Abstract
We demonstrate the versatility of a collection of insertions of the transposon Minos mediated integration cassette (MiMIC), in Drosophila melanogaster. MiMIC contains a gene-trap cassette and the yellow+ marker flanked by two inverted bacteriophage ΦC31 attP sites. MiMIC integrates almost at random in the genome to create sites for DNA manipulation. The attP sites allow the replacement of the intervening sequence of the transposon with any other sequence through recombinase mediated cassette exchange (RMCE). We can revert insertions that function as gene traps and cause mutant phenotypes to wild type by RMCE and modify insertions to control GAL4 or QF overexpression systems or perform lineage analysis using the Flp system. Insertions within coding introns can be exchanged with protein-tag cassettes to create fusion proteins to follow protein expression and perform biochemical experiments. The applications of MiMIC vastly extend the Drosophila melanogaster toolkit.
INTRODUCTION
Different types of transposons have been used to manipulate the Drosophila genome and to assess the function of genes, but each is designed for a specific purpose, and none is truly multi-facetted.. The most commonly used transposons are the P-element, piggyBac and Minos1–3. P-elements mobilize efficiently and often excise imprecisely. However, they exhibit a strong insertional bias for the 5' ends of genes4, 5. piggyBac elements show much less bias5, but mobilize less efficiently than P-elements and only excise precisely6. Minos elements have very little insertional bias5, 7, 8, transpose stably and efficiently in numerous organisms9, and excise imprecisely at a useful frequency6, 8.
The most popular application of transposons is to create mutations directly by insertion or by imprecise excision10. Transposons have been engineered to allow controlled misexpression of genes via upstream activating sequence (UAS) sites in the transposon vector4, 11 or to promote activation of reporters like GAL4 or β-galactosidase via nearby enhancers12, 13. Transposons can also function as gene traps if they carry a splice acceptor site (SA) followed by stop codons in all three reading frames and a polyadenylation site so that intronic insertions can interrupt transcription and translation14. Transposons containing a protein trap harbor a SA followed by a coding sequence tag and a splice donor site (SD). When the protein trap is inserted in a coding intron in the appropriate orientation and reading frame, it reveals the protein's expression pattern15, 16. Unfortunately, the frequency of P-element insertions in introns is low4, 5, 17, and only 1/6th of insertions within introns have the appropriate orientation and reading frame to function as protein traps. Hence, only about 2.5% of Drosophila genes have been tagged with a protein trap, even when a piggyBac having a lesser insertional bias was used18, 19. Each different application of transposons requires the generation and maintenance of thousands of single-insertion stocks. The burden of stock keeping has limited the availability of these different tools: less than 5% of the transposon stocks that have been generated in the past 25 years are still available5. For the vast majority of Drosophila genes, only one type of transposon insertion is still available.
Transposons can be engineered to include target sequences recognized by recombinases or integrases3, 20, 21 such as Flp22 and ΦC3123, 24, respectively. These enzymes can replace sequences within transposons via recombinase mediated cassette exchange (RMCE)25, 26. RMCE has been demonstrated in Drosophila with both Flp and ΦC3127, 28. ΦC31 integrase is the preferred enzyme due to its higher efficiency in transgenesis and unidirectional integration23, 24.
Here, we describe a new mutagenesis and genome manipulation system called Minos Mediated Integration Cassette (MiMIC). MiMIC is a Minos transposon that carries a dominant marker and a gene trap cassette flanked by two inverted ΦC31 attP sites. This transposon combines unbiased insertional mutagenesis with the ability to replace the gene trap cassette by RMCE. Using MiMIC insertions, virtually limitless gene modification and genome engineering can be performed. We illustrate the utility of this novel system through gene- and protein-trap experiments, and reversion of lethal phenotypes.
RESULTS
The MiMIC transposon
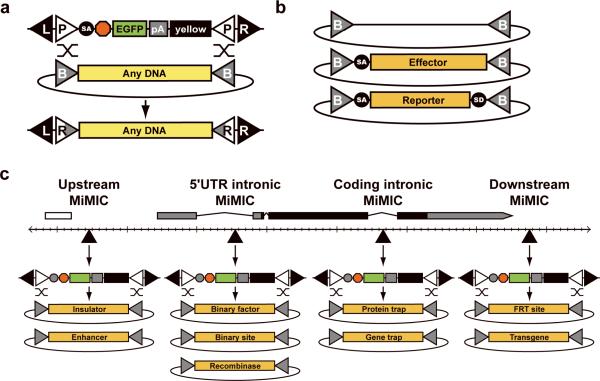
We engineered a new transposon vector, Minos Mediated Integration Cassette (MiMIC). Between the Minos 255-nt inverted repeats (Fig.1a), we included the yellow+ dominant body-color marker for identifying insertions, flanked by two inverted ΦC31 attP sites. We also included a mutagenic gene-trap cassette consisting of a SA followed by stop codons in all three reading frames, the coding sequence of the fluorescent protein EGFP, and an SV40 polyadenylation signal sequence. The sequences between the attP sites are replaceable through RMCE in vivo with any DNA cassette flanked by two inverted ΦC31 attB sites28. This replaces the yellow+ marker, so RMCE events can be identified by loss of body pigmentation. To demonstrate the versatility of MiMIC, we engineered three replacement cassettes: a neutral correction plasmid for removing the mutagenic gene trap in MiMIC, a gene-trap plasmid for introducing protein-coding sequences under control of the host gene promoter, and a protein-trap plasmid for incorporating reporter tags into the coding sequence of the host gene (Fig.1b and Supplementary Table 1).
Figure 1. The MiMIC transposon system.
(a) MiMIC consists of two Minos inverted repeats (L and R), two inverted ΦC31 attP sites (P), a gene trap cassette consisting of a splice acceptor site (SA) followed by stop codons in all three reading frames, and the EGFP coding sequence with a polyadenylation signal (pA), and the yellow+ marker. The sequence between the attP sites can be replaced via RMCE, resulting in two attR sites (R). (b) Three attB plasmids for RMCE: a correction plasmid consisting of a multiple cloning site, a gene-trap plasmid consisting of a SA fused to a downstream effector, and a protein-trap plasmid consisting of a reporter flanked by SA and SD sites. (c) Various MiMIC insertions in a hypothetical gene with regulatory element (white), 5' and 3' untranslated regions (grey), and coding regions (black), that can be used for several applications as indicated.
Since donor cassettes can contain any DNA fragment, MiMIC provides enormous flexibility (Fig.1c). MiMIC insertions near the 5' and 3' ends of genes allow the integration of regulatory elements such as enhancers or insulators to direct or restrict expression, respectively. Such insertions can also be used to integrate an FRT site for creating Flp-based chromosomal rearrangements3. Insertions in 5' UTR introns allow the incorporation of binary expression components, such as GAL4/UAS29 and QF/QUAS30 and recombinases such as Flp22. Insertions in coding introns allow integration of protein tags, including an ever-expanding repertoire of fluorescent markers and conditional protein destruction tags, and other gene-trap mutator cassettes. Finally, any insertion can be used as a generic docking site for integrating transgenes.
A MiMIC insertion screen
We created a collection of 4,464 single-insertion MiMIC lines, determined unique insertion sites for 3,633 insertions (81.3%) by inverse PCR, and associated the mapped insertions with features of annotated genes (Online Methods). Interestingly, 2,293 (63%) of the insertions map within 1,541 annotated genes, and 72% of these intragenic insertions map within introns, including 5' UTR introns and coding introns, both of which are valuable targets for RMCE-based gene manipulation (Supplementary Table 2). We have deposited 1,269 selected insertion lines in the Bloomington Drosophila Stock Center (BDSC) as part of the Drosophila Gene Disruption Project (GDP) collection5. Additional MiMIC lines will be selected for the GDP collection on a regular basis. We aim to deposit over 6,000 lines during the next four years. The GDP maintains an online database (http://flypush.imgen.bcm.tmc.edu/pscreen/) of lines that are available from the BDSC as well as lines that are in the process of being balanced, which may be obtained directly from the GDP.
MiMIC insertion mutants can be reverted by RMCE
The MiMIC transposon contains a gene-trap cassette. Hence, insertions in coding introns should truncate transcripts if MiMIC is inserted in the proper orientation. We selected four MiMIC transposons inserted in the proper orientation to be mutator gene traps: Mi{MIC}RfxMI00053, Mi{MIC}tutlMI00290, Mi{MIC}commMI00380, and Mi{MIC}wndMI00494 inserted in Rfx, tutl, comm and wnd, respectively. All four alleles are associated with a lethal phenotype. In three cases, the insertion failed to complement previously reported mutations of these genes (Supplementary Table 3), indicating that the lethality is indeed associated with the insertion. Mi{MIC}wndMI00494 is the exception. However, complementation data indicate that Mi{MIC}wndMI00494 and the previously reported alleles31 all contain second-site mutations responsible for the lethality, and that none of the transheterozygous wnd allelic combinations cause lethality. For Mi{MIC}RfxMI00053, complementation data indicate the presence of uncoordinated escapers for all allelic combinations, a phenotype that has been previously described for Rfx loss-of-function mutations32.
We then removed the gene-trap cassette from Mi{MIC}RfxMI00053, Mi{MIC}tutlMI00290 and Mi{MIC}commMI00380 by RMCE with a correction plasmid (Fig 1b), screened for loss of yellow+ (Supplementary Fig.1), and established that the lethality was reverted. Hence, intronic MiMIC insertions are mutagenic, and the mutation can be reverted through simple microinjection of a plasmid. This demonstrates that MiMIC is the cause of the lethal phenotypes in these three insertion alleles.
Binary expression and lineage analysis with MiMIC
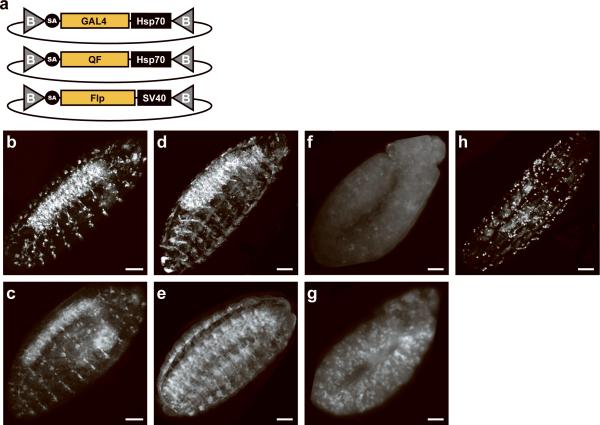
A substantial portion (20.4%) of intragenic MiMICs are localized to 5'UTR introns (Supplementary Table 2). Introduction of exogenous protein-coding sequences into these insertions allows expression under control of the endogenous gene regulatory elements. Hence, we constructed three gene-trap plasmids that encode GAL4, QF, and Flp (Fig.2a). We selected five 5'UTR intronic insertions: Mi{MIC}gogoMI00065, Mi{MIC}TlMI00181, Mi{MIC}capsMI00249, Mi{MIC}MYPT-75DMI00314 and Mi{MIC}BM-40-SPARCMI00329, inserted in gogo, Tl, caps, MYPT-75D, and BM-40-SPARC, respectively. We used RMCE to incorporate each of the three gene-trap cassettes into these insertions.
Figure 2. Binary expression and lineage analysis with MiMIC insertions.
(a) Gene-trap cassettes that incorporate the GAL4 or QF transactivators for binary activation, and the Flp recombinase for fate mapping. Live imaging (b, d) and confocal microscopy analysis using an anti-Cherry antibody (c, e) of the expression domain revealed by GAL 4 inserted in gogo (b,c ) or the caps locus (d,e). The expression domain of MYPT-75D revealed by GAL4 (f) or QF (g) integrated in MYPT-75D. Live imaging of the GAL4 expression pattern revealed by GAL4 inserted in BM-40-SPARC (h). Scale bars are 50 μm.
We tested the GAL4 insertions using a 10×UAS-mCherry cytoplasmic reporter (Online Methods), the QF insertions with a 5×QUAS-mtdTomato-3×HA membrane reporter30, and the Flp insertions with an act>y+>GAL4; UAS-GFP cytoplasmic Flp-out detector (Online Methods). GAL4 incorporated into the gogo insertion revealed expression in the embryonic peripheral and central nervous system, in agreement with RNA in situ hybridization data (Fig.2b,c) (Takashi Suzuki, personal communication). GAL4 inserted into caps faithfully recapitulated the known expression pattern33 (Fig.2d,e). The unknown expression pattern of MYPT-75D is revealed by both GAL4, QF and Flp integrated in Mi{MIC}MYPT-75DMI00314. GAL4 analysis reveals numerous scattered cells labeled during germ band extension (Fig.2f), some of which appear to be muscle precursors. This expression pattern was also revealed by QF analysis (Fig.2g), but resulted in stronger labeling due to the membrane marker driven by QUAS elements, instead of the cytoplasmic marker driven by UAS elements. Surprisingly, Flpout analysis revealed a much smaller subset of labeled cells, suggesting inefficient Flp-out (data not shown). Finally, GAL4 analysis of BM40-SPARC revealed an expression pattern very similar to that revealed by antibodies generated against the endogenous protein, including expression in hemocytes and fat body (Fig.2h)34, 35.
We performed PCR analysis to confirm the molecular nature of the RMCE events. In each case, PCR demonstrated that a productive binary activation or recombination activity is observed only when the cassette is integrated in the appropriate orientation for expression of the reporter (Supplementary Fig.2). Since RMCE can occur in either orientation, we expected a 50% chance of a productive exchange. However, only 25% of the gene-trap RMCE events are in the productive orientation for expression (Supplementary Table 4). Although we do not understand the cause, this suggests selection against productive reporter expression.
Protein trapping with MiMIC insertions
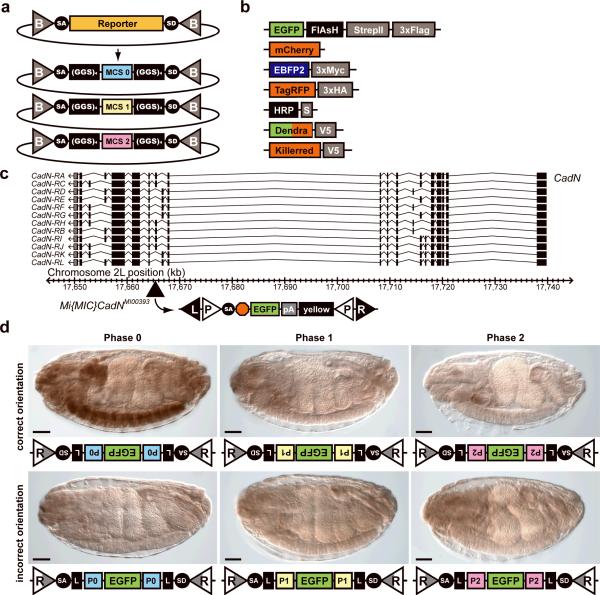
To determine the expression pattern of the protein product of a gene, including its subcellular localization, one can tag with an epitope for which antibodies are available or by live imaging. More than 51% of intragenic MiMIC insertions are in coding introns and permit protein trapping (Supplementary Table 2). We constructed protein-trap cassette plasmids with SA and SD sites flanking synthetic exons encoding a series of protein tags in three versions (Fig.3a,b and Supplementary Fig.3) so as to be able to convert any MiMIC insertion in a coding intron, regardless of its orientation or splicing phase (0,1, or 2), into a protein trap. We flanked the protein tag on both sides with a linker sequence encoding a quadruple GlyGlySer repeat to increase flexibility between the tag and the host protein. We engineered seven multi-tag cassettes for different applications. Most consist of a fluorescent tag and a peptide tag so that if in vivo fluorescence imaging is not possible due to low expression levels, the tag can still be detected by antibodies against the tags.
Figure 3. Protein trapping with MiMIC insertions.
(a) For each protein-trap cassette, three versions were constructed corresponding to the three intron phases (0, 1 and 2). (GGS)4, flexible peptide linker sequence encoding a GlyGlySer quadruplet tandem repeat. (b) Tag and multi-tag cassettes expressing the indicated reporters are shown. (c) A 100 kb genomic region containing CadN is shown. The location of the Mi{MIC}CadNMI00393 insertion in a phase 0 coding intron is indicated. (d) Integration of a phase 0 EGFP-FlAsH-StrepII-3×Flag cassette (EGFP) in the indicated orientation and intron phase. L, (GGS)4 linker; R, (attR); P0, splice phase 0; P1, splice phase 1; P2, splice phase 2. Scale bars are 50 μm.
In a first protein trapping test, we introduced the EGFP-FlAsH-StrepII-3×Flag multi-tag in all three phases into Mi{MIC}CadNMI00393, which is inserted into a phase 0 intron of CadN (Fig.3c) and used PCR to identify integration events for each orientation and phase. As expected, only the phase 0 cassette integrated in the correct orientation recapitulated the expected expression pattern, and none of the other five classes of events resulted in detectable expression (Fig.3d).
Protein expression analysis with multi-tag cassettes
Next, we evaluated expression patterns using seven different tags in six different genes in which MiMIC inserted in a coding intron: Mi{MIC}RfxMI00053, Mi{MIC}tutlMI00290, Mi{MIC}rheaMI00296, Mi{MIC}commMI00380, Mi{MIC}CadNMI00393, and MI{MIC}wndMI00494. These are inserted in Rfx (phase 1), tutl (phase 1), rhea (also known as talin) (phase 0), comm (phase 1), CadN (phase 0), and wnd (phase 2), respectively. We introduced the seven different tag cassettes (Fig.3b) with the proper intron phase (Fig.3a) into each of the six insertions (Table 1). We then performed PCR analysis to determine the orientation of each RMCE event and established that 48% of the integration events are in the desired orientation. This is in agreement with the 50% frequency expected by chance, suggesting that these events are not detrimental to host gene function. Indeed, the lethality associated with each of the original gene-trap insertions was often reverted upon RMCE with protein-trap tags. For Rfx and tutl, the lethality of the MiMIC gene trap was reverted in 69% and 86% of the protein-trap lines, respectively (Table 1). This demonstrates that in most cases protein function has been at least partially restored when the gene trap is removed and replaced by a protein trap. The reverted lines may be partial loss-of-function mutations or full revertants. Note that reversion of lethality did not occur in most RMCE events for the insertions in comm (15%) or wnd (9%).
Table 1. Protein-trapping experiments.
MiMIC insertions in six genes were tagged with different protein-trap cassettes using RMCE. For each insertion,gene, MiMIC line, gene-trap status, associated lethality and intron phase are indicated. For each RMCE experiment, tag component, total lines, number of expressing lines, percentage of expressing lines, and lethality (viable (V), >5% viable homozygotes), lethal (L), not applicable (NA)) are indicated.
| Insertion | RMCE | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Gene | MiMIC | Gene trap | Lethality | Phase | Taga | Total | Expression | % | Lethality |
|
| |||||||||
| Rfx | MI00053 | YES | Lethal | 1 | EGFP | 1 | 0 | 0% | NA |
| mCherry | 2 | 1 | 50% | L | |||||
| EBFP | 2 | 1 | 50% | V | |||||
| 3xHA | 2 | 1 | 50% | V | |||||
| S peptide | 5 | 2 | 40% | 2L | |||||
| Dendra | 6 | 5 | 83% | 5V | |||||
| V5 | 3 | 3 | 100% | 2V/L | |||||
|
| |||||||||
| tutl | MI00290 | YES | Lethal | 1 | EGFP | 4 | 3 | 75% | 3V |
| mCherry | 6 | 2 | 33% | 2V | |||||
| EBFP | 3 | 2 | 67% | 2V | |||||
| 3×HA | 1 | 0 | 0% | NA | |||||
| S peptide | 5 | 3 | 60% | 3V | |||||
| Dendra | 5 | 2 | 40% | 2V | |||||
| V5 | 5 | 2 | 40% | 2L | |||||
|
| |||||||||
| rhea | MI00296 | NO | Viable | 0 | EGFP | 5 | 3 | 60% | V/2L |
| mCherry | 4 | 2 | 50% | V/L | |||||
| EBFP | 6 | 3 | 50% | 2V/L | |||||
| 3×HA | 5 | 2 | 40% | 2V | |||||
| S peptide | 5 | 0 | 0% | NA | |||||
| Dendra | 6 | 4 | 67% | 3V/L | |||||
| V5 | 6 | 4 | 67% | 4V | |||||
|
| |||||||||
| comm | MI00380 | YES | Lethal | 1 | EGFP | 7 | 2 | 29% | V/L |
| mCherry | 3 | 2 | 67% | V/L | |||||
| EBFP | 5 | 2 | 40% | 2L | |||||
| 3×HA | 2 | 1 | 50% | L | |||||
| S peptide | 5 | 1 | 20% | L | |||||
| Dendra | 5 | 1 | 20% | L | |||||
| V5 | 5 | 4 | 80% | 4L | |||||
|
| |||||||||
| CadN | MI00393 | NO | Viable | 0 | EGFP | 5 | 3 | 60% | 3V |
| mCherry | 2 | 1 | 50% | V | |||||
| EBFP | 4 | 2 | 50% | V/L | |||||
| 3×HA | 4 | 1 | 25% | V | |||||
| S peptide | 3 | 1 | 33% | V | |||||
| Dendra | 4 | 2 | 50% | 2V | |||||
| V5 | 3 | 1 | 33% | V | |||||
|
| |||||||||
| wnd | MI00494 | YES | Lethal | 2 | EGFP | 5 | 5 | 100% | 5L |
| mCherry | 3 | 1 | 33% | L | |||||
| EBFP | 3 | 1 | 33% | L | |||||
| 3×HA | 3 | 0 | 0% | NA | |||||
| S peptide | 4 | 3 | 75% | 3L | |||||
| Dendra | 3 | 1 | 33% | V | |||||
| V5 | 1 | 0 | 0% | NA | |||||
|
| |||||||||
| Total | 166 | 80 | 48% | ||||||
Tag components of multi-tags used for expression analysis: EGFP (eGFP-FlAsH-StrepII-3×Flag), mCherry (mCherry), EBFP (eBFP-3×Myc), 3×HA (TagRFP-3×HA), S peptide (HRP-S peptide), Dendra (Dendra-V5), V5 (Killerred-V5).
Failure to revert the lethality of a MiMIC insertion by RMCE with a protein-trap cassette may result from the effect of the tags on protein function. Alternatively, some of the MiMIC-bearing chromosomes may contain second-site lethal mutations as previously observed in P-element stocks exposed to transposase13, or mutations could be induced during the RMCE procedure since ΦC31 integrase has been shown to induce DNA damage and chromosome rearrangements36–38. Both issues can be obviated by removing the second-site mutations by recombination. To test these possibilities, protein-trap alleles generated by RMCE that failed to revert the lethality of the gene-traps in comm (6 lines), wnd (4 lines), rhea (6 lines), and CadN (4 lines) were crossed to previously described alleles and deficiencies uncovering the corresponding loci. All of the protein-trap alleles of all the genes tested complemented the established lethal allele or deletion chromosome, providing evidence that the tagged fusion proteins indeed supply sufficient gene function to revert the lethality.
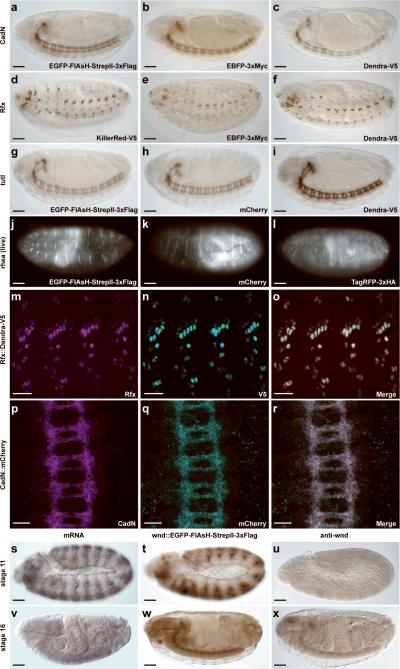
To determine the expression pattern and subcellular localization of the tagged proteins generated by RMCE, we first stained a large sample of tagged proteins using antibodies against GFP, V5, mCherry, and Dendra. We compared the expression of the same protein fused to different tags. Detection of CadN with EGFP-FlAsH-StrepII-3×Flag, EBFP2-3×Myc or Dendra-V5 shows very similar expression patterns (Fig.4a–c). Similarly, different tags integrated into Rfx (Fig.4d–f) and tutl (Fig.4g–i) show highly reproducible expression patterns. In the case of rhea, protein trapping allowed live imaging of three different fluorescent tags (Fig.4j–l). Furthermore, the observed expression patterns faithfully recapitulate the previously described expression patterns of CadN39, Rfx40, tutl41 and rhea42. To determine in more detail whether the fusion protein expression patterns faithfully report the cellular and subcellular localization of the endogenous proteins, we performed co-labeling experiments with antibodies against different tags and the endogenous protein for Rfx (Fig.4m–o) and CadN (Fig.4p–r). These experiments showed fully overlapping expression patterns in transheterozygotes that express both the tagged and the untagged proteins.
Figure 4. Expression analyses of tagged proteins.
(a–i) Detection of different protein-trap alleles of three genes using antibodies against several epitopes and diaminobenzidine/peroxidase (DAB) staining. In each panel, the tag or multi-tag is indicated in the lower right; the portion of the tag that was used as the antigen is italicized. (a–c) Expression of tagged CadN during embryonic stage 15: staining with (a) anti-EGFP, (b) anti-EBFP (c) anti-Dendra. (d-f) Expression of tagged Rfx in stage 15 embryos: staining with (d) anti-V5, (e) anti-EBFP (f) anti-Dendra. (g–i) Expression of tagged Tutl in the ventral nerve cord of stage 15 embryos: staining with (g) anti-EGFP, (h) anti-mCherry, (i) anti-Dendra. (j–l) Fluorescent detection of different protein-trap alleles in live stage 17 embryos expressing tagged Rhea: (j) EGFP (k) mCherry (l) TagRFP. (m–r) Co-localization of protein traps and endogenous proteins in stage 15 embyos for Rfx (m–o) and CadN (p–r). Anti-Rfx staining (m), anti-V5 staining (n), and co-staining (o) in an Rfx∷Dendra-V5 trap. Anti-CadN staining (p), anti-mCherry staining (q), and co-staining (r) in a CadN∷mCherry trap. (s–x) Novel expression detected using protein traps. (s,v) Expression of wnd detected using mRNA in situ hybridization. (t,w) Expression of Wnd detected by anti-EGFP staining of an EGFP-FlAsH-StrepII-3×Flag trap and (u,x) anti-Wnd staining during embryonic stages 11 (s–u) and 16 (v–x). Scale bars are 50 μm (a–l, s–x) and 20 μm (m–r).
In total, we tested 166 independent tagged fusion proteins generated by RMCE for 6 MiMIC insertions and observed that less than 3% (5/166) exhibited a different expression pattern than was anticipated. They were not associated with any particular gene or tag, suggesting that they were due to a faulty RMCE event, and this was confirmed by an aberrant PCR pattern. These data indicate that RMCE-based protein-trapping results in the precise incorporation of tags and will permit the determination of the expression pattern and subcellular distribution of numerous uncharacterized proteins.
Detection of new expression patterns
While analyzing the expression patterns of the different tags integrated into Mi{MIC}wndMI00494, we observed an expression pattern that was much broader and more complex than anticipated (Fig.4s–x and Supplementary Fig.4 online). Antibody staining31 showed weak expression of Wnd in the embryonic nervous system in stage 16 embryos but not in other tissues (Fig.4u,x). However, the Wnd fusion protein generated by RMCE has a very dynamic expression pattern that is complex (Fig.4s,v) and in agreement with the pattern revealed by RNA in situ hybridization (Fig.4t,w). These and many other immunohistochemical staining experiments on the 166 tagged proteins (data not shown) revealed that well characterized antibodies raised against tags that are integrated in fusion proteins are often superior to custom antibodies raised against the endogenous protein.
DISCUSSION
MiMIC permits a wide range of manipulations. Insertions in 5' UTR introns allow the expression of transcription factors such as GAL4 and QF, and recombinases such as Flp to generate gene-specific binary expression and recombination systems, respectively. Moreover, these insertions allow any current or future effector to be placed under the control of the endogenous gene's regulation. Genes that are tagged by insertions in coding introns can be manipulated in numerous ways. One can determine gene expression and subcellular protein distribution using light microscopy (Fig.4) and likely immuno-electron microscopy43. Tags inserted in transcription factors can be used for chromatin immunoprecipitation44. Other applications such as coimmunoprecipitation followed by mass spectrometry, and identification of RNA binding partners can also be performed. A complementary in vivo swapping approach was recently developed for enhancer trapping45.
The widespread adoption of the MiMIC system by the Drosophila research community will depend on substantially increasing the number of insertion lines that are available for public distribution. Currently, 1,269 unique MiMIC insertions are available from the BDSC and about 900 are being balanced and validated by re-sequencing. In the GDP, we plan to generate about 6,000 additional insertions during the next four years. Since Minos integrates almost at random in the genome5, 8, and about 33% of MiMIC insertions are in coding introns (Supplementary Table 2), the manipulations documented here will become feasible for many more Drosophila genes. The ability to assess gene expression patterns and subcellular protein distributions with high resolution (Fig.3 and 4) will vastly expand the number and quality of expression patterns of Drosophila genes.
Finally, both attP sites present in MiMIC can be used as docking sites for integration of gene targeting constructs46 that can be used to engineer the genome within the vicinity of the transposon insertion. A collection of about 6,000 insertions spaced about 40 kb apart5 should allow the manipulation of most genes by engineering and integrating large genomic constructs using the P[acman] system47, 48, recombineering methods3, and RMCE28.
ONLINE METHODS
Molecular biology
Primers were obtained from Operon or Sigma. PCR for cloning was performed with proofreading enzymes Pfu (Stratagene) or iProof (Biorad). Bacterial colony PCR was done with Qiagen HotStarTaq DNA Polymerase (QIAGEN) or Hot MultiTaq™ DNA polymerase (US DNA). PCR purification and gel extraction were performed with the QIAquick PCR Purification and QIAquick Gel Extraction Kits (QIAGEN), respectively. Restriction enzymes and T4 DNA ligase were from NEB. The SURE or SURE2 bacterial strains (Stratagene) were used for bacterial transformation experiments. Bacteria were grown in LB broth containing 1% NaCl for plasmid isolation and ampicilin (USB) at a final concentration of 100 μg/ml. Plasmid purifications were performed using the QIAprep Spin Miniprep Kit (QIAGEN) or the PureLink™ HiPure Plasmid Kit (Invitrogen), as indicated. All cloning experiments were verified by DNA sequencing.
Construction of the MiMIC transposon
pMiLRTetR was used as the Minos transposon backbone (gift of Stefan Oehler and Charalambos Savakis)49. pMiLRTetR was digested with HindIII and PacI and ligated with two annealed oligos, pMiLR-Correction-TOP and pMiLR-Correction-BOTTOM (Supplementary Table 5 for all primer sequences), resulting in the mini Minos plasmid, pMiLR-Correction.
Next, two 100-bp attP sites were obtained by PCR from pTA-attP (gift of Michelle Calos)50, the first one amplified with primers attP1-pMiLR-F and attP1-pMiLR-R, the second one amplified with primers attP2-pMiLR-F and attP2-pMiLR-R. Both attP amplicons, the first cut with HindIII and XhoI, and the second cut with XhoI and SacII, were ligated together into pMiLR-Correction cut with HindIII and SacII, resulting in the mini-Minos-attP plasmid, pMiLR-attP1-2. This plasmid has a stuffer fragment between each attP site and Minos inverted repeat that allows a specific inverse PCR reaction at either end (see below), as well as a multiple cloning site between the two inverted attP sites.
Subsequently, the intronless dominant body color marker yellow+ (y[+mDint2]) obtained from EPgy24 was subcloned as a SalI fragment into the MCS of pMiLR-attP1-2, resulting in pMiLR-attP1-2-yellow.
Finally, a gene-trap cassette was constructed, consisting of the Mhc intron 18 SA site51 obtained from pP-GC (gift of Xavier Morin and William Chia)15 and amplified with primers MHC-SA-XmaI-F and MHC-SA-EGFP-R, and an EGFP with SV40 polyadenylation signal obtained from pCA-GAP-Mut4-EGFP (gift of Ami Okada)52 and amplified with primers MHC-SA-EGFP-F and EGFP-SpeI-R. The two fragments were fused together using hybrid PCR53 and subcloned as an XmaI-SpeI fragment into pMiLR-attP1-2-yellow, resulting in the final transposon plasmid pMiLR-attP1-2-yellow-SA-EGFP, abbreviated to MiMIC or Mi{MIC}.
Generation of single-insertion MiMIC lines
Initial MiMIC transposition experiments were performed by co-injection of the MiMIC plasmid purified using the PureLink™ HiPure Plasmid Kit (Invitrogen) and mRNA encoding the Minos transposase generated from pBlueSKMimRNA (gift of Anastasios Pavlopoulos)54 by in vitro transcription after NotI linearization using the mMessage mMachine T7 Kit (Ambion) as described47, 54. Co-injection titration experiments were performed with plasmid/transposase concentrations (ng/μl) of 300/300, 300/100, 100/300 and 100/100. Injections were performed into a y* w* strain. Transgenic lines were mapped by genetic crosses using y* w*; T(2;3)apXa/ SM5;TM3, Sb and balanced with P{w[+mW.Scer\FRT.hs]=RS3}l(1)CB-6411-31, w1118/FM7h (BL6878) for the X chromosome, y1 w67c23; In(2LR)Gla, wgGla-1/SM6a (BL6600) for chromosome 2, y* w*; D/TM6b, Hu, Tb (Bellen lab) for chromosome 3 or y*; ry506/+; CiD/eyD for chromosome 4 (Bellen lab).
Subsequently, five MiMIC insertions on the X chromosome (Mi{MIC}MI00019, Mi{MIC}MI00030, Mi{MIC}MI00039, Mi{MIC}MI00040 and Mi{MIC}MI00069), obtained form the co-injection experiments, were remobilized to the autosomes using a transgenic source of transposase under the control of a heat-shock promoter (P{hsILMiT}, FlyBase ID FBtp0021508) inserted on a second chromosome balancer (P{hsILMiT}2.4; FlyBase ID FBti0073645) (gift of Charalambos Savakis)8. To allow the identification of new MiMIC remobilization events by screening for y+, this transposase source was moved into a y w background with y1 w*; nub2 b1 nocSco pr1cn1/CyO (BL3628) resulting in y w; nub2 b1 nocSco pr1 cn1/SM6a, P{hsILMiT}2.4. Heat shocks were performed for 2 hours in a 37° water bath on 5 consecutive days. Transposition efficiencies were initially as high as 43% for Mi{MIC}MI00040, but eventually dropped to about 7% for all donor insertions tested, for unknown reasons. Insertions in males were balanced with y* w*; T(2;3)apXa/ SM5;TM3, Sb. Insertions on the 4th chromosome were balanced with y*; ry506/+; ciD/eyD as described above.
During a second phase, three MiMIC insertions on the X chromosome (Mi{MIC}MI00019, Mi{MIC}MI00030, and Mi{MIC}MI00040) were remobilized to the TM3, Sb balancer chromosome, resulting in two independent 3rd chromosome transposon-donor balancer chromosomes, which we named MI00000A and MI00000B. These donor chromosomes were used to remobilize MiMIC to the X chromosome and autosomes using the y w; nocSco nub2 b1 nocSco pr1 cn1/SM6a, P{hsILMiT}2.4 stock described above. In addition Mi{MIC}MI00827 located on the X chromosome was mobilized to the autosomes.
Mapping and annotation of MiMIC insertion sites
We generated 4,464 strains containing insertions of the MiMIC transposon (nearly always single insertions) and mapped 3,633 insertions to a unique site in the reference genome sequence (Release 5; http://www.fruitfly.org). Sequences flanking MiMIC insertions were determined by inverse PCR and DNA sequencing, and mapped by alignment to the genome sequence, as described for MB lines in5 with one modification: genomic DNA was digested with Sau3A I or MboI (which are isoschizomers). A detailed protocol is available on the GDP website (http://flypush.imgen.bcm.tmc.edu/pscreen/). Lines that were selected for the GDP collection were balanced, and their insertion sites verified by re-sequencing before delivery to the BDSC.
We associated MiMIC insertions with annotated genes and gene features (FlyBase r5.32). Annotated features of one gene transcript often overlap those of another transcript, so we adopted a progressive strategy for associating MiMIC insertions with gene features, such that each insertion was assigned to only one feature. We first associated insertions with coding exons, followed by 5' UTR exons, 3' UTR exons, coding introns, 5' UTR introns, and 3' UTR introns. The remaining insertions were classified as intergenic. Note that this is a conservative approach that underestimates the number of insertions associated with lower ranked gene features.
Construction of correction cassettes for RMCE
Two 100-bp fragments containing attB sites were obtained by PCR from pTA-attB (gift of Michelle Calos)50, the first amplified with primers attB1-pBS-F and attB1-pBS-R, and the second amplified with primers attB2-pBS-F and attB2-pBS-R. The attB amplicons, the first cut with SacI and EcoRI, and the second cut with EcoRI and KpnI, were ligated together into pBS-KS and pBS-SK cut with SacI and KpnI, resulting in the mini-attB-RMCE plasmids, pBS-KS-attB1-2 and pBS-SK-attB1-2. These plasmids have two inverted attB sites flanking a multiple cloning site (XbaI, SpeI, PstI, EcoRI, XhoI, BamHI and HindIII).
Construction of gene-trap cassettes for RMCE
A gene-trap cassette incorporating the Mhc intron 18 SA site51 was PCR amplified from pP-GC (gift of Xavier Morin and William Chia)15 with primers SA-XbaI-F and SA-PstI-R. The resulting PCR fragment was cut with XbaI and PstI and subcloned into pBS-KS-attB1-2, cut with XbaI and PstI, resulting in the mini gene-trap plasmid, pBS-KS-attB1-2-GT-SA. The SA is followed by a multiple cloning site (PstI, EcoRI, XhoI, BamHI and HindIII).
A mutagenic GAL4 gene-trap cassette, encompassing the GAL4 coding sequence and Hsp70 polyadenylation signal, was obtained from plasmid pChs-GAL4 (Drosophila Genomics Resource Center)55, and PCR amplified with primers GAL4-Hsp70-EcoRI-F and GAL4-Hsp70-BamHI-R. A mutagenic QF gene-trap cassette, encompassing the QF coding sequence and Hsp70 polyadenylation signal, was obtained from plasmid pattB-QF-Hsp70 (Addgene)30, and PCR amplified with primers QF-SV40-EcoRI-F and QF-SV40-BamHI-R. A mutagenic Flp fate mapping gene-trap cassette, encompassing the FLPo56 coding sequence and SV40 polyadenylation signal, was obtained from plasmid pQUAS-DSCP-Flpo (Addgene)30, and PCR amplified with primers Flpo-SV40-EcoRI-F and Flpo-SV40-BamHI-R. The resulting PCR fragments were cut with EcoRI and BamHI and subcloned into pBS-KS-attB1-2-GT-SA, cut with EcoRI and BamHI, resulting in the plasmids pBS-KS-attB1-2-GT-SA-GAL4-Hsp70, pBS-KS-attB1-2-GT-SA-Flp-SV40, and pBS-KS-attB1-2-GT-SA-QF-Hsp70 respectively.
Construction of protein-trap cassettes for RMCE
Protein-trap cassettes were constructed for the three intron phases (0, 1, 2). The Mhc intron 18 SA and intron 17 SD sites51 were obtained from pP-GC (gift of Xavier Morin and William Chia)15. The protein trap with splice phase 0 was generated from two PCR fragments, the SA site amplified with primers SA-XbaI-F and SA-SD-Phase-0-R, and the SD site amplified with primers SA-SD-Phase-0-F and SD-HindIII-R. The protein trap with splice phase 1 was generated from two PCR fragments, the SA site amplified with primers SA-XbaI-F and SA-SD-Phase-1-R, and the SD site amplified with primers SA-SD-Phase-1-F and SD-HindIII-R. The protein trap with splice phase 2 was generated from two PCR fragments, the SA site amplified with primers SA-XbaI-F and SA-SD-Phase-2-R, and the SD site amplified with primers SA-SD-Phase-2-F and SD-HindIII-R. For each intron phase construct, the SA PCR fragment was cut with XbaI and BamHI, the SD PCR fragment was cut with BamHI and HindIII, and the digested fragments were subcloned in a three-way ligation into pBS-KS-attB1-2, cut with XbaI and HindIII, resulting in pBS-KS-attB1-2-PT-SA-SD-0, pBS-KS-attB1-2-PT-SA-SD-1 and pBS-KS-attB1-2-PT-SA-SD-2. These plasmids contain the SA site, a phase linker for phase 0, 1 or 2, and the SD site, between two inverted attB sites. The phase linker consists of a BamHI site used to sublone protein-trap tags (see below) between two (GlyGlySer)4 peptide-encoding linkers that may provide flexibility between the protein trap tag and the endogenous protein sequences (see below) (Supplementary Figure 3).
The fluorescent protein tag mCherry (gift of Roger Tsien)57 was used without codon optimization. The following protein and peptide tags were generated by gene synthesis by GENEART (http://www.geneart.com/) with codon usage biased toward Drosophila melanogaster: superfolder GFP58, enhanced blue fluorescent protein 259, TagRFP-T60, HRP61, Dendra262, 63, KillerRed64, optimized FlAsH peptide65, StrepII peptide66, 3×Flag peptide67, 3×Myc peptide68, 3×HA peptide69 and the TEV protease site70. The following peptide tags were generated by PCR and primer addition with codon usage biased towards Drosophila melanogaster based on the Codon Usage Database (http://www.kazusa.or.jp/codon/): S peptide71 and V5 peptide72. The following mult-itags were generated through hybrid PCR53 or PCR and primer addition, and subcloned in customized vector backbones (KJTV, unpublished): EGFP-FlAsH-StrepII-TEV-3×Flag, EBFP2-3×Myc, TagRFP-T-3×HA, HRP-S, Dendra-V5, KillerRed-V5.
The tags were then amplified by PCR as BamHI-Insert-GGC-BamHI fragments, to correct for cloning and reconstitution of the (GlyGlySer)4 linker (Supplementary Figure 3), and subcloned into the three-phase protein-trap plasmids described above. PCR amplification of EGFP-FlAsH-StrepII-TEV-3×Flag was performed with primers EGFPmultiFINAL-F and EGFPmultiFINAL-R, resulting in pBS-KS-attB1-2-PT-SA-SD-0-EGFP-FlAsH-StrepII-TEV-3×Flag, pBS-KS-attB1-2-PT-SA-SD-1-EGFP-FlAsH-StrepIITEV-3×Flag and pBS-KS-attB1-2-PT-SA-SD-2-EGFP-FlAsH-StrepII-TEV-3×Flag. PCR amplification of mCherry was performed with primers Cherry-F and Cherry-R, resulting in pBS-KS-attB1-2-PT-SA-SD-0-mCherry, pBS-KS-attB1-2-PT-SA-SD-1-mCherry and pBS-KS-attB1-2-PT-SA-SD-2-mCherry. PCR amplification of EBFP2-3×Myc was performed with primers EBFP2-Myc-F and EBFP2-Myc-R resulting in pBS-KS-attB1-2-PT-SA-SD-0-EGFP-EBFP2-3×Myc, pBS-KS-attB1-2-PT-SA-SD-1-EBFP2-3×Myc and pBS-KS-attB1-2-PT-SA-SD-2-EBFP2-3×Myc. PCR amplification of TagRFP-T-3×HA was performed with primers TagRFP-HA-F and TagRFP-HA-R, resulting in pBS-KS-attB1-2-PT-SA-SD-0-TagRFP-T-3×HA, pBS-KS-attB1-2-PT-SA-SD-1-TagRFP-T-3×HA and pBS-KS-attB1-2-PT-SA-SD-2-TagRFP-T-3×HA. PCR of HRP-S was performed with primers HRP-S-F, HRP-S-R1 and HRP-S-R2, resulting in pBS-KS-attB1-2-PT-SA-SD-0-HRP-S, pBS-KS-attB1-2-PT-SA-SD-1-HRP-S and pBS-KS-attB1-2-PT-SA-SD-2-HRPS. PCR of Dendra-V5 was performed with primers Dendra-V5-F, Dendra-V5-R1 and Dendra-V5-R2, resulting in pBS-KS-attB1-2-PT-SA-SD-0-Dendra-V5, pBS-KS-attB1-2-PT-SA-SD-1-Dendra-V5 and pBS-KS-attB1-2-PT-SA-SD-2-Dendra-V5. PCR of KillerRed-V5 was performed with primers KillerRed-V5-F, KillerRed-V5-R1 and KillerRed-V5-R2 resulting in pBS-KS-attB1-2-PT-SA-SD-0-KillerRed-V5, pBS-KS-attB1-2-PT-SA-SD-1-KillerRed-V5 and pBS-KS-attB1-2-PT-SA-SD-2-KillerRed-V5.
ΦC31-mediated RMCE
Initial RMCE tests were performed with pBS-KS-attB1-2 to ensure functionality of the plasmid backbone, since this plasmid is the progenitor of all constructs for protein-trap cassettes and other cassettes. pBS-KS-attB1-2 DNA was purified and co-injected with ΦC31 integrase mRNA, obtained from pET11ΦC31pA (a gift from Michelle Calos)23 by in vitro transcription after BamHI linearization using the mMessage mMachine T7 Kit (Ambion) as described23, 47. Microinjections were performed using the RMCE landing site 25C (gift of Jack Bateman and Ting Wu)28 at a plasmid concentration of 123 ng/μl and an mRNA concentration of 600 ng/μl. A transgenesis efficiency of 17.5% was obtained.
Subsequent injections were performed with transgenic ΦC31 integrase sources driven by vasa promoter elements located on the X chromosome (y1M{vas-int.B}ZH-2A w*) or the 4th chromosome (y1w*; M{vas-int.B}ZH-102D) (gifts of Johannes Bischof, Francois Karch and Konrad Basler)24. Plasmid was generally diluted to a concentration between 30 and 100 ng/μl. Microinjections were performed using the following MiMIC insertion lines: Mi{MIC}tutlMI00290 and Mi{MIC}CadNMI00393 on chromosome 2, and Mi{MIC}RfxMI00053, Mi{MIC}gogoMI00065, Mi{MIC}TlMI00181, Mi{MIC}capsMI00249, Mi{MIC}rheaMI00296, Mi{MIC}MYPT-75DMI00314, Mi{MIC}BM-40-SPARCMI00329, Mi{MIC}commMI00380, and Mi{MIC}wndMI00494 on chromosome 3. When lines contained a gene-trap insertion, they were injected as heterozygous balanced stocks. Microinjections were performed by crossing males from appropriate MiMIC lines to virgin females containing the ΦC31 integrase source. Since RMCE results in a genetically unmarked chromosome due to the removal of the y[+mDint2] marker, fly stocks were generated that contained the ΦC31 integrase source in a balanced background for the 2nd or 3rd chromosome to maintain the MiMIC insertion balanced in G0 animals: y1M{vas-int.B}ZH-2A w*; nocSco/CyO and y1M{vas-int.B}ZH-2A w*; Sb/TM6b, Hu, Tb. Appropriate G0 animals were crossed to balancer virgins: y1w67c23; In(2LR)Gla, wgGla-1/SM6a (BL6600) for chromosome 2 RMCE experiments, or y* w*; D/TM6b, Hu, Tb for chromosome 3 RMCE experiments. Transgenic G1 flies were scored for the absence of a yellow+ phenotype (loss of the y[+mDint2] marker) over a balancer or dominantly marked chromosome appropriate for the chromosome, and crossed to balancer virgins: y1w67c23; In(2LR)Gla, wgGla-1/SM6a (BL6600) (chromosome 2), or y* w*; D/TM6b, Hu, Tb (chromosome 3). Balanced transgenic G2 flies were intercrossed to establish stocks. A list with all RMCE efficiencies is available in Supplementary Table 6.
Molecular characterization of integration events
For PCR verification of RMCE integration events, DNA was extracted from 10 to 15 adult flies using the PureLink™ Genomic DNA Mini Kit (Invitrogen). PCR was performed with tag-specific primers and MiMIC specific primers. Tag-specific primers (Tag-F and Tag-R) are mCherry-Seq-F and mCherry-Seq-R for mCherry, EGFPdo-Seq-F and EGFPdo-Seq-R for EGFP, EBFP2do-Seq-F and EBFP2do-Seq-R for EBFP2, TagRFPdo-Seq-F and TagRFPdo-Seq-R for TagRFP, Hrpdo-Seq-F and Hrpdo-Seq-R for HRP, Dendrado-Seq-F and Dendrado-Seq-R for Dendra, Killerreddo-Seq-F and Killerreddo-Seq-R for Killerred, GAL4-1R and GAL4-5F for GAL4, FLP0-Seq-R and SV40pA-Long-F for Flpo, and QF-Seq-R1 and Hsp70-pA-Alt-F for QF. MiMIC specific primers are Orientation-MiL-F and Orientation-MiL-R. PCR reaction conditions were: 1 μl DNA, 1 μl primer 1, 1 μl primer 2, 2 μl 10× Buffer, 0.16 μl dNTPs (25 mM each), 0.08 μl Qiagen HotStarTaq DNA Polymerase (QIAGEN), 14.76 μl milliQ water. PCR cycling conditions in PTC-225 or DNA Engine (MJ Research) were: denaturation at 94° for 10 minutes, 40 cycles at 94° for 30 seconds, 60° for 30 seconds and 72° for 60 seconds, and post-amplification extension at 72° for 10 minutes.
For each RMCE event, 4 PCR reactions were performed: a first PCR reaction with primers Orientation-MiL-F and Tag-R, a second PCR reaction with primers Orientation-MiL-F and Tag-F, a third PCR reaction with primers Orientation-MiL-R and Tag-R, and a fourth PCR reaction with primers Orientation-MiL-R and Tag-F. Since the transposon integrates one or two orientations relative to the gene, only one in two RMCE events is productive with respect to creating a gene trap or protein trap, which is reflected in a positive PCR for reactions 1 and 4, or 2 and 3. A “1/4” PCR pattern is always desired for a productive RMCE event (for example a gene or protein trap), when the gene/transposon configuration is 1/1 or −1/−1. Conversely, a “2/3” PCR pattern is diagnostic of a productive RMCE event, when the gene/transposon configuration is 1/−1 or −1/1. The reverse holds for unproductive RMCE events (Supplementary Figure 2).
Genetic complementation testing
Genetic complementation tests were performed between lethal MiMIC insertion lines, or lethal RMCE derivatives of both lethal and viable MiMIC lines, and previously described mutant alleles. Mi{MIC}RfxMI00053 was tested with Rfx49 and Rfx253 (gifts of Anne Laurençon and Benedicte Durand)32, and Df(3R)Exel6157 (BL7636)73. Mi{MIC}tutlMI00290 was tested with P{ry+t7.2=PZ}tutl01085 (Bloomington stock number BL10979), tutl4 (gift of Kendal Broadie)41, tutl23 and tutlGAL4 (gifts of Yong Rao)74, tutlex383 (gift of Bader Al-Anzi)75 and Df(2L)ed-dp (BL702). Mi{MIC}rheaMI00296 was tested with rhea1 (BL2296) and Df(3L)W10 (BL2608). Mi{MIC}commMI00380 was tested with commA490 and commΔe39 (gifts of Guy Tear)76, and P{w+mCy+mDint2=EPgy2}commEY10154 (BL17644), Df(3L)BK10 (BL2992) and Df(3L)fz-M21 (BL5461). Mi{MIC}CadNMI00393 was tested with CadNM19FRT40A and CadNΔ18A FRT40A (gifts of Larry Zipursky)39, 77, and Df(2L)Exel7069 (BL7837). Mi{MIC}wndMI00494 was tested with wnd1, wnd2 and wnd3 (gifts of Aaron DiAntonio)31, and Df(3L)XS705 (BL5584) and Df(3L)Exel9007 (BL7942). Complementation tests for lethal protein-trap events were performed similarly.
GAL4/UAS, QF/QUAS and Flp experiments
RMCE experiments using the binary expression factors GAL4 and QF, and the Flp recombinase system were tested as follow: GAL4 swaps were crossed to y w; VK19∷10×UAS-mCherry-SV40 which provides strong mCherry overexpression driven by 10×UAS in a customized P[acman] construct (KJTV, unpublished), and were analyzed as described below. QF swaps were crossed to y1w1118; P{w+mC=QUAS-mtdTomato-3×HA}26 (BL30005)30 and analyzed as described below. Flp swaps were crossed to a customized actin-GAL4-Flp-out line driving UAS-EGFP (H.J.B., unpublished) and analyzed as described below.
Expression analysis
The following antibodies were used for expression analysis:, mouse anti-CadN (DN-Ex#8) at 1:20039 Developmental Studies Hybridoma Bank), rabbit anti-RFX at 1:5000 (a gift from Anne Laurençon and Benedicte Durand)40, rabbit anti-Wnd at 1:500 (a gift from Aaron DiAntonio)31, rabbit anti-GFP at 1:250 (Invitrogen), mouse anti-DsRed at 1:250 (Clontech), rabbit anti-TagRFP at 1:500 (Evrogen), rabbit anti-Dendra2 at 1:5000 (Evrogen), rabbit anti-Killerred at 1:1000 (Evrogen), mouse anti-Flag at 1:250 (Sigma-Aldrich), mouse anti-StrepII at 1:200 (Thermo Scientific), mouse anti-S at 1:100 (Thermo Scientific), mouse anti-V5 at 1:2000 (Invitrogen), mouse anti-c-Myc at 1:250 (Abcam) and mouse anti-HA at 1:200 (Covance).
Drosophila embryos (0 to 24 hours) were collected on grape-agar plates and were subsequently fixed for 20 minutes in a 1:1 mixture of 0.38% formaldehyde in PBS and heptane. The fixative was then removed and methanol added. After vigorously shaking, the heptane-methanol mixture was replaced by methanol, whereupon methanol was replaced by ethanol. Upon rehydration in PBS/0.2% Triton, embryos were blocked for 1 hour in PBS, 10% normal goat serum and incubated overnight with primary antibodies. Fluorescently labeled or HRP-conjugated secondary antibodies were obtained from Jackson ImmunoResearch and were used at a 1:250 dilution.
mRNA in situ hybridization
A 1 kb PCR fragment was obtained with primers Wnd-F and Wnd-R from the Wnd cDNA clone LD1485678 and subcloned into the pGemTeasy vector (Promega). In vitro transcription was performed according to standard procedures, using the DIG RNA labeling kit (Roche). Fixation and in situ hybridization were carried out following the Krause protocol79. The digoxigenin labeled RNA probes were detected by alkaline phosphatase reaction, using the Lehmann protocol80.
Supplementary Material
ACKNOWLEDGEMENTS
We thank B. Al-Anzi (Caltech), Konrad Basler (University of Zurich), J. Bateman (Bowdoin College), J. Bischof (University of Zurich), K. Broadie (Vanderbilt University), M. Calos (Stanford University), W. Chia (National University of Singapore), A. DiAntonio (Washington University), B. Durand (University of Lyon), F. Karch (University of Geneva), A. Laurençon (University of Lyon), L. Luo (Stanford University), X. Morin (Institute of Developmental Biology of Marseille), A. Nose (University of Tokyo), S. Oehler (University of Crete), A. Okada (Stanford University), A. Pavlopoulos (University of Cambridge), C. Potter (Johns Hopkins University), Y. Rao (McGill University), M. Ringuette (University of Toronto), C. Savakis (Biomedical Sciences Research Center Alexander Fleming), J. Shahab (University of Toronto), T. Suzuki (Max Planck Institute of Neurobiology), C. Tan (University of Missouri), G. Tear (King's College London), R. Tsien (University of California San Diego), T. Wu (Harvard University), L. Zipursky (University of California Los Angeles), the Bloomington Drosophila Stock Center (Indiana University), the Drosophila Genomics Resource Center (Indiana University), Addgene, and the Developmental Studies Hybridoma Bank for flies, plasmids, antibodies and communications. We thank S. Park and K. Wan at LBNL for assistance in mapping MiMIC insertions. We especially thank D. Bei, Y. Fang, J. Li, Z. Wang, X. Zheng and J. Yue for generating fly stocks. We thank T. Suzuki (Max Planck Institute of Neurobiology) for communication of unpublished results. This work was funded by NIH grant 2R01 GM067858 to A.C.S., R.A.H. and H.J.B. A.C.S. and H.J.B. are Investigators of the Howard Hughes Medical Institute. Plasmids are available through the Drosophila Genomics Resource Center (https://dgrc.cgb.indiana.edu/vectors/). Fly strains are available through the Bloomington Drosophila Stock Center.
Footnotes
ACCESSION NUMBERS GenBank: GU370067 (MiMIC vector), XXX, (pBS-KS-attB1-2), XXX (pBS-KS-attB1-2-GT-SA), XXX (pBS-KS-attB1-2-PT-SA-SD-0), XXX (pBS-KS-attB1-2-PT-SA-SD-1), XXX (pBS-KS-attB1-2-PT-SA-SD-2). Genomic sequences flanking MiMIC insertion sites in the 1,269 insertion lines selected for the GDP permanent collection have been submitted to GenBank, and supporting data have been deposited in FlyBase (http://flybase.org/).
REFERENCES
- 1.Ryder E, Russell S. Transposable elements as tools for genomics and genetics in Drosophila. Brief. Funct. Genomic. Proteomic. 2003;2:57–71. doi: 10.1093/bfgp/2.1.57. [DOI] [PubMed] [Google Scholar]
- 2.Venken KJ, Bellen HJ. Emerging technologies for gene manipulation in Drosophila melanogaster. Nat. Rev. Genet. 2005;6:167–178. doi: 10.1038/nrg1553. [DOI] [PubMed] [Google Scholar]
- 3.Venken KJ, Bellen HJ. Transgenesis upgrades for Drosophila melanogaster. Development. 2007;134:3571–3584. doi: 10.1242/dev.005686. [DOI] [PubMed] [Google Scholar]
- 4.Bellen HJ, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellen HJ, et al. The Drosophila Gene Disruption Project: Progress Using Transposons With Distinctive Site-Specificities. 2011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witsell A, Kane DP, Rubin S, McVey M. Removal of the bloom syndrome DNA helicase extends the utility of imprecise transposon excision for making null mutations in Drosophila. Genetics. 2009;183:1187–1193. doi: 10.1534/genetics.109.108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franz G, Savakis C. Minos, a new transposable element from Drosophila hydei, is a member of the Tc1-like family of transposons. Nucleic Acids Res. 1991;19:6646. doi: 10.1093/nar/19.23.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metaxakis A, Oehler S, Klinakis A, Savakis C. Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics. 2005;171:571–581. doi: 10.1534/genetics.105.041848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavlopoulos A, Oehler S, Kapetanaki MG, Savakis C. The DNA transposon Minos as a tool for transgenesis and functional genomic analysis in vertebrates and invertebrates. Genome Biol. 2007;8(Suppl 1):S2. doi: 10.1186/gb-2007-8-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spradling AC, et al. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rorth P, et al. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- 12.Bier E, et al. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- 13.Bellen HJ, et al. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- 14.Lukacsovich T, et al. Dual-tagging gene trap of novel genes in Drosophila melanogaster. Genetics. 2001;157:727–742. doi: 10.1093/genetics/157.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clyne PJ, Brotman JS, Sweeney ST, Davis G. Green fluorescent protein tagging Drosophila proteins at their native genomic loci with small P elements. Genetics. 2003;165:1433–1441. doi: 10.1093/genetics/165.3.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aleksic J, Lazic R, Muller I, Russell SR, Adryan B. Biases in Drosophila melanogaster protein trap screens. BMC. Genomics. 2009;10:249. doi: 10.1186/1471-2164-10-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quinones-Coello AT, et al. Exploring strategies for protein trapping in Drosophila. Genetics. 2007;175:1089–1104. doi: 10.1534/genetics.106.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buszczak M, et al. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev. Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 21.Wirth D, et al. Road to precision: recombinase-based targeting technologies for genome engineering. Curr. Opin. Biotechnol. 2007;18:411–419. doi: 10.1016/j.copbio.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 23.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage ϕC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific ϕC31 integrases. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlake T, Bode J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry. 1994;33:12746–12751. doi: 10.1021/bi00209a003. [DOI] [PubMed] [Google Scholar]
- 26.Baer A, Bode J. Coping with kinetic and thermodynamic barriers: RMCE, an efficient strategy for the targeted integration of transgenes. Curr. Opin. Biotechnol. 2001;12:473–480. doi: 10.1016/s0958-1669(00)00248-2. [DOI] [PubMed] [Google Scholar]
- 27.Horn C, Handler AM. Site-specific genomic targeting in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12483–12488. doi: 10.1073/pnas.0504305102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bateman JR, Lee AM, Wu CT. Site-Specific Transformation of Drosophila via ϕC31 Integrase-Mediated Cassette Exchange. Genetics. 2006;173:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 30.Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins CA, Wairkar YP, Johnson SL, Diantonio A. Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron. 2006;51:57–69. doi: 10.1016/j.neuron.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 32.Dubruille R, et al. Drosophila regulatory factor X is necessary for ciliated sensory neuron differentiation. Development. 2002;129:5487–5498. doi: 10.1242/dev.00148. [DOI] [PubMed] [Google Scholar]
- 33.Shishido E, Takeichi M, Nose A. Drosophila synapse formation: regulation by transmembrane protein with Leu-rich repeats, CAPRICIOUS. Science. 1998;280:2118–2121. doi: 10.1126/science.280.5372.2118. [DOI] [PubMed] [Google Scholar]
- 34.Martinek N, Zou R, Berg M, Sodek J, Ringuette M. Evolutionary conservation and association of SPARC with the basal lamina in Drosophila. Dev. Genes Evol. 2002;212:124–133. doi: 10.1007/s00427-002-0220-9. [DOI] [PubMed] [Google Scholar]
- 35.Martinek N, Shahab J, Saathoff M, Ringuette M. Haemocyte-derived SPARC is required for collagen-IV-dependent stability of basal laminae in Drosophila embryos. J. Cell Sci. 2008;121:1671–1680. doi: 10.1242/jcs.021931. [DOI] [PubMed] [Google Scholar]
- 36.Ehrhardt A, Engler JA, Xu H, Cherry AM, Kay MA. Molecular analysis of chromosomal rearrangements in mammalian cells after ϕC31-mediated integration. Hum. Gene Ther. 2006;17:1077–1094. doi: 10.1089/hum.2006.17.1077. [DOI] [PubMed] [Google Scholar]
- 37.Liu J, Jeppesen I, Nielsen K, Jensen TG. Phic31 integrase induces chromosomal aberrations in primary human fibroblasts. Gene Ther. 2006;13:1188–1190. doi: 10.1038/sj.gt.3302789. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Skjorringe T, Gjetting T, Jensen TG. PhiC31 integrase induces a DNA damage response and chromosomal rearrangements in human adult fibroblasts. BMC. Biotechnol. 2009;9:31. doi: 10.1186/1472-6750-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwai Y, et al. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 40.Vandaele C, Coulon-Bublex M, Couble P, Durand B. Drosophila regulatory factor X is an embryonic type I sensory neuron marker also expressed in spermatids and in the brain of Drosophila. Mech. Dev. 2001;103:159–162. doi: 10.1016/s0925-4773(01)00340-9. [DOI] [PubMed] [Google Scholar]
- 41.Bodily KD, Morrison CM, Renden RB, Broadie K. A novel member of the Ig superfamily, turtle, is a CNS-specific protein required for coordinated motor control. J. Neurosci. 2001;21:3113–3125. doi: 10.1523/JNEUROSCI.21-09-03113.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown NH, et al. Talin is essential for integrin function in Drosophila. Dev. Cell. 2002;3:569–579. doi: 10.1016/s1534-5807(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 43.Yao CK, et al. A synaptic vesicle-associated Ca2+ channel promotes endocytosis and couples exocytosis to endocytosis. Cell. 2009;138:947–960. doi: 10.1016/j.cell.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Negre N, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gohl DM, et al. A versatile in vivo system for directed dissection of gene expression patterns. Nat. Methods. 2011;8:231–237. doi: 10.1038/nmeth.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wesolowska N, Rong YS. The past, present and future of gene targeting in Drosophila. Fly. (Austin.) 2010;4:53–59. doi: 10.4161/fly.4.1.10993. [DOI] [PubMed] [Google Scholar]
- 47.Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 48.Venken KJ, et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klinakis AG, Loukeris TG, Pavlopoulos A, Savakis C. Mobility assays confirm the broad host-range activity of the Minos transposable element and validate new transformation tools. Insect Mol. Biol. 2000;9:269–275. doi: 10.1046/j.1365-2583.2000.00183.x. [DOI] [PubMed] [Google Scholar]
- 50.Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hodges D, Bernstein SI. Suboptimal 5' and 3' splice sites regulate alternative splicing of Drosophila melanogaster myosin heavy chain transcripts in vitro. Mech. Dev. 1992;37:127–140. doi: 10.1016/0925-4773(92)90075-u. [DOI] [PubMed] [Google Scholar]
- 52.Okada A, Lansford R, Weimann JM, Fraser SE, McConnell SK. Imaging cells in the developing nervous system with retrovirus expressing modified green fluorescent protein. Exp. Neurol. 1999;156:394–406. doi: 10.1006/exnr.1999.7033. [DOI] [PubMed] [Google Scholar]
- 53.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 54.Pavlopoulos A, Berghammer AJ, Averof M, Klingler M. Efficient transformation of the beetle Tribolium castaneum using the Minos transposable element: quantitative and qualitative analysis of genomic integration events. Genetics. 2004;167:737–746. doi: 10.1534/genetics.103.023085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Apitz H, et al. Identification of regulatory modules mediating specific expression of the roughest gene in Drosophila melanogaster. Dev. Genes Evol. 2004;214:453–459. doi: 10.1007/s00427-004-0423-3. [DOI] [PubMed] [Google Scholar]
- 56.Raymond CS, Soriano P. High-Efficiency FLP and ϕC31 Site-Specific Recombination in Mammalian Cells. PLoS. ONE. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 58.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 59.Ai HW, Shaner NC, Cheng Z, Tsien RY, Campbell RE. Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry. 2007;46:5904–5910. doi: 10.1021/bi700199g. [DOI] [PubMed] [Google Scholar]
- 60.Shaner NC, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith AT, et al. Expression of a synthetic gene for horseradish peroxidase C in Escherichia coli and folding and activation of the recombinant enzyme with Ca2+ and heme. J. Biol. Chem. 1990;265:13335–13343. [PubMed] [Google Scholar]
- 62.Gurskaya NG, et al. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat. Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 63.Chudakov DM, Lukyanov S, Lukyanov KA. Using photoactivatable fluorescent protein Dendra2 to track protein movement. Biotechniques. 2007;42:553, 555, 557. doi: 10.2144/000112470. [DOI] [PubMed] [Google Scholar]
- 64.Bulina ME, et al. A genetically encoded photosensitizer. Nat. Biotechnol. 2006;24:95–99. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- 65.Martin BR, Giepmans BN, Adams SR, Tsien RY. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat. Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 66.Schmidt TG, Koepke J, Frank R, Skerra A. Molecular interaction between the Strep-tag affinity peptide and its cognate target, streptavidin. J. Mol. Biol. 1996;255:753–766. doi: 10.1006/jmbi.1996.0061. [DOI] [PubMed] [Google Scholar]
- 67.Terpe K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2003;60:523–533. doi: 10.1007/s00253-002-1158-6. [DOI] [PubMed] [Google Scholar]
- 68.Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson IA, et al. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 70.Dougherty WG, Cary SM, Parks TD. Molecular genetic analysis of a plant virus polyprotein cleavage site: a model. Virology. 1989;171:356–364. doi: 10.1016/0042-6822(89)90603-x. [DOI] [PubMed] [Google Scholar]
- 71.Hackbarth JS, et al. S-peptide epitope tagging for protein purification, expression monitoring, and localization in mammalian cells. Biotechniques. 2004;37:835–839. [PubMed] [Google Scholar]
- 72.Southern JA, Young DF, Heaney F, Baumgartner WK, Randall RE. Identification of an epitope on the P and V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. J. Gen. Virol. 1991;72:1551–1557. doi: 10.1099/0022-1317-72-7-1551. [DOI] [PubMed] [Google Scholar]
- 73.Parks AL, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 74.Ferguson K, Long H, Cameron S, Chang WT, Rao Y. The conserved Ig superfamily member Turtle mediates axonal tiling in Drosophila. J. Neurosci. 2009;29:14151–14159. doi: 10.1523/JNEUROSCI.2497-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Al-Anzi B, Wyman RJ. The Drosophila immunoglobulin gene turtle encodes guidance molecules involved in axon pathfinding. Neural Dev. 2009;4:31. doi: 10.1186/1749-8104-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tear G, et al. commissureless controls growth cone guidance across the CNS midline in Drosophila and encodes a novel membrane protein. Neuron. 1996;16:501–514. doi: 10.1016/s0896-6273(00)80070-7. [DOI] [PubMed] [Google Scholar]
- 77.Nern A, et al. An isoform-specific allele of Drosophila N-cadherin disrupts a late step of R7 targeting. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12944–12949. doi: 10.1073/pnas.0502888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stapleton M, et al. A Drosophila full-length cDNA resource. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0080. RESEARCH0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lecuyer E, Parthasarathy N, Krause HM. Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. Methods Mol. Biol. 2008;420:289–302. doi: 10.1007/978-1-59745-583-1_18. [DOI] [PubMed] [Google Scholar]
- 80.Lehmann R, Tautz D. In situ hybridization to RNA. Methods Cell Biol. 1994;44:575–598. doi: 10.1016/s0091-679x(08)60933-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.