of good embryo development and implantation potential (Katz-Jaffe et al., 2006a). Furthermore, by using bioinformatics to create networks linking proteins found to be differentially regulated between biological samples, functional pathways can be identified (Dominguez et al., 2010). However, analysis of the proteome in a research setting requires extraction of proteins from lysed blastocysts, which limits the use of this technology in routine embryo assessment.
Analysis of the proteins contained in the surrounding embryo culture medium may provide a non-invasive method of using proteomic techniques to identify novel biomarkers of embryo viability (Katz-Jaffe et al., 2006b). Domínguez et al. have used protein array analysis to characterize pooled spent sequential culture media prior to SET and to compare it with control media. A lower abundance of CXCL13, stem cell factor and macrophage-stimulating protein--α, but a higher abundance of soluble TNF receptor 1 was detected in the sequential culture media containing a blastocyst relative to the control (Domínguez et al., 2008). Furthermore, blastocysts that went on to successfully implant were found to have a relatively lower abundance of CXCL13 and granulocyte-macrophage colony-stimulating factor in their culture media than those that did not implant; these proteins may be markers of successful implantation ability (Domínguez et al., 2008).
Domínguez et al. have also used co-culture studies to explore the role of the endometrium in implantation. This technique involves endometrial biopsy, followed by separation and culture of the endometrial epithelial cells (EEC). A total of 32 proteins were found to be up- or down-regulated in blastocysts that went on to successfully implant and were co-cultured with EEC relative to those cultured in sequential media; of these proteins, interleukin 6 (IL-6) was found to be the most abundantly secreted in the EEC culture. Furthermore, the IL-6 concentration in sequential media from blastocysts that implanted was significantly lower than that of blastocysts that did not implant (Dominguez et al., 2010). The proteomics of endometrial receptivity have also been studied by analysis of endometrial biopsies taken during the pre-receptive and receptive phases of the menstrual cycle (DeSouza et al., 2005; Domínguez et al., 2009; Haouzi et al., 2009a, b). Two proteins found to be differentially expressed in pre-receptive and receptive endometria were stathmin 1 and annexin A2 (down- and up-regulated in the receptive endometrium, respectively) (Domínguez et al., 2009). Support for the involvement of these proteins in endometrial receptivity was provided by the pre-receptive pattern of stathmin 1 and annexin A2 expression observed when an intrauterine device was used to model the non-receptive endometrium (Domínguez et al., 2009). In previous studies, a quantitative approach for proteomic assessment using isotope-coded affinity tags, affinity purification and online tandem mass spectrometry has been used to study differences between proliferative and secretory endometria (DeSouza et al., 2005). Only five proteins with significant differential expression were found; the glutamate NMDA receptor subunit zeta 1 precursor and FRAT1 were of greatest interest.
Endometrial receptivity has also been studied by other groups using non-invasive techniques. Boomsma et al. (2009) have used a multiplex immunoassay to analyse endometrial secretions, aspirated immediately prior to embryo transfer during IVF. Using this approach, cytokine profiles predictive of implantation and clinical pregnancy have been identified (Boomsma et al., 2009). The data obtained provide support for associations between MCP-I and IP-10 levels with implantation, and IL-1β and TNF-α levels with clinical pregnancy (Boomsma et al., 2009).
Recent studies have investigated discriminating signatures between individual euploid and aneuploid blastocysts. Microdrops of spent IVF culture medium from individual blastocysts of transferable quality were processed and analysed by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry to determine a blastocyst secretome fingerprint. Each individual blastocyst was then subjected to CGH for comprehensive analysis of all chromosomes (Fragouli et al., 2008). Of the 14 aneuploid blastocysts analysed, nine had a single chromosomal aneuploidy and five had chaotic changes involving more than two chromosomes. Secretome fingerprints from individual blastocysts identified protein signatures that allowed discrimination between euploid and aneuploid chromosomal constitutions (Katz-Jaffe et al., 2008).
Metabolomics
Metabolites represent the end products of cell regulatory processes and can be used to study the response of biological systems to genetic, nutritional and environmental influences; the metabolome (i.e. the complete set of small molecule metabolites found within a biological sample) therefore provides a good indicator of cellular activities.
High-resolution nuclear magnetic resonance (NMR) has been used to analyse the metabolic profile of follicular fluid samples from oocyte donors and has identified metabolites that could be useful as biomarkers of the follicular maturation state (Pinero-Sagredo et al., 2010). NMR spectroscopy has also been used for compositional analysis of other areas of the reproductive tract, including cervical mucus, ovarian tissue, fallopian tubes and uterine matter (Baskind et al., 2011). Incorporation of such techniques into female fertility research may be valuable for understanding subfertility and for predicting outcomes of assisted conception treatments (Baskind et al., 2011).
Metabolomic technology is being used in the development of a new method for aneuploidy detection. This approach is based on the assumption that an embryo with a missing or extra chromosome may have modified metabolism. Given the drawbacks of the use of PGS to detect chromosomal abnormalities, alternative methods would be of value. Analysis of the global metabolome of the embryo (using a combination of ultra-performance liquid chromatography and mass spectroscopy) to analyse low-molecular-weight molecules in spent culture media allows study of the embryo without invasive techniques (unlike PGS). By correlating metabolomic profiles with the results of subsequently performed PGS, it has been possible to differentiate between embryos with normal and abnormal chromosome numbers, and even classify embryos according to their chromosomal anomaly (Sanchez-Ribas et al., 2008). Although this technique may be limited by the low quantity of metabolites in spent culture media, preliminary results are promising and encourage further development of this non-invasive tool for embryo assessment.
Aneuploidy screening represents a unique and valuable application of metabolomics. Analysis of the metabolomic profile of embryo culture media using infra-red techniques is also being explored as an embryo assessment strategy (Seli et al., 2007, 2010; Scott et al., 2008).
Translating ‘omics’ technologies to the ART clinic
As with any ART procedure, the easier the technique, the more likely that it can be used to produce reliable results in routine procedures in ART clinics. Most ART laboratories have, or have easy access to, enzyme-linked immunosorbent assay platforms for routine hormone testing. However, few laboratories have sophisticated equipment for performing quantitative PCR or mass spectrometry. If the cost of this equipment falls over time, or as more focused assays become available, one or more of these technologies may become used routinely in clinical practice. Another possibility is that as procedures become routine and as key genes, proteins or small molecules are identified that correlate with pregnancy outcomes, clinical samples (cumulus cells, media from embryo cultures or blastomeres) could be collected and stored in specialized solutions or containers, and testing could be performed at a commercial facility. Such a process could allow cost-effective performance of the assays and standardization of the technologies. In addition, this practice could allow selection of high-quality oocytes based on cumulus cell profiles after the oocytes have been vitrified. Under such circumstances, there would not be immediate time pressures to obtain the results and oocytes could be assessed before transfer in a subsequent cycle. In the future, it is likely that a combination of multiple technologies will be used with the goal of obtaining a healthy newborn on the first attempted embryo transfer.
Conclusions
In conclusion, our knowledge of ovarian genetics is rapidly expanding and this review provides an important update of those genes identified to have a role in infertility, particularly POI. To date, the search for a candidate gene for PCOS has produced limited positive findings despite involvement of numerous genes. In the field of endometriosis, it has been proposed that the condition results from a series of multiple hits within target genes and gene expression microarray studies have identified a number of candidate gene families that may be differentially regulated. The members of the Fifth EVAR Workshop Group agreed that the way forward in advancing the knowledge of genes involved in reproduction and fertility-associated pathologies was through GWAS involving large numbers of patients. As GWAS are expensive to conduct, it was suggested that pooling of data from different research groups may be the way forward.
Future challenges for genomics and genetic screening of patients include identification of high-throughput strategies and how to apply findings to patients being seen in the ART clinic. The identification of high-quality embryos in ART remains a priority in order to select those with high potential development and implantation capability for transfer. Gene expression profiling of cumulus cells and proteomic and metabolomic technologies show promise for non-invasive oocyte and embryo assessment.
Authors' roles
B.C.J.M. Fauser: developed the Fifth EVAR Workshop scientific programme, participated as a Chairman, was involved in drafting the article and revising it critically for important intellectual content and approved the final version of the manuscript. K. Diedrich: participated in the Fifth EVAR Workshop as a Chairman, was involved in drafting the article and revising it critically for important intellectual content and approved the final version of the manuscript. P. Bouchard, F. Domínguez, M. Matzuk, S. Franks, S. Hamamah, C. Simón, D. Ezcurra and C.M. Howles: participated in the Fifth EVAR Workshop, were involved in drafting the article and revising it critically for important intellectual content and approved the final version of the manuscript. P. Devroey: was involved in the planning of the Fifth EVAR Workshop and was involved in drafting the article and revising it critically for important intellectual content and approved the final version of the manuscript.
Funding
The workshop and preparation of the manuscript were sponsored by an unrestricted educational grant from Merck Serono S.A., Geneva, Switzerland, an affiliate of Merck KGaA, Darmstadt, Germany.
Conflict of interest
B.C.J.M. Fauser has received fees and grant support from the following companies: Andromed, Ardana, Ferring Pharmaceuticals, Genovum, Glycotope, Merck Serono, MSD, Organon, Pantarhei Bioscience, Philips, PregLem, Schering, Schering-Plough, Serono and Wyeth. C.S. is an International Scientific Advisor for Merck Serono S.A., Geneva, Switzerland, an affiliate of Merck KGaA, Darmstadt, Germany. D. Ezcurra and C.M. Howles are employees of Merck Serono S.A., Geneva, Switzerland, an affiliate of Merck KGaA, Darmstadt, Germany.
Acknowledgements
The authors thank Olga Salvidio and Manuel Ortega of Merck Serono S.A., Geneva, Switzerland (an affiliate of Merck KGaA, Darmstadt, Germany) for their participation in the Fifth Evian Annual Reproduction Workshop Meeting. The authors also thank Jane Davies (J.D.), Mailee Wong (M.W.) and Carol Cooper (C.C.) of Caudex Medical (supported by Merck Serono S.A., Geneva, Switzerland, an affiliate of Merck KGaA, Darmstadt, Germany) for assistance in the preparation of this manuscript. J.D. and M.W. assisted with the writing of the first draft of the manuscript under the direction of the authors and incorporated their feedback. C.C. provided editorial assistance.
References
- Abbott DH, Dumesic DA, Franks S. Developmental origin of polycystic ovary syndrome—a hypothesis. J Endocrinol. 2002;174:1–5. doi: 10.1677/joe.0.1740001. [DOI] [PubMed] [Google Scholar]
- Aboura A, Dupas C, Tachdjian G, Portnoi MF, Bourcigaux N, Dewailly D, Frydman R, Fauser B, Ronci-Chaix N, Donadille B, et al. Array comparative genomic hybridization profiling analysis reveals deoxyribonucleic acid copy number variations associated with premature ovarian failure. J Clin Endocrinol Metab. 2009;94:4540–4546. doi: 10.1210/jc.2009-0186. [DOI] [PubMed] [Google Scholar]
- Ahonen P, Myllarniemi S, Sipila I, Perheentupa J. Clinical variation of autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) in a series of 68 patients. N Engl J Med. 1990;322:1829–1836. doi: 10.1056/NEJM199006283222601. [DOI] [PubMed] [Google Scholar]
- Aittomaki K, Lucena JL, Pakarinen P, Sistonen P, Tapanainen J, Gromoll J, Kaskikari R, Sankila EM, Lehvaslaiho H, Engel AR, et al. Mutation in the follicle-stimulating hormone receptor gene causes hereditary hypergonadotropic ovarian failure. Cell. 1995;82:959–968. doi: 10.1016/0092-8674(95)90275-9. [DOI] [PubMed] [Google Scholar]
- Aki T, Funakoshi T, Nishida-Kitayama J, Mizukami Y. TPRA40/GPR175 regulates early mouse embryogenesis through functional membrane transport by Sjogren's syndrome-associated protein NA14. J Cell Physiol. 2008;217:194–206. doi: 10.1002/jcp.21492. [DOI] [PubMed] [Google Scholar]
- Andersen AN, Goossens V, Ferraretti AP, Bhattacharya S, Felberbaum R, de Mouzon J, Nygren KG. Assisted reproductive technology in Europe, 2004: results generated from European registers by ESHRE. Hum Reprod. 2008;23:756–771. doi: 10.1093/humrep/den014. [DOI] [PubMed] [Google Scholar]
- Anderson RA, Sciorio R, Kinnell H, Bayne RA, Thong KJ, de Sousa PA, Pickering S. Cumulus gene expression as a predictor of human oocyte fertilisation, embryo development and competence to establish a pregnancy. Reproduction. 2009;138:629–637. doi: 10.1530/REP-09-0144. [DOI] [PubMed] [Google Scholar]
- Assou S, Anahory T, Pantesco V, Le Carrour T, Pellestor F, Klein B, Reyftmann L, Dechaud H, De Vos J, Hamamah S. The human cumulus–oocyte complex gene-expression profile. Hum Reprod. 2006;21:1705–1719. doi: 10.1093/humrep/del065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S, Haouzi D, Mahmoud K, Aouacheria A, Guillemin Y, Pantesco V, Reme T, Dechaud H, De Vos J, Hamamah S. A non-invasive test for assessing embryo potential by gene expression profiles of human cumulus cells: a proof of concept study. Mol Hum Reprod. 2008;14:711–719. doi: 10.1093/molehr/gan067. [DOI] [PubMed] [Google Scholar]
- Assou S, Haouzi D, De Vos J, Hamamah S. Human cumulus cells as biomarkers for embryo and pregnancy outcomes. Mol Hum Reprod. 2010;16:531–538. doi: 10.1093/molehr/gaq032. [DOI] [PubMed] [Google Scholar]
- Assou S, Boumela I, Haouzi D, Anahory T, Dechaud H, De Vos J, Hamamah S. Dynamic changes in gene expression during human early embryo development: from fundamental aspects to clinical applications. Hum Reprod Update. 2011;17:272–290. doi: 10.1093/humupd/dmq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attaoua R, Ait El Mkadem S, Radian S, Fica S, Hanzu F, Albu A, Gheorghiu M, Coculescu M, Grigorescu F. FTO gene associates to metabolic syndrome in women with polycystic ovary syndrome. Biochem Biophys Res Commun. 2008;373:230–234. doi: 10.1016/j.bbrc.2008.06.039. [DOI] [PubMed] [Google Scholar]
- Attar E, Bulun SE. Aromatase and other steroidogenic genes in endometriosis: translational aspects. Hum Reprod Update. 2006;12:49–56. doi: 10.1093/humupd/dmi034. [DOI] [PubMed] [Google Scholar]
- Barber TM, Bennett AJ, Groves CJ, Sovio U, Ruokonen A, Martikainen H, Pouta A, Hartikainen AL, Elliott P, Lindgren CM, et al. Association of variants in the fat mass and obesity associated (FTO) gene with polycystic ovary syndrome. Diabetologia. 2008;51:1153–1158. doi: 10.1007/s00125-008-1028-6. [DOI] [PubMed] [Google Scholar]
- Baskind NE, McRae C, Sharma V, Fisher J. Understanding subfertility at a molecular level in the female through the application of nuclear magnetic resonance (NMR) spectroscopy. Hum Reprod Update. 2011;17:228–241. doi: 10.1093/humupd/dmq039. [DOI] [PubMed] [Google Scholar]
- Bione S, Sala C, Manzini C, Arrigo G, Zuffardi O, Banfi S, Borsani G, Jonveaux P, Philippe C, Zuccotti M, et al. A human homologue of the Drosophila melanogaster diaphanous gene is disrupted in a patient with premature ovarian failure: evidence for conserved function in oogenesis and implications for human sterility. Am J Hum Genet. 1998;62:533–541. doi: 10.1086/301761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F, Simpson JL. Genetic basis of endometriosis. Ann N Y Acad Sci. 2004;1034:284–299. doi: 10.1196/annals.1335.030. [DOI] [PubMed] [Google Scholar]
- Boomsma CM, Kavelaars A, Eijkemans MJ, Lentjes EG, Fauser BC, Heijnen CJ, Macklon NS. Endometrial secretion analysis identifies a cytokine profile predictive of pregnancy in IVF. Hum Reprod. 2009;24:1427–1435. doi: 10.1093/humrep/dep011. [DOI] [PubMed] [Google Scholar]
- Borgstrom B, Hreinsson J, Rasmussen C, Sheikhi M, Fried G, Keros V, Fridstrom M, Hovatta O. Fertility preservation in girls with turner syndrome: prognostic signs of the presence of ovarian follicles. J Clin Endocrinol Metab. 2009;94:74–80. doi: 10.1210/jc.2008-0708. [DOI] [PubMed] [Google Scholar]
- Broekmans FJ, Soules MR, Fauser BC. Ovarian ageing: mechanisms and clinical consequences. Endocr Rev. 2009;30:465–493. doi: 10.1210/er.2009-0006. [DOI] [PubMed] [Google Scholar]
- Bruysters M, Christin-Maitre S, Verhoef-Post M, Sultan C, Auger J, Faugeron I, Larue L, Lumbroso S, Themmen AP, Bouchard P. A new LH receptor splice mutation responsible for male hypogonadism with subnormal sperm production in the propositus, and infertility with regular cycles in an affected sister. Hum Reprod. 2008;23:1917–1923. doi: 10.1093/humrep/den180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulun SE, Utsunomiya H, Lin Z, Yin P, Cheng YH, Pavone ME, Tokunaga H, Trukhacheva E, Attar E, Gurates B, et al. Steroidogenic factor-1 and endometriosis. Mol Cell Endocrinol. 2009;300:104–108. doi: 10.1016/j.mce.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Burney RO, Talbi S, Hamilton AE, Vo KC, Nyegaard M, Nezhat CR, Lessey BA, Giudice LC. Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology. 2007;148:3814–3826. doi: 10.1210/en.2006-1692. [DOI] [PubMed] [Google Scholar]
- Carey AH, Chan KL, Short F, White D, Williamson R, Franks S. Evidence for a single gene effect causing polycystic ovaries and male pattern baldness. Clin Endocrinol (Oxf) 1993;38:653–658. doi: 10.1111/j.1365-2265.1993.tb02150.x. [DOI] [PubMed] [Google Scholar]
- Cervelló I, Gil-Sanchis C, Mas A, Delgado-Rosas F, Martinez-Conejero JA, Galan A, Martinez-Romero A, Martinez S, Navarro I, Ferro J, et al. Human endometrial side population cells exhibit genotypic, phenotypic and functional features of somatic stem cells. PLoS One. 2010;5:e10964. doi: 10.1371/journal.pone.0010964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Brown CW, Matzuk MM. Genetic analysis of the mammalian transforming growth factor-beta superfamily. Endocr Rev. 2002;23:787–823. doi: 10.1210/er.2002-0003. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Zhao H, He L, Shi Y, Qin Y, Shi Y, Li Z, You L, Zhao J, Liu J, et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011;43:55–59. doi: 10.1038/ng.732. [DOI] [PubMed] [Google Scholar]
- Choi Y, Rajkovic A. Characterization of NOBOX DNA binding specificity and its regulation of Gdf9 and Pou5f1 promoters. J Biol Chem. 2006;281:35747–35756. doi: 10.1074/jbc.M604008200. [DOI] [PubMed] [Google Scholar]
- Choi Y, Ballow DJ, Xin Y, Rajkovic A. Lim homeobox gene, lhx8, is essential for mouse oocyte differentiation and survival. Biol Reprod. 2008;79:442–449. doi: 10.1095/biolreprod.108.069393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopoulos P, Mastorakos G, Gazouli M, Panidis D, Deligeoroglou E, Katsikis I, Papadias K, Diamandi-Kandarakis E, Creatsas G. Genetic variants in TCF7L2 and KCNJ11 genes in a Greek population with polycystic ovary syndrome. Gynecol Endocrinol. 2008;24:486–490. doi: 10.1080/09513590802196379. [DOI] [PubMed] [Google Scholar]
- Chrzanowska KH, Szarras-Czapnik M, Gajdulewicz M, Kalina MA, Gajtko-Metera M, Walewska-Wolf M, Szufladowicz-Wozniak J, Rysiewski H, Gregorek H, Cukrowska B, et al. High prevalence of primary ovarian insufficiency in girls and young women with Nijmegen breakage syndrome: evidence from a longitudinal study. J Clin Endocrinol Metab. 2010;95:3133–3140. doi: 10.1210/jc.2009-2628. [DOI] [PubMed] [Google Scholar]
- Chun SY, McGee EA, Hsu SY, Minami S, LaPolt PS, Yao HH, Bahr JM, Gougeon A, Schomberg DW, Hsueh AJ. Restricted expression of WT1 messenger ribonucleic acid in immature ovarian follicles: uniformity in mammalian and avian species and maintenance during reproductive senescence. Biol Reprod. 1999;60:365–373. doi: 10.1095/biolreprod60.2.365. [DOI] [PubMed] [Google Scholar]
- Ciampelli M, Fulghesu AM, Cucinelli F, Pavone V, Ronsisvalle E, Guido M, Caruso A, Lanzone A. Impact of insulin and body mass index on metabolic and endocrine variables in polycystic ovary syndrome. Metabolism. 1999;48:167–172. doi: 10.1016/s0026-0495(99)90028-8. [DOI] [PubMed] [Google Scholar]
- Cillo F, Brevini TA, Antonini S, Paffoni A, Ragni G, Gandolfi F. Association between human oocyte developmental competence and expression levels of some cumulus genes. Reproduction. 2007;134:645–650. doi: 10.1530/REP-07-0182. [DOI] [PubMed] [Google Scholar]
- Cooper HE, Spellacy WN, Prem KA, Cohen WD. Hereditary factors in the Stein–Leventhal syndrome. Am J Obstet Gynecol. 1968;100:371–387. doi: 10.1016/s0002-9378(15)33704-2. [DOI] [PubMed] [Google Scholar]
- Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, Amati P, Bisceglia L, Zelante L, Nagaraja R, Porcu S, et al. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat Genet. 2001;27:159–166. doi: 10.1038/84781. [DOI] [PubMed] [Google Scholar]
- De Baere E, Dixon MJ, Small KW, Jabs EW, Leroy BP, Devriendt K, Gillerot Y, Mortier G, Meire F, Van Maldergem L, et al. Spectrum of FOXL2 gene mutations in blepharophimosis-ptosis-epicanthus inversus (BPES) families demonstrates a genotype–phenotype correlation. Hum Mol Genet. 2001;10:1591–1600. doi: 10.1093/hmg/10.15.1591. [DOI] [PubMed] [Google Scholar]
- de Boer EJ, den Tonkelaar I, te Velde ER, Burger CW, van Leeuwen FE. Increased risk of early menopausal transition and natural menopause after poor response at first IVF treatment. Hum Reprod. 2003;18:1544–1552. doi: 10.1093/humrep/deg278. [DOI] [PubMed] [Google Scholar]
- De Vos M, Devroey P, Fauser BC. Primary ovarian insufficiency. Lancet. 2010;376:911–921. doi: 10.1016/S0140-6736(10)60355-8. [DOI] [PubMed] [Google Scholar]
- DeSouza L, Diehl G, Yang EC, Guo J, Rodrigues MJ, Romaschin AD, Colgan TJ, Siu KW. Proteomic analysis of the proliferative and secretory phases of the human endometrium: protein identification and differential protein expression. Proteomics. 2005;5:270–281. doi: 10.1002/pmic.200400920. [DOI] [PubMed] [Google Scholar]
- Di Pasquale E, Beck-Peccoz P, Persani L. Hypergonadotropic ovarian failure associated with an inherited mutation of human bone morphogenetic protein-15 (BMP15) gene. Am J Hum Genet. 2004;75:106–111. doi: 10.1086/422103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz FJ, Wigglesworth K, Eppig JJ. Oocytes determine cumulus cell lineage in mouse ovarian follicles. J Cell Sci. 2007;120:1330–1340. doi: 10.1242/jcs.000968. [DOI] [PubMed] [Google Scholar]
- Dixit H, Deendayal M, Singh L. Mutational analysis of the mature peptide region of inhibin genes in Indian women with ovarian failure. Hum Reprod. 2004;19:1760–1764. doi: 10.1093/humrep/deh342. [DOI] [PubMed] [Google Scholar]
- Dixit H, Rao LK, Padmalatha V, Kanakavalli M, Deenadayal M, Gupta N, Chakravarty B, Singh L. Mutational screening of the coding region of growth differentiation factor 9 gene in Indian women with ovarian failure. Menopause. 2005;12:749–754. doi: 10.1097/01.gme.0000184424.96437.7a. [DOI] [PubMed] [Google Scholar]
- Dixit H, Rao KL, Padmalatha VV, Kanakavalli M, Deenadayal M, Gupta N, Chakrabarty BN, Singh L. Mutational analysis of the betaglycan gene-coding region in susceptibility for ovarian failure. Hum Reprod. 2006;21:2041–2046. doi: 10.1093/humrep/del107. [DOI] [PubMed] [Google Scholar]
- Doherty E, Pakarinen P, Tiitinen A, Kiilavuori A, Huhtaniemi I, Forrest S, Aittomaki K. A novel mutation in the FSH receptor inhibiting signal transduction and causing primary ovarian failure. J Clin Endocrinol Metab. 2002;87:1151–1155. doi: 10.1210/jcem.87.3.8319. [DOI] [PubMed] [Google Scholar]
- Domínguez F, Gadea B, Esteban FJ, Horcajadas JA, Pellicer A, Simon C. Comparative protein-profile analysis of implanted versus non-implanted human blastocysts. Hum Reprod. 2008;23:1993–2000. doi: 10.1093/humrep/den205. [DOI] [PubMed] [Google Scholar]
- Domínguez F, Garrido-Gomez T, Lopez JA, Camafeita E, Quinonero A, Pellicer A, Simon C. Proteomic analysis of the human receptive versus non-receptive endometrium using differential in-gel electrophoresis and MALDI-MS unveils stathmin 1 and annexin A2 as differentially regulated. Hum Reprod. 2009;24:2607–2617. doi: 10.1093/humrep/dep230. [DOI] [PubMed] [Google Scholar]
- Dominguez F, Gadea B, Mercader A, Esteban FJ, Pellicer A, Simon C. Embryologic outcome and secretome profile of implanted blastocysts obtained after coculture in human endometrial epithelial cells versus the sequential system. Fertil Steril. 2010;93:774–782. doi: 10.1016/j.fertnstert.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk MM. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature. 1996;383:531–535. doi: 10.1038/383531a0. [DOI] [PubMed] [Google Scholar]
- Edson MA, Nagaraja AK, Matzuk MM. The mammalian ovary from genesis to revelation. Endocr Rev. 2009;30:624–712. doi: 10.1210/er.2009-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edson MA, Lin YN, Matzuk MM. Deletion of the novel oocyte-enriched gene, Gpr149, leads to increased fertility in mice. Endocrinology. 2010;151:358–368. doi: 10.1210/en.2009-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvin JA, Clark AT, Wang P, Wolfman NM, Matzuk MM. Paracrine actions of growth differentiation factor-9 in the mammalian ovary. Mol Endocrinol. 1999a;13:1035–1048. doi: 10.1210/mend.13.6.0310. [DOI] [PubMed] [Google Scholar]
- Elvin JA, Yan C, Wang P, Nishimori K, Matzuk MM. Molecular characterization of the follicle defects in the growth differentiation factor 9-deficient ovary. Mol Endocrinol. 1999b;13:1018–1034. doi: 10.1210/mend.13.6.0309. [DOI] [PubMed] [Google Scholar]
- Elvin JA, Yan C, Matzuk MM. Growth differentiation factor-9 stimulates progesterone synthesis in granulosa cells via a prostaglandin E2/EP2 receptor pathway. Proc Natl Acad Sci USA. 2000;97:10288–10293. doi: 10.1073/pnas.180295197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis S, Ward D, Murray A. Nonlinear association between CGG repeat number and age of menopause in FMR1 premutation carriers. Eur J Hum Genet. 2006;14:253–255. doi: 10.1038/sj.ejhg.5201510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JJ, Pendola FL, Wigglesworth K, Pendola JK. Mouse oocytes regulate metabolic cooperativity between granulosa cells and oocytes: amino acid transport. Biol Reprod. 2005;73:351–357. doi: 10.1095/biolreprod.105.041798. [DOI] [PubMed] [Google Scholar]
- Escobar-Morreale HF, Luque-Ramirez M, San Millan JL. The molecular-genetic basis of functional hyperandrogenism and the polycystic ovary syndrome. Endocr Rev. 2005;26:251–282. doi: 10.1210/er.2004-0004. [DOI] [PubMed] [Google Scholar]
- Farhi J, Homburg R, Ferber A, Orvieto R, Ben RZ. Non-response to ovarian stimulation in normogonadotrophic, normogonadal women: a clinical sign of impending onset of ovarian failure pre-empting the rise in basal follicle stimulating hormone levels. Hum Reprod. 1997;12:241–243. doi: 10.1093/humrep/12.2.241. [DOI] [PubMed] [Google Scholar]
- Ferriman D, Purdie AW. The inheritance of polycystic ovarian disease and a possible relationship to premature balding. Clin Endocrinol (Oxf) 1979;11:291–300. doi: 10.1111/j.1365-2265.1979.tb03077.x. [DOI] [PubMed] [Google Scholar]
- Feuerstein P, Cadoret V, Dalbies-Tran R, Guerif F, Bidault R, Royere D. Gene expression in human cumulus cells: one approach to oocyte competence. Hum Reprod. 2007;22:3069–3077. doi: 10.1093/humrep/dem336. [DOI] [PubMed] [Google Scholar]
- Fogli A, Gauthier-Barichard F, Schiffmann R, Vanderhoof VH, Bakalov VK, Nelson LM, Boespflug-Tanguy O. Screening for known mutations in EIF2B genes in a large panel of patients with premature ovarian failure. BMC Womens Health. 2004;4:8. doi: 10.1186/1472-6874-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragouli E, Lenzi M, Ross R, Katz-Jaffe M, Schoolcraft WB, Wells D. Comprehensive molecular cytogenetic analysis of the human blastocyst stage. Hum Reprod. 2008;23:2596–2608. doi: 10.1093/humrep/den287. [DOI] [PubMed] [Google Scholar]
- Franks S, McCarthy M. Genetics of ovarian disorders: polycystic ovary syndrome. Rev Endocr Metab Disord. 2004;5:69–76. doi: 10.1023/B:REMD.0000016125.05878.96. [DOI] [PubMed] [Google Scholar]
- Franks S, McCarthy MI, Hardy K. Development of polycystic ovary syndrome: involvement of genetic and environmental factors. Int J Androl. 2006;29:278–285. doi: 10.1111/j.1365-2605.2005.00623.x. [DOI] [PubMed] [Google Scholar]
- Franks S, Webber LJ, Goh M, Valentine A, White DM, Conway GS, Wiltshire S, McCarthy MI. Ovarian morphology is a marker of heritable biochemical traits in sisters with polycystic ovaries. J Clin Endocrinol Metab. 2008;93:3396–3402. doi: 10.1210/jc.2008-0369. [DOI] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaasenbeek M, Powell BL, Sovio U, Haddad L, Gharani N, Bennett A, Groves CJ, Rush K, Goh MJ, Conway GS, et al. Large-scale analysis of the relationship between CYP11A promoter variation, polycystic ovarian syndrome, and serum testosterone. J Clin Endocrinol Metab. 2004;89:2408–2413. doi: 10.1210/jc.2003-031640. [DOI] [PubMed] [Google Scholar]
- Galloway SM, McNatty KP, Cambridge LM, Laitinen MP, Juengel JL, Jokiranta TS, McLaren RJ, Luiro K, Dodds KG, Montgomery GW, et al. Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat Genet. 2000;25:279–283. doi: 10.1038/77033. [DOI] [PubMed] [Google Scholar]
- Gargett CE, Masuda H. Adult stem cells in the endometrium. Mol Hum Reprod. 2010;16:818–834. doi: 10.1093/molehr/gaq061. [DOI] [PubMed] [Google Scholar]
- Gargett CE, Cervello I, Hubbard S, Simon C. Adult stem cells in the human endometrium. In: Simon C, Pellicer A, editors. Stem Cells in Human Reproduction: Basic Science and Therapeutic Potential (Reproductive Medicine & Assisted Reproductive Techniques) London: Informa Healthcare; 2009. pp. 160–176. [Google Scholar]
- Gilchrist RB, Lane M, Thompson JG. Oocyte-secreted factors: regulators of cumulus cell function and oocyte quality. Hum Reprod Update. 2008;14:159–177. doi: 10.1093/humupd/dmm040. [DOI] [PubMed] [Google Scholar]
- Givens JR. Familial polycystic ovarian disease. Endocrinol Metab Clin North Am. 1988;17:771–783. [PubMed] [Google Scholar]
- Gosden RG, Faddy MJ. Ovarian aging, follicular depletion, and steroidogenesis. Exp Gerontol. 1994;29:265–274. doi: 10.1016/0531-5565(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Gosden RG, Faddy MJ. Biological bases of premature ovarian failure. Reprod Fertil Dev. 1998;10:73–78. doi: 10.1071/r98043. [DOI] [PubMed] [Google Scholar]
- Goswami D, Conway GS. Premature ovarian failure. Hum Reprod Update. 2005;11:391–410. doi: 10.1093/humupd/dmi012. [DOI] [PubMed] [Google Scholar]
- Guo SW. Emerging drugs for endometriosis. Expert Opin Emerg Drugs. 2008;13:547–571. doi: 10.1517/14728210802548360. [DOI] [PubMed] [Google Scholar]
- Gurates B, Amsterdam A, Tamura M, Yang S, Zhou J, Fang Z, Amin S, Sebastian S, Bulun SE. WT1 and DAX-1 regulate SF-1-mediated human P450arom gene expression in gonadal cells. Mol Cell Endocrinol. 2003;208:61–75. doi: 10.1016/s0303-7207(03)00198-9. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Mateo C, Colls P, Sánchez-Garcia J, Escudero T, Prates R, Ketterson K, Wells D, Munné S. Validation of microarray comparative genomic hybridization for comprehensive chromosome analysis of embryos. Fertil Steril. 2011;95:953–958. doi: 10.1016/j.fertnstert.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Hague WM, Adams J, Reeders ST, Peto TE, Jacobs HS. Familial polycystic ovaries: a genetic disease? Clin Endocrinol (Oxf) 1988;29:593–605. doi: 10.1111/j.1365-2265.1988.tb03707.x. [DOI] [PubMed] [Google Scholar]
- Hamamah S, Matha V, Berthenet C, Anahory T, Loup V, Dechaud H, Hedon B, Fernandez A, Lamb N. Comparative protein expression profiling in human cumulus cells in relation to oocyte fertilization and ovarian stimulation protocol. Reprod Biomed Online. 2006;13:807–814. doi: 10.1016/s1472-6483(10)61028-0. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Hamel M, Dufort I, Robert C, Gravel C, Leveille MC, Leader A, Sirard MA. Identification of differentially expressed markers in human follicular cells associated with competent oocytes. Hum Reprod. 2008;23:1118–1127. doi: 10.1093/humrep/den048. [DOI] [PubMed] [Google Scholar]
- Hamel M, Dufort I, Robert C, Leveille MC, Leader A, Sirard MA. Genomic assessment of follicular marker genes as pregnancy predictors for human IVF. Mol Hum Reprod. 2010;16:87–96. doi: 10.1093/molehr/gap079. [DOI] [PubMed] [Google Scholar]
- Hanley NA, Rainey WE, Wilson DI, Ball SG, Parker KL. Expression profiles of SF-1, DAX1, and CYP17 in the human fetal adrenal gland: potential interactions in gene regulation. Mol Endocrinol. 2001;15:57–68. doi: 10.1210/mend.15.1.0585. [DOI] [PubMed] [Google Scholar]
- Hao J, Yamamoto M, Richardson TE, Chapman KM, Denard BS, Hammer RE, Zhao GQ, Hamra FK. Sohlh2 knockout mice are male-sterile because of degeneration of differentiating type A spermatogonia. Stem Cells. 2008;26:1587–1597. doi: 10.1634/stemcells.2007-0502. [DOI] [PubMed] [Google Scholar]
- Haouzi D, Assou S, Mahmoud K, Tondeur S, Reme T, Hedon B, De Vos J, Hamamah S. Gene expression profile of human endometrial receptivity: comparison between natural and stimulated cycles for the same patients. Hum Reprod. 2009a;24:1436–1445. doi: 10.1093/humrep/dep039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haouzi D, Mahmoud K, Fourar M, Bendhaou K, Dechaud H, De Vos J, Reme T, Dewailly D, Hamamah S. Identification of new biomarkers of human endometrial receptivity in the natural cycle. Hum Reprod. 2009b;24:198–205. doi: 10.1093/humrep/den360. [DOI] [PubMed] [Google Scholar]
- Harris SE, Chand AL, Winship IM, Gersak K, Aittomaki K, Shelling AN. Identification of novel mutations in FOXL2 associated with premature ovarian failure. Mol Hum Reprod. 2002;8:729–733. doi: 10.1093/molehr/8.8.729. [DOI] [PubMed] [Google Scholar]
- Hartge P. Genetics of reproductive lifespan. Nat Genet. 2009;41:637–638. doi: 10.1038/ng0609-637. [DOI] [PubMed] [Google Scholar]
- Hawkins SM, Buchold GM, Matzuk MM. Minireview: the roles of small RNA pathways in reproductive medicine. Mol Endocrinol. 2011a;25:1257–1279. doi: 10.1210/me.2011-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins SM, Creighton CJ, Han DY, Zariff A, Anderson ML, Gunaratne PH, Matzuk MM. Functional microRNA involved in endometriosis. Mol Endocrinol. 2011b;25:821–832. doi: 10.1210/me.2010-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horcajadas JA, Pellicer A, Simon C. Wide genomic analysis of human endometrial receptivity: new times, new opportunities. Hum Reprod Update. 2007;13:77–86. doi: 10.1093/humupd/dml046. [DOI] [PubMed] [Google Scholar]
- Huang HY. Medical treatment of endometriosis. Chang Gung Med J. 2008;31:431–440. [PubMed] [Google Scholar]
- Hull ML, Escareno CR, Godsland JM, Doig JR, Johnson CM, Phillips SC, Smith SK, Tavare S, Print CG, Charnock-Jones DS. Endometrial–peritoneal interactions during endometriotic lesion establishment. Am J Pathol. 2008;173:700–715. doi: 10.2353/ajpath.2008.071128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong HJ, Kim HJ, Lee SH, Kwack K, Ahn SY, Choi YJ, Kim HG, Lee KW, Lee CN, Cha KY. Gene expression profiling of the pre-implantation mouse embryo by microarray analysis: comparison of the two-cell stage and two-cell block. Theriogenology. 2006;66:785–796. doi: 10.1016/j.theriogenology.2006.01.059. [DOI] [PubMed] [Google Scholar]
- Jeyasuria P, Ikeda Y, Jamin SP, Zhao L, De Rooij DG, Themmen AP, Behringer RR, Parker KL. Cell-specific knockout of steroidogenic factor 1 reveals its essential roles in gonadal function. Mol Endocrinol. 2004;18:1610–1619. doi: 10.1210/me.2003-0404. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Gemelos G, Baner J, Ryan A, Cinnioglu C, Banjevic M, Ross R, Alper M, Barrett B, Frederick J, et al. Preclinical validation of a microarray method for full molecular karyotyping of blastomeres in a 24-h protocol. Hum Reprod. 2010;25:1066–1075. doi: 10.1093/humrep/dep452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris H, Van den Abbeel E, Vos AD, Van Steirteghem A. Reduced survival after human embryo biopsy and subsequent cryopreservation. Hum Reprod. 1999;14:2833–2837. doi: 10.1093/humrep/14.11.2833. [DOI] [PubMed] [Google Scholar]
- Joshi S, Davies H, Sims LP, Levy SE, Dean J. Ovarian gene expression in the absence of FIGLA, an oocyte-specific transcription factor. BMC Dev Biol. 2007;7:67. doi: 10.1186/1471-213X-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao LC, Germeyer A, Tulac S, Lobo S, Yang JP, Taylor RN, Osteen K, Lessey BA, Giudice LC. Expression profiling of endometrium from women with endometriosis reveals candidate genes for disease-based implantation failure and infertility. Endocrinology. 2003;144:2870–2881. doi: 10.1210/en.2003-0043. [DOI] [PubMed] [Google Scholar]
- Kato K, Yoshimoto M, Kato K, Adachi S, Yamayoshi A, Arima T, Asanoma K, Kyo S, Nakahata T, Wake N. Characterization of side-population cells in human normal endometrium. Hum Reprod. 2007;22:1214–1223. doi: 10.1093/humrep/del514. [DOI] [PubMed] [Google Scholar]
- Kato K, Takao T, Kuboyama A, Tanaka Y, Ohgami T, Yamaguchi S, Adachi S, Yoneda T, Ueoka Y, Kato K, et al. Endometrial cancer side-population cells show prominent migration and have a potential to differentiate into the mesenchymal cell lineage. Am J Pathol. 2010;176:381–392. doi: 10.2353/ajpath.2010.090056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz-Jaffe MG, Linck DW, Schoolcraft WB, Gardner DK. A proteomic analysis of mammalian preimplantation embryonic development. Reproduction. 2005;130:899–905. doi: 10.1530/rep.1.00854. [DOI] [PubMed] [Google Scholar]
- Katz-Jaffe MG, Gardner DK, Schoolcraft WB. Proteomic analysis of individual human embryos to identify novel biomarkers of development and viability. Fertil Steril. 2006a;85:101–107. doi: 10.1016/j.fertnstert.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Katz-Jaffe MG, Schoolcraft WB, Gardner DK. Analysis of protein expression (secretome) by human and mouse preimplantation embryos. Fertil Steril. 2006b;86:678–685. doi: 10.1016/j.fertnstert.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Katz-Jaffe MG, Fagouli E, Fillipovits J, Wells D, Schoolcraft WB. Relationship between the human blastocyst secretome and chromosomal constitution. Fertil Steril. 2008;90:S80. [Google Scholar]
- Katz-Jaffe MG, McCallie BR, Janesch A, Filipovits JA, Schoolcraft WB, Gardner DK. Blastocysts from patients with polycystic ovaries exhibit altered transcriptome and secretome. Reprod Biomed Online. 2010;21:520–526. doi: 10.1016/j.rbmo.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Kehler J, Tolkunova E, Koschorz B, Pesce M, Gentile L, Boiani M, Lomeli H, Nagy A, McLaughlin KJ, Scholer HR, et al. Oct4 is required for primordial germ cell survival. EMBO Rep. 2004;5:1078–1083. doi: 10.1038/sj.embor.7400279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenigsberg S, Bentov Y, Chalifa-Caspi V, Potashnik G, Ofir R, Birk OS. Gene expression microarray profiles of cumulus cells in lean and overweight-obese polycystic ovary syndrome patients. Mol Hum Reprod. 2009;15:89–103. doi: 10.1093/molehr/gan082. [DOI] [PubMed] [Google Scholar]
- Keri RA, Nilson JH. A steroidogenic factor-1 binding site is required for activity of the luteinizing hormone beta subunit promoter in gonadotropes of transgenic mice. J Biol Chem. 1996;271:10782–10785. doi: 10.1074/jbc.271.18.10782. [DOI] [PubMed] [Google Scholar]
- Knauff EA, Franke L, van Es MA, van den Berg LH, van der Schouw YT, Laven JS, Lambalk CB, Hoek A, Goverde AJ, Christin-Maitre S, et al. Genome-wide association study in premature ovarian failure patients suggests ADAMTS19 as a possible candidate gene. Hum Reprod. 2009;24:2372–2378. doi: 10.1093/humrep/dep197. [DOI] [PubMed] [Google Scholar]
- Kovanci E, Simpson JL, Amato P, Rohozinski J, Heard MJ, Bishop CE, Carson SA. Oocyte-specific G-protein-coupled receptor 3 (GPR3): no perturbations found in 82 women with premature ovarian failure (first report) Fertil Steril. 2008;90:1269–1271. doi: 10.1016/j.fertnstert.2007.07.1373. [DOI] [PubMed] [Google Scholar]
- Laissue P, Christin-Maitre S, Touraine P, Kuttenn F, Ritvos O, Aittomaki K, Bourcigaux N, Jacquesson L, Bouchard P, Frydman R, et al. Mutations and sequence variants in GDF9 and BMP15 in patients with premature ovarian failure. Eur J Endocrinol. 2006;154:739–744. doi: 10.1530/eje.1.02135. [DOI] [PubMed] [Google Scholar]
- Laissue P, Christin-Maitre S, Bouchard P, Fellous M, Veitia RA. Mutations in the NOG gene are not a common cause of nonsyndromic premature ovarian failure. Clin Endocrinol (Oxf) 2007;66:900. doi: 10.1111/j.1365-2265.2007.02797.x. [DOI] [PubMed] [Google Scholar]
- Lavorgna G, Ueda H, Clos J, Wu C. FTZ-F1, a steroid hormone receptor-like protein implicated in the activation of fushi tarazu. Science. 1991;252:848–851. doi: 10.1126/science.1709303. [DOI] [PubMed] [Google Scholar]
- Lawson R, El-Toukhy T, Kassab A, Taylor A, Braude P, Parsons J, Seed P. Poor response to ovulation induction is a stronger predictor of early menopause than elevated basal FSH: a life table analysis. Hum Reprod. 2003;18:527–533. doi: 10.1093/humrep/deg101. [DOI] [PubMed] [Google Scholar]
- Layman LC. Editorial: BMP15—the first true ovarian determinant gene on the X-chromosome? J Clin Endocrinol Metab. 2006;91:1673–1676. doi: 10.1210/jc.2006-0548. [DOI] [PubMed] [Google Scholar]
- Legro RS, Driscoll D, Strauss JF, III, Fox J, Dunaif A. Evidence for a genetic basis for hyperandrogenemia in polycystic ovary syndrome. Proc Natl Acad Sci USA. 1998;95:14956–14960. doi: 10.1073/pnas.95.25.14956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legro RS, Bentley-Lewis R, Driscoll D, Wang SC, Dunaif A. Insulin resistance in the sisters of women with polycystic ovary syndrome: association with hyperandrogenemia rather than menstrual irregularity. J Clin Endocrinol Metab. 2002;87:2128–2133. doi: 10.1210/jcem.87.5.8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie ND, Immerman EB, Flach JE, Florez M, Fridovich-Keil JL, Elsas LJ. The human galactose-1-phosphate uridyltransferase gene. Genomics. 1992;14:474–480. doi: 10.1016/s0888-7543(05)80244-7. [DOI] [PubMed] [Google Scholar]
- Liang L, Soyal SM, Dean J. FIGalpha, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development. 1997;124:4939–4947. doi: 10.1242/dev.124.24.4939. [DOI] [PubMed] [Google Scholar]
- Lourenco D, Brauner R, Lin L, De Perdigo A, Weryha G, Muresan M, Boudjenah R, Guerra-Junior G, Maciel-Guerra AT, Achermann JC, et al. Mutations in NR5A1 associated with ovarian insufficiency. N Engl J Med. 2009;360:1200–1210. doi: 10.1056/NEJMoa0806228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde O, Magnus P, Sandvik L, Hoglo S. Familial clustering in the polycystic ovarian syndrome. Gynecol Obstet Invest. 1989;28:23–30. doi: 10.1159/000293493. [DOI] [PubMed] [Google Scholar]
- Luo X, Ikeda Y, Parker KL. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- Luoma P, Melberg A, Rinne JO, Kaukonen JA, Nupponen NN, Chalmers RM, Oldfors A, Rautakorpi I, Peltonen L, Majamaa K, et al. Parkinsonism, premature menopause, and mitochondrial DNA polymerase gamma mutations: clinical and molecular genetic study. Lancet. 2004;364:875–882. doi: 10.1016/S0140-6736(04)16983-3. [DOI] [PubMed] [Google Scholar]
- Malinak LR, Buttram VC, Jr, Elias S, Simpson JL. Heritage aspects of endometriosis. II. Clinical characteristics of familial endometriosis. Am J Obstet Gynecol. 1980;137:332–337. [PubMed] [Google Scholar]
- Mandon-Pepin B, Touraine P, Kuttenn F, Derbois C, Rouxel A, Matsuda F, Nicolas A, Cotinot C, Fellous M. Genetic investigation of four meiotic genes in women with premature ovarian failure. Eur J Endocrinol. 2008;158:107–115. doi: 10.1530/EJE-07-0400. [DOI] [PubMed] [Google Scholar]
- Masuda H, Matsuzaki Y, Hiratsu E, Ono M, Nagashima T, Kajitani T, Arase T, Oda H, Uchida H, Asada H, et al. Stem cell-like properties of the endometrial side population: implication in endometrial regeneration. PLoS One. 2010;5:e10387. doi: 10.1371/journal.pone.0010387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallie B, Schoolcraft WB, Katz-Jaffe MG. Aberration of blastocyst microRNA expression is associated with human infertility. Fertil Steril. 2010;93:2374–2382. doi: 10.1016/j.fertnstert.2009.01.069. [DOI] [PubMed] [Google Scholar]
- McKenzie LJ, Pangas SA, Carson SA, Kovanci E, Cisneros P, Buster JE, Amato P, Matzuk MM. Human cumulus granulosa cell gene expression: a predictor of fertilization and embryo selection in women undergoing IVF. Hum Reprod. 2004;19:2869–2874. doi: 10.1093/humrep/deh535. [DOI] [PubMed] [Google Scholar]
- Mehlmann LM, Saeki Y, Tanaka S, Brennan TJ, Evsikov AV, Pendola FL, Knowles BB, Eppig JJ, Jaffe LA. The Gs-linked receptor GPR3 maintains meiotic arrest in mammalian oocytes. Science. 2004;306:1947–1950. doi: 10.1126/science.1103974. [DOI] [PubMed] [Google Scholar]
- Mendelson CR, Kamat A. Mechanisms in the regulation of aromatase in developing ovary and placenta. J Steroid Biochem Mol Biol. 2007;106:62–70. doi: 10.1016/j.jsbmb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ME, Chatten J. Ovarian changes in ataxia telangiectasia. Acta Paediatr Scand. 1967;56:559–561. doi: 10.1111/j.1651-2227.1967.tb15424.x. [DOI] [PubMed] [Google Scholar]
- Murabito JM, Yang Q, Fox CS, Cupples LA. Genome-wide linkage analysis to age at natural menopause in a community-based sample: the Framingham Heart Study. Fertil Steril. 2005;84:1674–1679. doi: 10.1016/j.fertnstert.2005.05.046. [DOI] [PubMed] [Google Scholar]
- Murray A, Webb J, Dennis N, Conway G, Morton N. Microdeletions in FMR2 may be a significant cause of premature ovarian failure. J Med Genet. 1999;36:767–770. doi: 10.1136/jmg.36.10.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A, Bennett CE, Perry JR, Weedon MN, Jacobs PA, Morris DH, Orr N, Schoemaker MJ, Jones M, Ashworth A, et al. Common genetic variants are significant risk factors for early menopause: results from the Breakthrough Generations Study. Hum Mol Genet. 2011;20:186–192. doi: 10.1093/hmg/ddq417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaou D, Lavery S, Turner C, Margara R, Trew G. Is there a link between an extremely poor response to ovarian hyperstimulation and early ovarian failure? Hum Reprod. 2002;17:1106–1111. doi: 10.1093/humrep/17.4.1106. [DOI] [PubMed] [Google Scholar]
- Nisolle M, Donnez J. Peritoneal endometriosis, ovarian endometriosis, and adenomyotic nodules of the rectovaginal septum are three different entities. Fertil Steril. 1997;68:585–596. doi: 10.1016/s0015-0282(97)00191-x. [DOI] [PubMed] [Google Scholar]
- Nygren K, Andersen AN, Felberbaum R, Gianaroli L, de Mouzon J. On the benefit of assisted reproduction techniques, a comparison of the USA and Europe. Hum Reprod. 2006;21:2194. doi: 10.1093/humrep/del238. [DOI] [PubMed] [Google Scholar]
- Ohlsson Teague EM, Van der Hoek KH, Van der Hoek MB, Perry N, Wagaarachchi P, Robertson SA, Print CG, Hull LM. MicroRNA-regulated pathways associated with endometriosis. Mol Endocrinol. 2009;23:265–275. doi: 10.1210/me.2008-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson Teague EM, Print CG, Hull ML. The role of microRNAs in endometriosis and associated reproductive conditions. Hum Reprod Update. 2010;16:142–165. doi: 10.1093/humupd/dmp034. [DOI] [PubMed] [Google Scholar]
- Oldenburg RA, van Dooren MF, de Graaf B, Simons E, Govaerts L, Swagemakers S, Verkerk JM, Oostra BA, Bertoli-Avella AM. A genome-wide linkage scan in a Dutch family identifies a premature ovarian failure susceptibility locus. Hum Reprod. 2008;23:2835–2841. doi: 10.1093/humrep/den278. [DOI] [PubMed] [Google Scholar]
- Painter JN, Anderson CA, Nyholt DR, Macgregor S, Lin J, Lee SH, Lambert A, Zhao ZZ, Roseman F, Guo Q, et al. Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat Genet. 2011;43:51–54. doi: 10.1038/ng.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Luo X, Toloubeydokhti T, Chegini N. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol Hum Reprod. 2007;13:797–806. doi: 10.1093/molehr/gam063. [DOI] [PubMed] [Google Scholar]
- Pangas SA, Jorgez CJ, Matzuk MM. Growth differentiation factor 9 regulates expression of the bone morphogenetic protein antagonist gremlin. J Biol Chem. 2004;279:32281–32286. doi: 10.1074/jbc.M403212200. [DOI] [PubMed] [Google Scholar]
- Pangas SA, Choi Y, Ballow DJ, Zhao Y, Westphal H, Matzuk MM, Rajkovic A. Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8. Proc Natl Acad Sci USA. 2006;103:8090–8095. doi: 10.1073/pnas.0601083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks JC, McCallie BR, Janesch AM, Schoolcraft WB, Katz-Jaffe MG. Blastocyst gene expression correlates with implantation potential. Fertil Steril. 2011;95:1367–1372. doi: 10.1016/j.fertnstert.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Perez GI, Jurisicova A, Wise L, Lipina T, Kanisek M, Bechard A, Takai Y, Hunt P, Roder J, Grynpas M, et al. Absence of the proapoptotic Bax protein extends fertility and alleviates age-related health complications in female mice. Proc Natl Acad Sci USA. 2007;104:5229–5234. doi: 10.1073/pnas.0608557104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinero-Sagredo E, Nunes S, de Los Santos MJ, Celda B, Esteve V. NMR metabolic profile of human follicular fluid. NMR Biomed. 2010;23:485–495. doi: 10.1002/nbm.1488. [DOI] [PubMed] [Google Scholar]
- Platteau P, Staessen C, Michiels A, Van Steirteghem A, Liebaers I, Devroey P. Which patients with recurrent implantation failure after IVF benefit from PGD for aneuploidy screening? Reprod Biomed Online. 2006;12:334–339. doi: 10.1016/s1472-6483(10)61006-1. [DOI] [PubMed] [Google Scholar]
- Prueitt RL, Ross JL, Zinn AR. Physical mapping of nine Xq translocation breakpoints and identification of XPNPEP2 as a premature ovarian failure candidate gene. Cytogenet Cell Genet. 2000;89:44–50. doi: 10.1159/000015560. [DOI] [PubMed] [Google Scholar]
- Prueitt RL, Chen H, Barnes RI, Zinn AR. Most X;autosome translocations associated with premature ovarian failure do not interrupt X-linked genes. Cytogenet Genome Res. 2002;97:32–38. doi: 10.1159/000064052. [DOI] [PubMed] [Google Scholar]
- Qin Y, Choi Y, Zhao H, Simpson JL, Chen ZJ, Rajkovic A. NOBOX homeobox mutation causes premature ovarian failure. Am J Hum Genet. 2007;81:576–581. doi: 10.1086/519496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkovic A, Pangas SA, Ballow D, Suzumori N, Matzuk MM. NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science. 2004;305:1157–1159. doi: 10.1126/science.1099755. [DOI] [PubMed] [Google Scholar]
- Rius M, Daina G, Obradors A, Ramos L, Velilla E, Fernandez S, Martinez-Passarell O, Benet J, Navarro J. Comprehensive embryo analysis of advanced maternal age-related aneuploidies and mosaicism by short comparative genomic hybridization. Fertil Steril. 2011;95:413–416. doi: 10.1016/j.fertnstert.2010.07.1051. [DOI] [PubMed] [Google Scholar]
- Rocca WA, Bower JH, Maraganore DM, Ahlskog JE, Grossardt BR, de Andrade M, Melton LJ., III Increased risk of cognitive impairment or dementia in women who underwent oophorectomy before menopause. Neurology. 2007;69:1074–1083. doi: 10.1212/01.wnl.0000276984.19542.e6. [DOI] [PubMed] [Google Scholar]
- Rocca WA, Bower JH, Maraganore DM, Ahlskog JE, Grossardt BR, de Andrade M, Melton LJ., III Increased risk of parkinsonism in women who underwent oophorectomy before menopause. Neurology. 2008;70:200–209. doi: 10.1212/01.wnl.0000280573.30975.6a. [DOI] [PubMed] [Google Scholar]
- Russell DL, Robker RL. Molecular mechanisms of ovulation: co-ordination through the cumulus complex. Hum Reprod Update. 2007;13:289–312. doi: 10.1093/humupd/dml062. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ribas I, Dominguez F, Barr J, Castro A, Mato JM, Simon C. Metabolomic profile as a novel non-invasive tool for human embryo aneuploidy assessment. Fertil Steril. 2008;90:S100. [Google Scholar]
- Sasson IE, Taylor HS. Stem cells and the pathogenesis of endometriosis. Ann N Y Acad Sci. 2008;1127:106–115. doi: 10.1196/annals.1434.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Uehara S, Hashiyada M, Nabeshima H, Sugawara J, Terada Y, Yaegashi N, Okamura K. Genetic significance of skewed X-chromosome inactivation in premature ovarian failure. Am J Med Genet A. 2004;130A:240–244. doi: 10.1002/ajmg.a.30256. [DOI] [PubMed] [Google Scholar]
- Scott R, Seli E, Miller K, Sakkas D, Scott K, Burns DH. Noninvasive metabolomic profiling of human embryo culture media using Raman spectroscopy predicts embryonic reproductive potential: a prospective blinded pilot study. Fertil Steril. 2008;90:77–83. doi: 10.1016/j.fertnstert.2007.11.058. [DOI] [PubMed] [Google Scholar]
- Seli E, Sakkas D, Scott R, Kwok SC, Rosendahl SM, Burns DH. Noninvasive metabolomic profiling of embryo culture media using Raman and near-infrared spectroscopy correlates with reproductive potential of embryos in women undergoing in vitro fertilization. Fertil Steril. 2007;88:1350–1357. doi: 10.1016/j.fertnstert.2007.07.1390. [DOI] [PubMed] [Google Scholar]
- Seli E, Vergouw CG, Morita H, Botros L, Roos P, Lambalk CB, Yamashita N, Kato O, Sakkas D. Noninvasive metabolomic profiling as an adjunct to morphology for noninvasive embryo assessment in women undergoing single embryo transfer. Fertil Steril. 2010;94:535–542. doi: 10.1016/j.fertnstert.2009.03.078. [DOI] [PubMed] [Google Scholar]
- Shankar R, Gude N, Cullinane F, Brennecke S, Purcell AW, Moses EK. An emerging role for comprehensive proteome analysis in human pregnancy research. Reproduction. 2005;129:685–696. doi: 10.1530/rep.1.00524. [DOI] [PubMed] [Google Scholar]
- Shen WH, Moore CC, Ikeda Y, Parker KL, Ingraham HA. Nuclear receptor steroidogenic factor 1 regulates the mullerian inhibiting substance gene: a link to the sex determination cascade. Cell. 1994;77:651–661. doi: 10.1016/0092-8674(94)90050-7. [DOI] [PubMed] [Google Scholar]
- Simpson JL, Elias S, Malinak LR, Buttram VC., Jr. Heritable aspects of endometriosis. I. Genetic studies. Am J Obstet Gynecol. 1980;137:327–331. doi: 10.1016/0002-9378(80)90917-5. [DOI] [PubMed] [Google Scholar]
- Snieder H, MacGregor AJ, Spector TD. Genes control the cessation of a woman's reproductive life: a twin study of hysterectomy and age at menopause. J Clin Endocrinol Metab. 1998;83:1875–1880. doi: 10.1210/jcem.83.6.4890. [DOI] [PubMed] [Google Scholar]
- Su YQ, Wu X, O'Brien MJ, Pendola FL, Denegre JN, Matzuk MM, Eppig JJ. Synergistic roles of BMP15 and GDF9 in the development and function of the oocyte–cumulus cell complex in mice: genetic evidence for an oocyte-granulosa cell regulatory loop. Dev Biol. 2004;276:64–73. doi: 10.1016/j.ydbio.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Sugawara T, Kiriakidou M, McAllister JM, Kallen CB, Strauss JF., III Multiple steroidogenic factor 1 binding elements in the human steroidogenic acute regulatory protein gene 5′-flanking region are required for maximal promoter activity and cyclic AMP responsiveness. Biochemistry. 1997;36:7249–7255. doi: 10.1021/bi9628984. [DOI] [PubMed] [Google Scholar]
- Sullivan AK, Marcus M, Epstein MP, Allen EG, Anido AE, Paquin JJ, Yadav-Shah M, Sherman SL. Association of FMR1 repeat size with ovarian dysfunction. Hum Reprod. 2005;20:402–412. doi: 10.1093/humrep/deh635. [DOI] [PubMed] [Google Scholar]
- Sutton ML, Cetica PD, Beconi MT, Kind KL, Gilchrist RB, Thompson JG. Influence of oocyte-secreted factors and culture duration on the metabolic activity of bovine cumulus cell complexes. Reproduction. 2003;126:27–34. doi: 10.1530/rep.0.1260027. [DOI] [PubMed] [Google Scholar]
- Suzumori N, Yan C, Matzuk MM, Rajkovic A. Nobox is a homeobox-encoding gene preferentially expressed in primordial and growing oocytes. Mech Dev. 2002;111:137–141. doi: 10.1016/s0925-4773(01)00620-7. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Ozaki T, Okada M, Kurioka H, Kanasaki H, Miyazaki K. Increased prevalence of luteinizing hormone beta-subunit variant in patients with premature ovarian failure. Fertil Steril. 1999;71:96–101. doi: 10.1016/s0015-0282(98)00409-9. [DOI] [PubMed] [Google Scholar]
- Tan S, Scherag A, Janssen OE, Hahn S, Lahner H, Dietz T, Scherag S, Grallert H, Vogel CI, Kimmig R, et al. Large effects on body mass index and insulin resistance of fat mass and obesity associated gene (FTO) variants in patients with polycystic ovary syndrome (PCOS) BMC Med Genet. 2010;11:12. doi: 10.1186/1471-2350-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, et al. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci USA. 2000;97:9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempfer CB, Simoni M, Destenaves B, Fauser BC. Functional genetic polymorphisms and female reproductive disorders: part II—endometriosis. Hum Reprod Update. 2009;15:97–118. doi: 10.1093/humupd/dmn040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly JL. Commuting the death sentence: how oocytes strive to survive. Nat Rev Mol Cell Biol. 2001;2:838–848. doi: 10.1038/35099086. [DOI] [PubMed] [Google Scholar]
- Torgerson DJ, Thomas RE, Reid DM. Mothers and daughters menopausal ages: is there a link? Eur J Obstet Gynecol Reprod Biol. 1997;74:63–66. doi: 10.1016/s0301-2115(97)00085-7. [DOI] [PubMed] [Google Scholar]
- Toyoda S, Miyazaki T, Miyazaki S, Yoshimura T, Yamamoto M, Tashiro F, Yamato E, Miyazaki J. Sohlh2 affects differentiation of KIT positive oocytes and spermatogonia. Dev Biol. 2009;325:238–248. doi: 10.1016/j.ydbio.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Treloar SA, Do KA, Martin NG. Genetic influences on the age at menopause. Lancet. 1998;352:1084–1085. doi: 10.1016/S0140-6736(05)79753-1. [DOI] [PubMed] [Google Scholar]
- Tucci S, Futterweit W, Concepcion ES, Greenberg DA, Villanueva R, Davies TF, Tomer Y. Evidence for association of polycystic ovary syndrome in caucasian women with a marker at the insulin receptor gene locus. J Clin Endocrinol Metab. 2001;86:446–449. doi: 10.1210/jcem.86.1.7274. [DOI] [PubMed] [Google Scholar]
- Tung JY, Rosen MP, Nelson LM, Turek PJ, Witte JS, Cramer DW, Cedars MI, Reijo-Pera RA. Novel missense mutations of the Deleted-in-AZoospermia-Like (DAZL) gene in infertile women and men. Reprod Biol Endocrinol. 2006;4:40. doi: 10.1186/1477-7827-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uda M, Ottolenghi C, Crisponi L, Garcia JE, Deiana M, Kimber W, Forabosco A, Cao A, Schlessinger D, Pilia G. Foxl2 disruption causes mouse ovarian failure by pervasive blockage of follicle development. Hum Mol Genet. 2004;13:1171–1181. doi: 10.1093/hmg/ddh124. [DOI] [PubMed] [Google Scholar]
- Urbanek M. The genetics of the polycystic ovary syndrome. Nat Clin Pract Endocrinol Metab. 2007;3:103–111. doi: 10.1038/ncpendmet0400. [DOI] [PubMed] [Google Scholar]
- Urbanek M, Legro RS, Driscoll DA, Azziz R, Ehrmann DA, Norman RJ, Strauss JF, III, Spielman RS, Dunaif A. Thirty-seven candidate genes for polycystic ovary syndrome: strongest evidence for linkage is with follistatin. Proc Natl Acad Sci USA. 1999;96:8573–8578. doi: 10.1073/pnas.96.15.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanek M, Woodroffe A, Ewens KG, Diamanti-Kandarakis E, Legro RS, Strauss JF, III, Dunaif A, Spielman RS. Candidate gene region for polycystic ovary syndrome on chromosome 19p13.2. J Clin Endocrinol Metab. 2005;90:6623–6629. doi: 10.1210/jc.2005-0622. [DOI] [PubMed] [Google Scholar]
- van Asselt KM, Kok HS, Putter H, Wijmenga C, Peeters PH, van der Schouw YT, Grobbee DE, te Velde ER, Mosselman S, Pearson PL. Linkage analysis of extremely discordant and concordant sibling pairs identifies quantitative trait loci influencing variation in human menopausal age. Am J Hum Genet. 2004;74:444–453. doi: 10.1086/382136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Gaast MH, Eijkemans MJ, van der Net JB, de Boer EJ, Burger CW, van Leeuwen FE, Fauser BC, Macklon NS. Optimum number of oocytes for a successful first IVF treatment cycle. Reprod Biomed Online. 2006;13:476–480. doi: 10.1016/s1472-6483(10)60633-5. [DOI] [PubMed] [Google Scholar]
- van Dooren MF, Bertoli-Avellab AM, Oldenburg RA. Premature ovarian failure and gene polymorphisms. Curr Opin Obstet Gynecol. 2009;21:313–317. doi: 10.1097/gco.0b013e32832e0813. [DOI] [PubMed] [Google Scholar]
- van Montfoort AP, Geraedts JP, Dumoulin JC, Stassen AP, Evers JL, Ayoubi TA. Differential gene expression in cumulus cells as a prognostic indicator of embryo viability: a microarray analysis. Mol Hum Reprod. 2008;14:157–168. doi: 10.1093/molehr/gam088. [DOI] [PubMed] [Google Scholar]
- Vanderhyden BC, Armstrong DT. Role of cumulus cells and serum on the in vitro maturation, fertilization, and subsequent development of rat oocytes. Biol Reprod. 1989;40:720–728. doi: 10.1095/biolreprod40.4.720. [DOI] [PubMed] [Google Scholar]
- Varani S, Elvin JA, Yan C, DeMayo J, DeMayo FJ, Horton HF, Byrne MC, Matzuk MM. Knockout of pentraxin 3, a downstream target of growth differentiation factor-9, causes female subfertility. Mol Endocrinol. 2002;16:1154–1167. doi: 10.1210/mend.16.6.0859. [DOI] [PubMed] [Google Scholar]
- Verberg MF, Macklon NS, Nargund G, Frydman R, Devroey P, Broekmans FJ, Fauser BC. Mild ovarian stimulation for IVF. Hum Reprod Update. 2009;15:13–29. doi: 10.1093/humupd/dmn056. [DOI] [PubMed] [Google Scholar]
- Vercellini P, Somigliana E, Vigano P, Abbiati A, Barbara G, Crosignani PG. Endometriosis: current therapies and new pharmacological developments. Drugs. 2009;69:649–675. doi: 10.2165/00003495-200969060-00002. [DOI] [PubMed] [Google Scholar]
- Vinci G, Christin-Maitre S, Pasquier M, Bouchard P, Fellous M, Veitia RA. FOXO3a variants in patients with premature ovarian failure. Clin Endocrinol (Oxf) 2008;68:495–497. doi: 10.1111/j.1365-2265.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- Vink JM, Sadrzadeh S, Lambalk CB, Boomsma DI. Heritability of polycystic ovary syndrome in a Dutch twin-family study. J Clin Endocrinol Metab. 2006;91:2100–2104. doi: 10.1210/jc.2005-1494. [DOI] [PubMed] [Google Scholar]
- Voorhuis M, Onland-Moret NC, van der Schouw YT, Fauser BC, Broekmans FJ. Human studies on genetics of the age at natural menopause: a systematic review. Hum Reprod Update. 2010;16:364–377. doi: 10.1093/humupd/dmp055. [DOI] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell. 2004;6:133–144. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- Wathlet S, Adriaenssens T, Segers I, Verheyen G, Van de Velde H, Coucke W, Ron El R, Devroey P, Smitz J. Cumulus cell gene expression predicts better cleavage-stage embryo or blastocyst development and pregnancy for ICSI patients. Hum Reprod. 2011;26:1035–1051. doi: 10.1093/humrep/der036. [DOI] [PubMed] [Google Scholar]
- Watkins WJ, Umbers AJ, Woad KJ, Harris SE, Winship IM, Gersak K, Shelling AN. Mutational screening of FOXO3A and FOXO1A in women with premature ovarian failure. Fertil Steril. 2006;86:1518–1521. doi: 10.1016/j.fertnstert.2006.03.054. [DOI] [PubMed] [Google Scholar]
- Weck J, Mayo KE. Switching of NR5A proteins associated with the inhibin alpha-subunit gene promoter after activation of the gene in granulosa cells. Mol Endocrinol. 2006;20:1090–1103. doi: 10.1210/me.2005-0199. [DOI] [PubMed] [Google Scholar]
- Wehr E, Schweighofer N, Moller R, Giuliani A, Pieber TR, Obermayer-Pietsch B. Association of FTO gene with hyperandrogenemia and metabolic parameters in women with polycystic ovary syndrome. Metabolism. 2010;59:575–580. doi: 10.1016/j.metabol.2009.08.023. [DOI] [PubMed] [Google Scholar]
- Wells D, Fragouli E, Bianchi V, Borini A, Patrizio P. Identification of novel non-invasive biomarkers of oocyte aneuploidy. Fertil Steril. 2008;90:S35. [Google Scholar]
- Welt CK, Smith PC, Taylor AE. Evidence of early ovarian aging in fragile X premutation carriers. J Clin Endocrinol Metab. 2004;89:4569–4574. doi: 10.1210/jc.2004-0347. [DOI] [PubMed] [Google Scholar]
- Wittenberger MD, Hagerman RJ, Sherman SL, Conkie-Rosell A, Welt CK, Rebar RW, Corrigan EC, Simpson JL, Nelson LM. The FMR1 premutation and reproduction. Fertil Steril. 2007;87:456–465. doi: 10.1016/j.fertnstert.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Wu Y, Kajdacsy-Balla A, Strawn E, Basir Z, Halverson G, Jailwala P, Wang Y, Wang X, Ghosh S, Guo SW. Transcriptional characterizations of differences between eutopic and ectopic endometrium. Endocrinology. 2006;147:232–246. doi: 10.1210/en.2005-0426. [DOI] [PubMed] [Google Scholar]
- Xita N, Tsatsoulis A. Review: fetal programming of polycystic ovary syndrome by androgen excess: evidence from experimental, clinical, and genetic association studies. J Clin Endocrinol Metab. 2006;91:1660–1666. doi: 10.1210/jc.2005-2757. [DOI] [PubMed] [Google Scholar]
- Xue Q, Lin Z, Yin P, Milad MP, Cheng YH, Confino E, Reierstad S, Bulun SE. Transcriptional activation of steroidogenic factor-1 by hypomethylation of the 5′ CpG island in endometriosis. J Clin Endocrinol Metab. 2007;92:3261–3267. doi: 10.1210/jc.2007-0494. [DOI] [PubMed] [Google Scholar]
- Yan C, Wang P, DeMayo J, DeMayo FJ, Elvin JA, Carino C, Prasad SV, Skinner SS, Dunbar BS, Dube JL, et al. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol. 2001;15:854–866. doi: 10.1210/mend.15.6.0662. [DOI] [PubMed] [Google Scholar]
- Zhang X, Jafari N, Barnes RB, Confino E, Milad M, Kazer RR. Studies of gene expression in human cumulus cells indicate pentraxin 3 as a possible marker for oocyte quality. Fertil Steril. 2005;83:1169–1179. doi: 10.1016/j.fertnstert.2004.11.030. [DOI] [PubMed] [Google Scholar]
- Zhao H, Chen ZJ, Qin Y, Shi Y, Wang S, Choi Y, Simpson JL, Rajkovic A. Transcription factor FIGLA is mutated in patients with premature ovarian failure. Am J Hum Genet. 2008;82:1342–1348. doi: 10.1016/j.ajhg.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZZ, Croft L, Nyholt DR, Chapman B, Treloar SA, Hull ML, Montgomery GW. Evaluation of polymorphisms in predicted target sites for micro RNAs differentially expressed in endometriosis. Mol Hum Reprod. 2011;17:92–103. doi: 10.1093/molehr/gaq084. [DOI] [PMC free article] [PubMed] [Google Scholar]