Abstract
Ebp are endocarditis- and biofilm-associated pili of Enterococcus faecalis that are also important in experimental urinary tract infections (UTIs). Our analyses, using available genomes, found that the ebp locus is unique to enterococci. In E. faecalis, the ebp locus is very highly conserved and only 1/473 E. faecalis isolates tested lacked ebpABC, while only 1.2% had the bee pilus locus. No other pilus-encoding operon was identified in 55 available genomes, indicating that the vast majority of E. faecalis strains (unlike Enterococcus faecium and streptococci) have a single pilus locus. Surface expression studies showed that Ebp pili were produced in vitro by 91/91 brain heart infusion (BHI) plus serum-grown E. faecalis isolates and that strain OG1RF expressed pili at even higher levels in rat endocarditis vegetations. However, Ebp expression was restricted to 30 to 72% of E. faecalis cells, consistent with a bistability mode of expression. We also evaluated E. faecalis interactions with human platelets and found that growth of E. faecalis in BHI plus serum significantly enhanced adherence to human platelets and that sortase deletion mutants (the ΔsrtA, Δbps, and ΔbpsΔsrtA mutants) were markedly defective. Further studies identified that Ebp pili, but not the microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) Ace and Fss2, mediate adherence of E. faecalis to platelets. Taken together, our data show that the immunogenic (in human endocarditis patients) and commonly expressed Ebp pili, which are known to be important for experimental endocarditis, are highly conserved and mediate adherence to platelets, suggesting that Ebp pili may be a reasonable immunotherapeutic target for prevention or possibly treatment of endocarditis caused by this species.
INTRODUCTION
Enterococcus faecalis has been recognized as a causative agent of community-acquired infective endocarditis (IE) since the turn of the last century (41, 42), accounting for ∼5 to 20% of total cases of IE. Enterococci have also been reported as the second most common cause of health care-associated (HA) endocarditis (14, 17). The recent increase in HA enterococcal infections, especially those caused by multidrug-resistant strains, has created therapeutic problems, thus emphasizing the need for alternative strategies for prevention or therapy, such as immunoprophylaxis. Growing evidence from other Gram-positive pathogens suggests that sortase-assembled pilus subunits may serve as candidates for the development of novel immunotherapies. For example, it has been demonstrated in pneumococci, group A streptococci (GAS), and group B streptococci (GBS) that a combination of pilus subunit proteins elicits antibodies that are capable of inducing complement-dependent opsonophagocytic killing and conferring protective immunity (16, 35, 39).
Our previous efforts to identify surface-exposed virulence factors of E. faecalis predicted that 17 LPXTG-type cell wall-associated proteins of E. faecalis strain V583 likely encode microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) or pilus subunits (50, 64). Subsequent studies, including our companion paper (48), demonstrated that four of these proteins (the collagen adhesin Ace and fibrinogen adhesins Fss1, Fss2, and Fss3) and Ebp pili (endocarditis- and biofilm-associated pili) mediate adherence to host extracellular matrix (ECM) proteins (46, 50, 61). Further analyses found that both Ace and Ebp pili (which are assembled from three subunits, EbpA, -B, and -C) are ubiquitous among E. faecalis isolates (47, 50) and are antigenic during human infections, including IE (47, 50), with little or no expression in vitro except under specific growth conditions (e.g., growth medium supplemented with serum), at least by E. faecalis strain OG1RF (44, 50). Disruption of genes encoding either Ace or Ebp pili has resulted in attenuation in animal models of IE (50, 67) and urinary tract infection (UTI) (31, 66). While sequence variability and expression of Ace by diverse E. faecalis strains have been described (19, 31, 47, 73), no such reports are available on genes encoding Ebp pili.
In addition to adherence of circulating bacteria to ECM proteins likely exposed on the damaged vascular endocardial surface, the presence of platelets has also been shown to facilitate binding of bacteria to vegetations on heart valves, resulting in infective endocarditis, which may then lead to heart failure or to septic emboli, major complications of this disease, as well as death (15, 27, 40). Bacterial interactions with platelets generally occur either directly through a bacterial surface protein or indirectly by a plasma-bridging molecule (15, 27). For example, GspB and Hsa proteins of Streptococcus gordonii interact directly with the platelet membrane glycoprotein Ibα (2, 26, 60), while the Staphylococcus aureus MSCRAMM ClfA interacts indirectly via fibrinogen with the αIIbβ3 platelet receptor (34). Although some reports have shown variable platelet aggregation and variable adherence phenotypes (8–10, 24, 56, 59, 74) of E. faecalis and Enterococcus faecium strains, to our knowledge nothing is known about factors responsible for enterococcal adherence to platelets.
The present study was aimed at testing our hypotheses (i) that E. faecalis strains adhere to human platelets, (ii) that either MSCRAMMs or Ebp pili may facilitate adherence to platelets, (iii) that Ebp pili are expressed during experimental IE by OG1RF, and (iv) that the Ebp pili of diverse E. faecalis strains are produced under in vitro growth conditions that may mimic physiologically relevant environments. We also sought to determine if the Ebp pilus-encoding genes, which are part of the E. faecalis core genome, are highly conserved across clonal complexes, as this would have implications for potential immunologically directed strategies in the future.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A total of 518 E. faecalis isolates collected from five continents (from Argentina, Belgium, Canada, Chile, China, Colombia, France, Japan, Lebanon, New Zealand, Spain, Thailand, United Kingdom, and the United States) were included in this study. These E. faecalis isolates had been recovered (between 1926 and 2009) from human clinical specimens, stools of hospitalized patients, stools of community volunteers, and animals. The sources of clinically derived strains include blood, bile, bone, catheters, cervix, cerebrospinal fluid, placenta, peritoneal fluid, urine, and several types of wounds. E. faecalis isolates were identified to the species level by biochemical tests and by hybridization using an intragenic ace probe (47). Wild-type E. faecalis OG1RF (4), its isogenic deletion mutants, and the complementation construct used for platelet assays are listed in Table S1 in the supplemental material.
E. faecalis isolates were routinely grown in brain heart infusion (BHI) broth/agar (Difco Laboratories, Detroit, MI) and, for expression studies, in BHI broth containing 40% heat-inactivated horse serum (BHI-S) (Sigma, St. Louis, MO). For platelet assays, they were grown in BHI, BHI-S, or tryptic soy broth (Difco) supplemented with 0.25% glucose (TSBG).
Colony hybridization.
E. faecalis isolates were inoculated onto sterile nylon membranes that had been placed on BHI agar plates and grown overnight. The colonies were lysed, and the filters were hybridized under high-stringency conditions as previously described (65). Intragenic DNA probes were obtained by PCR amplification of E. faecalis OG1RF genomic DNA (for ebpA, ebpB, and ebpC) and E. faecalis TX0006 genomic DNA (for bee1, bee2, and bee3) by using the appropriate primers and conditions described earlier (50, 72). 32P-labeled DNA probes were prepared using the RadPrime DNA labeling system (Invitrogen, Carlsbad, CA) according to the protocol supplied by the manufacturer.
The ebp locus sequence analysis.
To study the extent of variability within the genes encoding individual Ebp pilus components, we analyzed the sequence of the ebp locus from 54 E. faecalis strains (see Table S2 in the supplemental material). Draft genomes of these E. faecalis strains (sequenced as part of a collaboration with George M. Weinstock at the Genome Sequencing Center at Washington University School of Medicine [25 isolates] and Baylor College of Medicine [4 isolates] and by other researchers at The Broad Institute [22 isolates] [51] and at the J. Craig Venter Institute [3 isolates]) are available at http://www.ncbi.nlm.nih.gov/genomes/MICROBES. For the 30 E. faecalis strains for which the multilocus sequence type (MLST) was not previously derived, the sequences of 7 housekeeping genes (57) were analyzed in accordance with the international E. faecalis MLST database available at http://efaecalis.mlst.net/. The 54 sequenced strains were differentiated into 33 distinct sequence types (STs) by MLST (see Table S2 in the supplemental material), including three new STs. We also analyzed the sequence of the ebp locus from 19 E. faecium, one Enterococcus gallinarum, and three Enterococcus casseliflavus genomes available at NCBI.
To identify the nucleotide (nt) variation within the individual ebp genes, sequences from different isolates were compared to the corresponding sequences in the well-studied E. faecalis strain OG1RF (4). Each ebp gene sequence differing by one or more nucleotides was considered a different allele (nucleotide type), and distinct allelic sequences were assigned an arbitrary number. Haplotypes (defined as a combination of ebpA, ebpB, and ebpC types) were determined by allelic variation in the three ebp genes.
Phylogenetic analysis and tests for selection and recombination.
Multiple sequence alignments of individual ebp genes were done by the ClustalW method using the MegAlign program of the DNASTAR software (Lasergene, Madison, WI). Phylogenetic trees (cladograms) based on the matrix of pairwise sequence divergence were constructed using the MegAlign program. Splits graphs were computed from the sequences for each individual gene and also from the complete ebp operon. SplitsTree4 (22) (available at http://www.splitstree.org) was used to evaluate split decomposition network methods. Split networks are combinatorial generalizations of phylogenetic trees and are designed to represent incompatibilities within data sets. Nonsynonymous (dN) and synonymous (dS) substitutions per (non)synonymous nucleotide site were used to estimate whether the genes encoding pilin subunits are under selection pressure, as described in reference 1. To identify specific amino acids undergoing positive or purifying selection, the rate of nonsynonymous to synonymous polymorphisms (Ka/Ks) for each codon position (70) was performed using an Internet-based selecton server (http://selecton.tau.ac.il/). Potential recombination events between ebp sequences were identified using the maximum chi-square test (68), Sawyer's runs test (58), and 1,000 resamplings. Both of these tests are nucleotide substitution distribution methods that test for clustering of polymorphisms along an alignment (54). The START2 (sequence type analysis and recombinational test) program (available at http://pubmlst.org) (23) was used to determine the ratio of dN/dS and recombination tests described above.
Whole-cell ELISA.
Surface expression of EbpA and EbpC by E. faecalis isolates grown for 10 h in BHI-S was detected by a whole-cell enzyme-linked immunosorbent assay (ELISA), as described in the companion paper (48). Of note, anti-rEbpB antibodies that react reasonably well with polymerized pili on Western blots, similar to anti-rEbpC and anti-rEbpA antibodies (50), showed relatively low reactivity with OG1RF cells when analyzed by flow cytometry (as well as by whole-cell ELISA) (25), suggesting that EbpB epitopes may be masked in the native Ebp pilus structure and thus not accessible to antibodies in these assays. Therefore, we did not use these antibodies in surface expression studies with bacteria. To confirm that whole cells of all strains were actually bound to the microtiter plates, antiserum raised against formalin-killed E. faecalis strain HH22 whole cells was used as a positive control.
Flow cytometry. (i) Bacteria.
For flow cytometry analysis, bacteria grown in BHI-S were blocked with newborn calf serum (NBCS; Sigma) before being incubated at 4°C for 2 h with 20 μg/ml preimmune or affinity-purified anti-rEbpA- or anti-rEbpC-specific antibodies (50) in dilution buffer (phosphate-buffered saline [PBS] containing 20% NBCS and 0.1% bovine serum albumin [BSA]). Bacteria were then labeled with goat anti-rabbit F(ab′)2 fragment conjugated with R-phycoerythrin (Jackson Immunoresearch, West Grove, PA) at 4°C for 2 h, fixed with 1% paraformaldehyde in PBS, and analyzed using a Coulter EPICS XL AB6064 flow cytometer (Beckman Coulter) and System II software.
(ii) Endocarditis vegetations.
Sterile and OG1RF-infected vegetations were produced in rats, as described earlier (50, 67). Vegetations harvested from heart valves 48 h after injection of bacteria were processed by sonication, filtration, and centrifugation to remove host tissue debris, using a method previously described by us (49). Processed samples were labeled with affinity-purified anti-rEbpC antibodies and with immunoglobulins purified from antiserum raised against formalin-killed E. faecalis HH22 whole cells (positive control). To assess possible cross-reactivity of anti-rEbpC antibody with host tissue, a noninfected vegetation sample was also probed with anti-rEbpC-specific antibodies. Forward scatter (for analysis of particle sizes in the sample) and side scatter (for analysis of cell granularity or internal complexity) of cells processed from the vegetation were analyzed and compared with in vitro-grown E. faecalis OG1RF cells.
Quantitative assay for adherence to human platelets.
Adherence of E. faecalis to human platelets was assessed quantitatively as described previously for Streptococcus mitis (37) with minor modifications. In brief, human platelets (prepared using the method described in reference 71) were immobilized in poly-l-lysine-coated tissue culture wells to produce platelet monolayers of 90% confluence. To reduce nonspecific adherence, coated wells were treated for 2 h at room temperature with a casein solution (blocking reagent; Roche Applied Sciences, Indianapolis, IN) in Dulbecco's phosphate-buffered saline (DPBS). After removing the blocking solution, the wells were inoculated with approximately 1 × 108 to 5 × 108 CFU of appropriately grown E. faecalis cells suspended in 1 ml of DPBS and incubated at 37°C for 2 h, with gentle rocking to enhance mixing. Unbound bacteria were removed by washing with DPBS, and bound bacteria were then recovered by scraping and resuspension in 1 ml DPBS. The number of organisms bound was determined by plating serial dilutions of the suspension onto BHI agar, and binding was expressed as a percentage of the inoculum. Assays were carried out in triplicate on three to four separate occasions, using platelets from a different human donor on each day. Differences in platelet binding were compared by the unpaired t test.
RESULTS AND DISCUSSION
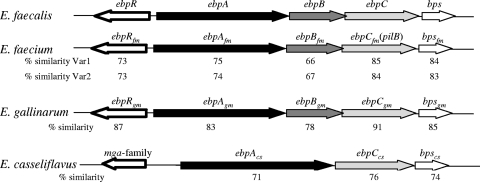
Distribution, organization, and conservation of pilus-encoding genes among diverse E. faecalis strains and other enterococci. (i) Distribution in E. faecalis.
Of the two E. faecalis pilus-encoding gene clusters described to date, the ebp locus (part of the core genome) was previously shown to be present in all 408 E. faecalis isolates tested (50). The second cluster, bee (for biofilm enhancer in Enterococcus), encoded on a conjugative plasmid, was restricted to 2 of 40 isolates tested (72). Here, we tested 64 additional isolates for ebp and 473 isolates for bee by colony hybridization. Our results showed that only a single (fecal) isolate (TX1346) lacked ebpA, ebpB, and ebpC. Consistent with a previous report (72), the bee locus (bee1, bee2, bee3) was present in 1% of 359 clinical isolates, 2% of 58 hospital-derived stool isolates, 6% of 33 community-derived stool isolates, and 0% of 23 animal isolates from our collection (1.2% overall). These results (along with genome analysis [see below]) suggest that most E. faecalis strains encode only one pilus-encoding locus (ebp), similar to GAS FCT (fibronectin- and collagen-binding protein and T antigen) pilus loci (55); however, unlike ebp, the FCT locus is highly variable in gene content and organization in different M serotypes (see below). In contrast, many isolates of GBS, E. faecium, and corynebacteria possess two or more pilus-encoding loci on variable genomic islands which, at times, are flanked by transposable elements (28, 36, 69).
(ii) ebp locus organization in E. faecalis.
To study the gene organization and extent of conservation within each gene encoding an Ebp pilus component (see below), we analyzed the sequence of this locus from 54 of 55 E. faecalis sequenced strains representing 33 MLSTs (see Table S1 in the supplemental material). The organization of the predicted backbone (ebpC) and the two predicted ancillary subunit-encoding genes (ebpA and ebpB) was ebpA-ebpB-ebpC-bps in all isolates tested (Fig. 1). Furthermore, our analysis identified a single pilus locus (ebp) in these E. faecalis genomes, except strain HIP11704, which also contained a partial bee locus in addition to the ebp locus. A draft genome of E. faecalis strain TX1346 (55th sequenced strain) was found to lack an ebp-like locus, confirming our hybridization results. Sequence alignments with the completed genomes of strains OG1RF and V583 enabled us to identify two contigs (Cont1322.1 and Cont123.1) in the TX1346 genome database that encode open reading frames (ORFs) surrounding the deleted ebp locus. Sequencing over this region suggested that TX1346 has undergone a 12-kb deletion (12,041 nt versus OG1RF and 12,035 nt versus V583) replaced by a 1,524-nt fragment encoding an IS1542 transposase, a member of the IS256 family of insertion sequence elements (12). The deleted region corresponds to 9 complete genes in V583 (ef1089 to ef1097), including the ebp locus (ef1090 to ef1094), as well as the 3′ end of ef1088.
Fig. 1.
Ebp-type pilus gene clusters (drawn to scale) identified in the chromosomes of four enterococcal species. Each cluster contains a predicted pilus-specific sortase gene (white arrows with thin borders), genes encoding a major subunit (light gray arrows), one or two minor subunits (black and dark gray arrows), and a gene encoding a cognate regulator (white arrows with thick borders). A second variant (Var 2) of the ebp locus found in E. faecium had an identical organization to variant one (Var 1). The percent similarity of each protein with its E. faecalis homolog is shown.
(iii) Each Ebp subunit-encoding gene of E. faecalis is highly conserved.
Comparison of the sequences of each individual subunit-encoding gene, ebpA, ebpB, and ebpC, from E. faecalis strains representing 33 STs revealed 98.4% to 99.99% identity at the DNA level and 97.5% to 100% identity at the protein level. The polymorphic sites in all three subunit-encoding genes were distributed randomly over the entire genes (data not shown). The numbers of polymorphic nucleotides ranged from 152 (128 point mutations and 24 insertion/deletion changes in ebpA) to 66 (ebpC) (Table 1). Insertion/deletion changes, which were found only in ebpA, were all in-frame changes (an 18-nt deletion in strain E1Sol and a 6-bp insertion in strains T8 and JH2-2). Most point mutations were found to be synonymous (silent) (e.g., EbpC, 41 of 66 mutations were synonymous), and several nonsynonymous mutations caused changes to a similar functional group of amino acids (aa) (e.g., EbpC, 8 of 25 mutations were nonsynonymous). Only one strain (DS5) had a nonsense transversion (C1967A in ebpA), which resulted in a premature stop codon (see expression studies below), while ebpB and ebpC were intact in all strains. The pilin motif and E box, shown to be important for assembly of pilus fibers in corynebacteria (36), were conserved in all strains.
Table 1.
Ebp pilin subunit gene characteristics in 54 E. faecalis isolates
| Gene | Length (nt) | No. of alleles (nt)a | No. of amino acid types | % identityb | No. of polymorphisms |
dN/dS ratioc | |
|---|---|---|---|---|---|---|---|
| nt | aa | ||||||
| ebpA | 3,414 to 3,438 | 32 | 29 | 98.8 to 99.99 | 152d | 57 | 0.13 |
| ebpB | 1,431 | 27 | 24 | 98.4 to 99.99 | 67e | 31 | 0.27 |
| ebpC | 1,884 | 23 | 20 | 98.8 to 99.99 | 66e | 25 | 0.13 |
| 7 housekeeping genes | 395 to 583f | 8 to 19 | NDg | ND | 7–20 | ND | 0 to 0.07 |
Nucleotide sequence types.
Percent identities between different nucleotide types (alleles).
dN/dS, a measure for estimating whether a particular sequence is under selection pressure: dN/dS < 1, nonsynonymous mutations are under negative (purifying) selection; dN/dS = 1, all changes are neutral; dN/dS > 1, substitutions are driven by positive (diversifying) selection.
One hundred twenty-eight point mutations and 24 insertion or deletion changes.
All point mutations.
Region used for MLST analysis.
ND, not determined.
A total of 32 alleles were observed for ebpA, 27 for ebpB, and 23 for ebpC, but the number of amino acid sequence types was slightly lower due to silent mutations; the majority of these sequence types differed only by 1 or 2 nt or aa (Table 1). Variation between individual amino acid sequence types of all three subunit proteins (Fig. 2) was in the range of 0.1% to 2.4%, which is low compared to that of another, presumably evolving, surface-exposed antigenic protein, Ace. The amino acid variation of Ace in this set of isolates was >7% in the A domain and was much higher for the complete protein due to variation in the number of B domain repeats.
Fig. 2.
Amino acid variation in Ebp pilin components of 54 E. faecalis strains representing 33 MLSTs. Each ebp sequence that differed by ≥1 nt was designated a different allele and assigned an arbitrary number, and the same numbering was retained for each amino acid type. Alleles that differed only by silent mutations and resulted in identical amino acid types are positioned below their identical amino acid type (for example, EbpA20 and EbpA21, which are identical to EbpA4, are placed below EbpA4). Amino acids present in each of the variable sites of E. faecalis OG1RF are shown. The position of each variable site within the sequenced fragment is shown by the number above each amino acid, read vertically.
A total of 35 unique Ebp haplotypes (which are likely inherited as a unit) were identified among the 33 MLSTs (see Table S2 in the supplemental material). While most isolates belonging to the same MLST had an identical Ebp haplotype (see Table S1 in the supplemental material), a few had 1 to 2 aa changes in one or more Ebp subunit proteins (details in Table S3 in the supplemental material). Similarly, related MLSTs (ST25 and ST364) had identical Ebp haplotypes. Also, two common Ebp haplotypes were identical in several unrelated STs (see Table S4 in the supplemental material). The incongruent phylogenetic trees of individual ebp genes and split decomposition analysis indicate remnants of horizontal gene transfer or recombination. Analysis by the maximum chi-square test on all possible pairwise combinations of alleles (1,000 randomizations; significance value, P < 0.05) identified nt 477, 747, 768, and 1053 of ebpA as potential recombination sites. The ratio of the rate of nonsynonymous (Ka or dN) to synonymous (Ks or dS) polymorphisms (dN/dS values are shown in Table 1) suggests that none of the ebp pilin subunit genes are under positive selection pressure. Due to the small number of polymorphic sites, reliable prediction of specific amino acids under selection and calculation of associated posterior probabilities were not possible. Taken together, the same amino acid replacements of Ebp proteins that likely occurred independently in different STs may not reflect common ancestry but rather represent parallel, reverse, or convergent changes.
(iv) Ebp-type pili of other enterococcal species.
A subsequent search for Ebp-type pilus (defined as >70% aa similarity with all subunit proteins) genes in available genome databases identified that Ebp-type pili are unique to enterococci and present in 19 of 22 E. faecium genomes (20, 63), 1 of 1 E. gallinarum genome, and 3 of 3 E. casseliflavus genomes (Fig. 1). Their genetic organization is highly conserved in each species, but the E. casseliflavus isolates lack an ebpB homolog. Similar to E. faecalis, E. faecium and E. gallinarum also have ebpR (encoding a positive regulator of the ebp operon in E. faecalis [5, 6]) in the same position (Fig. 1). The gene immediately upstream of the E. casseliflavus ebp locus has little similarity to ebpR; however, sequence analyses predict that it, like ebpR, also codes for a member of the AtxA/Mga regulator family. Interestingly, sequence analysis of the E. faecium ebp operon found two variants of this locus with ∼90% identities between their subunit-encoding genes; however, both variants had almost similar amino acid similarities with their E. faecalis homolog (Fig. 1). As previously reported, a number of E. faecium isolates have one or more of three additional pilus loci (fms11-fms19-fms16, fms14-fms17-fms13, or fms21 [pilA]-pilE-fms20 [pilF]), and each of these loci also encodes a sortase C, predicted to be involved in pilus biogenesis (20, 21, 62).
Taken together, Ebp-type pili of four different species of enterococci have similar gene organizations, including two to three conserved pilin-encoding genes, a single sortase C-encoding gene (bps-like) located at the 3′ end of the ebp loci (downstream of the major pilin gene ebpC), and a divergently transcribed regulatory gene, ebpR, at the 5′ start of the ebp locus. This is in contrast to the pilus-encoding loci of some other Gram-positive bacteria, such as the FCT locus of GAS, which is highly variable in gene content and organization and possesses ≥1 sortases (28, 36). For example, FCT-3 pili of a GAS serotype M3 strain have a collagen adhesin Cpa as a minor pilin, FCT-1 pili of M6 have a fibronectin adhesin PrtF1, and FCT-4 pili of M12 have both Cpa and PrtF1 (30, 53).
Ebp pilus expression by diverse E. faecalis isolates in BHI-S in vitro and in experimental IE vegetations. (i) Most E. faecalis isolates express Ebp pili after growth in BHI-S.
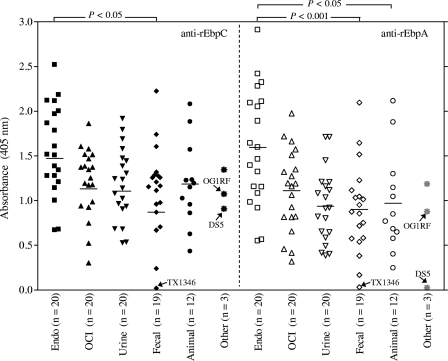
To investigate if the expression of Ebp pili observed with strain OG1RF after growth in BHI-S is a general property of E. faecalis, we analyzed Ebp pilus expression of 93 diverse E. faecalis isolates, including 25 sequenced strains by whole-cell ELISA using affinity-purified anti-rEbpC- or anti-rEbpA-specific antibodies. These isolates were grouped on the basis of their origin: 20 endocarditis; 20 other, clinical; 20 urine; 19 community, fecal; 12 animal; and 2 of unknown origin. OG1RF was used as a positive control. As shown in Fig. 3, all strains grown in BHI-S, except the one (fecal) isolate, TX1346, that lacks the ebp locus, were found to express both EbpA and EbpC on their surface. ST55 strain DS5 (of unknown origin), which has an internal stop codon in ebpA (see above), lacked EbpA, while it still expressed EbpC. Individual differences were observed in EbpA and EbpC expression levels (quantified by optical density at 405 nm [OD405] values) of different strains ranging from 0.31 to 2.5 for EbpC and 0.38 to 2.9 for EbpA (excluding TX1346). Statistical analysis, using Pearson's correlation test, confirmed that the amount of surface EbpC correlated strongly (r = 0.946; P < 0.0001) with the amount of surface EbpA (see Fig. S1 in the supplemental material).
Fig. 3.
Determination of EbpC and EbpA surface expression by diverse E. faecalis strains grown in BHI-S, grouped by source of isolation. Affinity-purified anti-rEbpC and anti-rEbpA antibodies were used in these whole-cell ELISAs. Each data point represents the mean absorbance at 405 nm ± SD from 9 wells. Results are from three independent experiments. Horizontal bars represent the means (absorbance) of each group of isolates. Endo, strains isolated from patients with E. faecalis endocarditis; OCI, strains isolated from E. faecalis nonendocarditis clinical infections; urine, strains isolated from urine; fecal, isolates from feces of community healthy volunteers; animal, isolates from animal origin; other, includes isolates of unknown origin and the control OG1RF. TX1346 that lacks the ebp locus, DS5, which has an internal stop codon in ebpA, and the control, OG1RF, are labeled with arrows. Mean Ebp pilin (both EbpC and EbpA) expression levels of strains from different sources were compared using ANOVA and Bonferroni's posttest; P = 0.02 and P = 0.0003 for EbpC and EbpA by ANOVA, respectively. Significant differences between individual groups assessed by the post hoc test are shown in the figure.
Although expression of Ebp pili after growth in BHI-S was detected in all categories of isolates, a statistically significant difference was observed between the Ebp pilin (both EbpC and EbpA) expression levels of strains from different sources (P = 0.02 and P = 0.0003 by analysis of variance [ANOVA], respectively). Multiple comparisons with Bonferroni's correction showed significant differences between the endocarditis group versus the community-derived fecal isolate group (P < 0.05 and P < 0.001, for EbpC and EbpA, respectively) and versus the animal isolate group for EbpA (P < 0.05).
(ii) Bistable surface expression of Ebp pili.
Using immunogold electron microscopy and fluorescence-activated cell sorting (FACS), we previously estimated that only a proportion of OG1RF cells (<10% of BHI-, <20% of TSBG-, and <35% of BHI-S-grown cells) had Ebp pili on their surface in vitro (25, 29, 50). Here, we quantified the surface-localized Ebp pilin proteins of 7 diverse endocarditis isolates using FACS and found that 30% (OG1RF) to 72% (TX0001) of cells of the investigated strains expressed EbpA or EbpC after growth in BHI-S (see Fig. S2 in the supplemental material); 6 of 7 strains were inducible (2- to 5-fold) in BHI-S versus in BHI (data not shown). In contrast to Ace (see Fig. S2J and K in the supplemental material), two different peaks (heterogeneity manifested by the bifurcation into two distinct subpopulations) could be distinguished among anti-Ebp-stained versus unstained cells for all strains (see Fig. S2A to H in the supplemental material), suggesting a bistable expression mode for Ebp. Growth in BHI also demonstrated bistable expression, albeit with fewer Ebp-expressing cells. Similar bistable expression was reported for FCT pili of GAS strains (43). While a biological benefit of bistability is currently unknown, it has been proposed to prepare a subpopulation of bacteria for rapidly changing environmental conditions (13, 75).
(iii) In vivo expression of Ebp pili during experimental infective endocarditis.
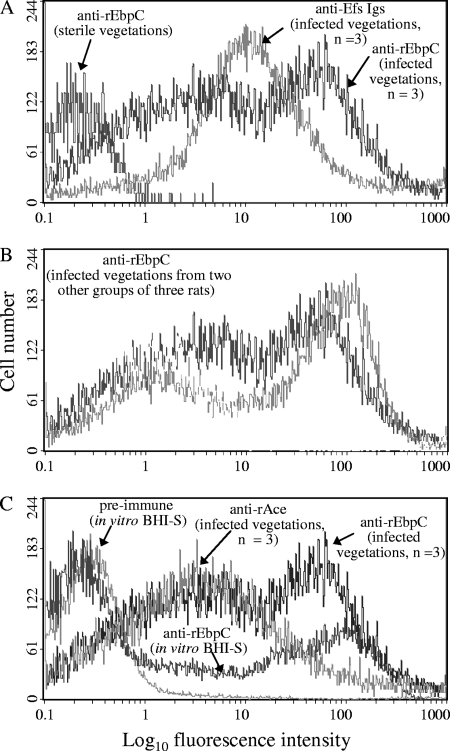
To quantify the percentage of Ebp-positive cells in vivo, flow cytometry analyses were carried out on extracts directly processed from IE vegetations infected with OG1RF grown in BHI. Processed sterile vegetations from noninfected rats probed with anti-rEbpC-specific antibodies, which served as a negative control, showed labeling of a minor fraction (<5%) of bacterium-sized particles (Fig. 4A). Immunoglobulins from an antiserum raised against formalin-killed E. faecalis strain HH22 whole cells (used as a positive control) bound 85% of bacterium-sized particles of processed infected vegetations, indicating that the majority of these particles were E. faecalis cells (Fig. 4A). Affinity-purified anti-EbpC-specific antibodies bound to ∼57 to 63% of bacterium-sized particles (∼67 to 74% of particles labeled by the general anti-E. faecalis HH22 antiserum) from multiple rat endocarditis vegetations infected with OG1RF (Fig. 4A and B). The higher percentage (Fig. 4C) of Ebp-expressing OG1RF cells from vegetations versus ∼30% of cells grown in BHI-S demonstrates that Ebp pili are actively expressed in host vegetations during IE. Furthermore, this result is consistent with our previous finding of anti-Ebp pilin antibodies in human endocarditis patient sera infected by diverse E. faecalis strains (64).
Fig. 4.
Flow cytometry analysis of Ebp pilus expression measured by EbpC on E. faecalis OG1RF cells derived from vegetations of rat experimental infective endocarditis. (A) Surface expression of Ebp pili by bacterium-sized particles derived from a mixture of vegetations of three rats; (B) expression of Ebp pili by bacterium-sized particles derived from a mixture of vegetations of two other groups of three rats each; (C) for comparison, OG1RF cells grown in vitro in BHI-S (the growth condition that exhibited the most in vitro Ebp pili expression), stained with preimmune immunoglobulins and anti-rEbpC, as well as surface expression of Ace (a positive control, which we previously reported [67]) by bacterium-sized particles derived from a mixture of vegetations from the three rats from panel A are shown. Vegetations were produced as described earlier (50, 67), and rats were intravenously (i.v.) injected with BHI-grown OG1RF. Processed sterile and infected vegetations were mixed and then divided into aliquots for labeling with (i) immunoglobulins purified from antiserum raised against formalin-killed E. faecalis HH22 whole cells (anti-Efs; positive control), (ii) affinity-purified anti-rAce-specific antibodies (positive control), and (iii) affinity-purified anti-rEbpC-specific antibodies, followed by incubation with R-phycoerythrin-conjugated antibodies. Specific binding by anti-rEbpC, anti-Efs, or anti-rAce antibodies is indicated as log fluorescence intensity on the x axis. Each histogram represents ∼5,000 (noninfected vegetations), ∼25,000 (infected vegetations), and 50,000 (in vitro-grown cultures) events of bacterium-sized particles.
The role of E. faecalis MSCRAMMs and pili on adherence to platelets.
Adherence of pathogenic bacteria to platelets is believed to play an important role in the pathogenesis of IE. Numerous endocarditis-associated pathogens have been shown to adhere to platelets directly or indirectly in vitro, through a variety of mechanisms (11, 15, 27). Adherence to platelets in vitro has been linked to virulence of common endocarditis-associated species, including S. aureus, S. gordonii, and Streptococcus sanguinis (11, 15, 27).
(i) Effect of growth conditions on adherence of E. faecalis strain OG1RF to immobilized human platelets.
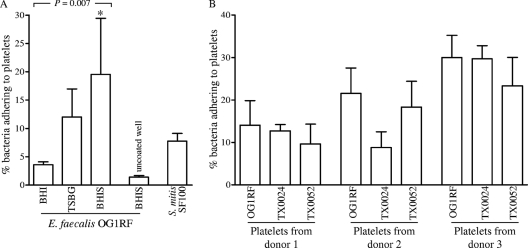
Since platelets are a major component of IE vegetations and because we found that E. faecalis MSCRAMMs and pilus mutants are attenuated in endocarditis (50, 67), we tested whether E. faecalis strains were able to adhere to human platelets. Pilot analyses of platelet adherence by E. faecalis strain OG1RF, grown in a standard laboratory medium (BHI) to three different growth phases, showed that after 2 h of incubation with platelet monolayers, bacterial adherence ranged from 0.9% to 4.2% of the inoculum (means ± standard deviations [SD] of 3.1% ± 1.0% with log-phase culture, 2.5% ± 0.9% with late-log-phase culture, and 1.1% ± 0.3% with overnight culture) (data not shown). Only 0.2 to 0.7% of the OG1RF inoculum bound to uncoated polylysine plastic wells (negative control), and BHI-grown OG1RF platelet adherence was 2- to 6-fold lower than that by the experimental control Streptococcus mitis strain SF100, previously shown to adhere efficiently to platelets (3, 37). Since growth medium has been found to influence the ability of E. faecalis strains to form biofilms (e.g., TSBG) (52) and enhance their adherence to several extracellular matrix proteins (e.g., BHI-S) (45), at least in part via increased expression of MSCRAMMs and pili (44, 48, 50), we next tested the adherence of OG1RF, grown to late logarithmic phase in TSBG and BHI-S, to human platelets derived from different donors. As shown in Fig. 5A, adherence to platelets by OG1RF from different culture conditions increased progressively, starting with cells grown in BHI, TSBG (3.4-fold versus BHI), and BHI-S (5.5-fold versus BHI and 1.6-fold versus TSBG) (P = 0.007 by ANOVA and P < 0.01 by the post hoc test for BHI versus BHI-S). The substantial variability in adherence to platelets from different donors (even cells grown in BHI-S showed no adherence to platelets from 3 of the 7 donors) likely explains the relatively high standard deviations of adherence data from independent experiments (e.g., 19.5 ± 10 for BHI-S); nevertheless, the adherence differences between independent experiments were highly reproducible. Subsequent experiments were carried out using platelets from the donors showing adherence.
Fig. 5.
Adherence of E. faecalis strains to immobilized human platelets. (A) Influence of growth conditions on the adherence of E. faecalis strain OG1RF; (B) adherence of OG1RF and two endocarditis-derived E. faecalis strains grown in BHI-S. Bars represent mean percentages ± SD of bacteria adhering to platelets (from three different donors) coated onto >9 wells from 3 to 4 independent experiments. S. mitis SF100, grown in Todd-Hewitt broth, as described in references 37 and 38, was used as a positive control. Means of adherence percentages of different strains were compared using ANOVA and Bonferroni's posttest. *, P < 0.01 by the post hoc test for BHI versus BHI-S.
We next tested the platelet adherence of two diverse endocarditis isolates. As shown in Fig. 5B, both strains grown in BHI-S were found to adhere to platelets obtained from three different donors, and the adherence percentages varied depending on the donor, similar to OG1RF. For TX0024 (also referred to as MC02152 [47]), the percentage of platelet-adhering cells ranged from 4.6% (donor 2) to 31.75% (donor 3), and that for TX0052 ranged from 6.4% (donor 1) to 30.63% (donor 3). These results indicate that human platelet adherence may be a common phenomenon among E. faecalis, albeit with considerable variability between individual E. faecalis strains and platelets from different donors. We speculate that the observed specificity of E. faecalis to platelets of some individuals may be due to genetic polymorphisms or differential expression of platelet receptors; similar heterogeneity was reported for the binding of Helicobacter pylori to human gastric MUC5AC mucin glycoforms (33). This suggests the possibility of a genetic predisposition to enterococcal infections.
(ii) Requirement of sortase activity for platelet adherence.
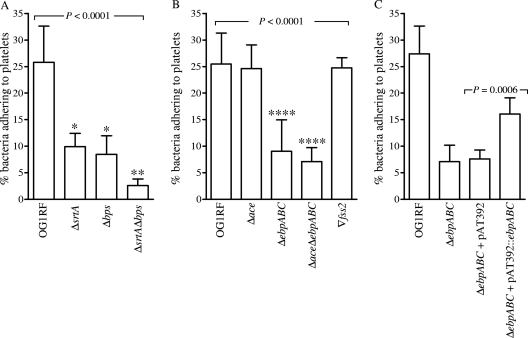
Since cell wall-anchored proteins of other pathogens (11, 27) are known to promote bacterial adhesion to human platelets, we evaluated the global impact of LPXTG-type surface-anchored proteins and pili using mutants of sortases. E. faecalis OG1RF contains two sortase genes (housekeeping srtA and pilus-associated bps) that are ubiquitous (25) among other E. faecalis isolates but lacks ef2524, a sortase A homolog found in the mobile genetic element efaC1 (ef2512 to ef2546) (7, 32). As shown in Fig. 6A, ΔsrtA, Δbps, and ΔbpsΔsrtA mutants grown in BHI-S were defective in adherence to platelets versus that of wild-type OG1RF (P < 0.0001 by ANOVA). The mean percentages ± SD of bacteria adhering of the ΔsrtA, Δbps, and ΔbpsΔsrtA mutants were 9.9% ± 2.5%, 8.4% ± 3.5%, and 2.6% ± 1.2%, respectively, versus 25.8% ± 6.8% for the wild type. The reduction in platelet binding seen with the double-deletion mutant (ΔbpsΔsrtA mutant) versus each of the single-deletion mutants was also significant (P < 0.001). These results indicate that binding of OG1RF to platelets is mediated predominantly by one or more surface-anchored sortase targets.
Fig. 6.
Effect of deletion of bps, srtA, ebpABC, or MSCRAMM-encoding genes and complementation constructs on the adherence of BHI-S-grown OG1RF to immobilized human platelets. (A) Comparison of platelet adherence of wild-type OG1RF and its sortase mutants (ΔsrtA, Δbps, and ΔsrtAΔbps mutants); (B) adherence by ebp, ace, and fss2 mutants; (C) adherence by complementation derivatives. Bars represent mean percentages ± SD of bacteria adhering to platelets (from three different donors) coated onto 6 to 9 wells from two to three independent experiments. Means of adherence percentages of different strains were compared using ANOVA and Bonferroni's posttest. Means of the complemented strain were compared with the appropriate vector-only control strain using the t test. *, P < 0.001 versus wild-type, P < 0.05 versus double deletion, and nonsignificant versus other single deletions; **, P < 0.001 versus wild type and P < 0.05 versus both single deletions; ****, P < 0.001 versus wild type and the Δace and Δfss2 mutants.
Adhesion to human platelets is directly mediated by Ebp pili of E. faecalis OG1RF.
The above-mentioned data indicated that the Ebp pili (assembly and cell wall anchoring of which are dependent on Bps and SrtA, respectively) (18, 25, 50) and possibly other SrtA targets (e.g., ace encoding a collagen adhesin, fss2 encoding a fibrinogen adhesin) (46, 61) contribute to adherence to human platelets of BHI-S-grown E. faecalis cells. To address this possibility, wild-type OG1RF and the Δace, ΔebpABC, ΔaceΔebpABC, and Δfss2 mutants were tested for this phenotype. As shown in Fig. 6B, OG1RF lacking ebpABC, but not ace or fss2 mutants, showed decreased platelet adherence (P < 0.0001 by ANOVA). The decrease for the ΔebpABC mutant was highly significant compared to that of the wild type or Δace or Δfss2 mutants (P < 0.001 by the post hoc test). This observation was further supported by the values obtained for the ΔaceΔebpABC mutant, which were similar to that of the ΔebpABC mutant and showed 3.5-fold reduction versus the wild type or Δace mutant (P < 0.001 by the post hoc test). Highly significant enhancement in platelet adherence was observed by the complemented strain (ΔebpABC mutant with pAT392::ebpABC) over the empty vector control (Fig. 6C; P = 0.0006); however, this restoration was only to 65% of the wild-type level. The higher levels of unpolymerized pilin components leading to shorter, incompletely polymerized pili (see companion paper [48]) could explain the lack of full restoration of the adherence phenotype by the complemented strain. Our preliminary analyses with OG1RF mutants of three additional genes encoding MSCRAMMs (fss1, ef2224, and ef1269) showed no effect on BHI-S-induced platelet adherence (data not shown). Thus, our data demonstrate that Ebp pili contribute to E. faecalis OG1RF platelet adherence under the conditions tested. While Ebp pili also mediate fibrinogen adherence, whether these pili interact directly with platelet receptors like GspB and Hsa proteins of S. gordonii (2, 26, 60) or indirectly via fibrinogens like S. aureus ClfA and FnbpA (26) will be subjects of our future studies. Furthermore, the Ebp expression levels of OG1RF, TX24, and TX52 did not parallel platelet adherence levels, indicating saturation or involvement of additional bacterial factors in this phenotype.
In summary, we have demonstrated that deletion of ebpABC results in significant attenuation in the ability of E. faecalis to adhere to platelets, which is likely one of the factors contributing to vegetation formation/infection and may help explain our previous experimental evidence that ebpABC is important for E. faecalis endocarditis (50). We have also shown that Ebp pili are actively expressed by the majority of strains after growth in medium supplemented with serum, a biological cue with in vivo relevance, and are also expressed within host vegetations during endocarditis produced by OG1RF. Finally, the highly conserved nature of Ebp pili, in combination with data showing that anti-Ebp antibodies inhibit fibrinogen adherence of E. faecalis (companion paper [48]), underscores the possibility of using Ebp pilin components as targets for immunoprophylaxis or possibly in combination with antibiotics for treating E. faecalis endocarditis. The observations that anti-rAce antibodies (polyclonal and monoclonal) inhibit adherence of Ace to collagen (19, 46) and to epithelial cell lines (19), in addition to their protective effect in the IE model (50, 67), point toward the prospect of using both Ace and the Ebp pilus components, in combination, as targets for antibody-based therapy against E. faecalis infections. Furthermore, since Ebp pilus-like surface structures (with >70% similarities between individual pilin components) and Ace homologs are also found in other enterococci, including the hospital-associated emerging pathogen E. faecium, future cross-protection studies may reveal opportunities for the development of immunotherapeutics that may be useful for the prevention and treatment of infective endocarditis caused by different enterococcal species.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grants R37 AI47923 and R01 AI47923 from the Division of Microbiology and Infectious Diseases, NIAID, to B.E.M. and the Department of Veteran Affairs and NIH grants R01 AI041513 and R01 AI057433 to P.M.S.
We thank Barbara Bensing (UCSF, CA) for helpful scientific discussions on platelet interactions. We gratefully acknowledge the many physicians and researchers (both active and retired) around the world for providing isolates for our 40-year strain collection.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 18 April 2011.
REFERENCES
- 1. Bambini S., et al. 2009. Distribution and genetic variability of three vaccine components in a panel of strains representative of the diversity of serogroup B meningococcus. Vaccine 27:2794–2803 [DOI] [PubMed] [Google Scholar]
- 2. Bensing B. A., Gibson B. W., Sullam P. M. 2004. The Streptococcus gordonii platelet binding protein GspB undergoes glycosylation independently of export. J. Bacteriol. 186:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bensing B. A., Rubens C. E., Sullam P. M. 2001. Genetic loci of Streptococcus mitis that mediate binding to human platelets. Infect. Immun. 69:1373–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bourgogne A., et al. 2008. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 9:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bourgogne A., et al. 2007. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. J. Bacteriol. 189:6490–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bourgogne A., Thomson L. C., Murray B. E. 2010. Bicarbonate enhances expression of the endocarditis and biofilm associated pilus locus, ebpR-ebpABC, in Enterococcus faecalis. BMC Microbiol. 10:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burrus V., Pavlovic G., Decaris B., Guedon G. 2002. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 48:77–97 [DOI] [PubMed] [Google Scholar]
- 8. Clawson C. C. 1973. Platelet interaction with bacteria. 3. Ultrastructure. Am. J. Pathol. 70:449–471 [PMC free article] [PubMed] [Google Scholar]
- 9. Clawson C. C., White J. G. 1971. Platelet interaction with bacteria. I. Reaction phases and effects of inhibitors. Am. J. Pathol. 65:367–380 [PMC free article] [PubMed] [Google Scholar]
- 10. Clawson C. C., White J. G. 1971. Platelet interaction with bacteria. II. Fate of the bacteria. Am. J. Pathol. 65:381–397 [PMC free article] [PubMed] [Google Scholar]
- 11. Clemetson K. J. 2010. Platelets and pathogens. Cell. Mol. Life Sci. 67:495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Darini A. L., Palepou M. F., Woodford N. 1999. Nucleotide sequence of IS1542, an insertion sequence identified within VanA glycopeptide resistance elements of enterococci. FEMS Microbiol. Lett. 173:341–346 [DOI] [PubMed] [Google Scholar]
- 13. Dubnau D., Losick R. 2006. Bistability in bacteria. Mol. Microbiol. 61:564–572 [DOI] [PubMed] [Google Scholar]
- 14. Fernandez-Guerrero M. L., Verdejo C., Azofra J., de Gorgolas M. 1995. Hospital-acquired infectious endocarditis not associated with cardiac surgery: an emerging problem. Clin. Infect. Dis. 20:16–23 [DOI] [PubMed] [Google Scholar]
- 15. Fitzgerald J. R., Foster T. J., Cox D. 2006. The interaction of bacterial pathogens with platelets. Nat. Rev. Microbiol. 4:445–457 [DOI] [PubMed] [Google Scholar]
- 16. Gianfaldoni C., et al. 2007. Streptococcus pneumoniae pilus subunits protect mice against lethal challenge. Infect. Immun. 75:1059–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giannitsioti E., et al. 2007. Nosocomial vs. community-acquired infective endocarditis in Greece: changing epidemiological profile and mortality risk. Clin. Microbiol. Infect. 13:763–769 [DOI] [PubMed] [Google Scholar]
- 18. Guiton P. S., et al. 2009. Contribution of autolysin and sortase A during Enterococcus faecalis DNA-dependent biofilm development. Infect. Immun. 77:3626–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hall A. E., et al. 2007. Monoclonal antibodies recognizing the Enterococcus faecalis collagen-binding MSCRAMM Ace: conditional expression and binding analysis. Microb. Pathog. 43:55–66 [DOI] [PubMed] [Google Scholar]
- 20. Hendrickx A. P., et al. 2008. Expression of two distinct types of pili by a hospital-acquired Enterococcus faecium isolate. Microbiology 154:3212–3223 [DOI] [PubMed] [Google Scholar]
- 21. Hendrickx A. P., et al. 2010. Differential PilA pilus assembly by a hospital-acquired and a community-derived Enterococcus faecium isolate. Microbiology 156:2649–2659 [DOI] [PubMed] [Google Scholar]
- 22. Huson D. H., Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267 [DOI] [PubMed] [Google Scholar]
- 23. Jolley K. A., Feil E. J., Chan M. S., Maiden M. C. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230–1231 [DOI] [PubMed] [Google Scholar]
- 24. Kahn R. A., Flinton L. J. 1974. The relationship between platelets and bacteria. Blood 44:715–721 [PubMed] [Google Scholar]
- 25. Kemp K. D., Singh K. V., Nallapareddy S. R., Murray B. E. 2007. Relative contributions of Enterococcus faecalis OG1RF sortase-encoding genes, srtA and bps (srtC), to biofilm formation and a murine model of urinary tract infection. Infect. Immun. 75:5399–5404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kerrigan S. W., et al. 2008. Molecular basis for Staphylococcus aureus-mediated platelet aggregate formation under arterial shear in vitro. Arterioscler. Thromb. Vasc. Biol. 28:335–340 [DOI] [PubMed] [Google Scholar]
- 27. Kerrigan S. W., Cox D. 2010. Platelet-bacterial interactions. Cell. Mol. Life Sci. 67:513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kline K. A., Dodson K. W., Caparon M. G., Hultgren S. J. 2010. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol. 18:224–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kline K. A., et al. 2009. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J. Bacteriol. 191:3237–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kratovac Z., Manoharan A., Luo F., Lizano S., Bessen D. E. 2007. Population genetics and linkage analysis of loci within the FCT region of Streptococcus pyogenes. J. Bacteriol. 189:1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lebreton F., et al. 2009. ace, which encodes an adhesin in Enterococcus faecalis, is regulated by Ers and is involved in virulence. Infect. Immun. 77:2832–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lepage E., et al. 2006. Comparative genomic hybridization analysis of Enterococcus faecalis: identification of genes absent from food strains. J. Bacteriol. 188:6858–6868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Linden S., et al. 2002. Strain- and blood group-dependent binding of Helicobacter pylori to human gastric MUC5AC glycoforms. Gastroenterology 123:1923–1930 [DOI] [PubMed] [Google Scholar]
- 34. Loughman A., et al. 2005. Roles for fibrinogen, immunoglobulin and complement in platelet activation promoted by Staphylococcus aureus clumping factor A. Mol. Microbiol. 57:804–818 [DOI] [PubMed] [Google Scholar]
- 35. Maione D., et al. 2005. Identification of a universal group B Streptococcus vaccine by multiple genome screen. Science 309:148–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mandlik A., Swierczynski A., Das A., Ton-That H. 2008. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 16:33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mitchell J., Siboo I. R., Takamatsu D., Chambers H. F., Sullam P. M. 2007. Mechanism of cell surface expression of the Streptococcus mitis platelet binding proteins PblA and PblB. Mol. Microbiol. 64:844–857 [DOI] [PubMed] [Google Scholar]
- 38. Mitchell J., Sullam P. M. 2009. Streptococcus mitis phage-encoded adhesins mediate attachment to {alpha}2-8-linked sialic acid residues on platelet membrane gangliosides. Infect. Immun. 77:3485–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mora M., et al. 2005. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. U. S. A. 102:15641–15646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moreillon P., Que Y. A., Bayer A. S. 2002. Pathogenesis of streptococcal and staphylococcal endocarditis. Infect. Dis. Clin. North Am. 16:297–318 [DOI] [PubMed] [Google Scholar]
- 41. Murray B. E. 1998. Enterococci, p. 1723–1730 In Gorbach S. L., Bartlett J. G., Blacklow N. R. (ed.), Infectious diseases, 2nd ed. W. B. Saunders Company, Philadelphia, PA [Google Scholar]
- 42. Murray B. E. 1990. The life and times of the Enterococcus. Clin. Microbiol. Rev. 3:46–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakata M., et al. 2009. Mode of expression and functional characterization of FCT-3 pilus region-encoded proteins in Streptococcus pyogenes serotype M49. Infect. Immun. 77:32–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nallapareddy S. R., Murray B. E. 2006. Ligand-signaled upregulation of Enterococcus faecalis ace transcription, a mechanism for modulating host-E. faecalis interaction. Infect. Immun. 74:4982–4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nallapareddy S. R., Murray B. E. 2008. Role played by serum, a biological cue, in the adherence of Enterococcus faecalis to extracellular matrix proteins, collagen, fibrinogen, and fibronectin. J. Infect. Dis. 197:1728–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nallapareddy S. R., Qin X., Weinstock G. M., Hook M., Murray B. E. 2000. Enterococcus faecalis adhesin, Ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. Infect. Immun. 68:5218–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nallapareddy S. R., Singh K. V., Duh R. W., Weinstock G. M., Murray B. E. 2000. Diversity of ace, a gene encoding a microbial surface component recognizing adhesive matrix molecules, from different strains of Enterococcus faecalis and evidence for production of Ace during human infections. Infect. Immun. 68:5210–5217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nallapareddy S. R., Singh K. V., Sillanpää J., Zhao M., Murray B. E. 2011. Relative contributions of Ebp pili and the collagen adhesin Ace to host extracellular matrix protein adherence and experimental urinary tract infection by Enterococcus faecalis OG1RF. Infect. Immun. 79:2901–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nallapareddy S. R., Singh K. V., Murray B. E. 2008. Contribution of the collagen adhesin Acm to pathogenesis of Enterococcus faecium in experimental endocarditis. Infect. Immun. 76:4120–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nallapareddy S. R., et al. 2006. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Invest. 116:2799–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Palmer K. L., et al. 2010. High-quality draft genome sequences of 28 Enterococcus sp. isolates. J. Bacteriol. 192:2469–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pillai S. K., et al. 2004. Effects of glucose on fsr-mediated biofilm formation in Enterococcus faecalis. J. Infect. Dis. 190:967–970 [DOI] [PubMed] [Google Scholar]
- 53. Podbielski A. 2007. Flexible architecture of the Streptococcus pyogenes FCT genome region: finally the clue for understanding purulent skin diseases and long-term persistence? J. Bacteriol. 189:1181–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Posada D. 2002. Evaluation of methods for detecting recombination from DNA sequences: empirical data. Mol. Biol. Evol. 19:708–717 [DOI] [PubMed] [Google Scholar]
- 55. Quigley B. R., Zahner D., Hatkoff M., Thanassi D. G., Scott J. R. 2009. Linkage of T3 and Cpa pilins in the Streptococcus pyogenes M3 pilus. Mol. Microbiol. 72:1379–1394 [DOI] [PubMed] [Google Scholar]
- 56. Rasmussen M., Johansson D., Sobirk S. K., Morgelin M., Shannon O. 2010. Clinical isolates of Enterococcus faecalis aggregate human platelets. Microbes Infect. 12:295–301 [DOI] [PubMed] [Google Scholar]
- 57. Ruiz-Garbajosa P., et al. 2006. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 44:2220–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sawyer S. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6:526–538 [DOI] [PubMed] [Google Scholar]
- 59. Scheld W. M., Zak O., Vosbeck K., Sande M. A. 1981. Bacterial adhesion in the pathogenesis of infective endocarditis. Effect of subinhibitory antibiotic concentrations on streptococcal adhesion in vitro and the development of endocarditis in rabbits. J. Clin. Invest. 68:1381–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Siboo I. R., Cheung A. L., Bayer A. S., Sullam P. M. 2001. Clumping factor A mediates binding of Staphylococcus aureus to human platelets. Infect. Immun. 69:3120–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sillanpaa J., et al. 2009. A family of fibrinogen-binding MSCRAMMs from Enterococcus faecalis. Microbiology 155:2390–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sillanpaa J., et al. 2008. Identification and phenotypic characterization of a second collagen adhesin, Scm, and genome-based identification and analysis of 13 other predicted MSCRAMMs, including four distinct pilus loci, in Enterococcus faecium. Microbiology 154:3199–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sillanpaa J., et al. 2010. Characterization of the ebp pilus-encoding operon of Enterococcus faecium and its role in biofilm formation and virulence in a murine model of urinary tract infection. Virulence 1:236–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sillanpaa J., Xu Y., Nallapareddy S. R., Murray B. E., Hook M. 2004. A family of putative MSCRAMMs from Enterococcus faecalis. Microbiology 150:2069–2078 [DOI] [PubMed] [Google Scholar]
- 65. Singh K. V., Coque T. M., Weinstock G. M., Murray B. E. 1998. In vivo testing of an Enterococcus faecalis efaA mutant and use of efaA homologs for species identification. FEMS Immunol. Med. Microbiol. 21:323–331 [DOI] [PubMed] [Google Scholar]
- 66. Singh K. V., Nallapareddy S. R., Murray B. E. 2007. Importance of the ebp (endocarditis- and biofilm-associated pilus) locus in the pathogenesis of Enterococcus faecalis ascending urinary tract infection. J. Infect. Dis. 195:1671–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Singh K. V., Nallapareddy S. R., Sillanpaa J., Murray B. E. 2010. Importance of the collagen adhesin Ace in pathogenesis and protection against Enterococcus faecalis experimental endocarditis. PLoS Pathog. 6:e1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Smith J. M. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126–129 [DOI] [PubMed] [Google Scholar]
- 69. Soriani M., Telford J. L. 2010. Relevance of pili in pathogenic streptococci pathogenesis and vaccine development. Future Microbiol. 5:735–747 [DOI] [PubMed] [Google Scholar]
- 70. Stern A., et al. 2007. Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 35:W506–W511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sullam P. M., Valone F. H., Mills J. 1987. Mechanisms of platelet aggregation by viridans group streptococci. Infect. Immun. 55:1743–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tendolkar P. M., Baghdayan A. S., Shankar N. 2006. Putative surface proteins encoded within a novel transferable locus confer a high-biofilm phenotype to Enterococcus faecalis. J. Bacteriol. 188:2063–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tomita H., Ike Y. 2004. Tissue-specific adherent Enterococcus faecalis strains that show highly efficient adhesion to human bladder carcinoma T24 cells also adhere to extracellular matrix proteins. Infect. Immun. 72:5877–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Usui Y., Ichiman Y., Suganuma M., Yoshida K. 1991. Platelet aggregation by strains of enterococci. Microbiol. Immunol. 35:933–942 [DOI] [PubMed] [Google Scholar]
- 75. Veening J. W., Smits W. K., Kuipers O. P. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62:193–210 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.