Abstract
Chronic lung infection is the major cause of morbidity and premature mortality in cystic fibrosis (CF) patients. Bacteria of the Burkholderia cepacia complex are the most threatening pathogens in CF, and a better understanding of how these bacteria adapt to the CF airway environment and resist the host defense mechanisms and therapeutically administered antibiotics is crucial. To provide clues to the adaptive strategies adopted by Burkholderia cenocepacia during long-term colonization, we carried out a phenotypic assessment of 11 clonal variants obtained at the major Portuguese CF Center in Lisbon from sputa of the same CF patient during 3.5 years of colonization of the lungs, until the patient's death with cepacia syndrome. Phenotypic characterization included susceptibility assays against different classes of antimicrobials and characterization of cell motility, cell hydrophobicity and zeta potential, colony and cell morphology, fatty acid composition, growth under iron limitation/load conditions, exopolysaccharide production, and size of the biofilms formed. The results suggest the occurrence of clonal expansion during long-term colonization. For a number of the characteristics tested, no isolation time-dependent consistent alteration pattern could be identified. However, the values for antimicrobial susceptibility and swarming motility for the first B. cenocepacia isolate, thought to have initiated the infection, were consistently above those for the clonal variants obtained during the course of infection, and the opposite was found for the zeta potential. The adaptive strategy for long-term colonization, described here for the first time, involved the alteration of membrane fatty acid composition, in particular a reduction of the degree of fatty acid saturation, in the B. cenocepacia variants retrieved, along with the deterioration of pulmonary function and severe oxygen limitation.
INTRODUCTION
Morbidity and mortality in cystic fibrosis (CF) relate to chronic airway infection with a variety of bacterial species, which contributes significantly to tissue destruction and continuous deterioration of lung function (39, 40). In particular, when Pseudomonas aeruginosa and Burkholderia cepacia complex (BCC) bacteria become established, these bacteria are difficult to eradicate from CF lungs due to their intrinsic resistance to multiple antibiotics and to antimicrobial peptides of the innate immunity and also to a rapid development of multidrug resistance. The BCC is a heterogeneous group that comprises at least 17 closely related species that are ubiquitous in the environment (28). A few recent studies have gained insights into the complexity of the strategies developed by P. aeruginosa cells in order to adapt to the stressing conditions to which they are exposed in CF airways (16, 39, 40). However, equivalent studies on BCC bacteria are still lacking, although infections involving these bacteria, especially Burkholderia cenocepacia, are particularly feared by CF patients because, in contrast to the case for P. aeruginosa, a subset of patients infected with these bacteria succumb to “cepacia syndrome,” which is characterized as a fatal necrotizing pneumonia with bacteremia (8, 28). Moreover, CF patients infected with B. cenocepacia have a substantially worse prognosis than those infected with P. aeruginosa only, and many centers refuse to perform lung transplantation on CF patients colonized with BCC bacteria (29, 41).
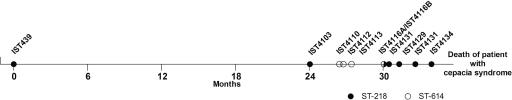
In many human infections, hosts and pathogens may coexist for years. During chronic colonization of a CF patient's airways, bacteria of the BCC experience changing selection pressures, in particular those resulting from challenges of the immune defenses, antimicrobial therapy, and oxygen limitation (21). These stressing conditions were found to lead to the adaptive evolution of P. aeruginosa, a major respiratory pathogen in CF (40). Over time, during long-term colonization, multiple phenotypic variants of the underlying clonal population emerge and become established in the patient's airways as the result of genetic adaptations (28). Clonal variants of BCC bacteria recovered from several chronically colonized CF patients at the major Portuguese CF treatment center, at Santa Maria Hospital (HSM), were found to exhibit various antimicrobial susceptibility levels (25, 27). In general, the more resistant variants were isolated following pulmonary exacerbation and aggressive antibiotic therapy (25). In order to obtain clues to the adaptive strategies developed by B. cenocepacia during long-term colonization of CF lungs, in the present study we carried out a phenotypic assessment of a number of relevant characteristics of 11 sequential isolates of B. cenocepacia (recA lineage III-A) obtained at HSM from the same CF patient during molecular epidemiological studies carried out by our research group (9, 10). This patient (patient J) was chronically colonized with the same B. cenocepacia strain for 3.5 years, from January 1999 to July 2002, until the patient's death with cepacia syndrome following progressive deterioration of pulmonary function (8, 9). The clonal nature of the B. cenocepacia isolates under study, as shown by the common ribopattern with EcoRI produced by these variants (9), was confirmed in the present work by their multilocus sequence typing (MLST) profiles (2). A preliminary phenotypic assessment of some of these clonal variants had been performed before by comparing antimicrobial susceptibility profiles (25). In the present study, this work was extended to all of the isolates retrieved from the patient and to a number of other relevant phenotypes in the context of persistent respiratory infections in CF patients.
MATERIALS AND METHODS
Bacterial isolates and culture conditions.
Eleven B. cenocepacia recA lineage III-A isolates (9), obtained at the HSM Cystic Fibrosis Center in Lisbon, Portugal, were used in this work (Table 1). Isolates were obtained from January 1998 to July 2002, as part of the hospital routine, from respiratory secretions of the same chronically infected CF patient during prolonged colonization. According to this routine, sputum samples are obtained from CF patients every 2 to 3 months, during periodic consultations to monitor their clinical status, or more often for patients showing clinical deterioration. Isolates IST4116A and IST4116B, with different colony morphologies, were obtained in the same isolation procedure. These isolates belong to the same clonal complex, as do all other isolates tested in this study (9). Bacterial cultures were stored at −80°C in 1:1 (vol/vol) glycerol. When in use, bacteria were cultivated on Luria-Bertani agar (LB agar; Difco, Sparks, MD) plates.
Table 1.
Clonal isolates of B. cenocepacia obtained from the same persistently colonized CF patienta
All isolates had the same ribopattern (ribopattern 11), as determined by Cunha et al. (9). The species and ribopattern of IST4134 were determined in the present work.
b Allelic profiles were determined as proposed by Baldwin et al. (2).
c Isolates IST4116A and IST4116B were obtained during the same isolation procedure.
MLST analysis.
Total genomic DNAs were extracted from B. cenocepacia isolates harvested from overnight growth in LB medium with orbital agitation at 37°C, using a cell and tissue kit (Gentra Systems, Qiagen, Germany). The concentrations of genomic DNA solutions were estimated using an ND-1000 spectrophotometer (NanoDrop). MLST analysis was performed using the primers and conditions described by Baldwin et al. (2) for seven conserved housekeeping genes (atpD, gltB, gyrB, recA, lepA, phaC, and trpB). Amplification reactions for all primers were carried out using an initial step of denaturation for 2 min at 94°C, followed by 30 cycles of 1 min at 94°C for denaturation, 1 min at 58°C for primer annealing, and 2 min at 72°C for polymerization, followed by a final extension step of 72°C for 7 min. The amplification mixtures contained the following, in a total volume of 50 μl: 75 ng of DNA template, 1 μl of 0.8 mM (each) deoxynucleoside triphosphates, 1.5 μl of 50 mM MgCl2, 1 μl of 20 pmol (each) forward and reverse primers, 1.25 U of Taq polymerase (Biotaq Taq polymerase; Bioline), and 5 μl of reaction buffer (10×; supplied by the polymerase manufacturer). The amplification products were confirmed following the separation of the PCR products by 0.7% (wt/vol) horizontal agarose gel electrophoresis, and the bands were excised, purified using a gel extraction kit (JETquick spin column technique; Genomed, Germany), and sequenced. The sequence data for each isolate were added to a group of known sequences, available in the Burkholderia cepacia complex MLST database (http://pubmlst.org/bcc), to simultaneously be aligned and edited to the correct sequence length using ClustalW software (www.clustal.org). Alleles for each of the seven loci were assigned, and the allelic profile (string of seven integers) was used to define the sequence type (ST), using the same database. A new ST number was assigned to a novel allelic combination that was submitted and added to the BCC MLST database under the designation ST-614.
Antimicrobial susceptibility assays.
The susceptibilities of the isolates under study to several antimicrobials of different classes were compared using the Etest and broth microdilution methods. Due to the high antibiotic resistance of the isolates tested, the antibiotic concentrations used in the Etest strips were not high enough to differentiate the MIC values for a number of isolates and antimicrobials (e.g., gentamicin and ceftazidime). Given this, the MIC values were also assessed using the broth microdilution method.
(i) Etest method.
Isolated colonies from LB agar plates were grown in LB broth (Difco) at 37°C until mid-exponential phase and then diluted to an optical density at 640 nm (OD640) of 0.2 in 0.9% NaCl (wt/vol), and 100 μl of this cell suspension was plated onto Mueller-Hinton (MH) agar (Difco) plates. Etest strips (AB Biodisk, Solna, Sweden) with the antimicrobial agents to be tested (ceftazidime, 0.016 to 256 μg ml−1; imipenem, 0.002 to 32 μg ml−1; meropenem, 0.002 to 32 μg ml−1; tobramycin, 0.064 to 1,024 μg ml−1; gentamicin, 0.064 to 1,024 μg ml−1; ciprofloxacin, 0.002 to 32 μg ml−1; and trimethoprim-sulfamethoxazole, 0.002 to 32 μg ml−1) were used, and results were interpreted according to CLSI guidelines (7). MIC values are mean values for at least three independent experiments.
(ii) Broth microdilution susceptibility test method.
Liquid cultures grown in LB medium at 37°C until the mid-exponential phase were harvested by centrifugation and then suspended in MH (Difco) broth and diluted to a standardized culture OD640 of 0.21. Of these cell suspensions, 190 μl was used to inoculate the wells of a 96-well polystyrene microtiter plate (Greiner Bio-One, Frickenhausen, Germany) containing 10 μl of the antibiotic solution to be tested. This test was performed in accordance with the recommendations of NCCLS (31), in at least three independent experiments. Ceftazidime, gentamicin, and tobramycin were obtained from Sigma-Aldrich (St. Louis, MO), and ciprofloxacin was obtained from ICN Biomedicals Inc. (OH); all antimicrobials were in powder form. The range of concentrations used varied depending on the antimicrobial tested, with ranges of 128 to 1,700 μg ml−1 for gentamicin, 1.75 to 10 μg ml−1 for ciprofloxacin, 128 to 512 μg ml−1 for tobramycin, 60 to 1,150 μg ml−1 for ceftazidime, and 12 to >32 μg ml−1 for meropenem. The microplates were incubated, and the OD640 values of the cultures in the wells were measured in a VERSAmax tunable microplate reader (Molecular Devices Corporation, Sunnyvale, CA) after 24 h of incubation at 37°C. The microplate reader was connected to a computer running SoftMax Pro 4.8 spectrophotometric software (Molecular Devices Corporation). Positive (without antibiotic) and negative (not inoculated) controls were included.
Colony morphotypes.
The comparison of colony morphologies was performed as described by Bernier et al. (3). The isolates were grown in LB (Difco) broth at 37°C for approximately 24 h with shaking (250 rpm), and after serial dilution into fresh LB broth, they were plated onto LB agar plates and incubated at 37°C for 48 h, followed by an additional period of 24 h at room temperature to more clearly distinguish the three different colony morphologies, identified as “smooth,” “rough,” and “semirough.” A Canon digital camera using a Stemi 2000-C stereomicroscope at a magnification of ×50 was used to capture pictures of the colonies.
Cell morphology.
Cell size and morphological parameters, such as equivalent diameter, circularity, and elongation, were determined by fluorescence microscopy. Cells were harvested and stained with a Live/Dead BacLight bacterial viability kit from Molecular Probes (Invitrogen Co., Spain). The samples were prepared according to the protocol provided by the manufacturer and were observed using an Olympus CX40 microscope with an Olympus U-RFL-T burner and a U-MWB mirror cube unit (BP450-480 excitation filter and BA515 barrier filter). Images were captured by an Evolution MP5.1 charge-coupled device (CCD) color camera using Image-Pro Plus software, both from Media Cybernetics, Inc. Image analysis was carried out using Visilog 5 software from Noesis (13). At least 10 images were taken for each sample. The error associated with image analysis was ±6.8%, based on the standard deviation and sample mean for 10 repeated images of the same sample, quoted for a confidence interval of 99.5%.
Zeta potential assay.
The zeta potential of cells collected in the mid-exponential phase, washed twice, and suspended in 10 mM KNO3 was calculated from the measured electrophoretic mobility by considering the Smoluchowski approximation by use of Zetasizer v6.12 software from Malvern Instruments, Ltd. (United Kingdom). The electrophoretic mobility was measured using a Zetasizer Nano ZS Doppler electrophoretic light scattering analyzer from Malvern Instruments, Ltd.
Determination of bacterial hydrophobicity and cell migration velocity.
The cell surface hydrophobicity of B. cenocepacia isolates under study was assessed by measuring the adhesion of cells to n-hexadecane, based on the method proposed by Rosenberg (38). Cells from cultures in the mid-exponential growth phase were washed twice with 50 mM phosphate-buffered saline (PBS; pH 7.0) and resuspended in the same buffer. Bacterial suspensions (1.2 ml) were overlaid with 0.2 ml of n-hexadecane (Sigma-Aldrich) in test tubes. These were agitated at full speed for 45 s on a vortex machine and allowed to stand still for 15 s. The migration of the cells was assessed by measuring the OD600 of the aqueous phase at 1-min intervals after removing the organic phase.
EPS production.
Exopolysaccharide (EPS) production was assessed by confluent growth of the different isolates on LB agar plates. Plates were inoculated with 100 μl of a suspension of cells harvested during the exponential phase of growth and resuspended to obtain a standardized OD640 of 0.2 ± 0.02. These plates were incubated for 5 days at 37°C and then scraped, and the material obtained was resuspended in 0.9% NaCl (wt/vol). The bacterial cells present in these suspensions were removed by centrifugation at 20,000 × g for 15 min. The EPS was precipitated from the cell-free supernatant by the addition of 2.5 volumes of cold ethanol, air dried, and redissolved in distilled water. The total sugar content was assessed by the phenol-sulfuric acid method (15), using the EPS produced by isolate B. cepacia IST408 as a standard. For this purpose, the EPS solution was further dialyzed against distilled water at 4°C for 24 h and then recovered by freeze-drying. The cell pellets obtained from each plate were washed once with 0.9% NaCl, and the protein content was quantified by the biuret method, using bovine serum albumin (BSA; Sigma-Aldrich) as a standard (22). EPS production was expressed in grams of total sugars per protein (g·g protein−1). The results are means for at least three independent cultivations and three determinations of the total sugar and protein contents for each sample.
Biofilm assays.
The biofilm formation assay was based on the methodology described by O'Toole and Kolter. Liquid cultures grown in LB medium at 37°C until the mid-exponential phase were diluted to a standardized culture OD640 of 0.5, and 20 μl of cell suspension was used to inoculate the wells of a 96-well polystyrene microtiter plate (Greiner Bio-One, Frickenhausen, Germany) containing 180 μl of LB medium. Wells containing sterile growth medium were used as negative controls. These microtiter plates were incubated at 37°C for 24 h without shaking. Quantification of the amount of biomass attached to the microtiter dish was carried out as described before, using crystal violet staining (33, 38).
Effect of iron availability on bacterial growth.
The effect of iron availability on the growth of the different B. cenocepacia isolates examined was assessed using an iron-poor chemically defined medium (CDM) (6), containing 48 mM glucose, 7.4 mM KCl, 6 mM NaCl, 48 mM (NH4)2SO4, 0.5 mM MgSO4·7H2O, 60 mM MOPS (morpholinepropanesulfonic acid), 3.8 mM K2HPO4, and 0.1% Casamino Acids (Bacto; Becton Dickinson and Company). Iron-loaded CDM was obtained by CDM supplementation with 100 μM FeCl3. Cells of the different clonal isolates in the mid-exponential phase of growth were inoculated, at an initial OD640 of 0.05, into CDM either supplemented or not supplemented with iron. Growth was followed by measuring the culture OD640 and, in certain cases, also the number of viable cells, determined based on the number of CFU. The values for CFU/ml shown are the mean values for three independent growth experiments.
Lipid extraction and determination of fatty acid composition.
Bacterial cells used to assess fatty acid composition were grown under both aerophilic and microaerophilic conditions in LB agar plates incubated for 24 h at 37°C. The microaerophilic atmosphere, containing 5 to 8% oxygen and 12 to 15% carbon dioxide, was generated in sealed jars by use of a GENbox microaerator (bioMérieux, Marcy L'Etoile, France). The extraction of cellular lipids was carried out as described by Findlay et al. (17). The fatty acids were derivatized into fatty acid methyl esters (FAME) and analyzed by gas chromatography on an Agilent 6890N gas chromatograph with a flame ionization detector (FID) and a 7683 B series injector equipped with a 30-m HP-5 capillary column from J&W Scientific, as described before (12). Peak identification was achieved using qualitative standard mixtures of bacterial FAME and polyunsaturated fatty acids, both from Supelco (Sigma-Aldrich), and a mixture of methyl cis-11-octadecenoate from Sigma-Aldrich. The average error associated with the GC quantification of each FAME, calculated based on seven independently prepared standard solutions, was ±5.1%, quoted for a confidence interval of 99.5%.
Motility assays. (i) Swimming assay.
Dry swim agar plates containing 30 ml of 1% (wt/vol) tryptone (Difco), 0.5% (wt/vol) NaCl, and 0.3% (wt/vol) agar (Oxoid, Cambridge, United Kingdom) were point inoculated by use of a sterile toothpick with bacterial cultures from isolated colonies grown on Pseudomonas isolation agar (PIA; Difco) for 24 h at 37°C. These swim agar plates were incubated at 37°C for 24 h, and the circular turbid zone formed by the bacterial cells migrating away from the point of inoculation was measured. Values are the means for at least three independent growth experiments.
(ii) Swarming assay.
Dry swarm agar plates containing 30 ml of 0.8% (wt/vol) nutrient broth (Difco), 0.5% (wt/vol) glucose, and 0.5% (wt/vol) agar (Oxoid) were point inoculated and incubated as described for the swimming assay. The diameter of the zone of growth around the point of inoculation was measured. Values are means for at least three independent growth experiments.
RESULTS
MLST analysis.
MLST of the 11 B. cenocepacia sequential isolates under study was performed through assessment of the allelic variation of seven housekeeping locus sequences proposed by Baldwin et al. (2). These genes were amplified and sequenced, and the allelic types of the seven loci were assigned (Table 1) and submitted to the Burkholderia cepacia complex MLST database. They are available publicly at http://pubmlst.org/bcc/. Six of the tested isolates (IST4103, IST4116B, IST4131, IST4129, IST4130, and IST4134) had the same allelic profile as the first isolate obtained from CF patient J (IST439) and were assigned to ST-218. This ST was already present in the BCC MLST database before this work, since isolate IST439 had been analyzed by Baldwin et al. (2). The above isolates are considered clonal because they are indistinguishable at the seven loci. Isolates IST4110, IST4112, IST4113, and IST4116A produced a new ST that was designated ST-614 and added to the BCC MLST database (Table 1). Although the isolates that exhibit sequence types ST-218 and ST-614 are not identical clones, they are considered part of the same clonal complex because ST-614 is a single-locus variant (SLV) of ST-218 (Table 1). This variation is registered at the gyrB locus (encoding subunit B of DNA gyrase) and involves 14 different nucleotides between gyrB1 and gyrB186 (Fig. 1). The differences registered in gyrB suggest that other mutations might have occurred in other loci during prolonged colonization in the airways of the CF patient and during aggressive antibiotic therapy. The MLST profiles of the isolates analyzed in this study confirmed that they all belong to the B. cenocepacia species and are consistent with the clonal nature of these isolates proposed before, based on other molecular methods (9). The species and ribopattern of the last isolate obtained from the CF patient (IST4134) were also determined in this study because this isolate was missing in the cohort of isolates examined previously (9).
Fig. 1.
Polymorphic sites within the gyrB locus of the BCC MLST scheme corresponding to the new sequence type, ST-614, described in this work, compared to ST-218. Nucleotide sites are numbered from the first nucleotide position in the gyrB gene.
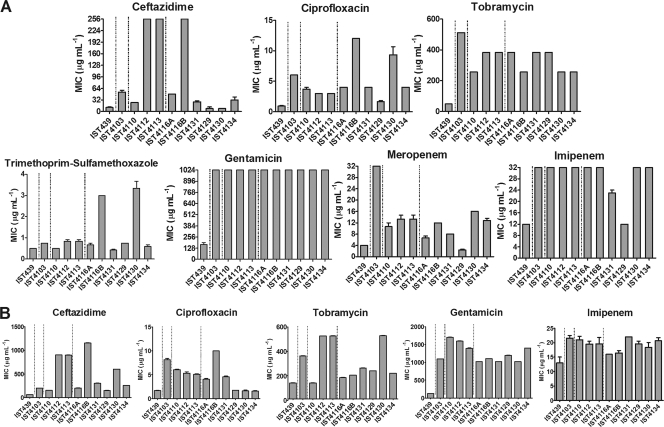
Antimicrobial susceptibility patterns of B. cenocepacia clonal isolates.
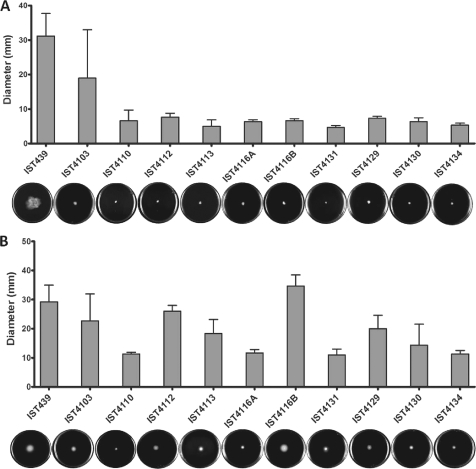
The susceptibility patterns of the sequential B. cenocepacia clonal isolates under study were determined by the Etest method, using seven antimicrobials of five distinct classes: cephalosporins (ceftazidime) and carbapenems (imipenem and meropenem), targeting cell wall biosynthesis; aminoglycosides, targeting protein synthesis by binding to the 30S ribosomal unit (gentamicin and tobramycin); fluoroquinolones, targeting DNA replication (ciprofloxacin); and the folate pathway inhibitor trimethoprim-sulfamethoxazole. The 11 clonal isolates under analysis showed different resistance profiles against all antibiotics tested (Fig. 2), consistent with previous results reported for the first 10 isolates retrieved from patient J for piperacillin-tazobactam, ceftazidime, and imipenem (25). The MIC values obtained based on the Etest method varied, with ranges of 192 to >1,024 μg ml−1 (gentamicin), 1 to 12 μg ml−1 (ciprofloxacin), 48 to 512 μg ml−1 (tobramycin), 0.5 to 3.5 μg ml−1 (trimethoprim-sulfamethoxazole), 12 to >32 μg ml−1 (imipenem), 2 to >32 μg ml−1 (meropenem), and 8 to >256 μg ml−1 (ceftazidime). The MIC values were also obtained based on the broth microdilution method for the 11 isolates, especially for those antimicrobials whose MIC values for the different isolates could not be differentiated by the Etest method due to these being the maximal concentration that could be tested. Values varied within wide ranges of concentrations: 128 to 1,700 μg ml−1 for gentamicin, 1.75 to 10 μg ml−1 for ciprofloxacin, 128 to 512 μg ml−1 for tobramycin, 12 to 22 μg ml−1 for imipenem, and 60 to 1,150 μg ml−1 for ceftazidime. The first isolate was found to be consistently more susceptible to all antimicrobials tested than all of the sequential clonal variants retrieved from the patient.
Fig. 2.
MIC values, determined using the Etest method (A) and the microdilution method (B), for the activities of the indicated antimicrobial agents of different classes against B. cenocepacia clonal isolates obtained during long-term colonization of CF patient J. Vertical lines indicate when the CF patient received intravenous antibiotic therapy with ceftazidime and gentamicin.
Colony morphotype, cell size and morphology, zeta potential, and hydrophobicity.
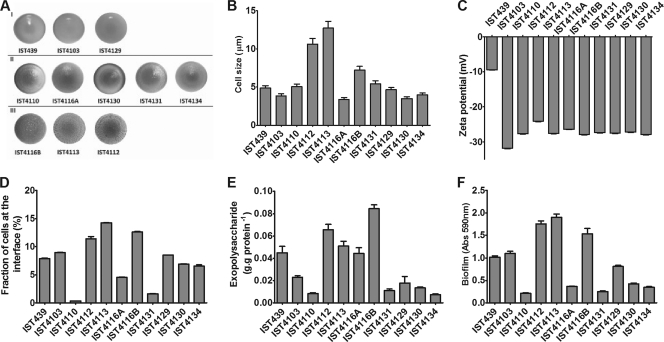
The B. cenocepacia clonal variants under study exhibited three different colony morphotypes (Fig. 3A). The colony morphotype of the initial isolate (IST439) was considered smooth, as well as the morphotype exhibited by isolates IST4103 and IST4129. The isolates IST4112, IST4113, and IST4116B were considered rough, while the remaining isolates, IST4110, IST4116A, IST4130, IST4131, and IST4134, which exhibited an intermediary morphotype, were considered semirough.
Fig. 3.
(A) Colony morphology. I, smooth; II, semirough; III, rough. (B) Cell size. (C) Zeta potential. (D) Fraction of cells at the interface of water and n-hexadecane. (E) Exopolysaccharide produced. (F) Sizes of biofilms formed by the sequential clonal isolates of B. cenocepacia retrieved from the same persistently colonized patient J.
The cell size of the B. cenocepacia clonal variants also varied, with isolates IST4112, IST4113, and IST4116B exhibiting cell sizes 2.4-, 2.9-, and 1.6-fold higher, respectively, than the average size of the remaining isolates (Fig. 3B). Curiously, these were the same isolates that formed rough colonies (Fig. 3A). The cells of these isolates had the shape of almost perfect circles (circularity of ca. 1 and elongation of ca. 1.1) (data not shown), while the other isolates exhibited more elongated forms (circularity and elongation, ca. 1.2 to 1.3). The value of the zeta potential (an indirect measure of the cell surface charge [13]) of the first isolate, IST439, was significantly below the values of all other clonal isolates (−9.5 mV, compared with values of around −30 mV) (Fig. 3C). The bacterial cell surface hydrophobicity was determined by the microbial adhesion to hydrocarbons (MATH) method (13, 18), using n-hexadecane as a solvent. This test has commonly been used to assess the hydrophobicity of bacterial cells, although cell migration toward the n-hexadecane layer may be the result of both hydrophobic and electrostatic interactions (18). The velocity of cell migration toward the n-hexadecane layer in the MATH test showed no significant differences between the tested isolates (data not shown). However, the fraction of cells at the interface suggested that isolate IST4113 was the most hydrophobic compared with the first isolate, IST439, while isolates IST4110, IST4116A, and IST4131 had the lowest surface hydrophobicity values (Fig. 3D).
Exopolysaccharide production and size of the biofilms formed.
The amount of EPS produced (Fig. 3E) and the size of the biofilm formed (Fig. 3F) in vitro by the different clonal variants were also found to vary along the colonization period. Isolates IST4112, IST4113, and IST4116B formed the largest biofilms, while the biofilms produced by IST4110, IST4116A, IST4131, IST4130, and IST4134 were much smaller, with the first isolate, IST439, exhibiting an intermediary value (Fig. 3F). In general, a correlation could be established between the amounts of EPS produced and the sizes of the biofilms formed by the different B. cenocepacia clonal isolates, with the exception of IST4103 and IST4116A. This observation is consistent with the concept that the EPS produced contributes to the development of thicker and more stable biofilms in bacteria of the B. cepacia complex (11).
Growth efficiency under iron limitation.
The growth curves of the isolates under study were compared using an iron-limited minimal CDM and the same medium supplemented with 100 μM Fe3+ (iron-loaded medium). Examples of the growth curves obtained for isolates IST439, IST4113, IST4116A, and IST4134 in both media are shown in Fig. 4A. As expected, the biomass attained (associated with the culture OD640) after the same incubation time (when the culture entered the stationary phase of growth after 24 h of incubation) under iron limitation was significantly below the levels attained under iron saturation conditions (Fig. 4A and B). The results also show that the first isolate, IST439, exhibited less efficient growth under iron limitation than the other isolates, while IST4113's growth performance under iron limitation was the best, reaching both the highest biomass and viable cell concentrations (Fig. 4A to D). However, in the iron-supplemented medium, the growth of the first isolate, IST439, was faster, reaching the highest biomass concentration (Fig. 4A to D). Differences in the growth performance of isolates IST439 and IST4113 in iron-limited or iron-loaded medium were confirmed based on the viable cell concentrations of the two cultures (Fig. 4B) and are more evident using a linear scale (Fig. 4C). The comparison of culture optical densities reached by all isolates after 24 h of growth in iron-limiting and iron-loaded CDM (Fig. 4D) indicates that the first isolate was consistently the best suited to grow under iron-loaded conditions but, in general, exhibited the worst performance under iron limitation conditions. Exceptions were registered for isolates that exhibited a poor growth performance even under iron-loaded conditions.
Fig. 4.
(A) Growth curves on a semilog scale based on the OD640 values of isolates IST439, IST4103, IST4110, and IST4113 in iron-limited CDM (○) and in the same medium supplemented with 100 μM Fe3+ (•). (B) Growth curves based on the concentrations of viable cells (expressed as numbers of CFU per ml) in the cultures of isolates IST439 (• and ○) and IST4113 (■ and □) in the same iron-limited CDM (○ and □) and in this medium supplemented with 100 μM Fe3+ (• and ■). (C) Growth curves of IST439 (• and ○) and IST4113 (■ and □), compared on a linear scale, in CDM (○ and □) and in CDM supplemented with 100 μM Fe3+ (• and ■). (D) Culture optical densities after 24 h of growth in CDM or CDM supplemented with 100 μM Fe3+ for all sequential clonal variants of B. cenocepacia obtained during long-term colonization of patient J.
Fatty acid composition.
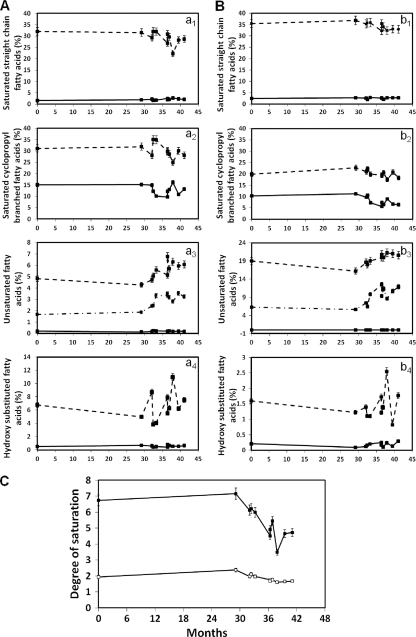
The most abundant fatty acids in the clonal variants examined were tetradecanoic (myristic) acid (C14:0), cis-9-hexadecenoic (palmitoleic) acid (C16:1ω7), methyl 3-hydroxytetradecanoic acid (C14:0 3-OH), hexadecanoic (palmitic) acid (C16:0), cis-9,1-methyleneoctadecanoic acid (cyC17:0), methyl 2-hydroxyhexadecanoic acid (C16:0 2-OH), cis-11-octadecenoic (vaccenic) acid (C18:1ω7cis), methyl trans-9- octadecenoate (elaidic) acid (C18:1ω9trans), and cis-9,10-methylenehexadecanoic acid (cyC19:0). These fatty acids, which have previously been identified in Burkholderia strains (5, 42), represented over 86% of the total amount of fatty acids determined for the isolates under study. In general, under both microaerophilic and aerophilic conditions, the content of saturated fatty acids decreased in the isolates retrieved along the colonization period in the CF patient's lungs (Fig. 5A and B), in particular the content of saturated cyclopropyl branched fatty acids in those isolates obtained during the last months of life. The decrease in the content of saturated fatty acids was followed by an increase in the percentage of unsaturated fatty acids registered from the first to the last isolates under both growth conditions, although the fatty acid contents were significantly different in cells grown under aerophilic and microaerophilic conditions.
Fig. 5.
Fatty acid compositions of sequential clonal variants of B. cenocepacia obtained during long-term colonization of patient J, grown on LB agar plates under aerophilic (A) and microaerophilic (B) conditions. The main fatty acids analyzed were saturated straight-chain fatty acids C14:0 (dashed line) and C16:0 (full line) (a1 and b1); saturated cyclopropyl branched fatty acids C17:0 cyclo (dashed line) and C19:0 cyclo (full line) (a2 and b2); unsaturated fatty acids C18:1ω9trans (full line), C18:1ω7cis (dashed line), and C16:1ω7 (dashed-dotted line) (a3 and b3); and hydroxy-substituted fatty acids C14:0 3-OH (full line) and C16:0 2-OH (dashed line) (a4 and b4). (C) Degrees of saturation of fatty acids in the same sequential clonal isolates of B. cenocepacia grown under aerophilic (■) or microaerophilic (□) conditions. The fatty acid compositions of these isolates are shown in panels A and B.
In general, the degree of saturation of the membrane fatty acids, defined as the ratio between the saturated fatty acids C16:0 and octadecanoic acid (C18:0) (Fig. 5, a1 and b1) and the unsaturated fatty acids methyl cis-9-hexadecenoate (palmitoleic) acid (C16:1cis9), C18:1ω7cis, and C18:1ω9trans (Fig. 5, a2 and b2), was found to be significantly lower in cells grown under microaerophilic conditions than under aerophilic conditions, decreasing in cells of the sequential clonal variants as the colonization time increased, approaching the date of the patient's death (Fig. 5C). The decrease of the saturation degree was accompanied by an increase of the percentage of unsaturated fatty acids (Fig. 5, a3 and b3). The cells grown under microaerophilic conditions exhibited a higher percentage of unsaturated fatty acids than the cells grown under aerophilic conditions. In the last isolates retrieved from the patient, the decrease of the saturation degree was related mainly to a decrease in the amount of C18:0 and C16:0 and to an increase in the amount of cis-11-octadecenoic acid (C18:1ω7cis). Since the presence of cis-11-octadecenoic acid in cell membranes is related to anaerophilic growth (20), the higher concentration of this fatty acid (Fig. 5, a3 and b3) registered in bacterial isolates obtained during the last stages of CF infection, when the level of oxygen in the airways was very limited, is also consistent with the hypothesis that fatty acid alteration is the result of bacterial adaptation to the selective pressure faced in CF lungs.
Swarming and swimming motility.
Compared with the first isolate, IST439, all of the other sequential clonal variants obtained during the 3.5 years of bacterial persistence in the patient's lungs exhibited lower swarming motilities (Fig. 6A). However, the variation in swimming motility values did not exhibit a specific pattern (Fig. 6B).
Fig. 6.
Swarming (A) and swimming (B) motilities of sequential clonal variants of B. cenocepacia retrieved during long-term colonization of patient J.
DISCUSSION
Bacteria of the BCC are ubiquitous in the environment, have very high metabolic versatility, and can cause chronic opportunistic infections in immunocompromised patients, in particular in patients with CF (28). Although these bacteria can colonize the lungs while causing no symptoms and having no long-term effect, in general they lead to chronic infection and to a continuous decline in lung function (28). In the worst-case scenario, they can cause the fatal “cepacia syndrome” (8, 28). Chronic infections with BCC bacteria are very difficult to treat due to their intrinsic resistance to a large number of antimicrobials and their ability to develop high-level resistance during antibiotic treatment and to adapt to and resist other adverse environmental conditions (4, 25). This fact severely limits the effective treatment of respiratory infections, making BCC bacteria very difficult (if it is even possible) to eradicate from the CF lung (8, 25, 28). In general, B. cenocepacia is the most common BCC species recovered in CF centers worldwide, and it is frequently associated with severe infections (26, 36). However, there are documented exceptions, in particular in the major Portuguese Cystic Fibrosis Center at HSM, in Lisbon, where the species B. cepacia dominates (8, 10). The first epidemiological survey of BCC bacteria involved in respiratory infections among the Portuguese CF population under surveillance at this center was reported by our laboratory in 2000 (37). This study was followed by others, covering isolates of B. cenocepacia, B. cepacia, Burkholderia multivorans, and Burkholderia stabilis obtained from 1995 to 2006 (9, 10, 11, 25). The present study was designed to try to obtain clues to the adaptive strategies adopted by B. cenocepacia during chronic infection through the systematic assessment of a number of relevant phenotypic characteristics, in the context of CF infection, of 11 serial clonal variants obtained at HSM from a CF patient who was chronically infected for 3.5 years, until the patient's death with cepacia syndrome. When this work was started, it was known that the recA restriction fragment length polymorphism (RFLP) and EcoRI ribopattern profiles of these B. cenocepacia isolates are indistinguishable (9). The MLST performed during this study revealed that four of the isolates exhibited a novel allelic profile, the sequence type ST-614, while the other isolates, including IST439, considered the variant that started the infection, belonged to the already described group ST-218. Although they are not identical clones, all of the isolates are part of the same clonal based upon related sequence types (BURST) group, since only one of the seven loci (the gyrB locus) tested exhibited an alteration (in 14 nucleotides). This result is in agreement with the idea that a CF patient's lungs can be infected chronically for years by one or a few lineages of P. aeruginosa or BCC species (9, 10, 14, 19). Together with other results obtained during this study, this observation also supports the concept of the occurrence of B. cenocepacia clonal expansion during chronic lung colonization, presumably as the result of mutations and selective pressures occurring in the CF lung environment, in particular those exerted by the immune defenses, antibiotic therapy, and oxygen limitation, as described for P. aeruginosa (32).
Concerning several of the phenotypic characteristics of the clonal variants under study, no colonization time-dependent alteration patterns could be identified. This fact could be the result of the heterogeneity of the colonizing clonal population and the changes of the selection pressures occurring during long-term colonization, in particular those related to the application of intravenous antibiotic therapy and the continuous or rapid deterioration of lung function. In fact, with only one exception, i.e., isolates IST4116A and IST4116B, obtained during the same isolation procedure and known to exhibit different colony morphologies, only a single isolate picked up at random per isolation date was tested. Remarkably, the phenotypic traits that did not show a clear time-dependent variation pattern were also found to be divergent in isolates IST4116A and IST4116B, obtained during the some isolation procedure. However, in general, the properties of the isolate considered to have initiated the infection appeared to differ significantly from those exhibited by the isolates obtained during the course of the infection during a period ranging from 29 to 41 months after the isolation of the first B. cenocepacia isolate. Indeed, the antimicrobial susceptibility of the first isolate was significantly higher than the susceptibilities of the other clonal variants toward all tested antimicrobials. This pattern is absolutely clear for the aminoglycoside gentamicin, used as intravenous therapy during specific periods of the patient's hospitalization due to pulmonary exacerbation. The ability of B. cenocepacia to develop high resistance levels during antibiotic treatment is in agreement with the generalized idea that this adaptive mechanism is among the important features contributing to persistent infection. Although bacterial resistance to antibiotics seems to be linked to the growth of bacteria in communities embedded in a protective polysaccharide matrix or biofilm, no correlation could be established for the different variants between antibiotic resistance and the size of the biofilm formed, indicating that other relevant mechanisms also contribute to the increased resistance registered toward several antimicrobials of different classes. Indeed, a number of underlying mechanisms were recently hypothesized based on a quantitative proteomic analysis of the first isolate, IST439, compared with the highly resistant variant IST4113 (27). In general, a correlation was found between the size of the biofilm and the amount of the exopolysaccharide cepacian synthesized by each variant, consistent with the proposed role of cepacian in the formation of larger biofilms in isogenic strains (11).
The swarming motility of the first isolate was found to be consistently higher than the swarming motility values of all subsequent clonal isolates, while a less negative value for the zeta potential was registered for cells of isolate IST439 than for the other clonal variants. These results suggest that the alteration of these two traits may be among the strategies used by B. cenocepacia to persist during progressive CF lung disease. Swarming motility involves the coordinated and rapid movement of a bacterial population across a semisolid surface, playing a crucial role in the establishment of a number of infections, in particular those in a CF patient's lungs (43). Although at this time there is no supported explanation for the higher level of swarming motility exhibited by the first isolate than by the subsequent isolates, we are tempted to hypothesize that this could be related to differences at the level of fatty acid metabolism that may affect biosurfactant production. Indeed, the swarming process involves the release of biosurfactants which act by reducing the local surface tension; rhamnolipids are the biosurfactant involved in swarming motility in P. aeruginosa (24). The less negative value of the zeta potential of IST439, which is associated with a less negative cell surface net charge, could also be related to higher values for swarming motility, since the cell surface charge has been proven to be important for cell-cell or cell-surface interactions essential in swarming motility (30).
During this study, we also compared the growth efficiencies of the clonal variants of B. cenocepacia under iron-limiting conditions. The main conclusion was that although the concentration of biomass produced by the first isolate, IST439, was higher than the concentration of biomass produced by all other isolates when cultures entered the stationary phase of growth after the same time of incubation in iron-loaded minimal medium, this isolate was less suited to growth under iron limitation conditions than most of the subsequent isolates. Pathogenic bacteria require iron as a cofactor for numerous metabolic enzymes, including those involved in aerobic respiration. The free iron concentration in living organisms is usually too low to be sufficient for bacterial growth, although higher concentrations may be present in the CF lung (34). Increased iron in the cystic fibrosis airway was proposed to play an important role in facilitating P. aeruginosa infection and contributing to anaerobic biofilm growth (35). Moreover, the importance of iron homeostasis in the CF lung and its role in determining the success and chronicity of P. aeruginosa infection are well documented (34, 35). Bacteria have evolved multiple metabolic pathways for efficient iron acquisition to successfully become established in the lungs, such as the synthesis of siderophores. Based on results from a quantitative proteomic analysis carried out recently in our laboratory, it is known that the clonal variant that exhibited the highest growth efficiency under iron limitation conditions (IST4113) has a higher content of proteins involved in binding and transport of iron ions than the first isolate, IST439, which showed one of the lowest growth performances under iron limitation conditions (27). These proteins include the TonB-dependent siderophore receptor, the TonB-dependent copper receptor, and the FAD-binding 9 siderophore-interacting protein (MxcB) (27).
The results of the present work also strongly suggest that the alteration of B. cenocepacia's ability to synthesize membranes with a different fatty acid composition, in particular at the level of fatty acid saturation, constitutes an important adaptation strategy to long-term colonization, especially at the end stage of CF lung disease. Indeed, the isolates recovered during the last 10 months of the patient's life exhibited a very evident time-dependent decrease of fatty acid saturation. At first, this adaptive response was considered intriguing because oxidative stress is among the stresses that bacteria are expected to be exposed to during colonization and because unsaturated fatty acids are more susceptible than saturated fatty acids to oxy-radical-mediated lipid peroxidation (44). Given that a minimum level of saturation was registered for the isolates obtained during the last months of life of patient J, we hypothesized that this adaptive trait could be due to severe oxygen depletion in the CF lungs. Consistent with this proposal, the fatty acid saturation degree and the amount of saturated cyclopropyl branched fatty acids in the isolates grown under microaerophilic conditions were significantly below those in isolates grown under aerophilic conditions. According to the hospital records, when isolate IST439 was obtained, the forced expiratory value in the first second (FEV1) was 22%, below the value obtained prior to its isolation (27%). No further values of FEV1 were registered during the later stages of the patient's life due to the highly severe deterioration of pulmonary function (8). It is known that in the lungs of CF patients, bacteria grow to high densities in mucopurulent material that is limited or depleted in oxygen. Although there is a generalized idea that growth in these circumstances is dependent on anaerobic nitrate respiration, microaerobic respiration appears to be the predominant mode of P. aeruginosa growth in CF lungs (1). Remarkably, during chronic infection of the CF lungs, oxygen-limiting conditions seem to contribute to persistent infection with P. aeruginosa (42). Despite the importance of bacterial metabolism under microaerophilic or anaerophilic conditions, little is known about the underlying mechanisms and how anaerobic metabolism contributes to a persistent infection, even for P. aeruginosa. However, it appears that the adaptation process affects predominantly metabolic pathways in this species, in particular those involving fatty acids, amino acids, and energy generation (23). Although it is too early to make firm conclusions, the adaptive phenomenon described here for the first time, involving the reduction of fatty acid saturation in B. cenocepacia along with the deterioration of pulmonary function, appears to constitute an adaptive shift to a fatty acid metabolism more suited to severely oxygen-limited conditions.
ACKNOWLEDGMENTS
This work was supported by FEDER and by the Portuguese Foundation for Science and Technology, FCT (contracts PTDC/SAU-MII/69591/2006 and ERA-PTG/SAU/0001/2008, in the context of the ADHRES Signature Project of EraNet Pathogenomics; Ph.D. grants SFRH/BD/32729/2006 [C.P.C.] and SFRH/BD/37012/2007 [A.M.]; and a contract under the program Ciência2007 awarded to C.C.C.R.D.C.). This study was performed under COST Action BM1003, Microbial Cell Surface Determinants of Virulence as Targets for New Therapeutics in Cystic Fibrosis.
Footnotes
Published ahead of print on 2 May 2011.
REFERENCES
- 1. Alvarez-Ortega C., Harwood C. S. 2007. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65:153–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baldwin A., et al. 2005. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J. Clin. Microbiol. 43:4665–4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bernier S. P., Nguyen D. T., Sokol P. A. 2008. A LysR-type transcriptional regulator in Burkholderia cenocepacia influences colony morphology and virulence. Infect. Immun. 76:38–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bevivino A., et al. 2002. Burkholderia cepacia complex bacteria from clinical and environmental sources in Italy: genomovar status and distribution of traits related to virulence and transmissibility. J. Clin. Microbiol. 40:846–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bramer C. O., Vandamme P., da Silva L. F., Gomez J. G. C., Steinbuchel A. 2001. Burkholderia sacchari sp. nov., a polyhydroxyalkanoate-accumulating bacterium isolated from soil of a sugar-cane plantation in Brazil. Int. J. Syst. Evol. Microbiol. 51:1709–1713 [DOI] [PubMed] [Google Scholar]
- 6. Buhler T., Ballestero S., Desai M., Brown M. R. 1998. Generation of a reproducible nutrient-depleted biofilm of Escherichia coli and Burkholderia cepacia. J. Appl. Microbiol. 85:457–462 [DOI] [PubMed] [Google Scholar]
- 7. CLSI 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. CLSI/NCCLS document M100-S15. CLSI, Wayne, PA [Google Scholar]
- 8. Correia S., et al. 2008. The clinical course of Burkholderia cepacia complex bacteria respiratory infection in cystic fibrosis patients. Rev. Port. Pneumol. 14:5–26 [PubMed] [Google Scholar]
- 9. Cunha M. V., et al. 2003. Molecular analysis of Burkholderia cepacia complex isolates from a Portuguese cystic fibrosis center: a 7-year study. J. Clin. Microbiol. 41:4113–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cunha M. V., et al. 2007. Exceptionally high representation of Burkholderia cepacia among B. cepacia complex isolates recovered from the major Portuguese cystic fibrosis center. J. Clin. Microbiol. 45:1628–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cunha M. V., et al. 2004. Studies on the involvement of the exopolysaccharide produced by cystic fibrosis-associated isolates of the Burkholderia cepacia complex in biofilm formation and in persistence of respiratory infections. J. Clin. Microbiol. 42:3052–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Carvalho C. C. C. R., Fatal V., Alves S. S., da Fonseca M. M. R. 2007. Adaptation of Rhodococcus erythropolis cells to high concentrations of toluene. Appl. Microbiol. Biotechnol. 76:1423–1430 [DOI] [PubMed] [Google Scholar]
- 13. de Carvalho C. C. C. R., Pons M. N., da Fonseca M. M. R. 2003. Principal components analysis as a tool to summarise biotransformation data: influence on cells of solvent type and phase ratio. Biocatal. Biotransformation 21:305–314 [Google Scholar]
- 14. Drevinek P., Mahenthiralingam E. 2010. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin. Microbiol. Infect. 16:821–830 [DOI] [PubMed] [Google Scholar]
- 15. Dubois M., Gilles K. A., Hamilton J. K., Rebers P. A., Smith F. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350–356 [Google Scholar]
- 16. Feliziani S., et al. 2010. Mucoidy, quorum sensing, mismatch repair and antibiotic resistance in Pseudomonas aeruginosa from cystic fibrosis chronic airways infections. PLos One 5:e12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Findlay R. H., King G. M., Watling L. 1989. Efficacy of phospholipid analysis in determining microbial biomass in sediments. Appl. Environ. Microbiol. 55:2888–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geertsema-Doornbusch G. I., van der Mei H.C., Busscher H. J. 1993. Microbial cell surface hydrophobicity: the involvement of electrostatic interactions in microbial adhesion to hydrocarbons (MATH). J. Microbiol. Methods 18:61–68 [Google Scholar]
- 19. Govan J. R., Brown A. R., Jones A. M. 2007. Evolving epidemiology of Pseudomonas aeruginosa and the Burkholderia cepacia complex in cystic fibrosis lung infection. Future Microbiol. 2:153–164 [DOI] [PubMed] [Google Scholar]
- 20. Guerzoni M. E., Lanciotti R., Cocconcelli P. S. 2001. Alteration in cellular fatty acid composition as a response to salt, acid, oxidative and thermal stresses in Lactobacillus helveticus. Microbiology 147:2255–2264 [DOI] [PubMed] [Google Scholar]
- 21. Harrison F. 2007. Microbial ecology of the cystic fibrosis lung. Microbiology 153:917–923 [DOI] [PubMed] [Google Scholar]
- 22. Herbert D., Phipps P. J., Stange R. E. 1971. Chemical analysis of microbial cells, vol. 5B Academic Press, London, United Kingdom [Google Scholar]
- 23. Hoboth C., et al. 2009. Dynamics of adaptive microevolution of hypermutable Pseudomonas aeruginosa during chronic pulmonary infection in patients with cystic fibrosis. J. Infect. Dis. 200:118–130 [DOI] [PubMed] [Google Scholar]
- 24. Inoue T., Shingaki R., Fukui K. 2008. Inhibition of swarming motility of Pseudomonas aeruginosa by branched-chain fatty acids. FEMS Microbiol. Lett. 281:81–86 [DOI] [PubMed] [Google Scholar]
- 25. Leitão J., et al. 2008. Variation of the antimicrobial susceptibility profiles of Burkholderia cepacia complex clonal isolates obtained from chronically infected cystic fibrosis patients: a five-year survey in the major Portuguese treatment center. Eur. J. Clin. Microbiol. Infect. Dis. 27:1101–1111 [DOI] [PubMed] [Google Scholar]
- 26. LiPuma J. J., et al. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit. Care Med. 164:92–96 [DOI] [PubMed] [Google Scholar]
- 27. Madeira A., Santos P. M., Coutinho C. P., Pinto-de-Oliveira A., Sá-Correia I. 2010. Quantitative proteomics (2D-DIGE) reveals molecular strategies employed by Burkholderia cenocepacia to adapt to the airways of cystic fibrosis patients under antimicrobial therapy. Proteomics 11:1313–1328 [DOI] [PubMed] [Google Scholar]
- 28. Mahenthiralingam E., Baldwin A., Vandamme P. 2002. Burkholderia cepacia complex infection in patients with cystic fibrosis. J. Clin. Microbiol. 51:533–538 [DOI] [PubMed] [Google Scholar]
- 29. Marolda C. L., Hauroder B., John M. A., Michel R., Valvano M. A. 1999. Intracellular survival and saprophytic growth of isolates from the Burkholderia cepacia complex in free-living amoebae. Microbiology 145:1509–1517 [DOI] [PubMed] [Google Scholar]
- 30. McCoy A. J., Liu H. J., Falla T. J., Gunn J. S. 2001. Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides. Antimicrob. Agents Chemother. 45:2030–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. NCCLS 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed National Committee for Clinical Laboratory Standards, Wayne, PA [Google Scholar]
- 32. Oliver A., Canton R., Campo P., Baquero F., Blazquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1253 [DOI] [PubMed] [Google Scholar]
- 33. O'Toole G. A., Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 34. Reid D. W., Anderson G. J., Lamont I. L. 2009. Role of lung iron in determining the bacterial and host struggle in cystic fibrosis. Am. J. Physiol. Lung C 297:L795–L802 [DOI] [PubMed] [Google Scholar]
- 35. Reid D. W., Carroll V., O'May C., Champion A., Kirov S. M. 2007. Increased airway iron as a potential factor in the persistence of Pseudomonas aeruginosa infection in cystic fibrosis. Eur. Respir. J. 30:286–292 [DOI] [PubMed] [Google Scholar]
- 36. Reik R., Spilker T., LiPuma J. J. 2005. Distribution of Burkholderia cepacia complex species among isolates recovered from persons with or without cystic fibrosis. J. Clin. Microbiol. 43:2926–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Richau J. A., et al. 2000. Molecular typing and exopolysaccharide biosynthesis of Burkholderia cepacia isolates from a Portuguese cystic fibrosis center. J. Clin. Microbiol. 38:1651–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rosenberg M. 1981. Bacterial adherence to polystyrene: a replica method of screening for bacterial hydrophobicity. Appl. Environ. Microbiol. 42:375–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schobert M., Tielen P. 2010. Contribution of oxygen-limiting conditions to persistent infection of Pseudomonas aeruginosa. Future Microbiol. 5:603–621 [DOI] [PubMed] [Google Scholar]
- 40. Smith E. E., et al. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103:8487–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Snell G. I., Dehoyos A., Krajden M., Winton T., Maurer J. R. 1993. Pseudomonas-cepacia in lung-transplant recipients with cystic-fibrosis. Chest 103:466–471 [DOI] [PubMed] [Google Scholar]
- 42. Vandamme P., et al. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188–1200 [DOI] [PubMed] [Google Scholar]
- 43. Verstraeten N., et al. 2008. Living on a surface: swarming and biofilm formation. Trends Microbiol. 16:496–506 [DOI] [PubMed] [Google Scholar]
- 44. Wagner B. A., Buettner G. R., Burns C. P. 1994. Free radical-mediated lipid-peroxidation in cells—oxidizability is a function of cell lipid bis-allylic hydrogen content. Biochemistry 33:4449–4453 [DOI] [PubMed] [Google Scholar]