Abstract
Pseudomonas aeruginosa, an opportunistic pathogen of clinical importance, causes chronic airway infections in patients with cystic fibrosis (CF). Current literature suggests that pockets with reduced oxygen tension exist in the CF airway mucus. However, virulence features of this opportunistic pathogen under such conditions are largely unknown. Cell-free supernatant of the standard laboratory P. aeruginosa strain PAO1 obtained from anaerobic culture, but not aerobic culture, failed to kill A549 human airway epithelial cells. Further investigation revealed that this reduced cytotoxicity upon anaerobiosis was due to the suppressed secretion of elastase, a virulence factor controlled by P. aeruginosa quorum sensing (QS). Both a lacZ-reporter fusion assay and quantitative real-time PCR (RT-PCR) analysis demonstrated that transcription of the elastase-encoding lasB gene was substantially decreased during anaerobic growth compared with aerobic growth. Moreover, transcription of other genes controlled by the LasI/R QS system, such as rhlR, vqsR, mvfR, and rsaL, was also repressed under the same anaerobic growth conditions. Importantly, synthesis of 3-oxo-C12-HSL (PAI-1), an autoinducer molecule that mediates induction of the LasI/R QS system, was >22-fold decreased during anaerobic growth while C4-HSL (PAI-2), which mediates RhlI/R QS, was nondetectable under the same growth conditions. Transcription of the lasB gene was restored by exogenous supplementation with autoinducers, with PAI-2 more effective than PAI-1 or Pseudomonas quinolone signal (PQS) at restoring transcription of the lasB gene. Together, these results suggest that anaerobiosis deprives P. aeruginosa of the ability to regulate its virulence via QS and this misregulation attenuates the pathogenic potential of this important pathogen.
INTRODUCTION
Pseudomonas aeruginosa is a clinically important Gram-negative bacterium that is the causative agent of chronic airway infections in patients suffering from pneumonia and bronchiectasis, including cystic fibrosis (CF) (57). P. aeruginosa has developed highly sophisticated virulence mechanisms and secretes a wide range of extracellular virulence factors, such as proteases (50), exotoxin A (53), rhamnolipids (23), pyocyanin (24), and siderophores (11). Production of these virulence factors is regulated to a large extent by a cell density-dependent gene regulatory mechanism termed quorum sensing (QS) (35). The importance of QS in P. aeruginosa virulence has been clearly elucidated in studies using a range of infection models (10, 33, 48) and cultured host cells (8, 41).
There are three well-characterized QS systems in P. aeruginosa: the las, rhl, and pqs systems, each of which plays a distinct role in orchestrating the expression of numerous virulence-associated genes (44). The las and rhl systems were initially identified to be essential for elastase and rhamnolipid production, respectively (35). Each system is composed of a transcriptional activator protein (LasR or RhlR) and a cognate autoinducer synthase, LasI or RhlI, that produces N-(3-oxododecanoyl)-l-homoserine lactone (3-oxo-C12-HSL, PAI-1) and N-butyryl-l-homoserine (C4-HSL, PAI-2), respectively. Upon binding to its cognate signal molecule, LasR or RhlR activates the transcription of target genes (35). P. aeruginosa QS is also regulated by another system, which involves potentiation of transcriptional activation by MvfR (also known as PqsR) upon binding of a Pseudomonas quinolone signal (PQS) (55). The PQS/MvfR complex actively participates in the intertwined P. aeruginosa QS network, and accumulating evidence now suggests that PQS-mediated QS is absolutely required for the uninterrupted production of elastase (29, 36).
In the CF lung, the lack of a functional cystic fibrosis transmembrane conductance regulator (CFTR) channel results in the overproduction of a viscous and stagnant mucus layer (26), on which P. aeruginosa becomes established as a microbial community known as a biofilm. This abnormally altered CF airway has been reported to harbor regions with a steep oxygen gradient ranging from aerobic to anaerobic (40, 54). Given the fact that P. aeruginosa is able to grow anaerobically in the presence of alternative electron acceptors such as nitrate (NO3−) or nitrite (NO2−) that are present in sufficient quantity in a CF mucus layer (21, 31, 59), further research on bacterial responses to an anaerobic environment should be pursued for an integrated understanding of its virulence mechanisms. From this perspective, it is of particular interest that P. aeruginosa growing by anaerobic respiration forms a significantly more robust biofilm than that formed during aerobic growth, allowing the establishment of a resistant mode of bacterial proliferation (32, 59). Moreover, bactericidal activity of polymorphonuclear neutrophils (PMN) was significantly decreased under conditions of low oxygen tension due to the impaired production of hydrogen peroxide (30). Together, these results suggest that long-term survival of P. aeruginosa can be facilitated by the growth under reduced oxygen tension in the CF airway.
Recent reports revealed that a considerable proportion of P. aeruginosa isolates from CF patients possess mutations in the lasR gene (12, 19). Being contradictory to the current view that the lasR-mediated QS system is essential for P. aeruginosa virulence, these findings suggest that (i) QS in the CF airway may not be required for bacterial survival, especially at the chronic stage, and (ii) QS regulation may occur differently under conditions with reduced oxygen tension. Although a study of elastase production in response to various degrees of oxygen potential was reported elsewhere (39), no in-depth understanding of anaerobiosis-induced modulation of QS has been achieved. We undertook the present study to gain insight into how QS regulation is modulated upon growth under anaerobic conditions and to determine the effect of this modulation on bacterial virulence. Understanding the mode of QS regulation under such conditions will aid the development of evidence-based clinical guidelines for the management of P. aeruginosa airway infections.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. aeruginosa laboratory strains (PAO1, PA14, PAK, and FRD1) and pneumonia patient isolates have been previously described (25, 47, 57, 58). The PAO1 ΔlasB mutant was purchased from a P. aeruginosa transposon mutant library (www.genome.washington.edu/UWGC/pseudomonas) and sequence verified. Unless otherwise indicated, the strains were routinely grown in Luria-Bertani broth (LB; 10 g tryptone, 5 g NaCl, 5 g yeast extract per liter) at 37°C. P. aeruginosa was grown anaerobically in an anaerobic chamber (Coylab Inc., Grass Lake, MI) that was filled with mixed gas (nitrogen, 90%; hydrogen, 5%; carbon dioxide, 5%) and maintained at a temperature of 37°C. Chamber operation to achieve and maintain anaerobic environments was performed according to the manufacturer's instructions. To enhance anaerobic growth, bacteria were inoculated in a flask with a stirrer bar in it and the flask was placed on top of the stirrer plate to allow homogeneous mixing. Anaerobic growth was supported by the addition of 0.4% KNO3 to the culture medium (60).
Cell viability assay.
To compare the cytotoxic potential of PAO1 grown aerobically with that grown anaerobically, cell-free culture supernatants were harvested from aerobic and anaerobic cultures that had grown to similar final cell densities. A549 human airway epithelial cells (4) were grown in minimum essential medium (MEM; Gibco/BRL, Rockville, MD) containing 10% fetal bovine serum and 1% penicillin-streptomycin (Gibco/BRL). The cells were placed in the wells of 96-well plates at a density of 1 × 104 cells/well, and the plates were incubated overnight under normal culture conditions (37°C and 5% CO2). After 1 h of adjustment with serum-free medium, bacterial culture supernatants reconstituted in the same serum-free medium were added to the A549 cells. After a 6-h treatment, A549 cell viability was assessed using an MTT assay kit (Sigma-Aldrich) following the instructions provided. A549 cell viability was also examined by trypan blue viability assay (49).
Construction of a PlasB::lacZ reporter strain and β-galactosidase assay.
The lasB promoter region was PCR amplified from the P. aeruginosa PAO1 chromosome using primers lasB-PF (5′-CATATACTAGTAACCTAGCTGCCACCTGCTTT-3′) and lasB-PR (5′-GTAAAGGATCCCTTGTTCAGTTCTCCTGGTTTTTTC-3′), and the lacZ-containing open reading frame (ORF) was amplified from the pTnKGL3 (61) vector using primers lacZ-F (5′-TATACGGATCCATGACCATGATTACGGATTCACTG-3′) and lacZ-R (5′-TGGTTCTCGAGACCTTTAATAGATTATATTACTAATTAATTGGGGA-3′). The lasB promoter region was double digested with SpeI/BamHI, and the lacZ ORF was digested with both BamHI and XhoI. The sequence-specific chromosomal delivery vector pUC18T mini-Tn7T-Gm (7) was also double digested with SpeI/XhoI, and the two digested PCR products were ligated into the cut vector. The constructed plasmid was then integrated into the P. aeruginosa PAO1 genome as described previously (6). The β-galactosidase activity assay was performed as described previously (61).
qRT-PCR analysis.
Total RNA was extracted from harvested cells using TRIzol (Invitrogen) and an RNeasy kit (Qiagen) according to the manufacturers' instructions. RNA quantification was performed using a Nanodrop spectrophotometer (model no. ASP2680; CellTAGen Inc., Seoul, South Korea). cDNA was synthesized using a Primescript reverse transcriptase kit (Takara Bio Inc., Shiga, Japan) with random primers (5′-NSNSNSNSNS-3′, where N = A, T, C, or G and S = C or G). Real-time PCRs were monitored using a StepOne real-time PCR system (Applied Biosystems, Carlsbad, CA). SYBR premix Ex Taq (Takara) was used for PCRs according to the manufacturer's instructions. Transcript levels of the rpoD gene were similar in cells grown under aerobic and anaerobic conditions, and transcript levels of rpoD were thus used to normalize the real-time PCR results. The primers used for quantitative real-time PCR (qRT-PCR) are listed in Table S1 in the supplemental material.
Quantification of PAI-1 and PAI-2 in cell-free culture supernatants.
Filter-passed bacterial culture supernatants were sequentially extracted with two equal volumes of ethyl acetate containing 0.01% (final concentration) glacial acetic acid. The ethyl acetate phase was collected and then evaporated to dryness. The dried residues were then dissolved in high-performance liquid chromatography (HPLC)-grade ethyl acetate and stored at −20°C. Quantification of PAI-1 was performed using gas chromatography-mass spectrometry (GC-MS) with commercially purchased PAI-1 (Sigma-Aldrich) as a standard. Gas chromatographic analyses of PAI-1 in the solvent extracts were carried out using an Agilent 6890 Plus gas chromatograph equipped with a DB-5 MS capillary column (30 m by 0.25-mm inside diameter [i.d.], 0.25-μm film thickness, 5% diphenyl-95% dimethylsiloxane phase; J&W Scientific, Folsom, CA). Mass spectra were obtained using a quadruple mass spectrometer system with a 5973N mass selective detector (Agilent Technologies Inc., Santa Clara, CA). Chromobacterium violaceum CV026 (27) was used to quantify PAI-2 present in the culture supernatants with commercially purchased PAI-2 (Sigma-Aldrich) as a standard. CV026 was inoculated in LB supplemented with PAI-2 of known concentration or the supernatant to be tested and grown for 16 to 18 h at room temperature with vigorous shaking. A 1-ml aliquot of each culture was centrifuged to precipitate the insoluble violacein. Then, 1 ml of dimethyl sulfoxide (DMSO) was added to dissolve the pellet. The absorbance of the completely solubilized violacein was measured with a ThermoMax microplate reader (Molecular Devices, Sunnyvale, CA) at a wavelength of 585 nm.
Western blot analysis and cellular fractionation.
One-milliliter aliquots of aerobic or anaerobic cultures of PAO1 grown to similar final cell densities (optical density at 600 nm [OD600] of ∼3.0) were centrifuged at 14,000 rpm for 5 min. Supernatants were passed through an 0.2-μm Acrodisc syringe filter (Pall Life Science Inc., Ann Arbor, MI) and saved as culture supernatants. Cell pellets were resuspended in 100 μl of B-PER protein extraction reagent (Thermo Fisher Scientific Inc., Rockford, IL) and incubated for 10 min at room temperature. Lysed cells were centrifuged at 14,000 rpm for 10 min, and supernatants were recovered (cell extract fractions). Insoluble precipitates were then resuspended with 50 μl of the same B-PER protein extraction reagent (membrane fractions). An antibody against P. aeruginosa elastase was a kind gift from Efrat Kessler (Tel Aviv University, Israel). Twenty microliters of culture supernatant and 20 μg of cell extract fraction and membrane fraction were loaded onto 12% polyacrylamide gels. Proteins separated on the gel by electrophoresis were transferred to nitrocellulose membranes (Hybond-ECL; GE Health Care), and membranes were blocked with 5% skim mile in Tris-buffered saline–Tween (TBST) buffer. Membranes were then probed with antielastase antibody (1:5,000) for 2 h and washed six times with TBST for 10 min each time. Membranes were then reprobed with anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:5,000) and washed three more times with TBST. Finally, the membranes were incubated with ECL solution for 2 min and signals were detected on X-ray film (Kodak).
Statistical analysis.
Data are expressed as means ± standard deviations (SD). An unpaired Student's t test was used to analyze the data. To compare differences among more than three groups, one-way analysis of variance (ANOVA) was used. A P value of <0.05 was considered statistically significant. All the experiments were repeated for reproducibility.
RESULTS
Cell-free supernatants of anaerobic cultures failed to kill human airway epithelial cells.
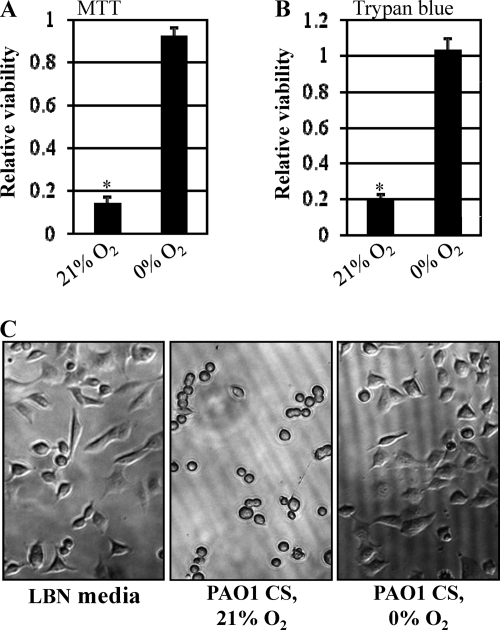
To compare the cytotoxicities of virulence factors secreted during aerobic and anaerobic growth, we treated A549 human airway epithelial cells with the culture supernatants from bacteria growing aerobically or anaerobically. Because production of virulence factors occurs in a cell density-dependent manner, cell-free supernatants were harvested from cultures that grew to similar final cell densities (i.e., OD600 of ∼3.75 for aerobic culture versus ∼3.59 for anaerobic culture). To support anaerobic growth, medium was supplemented with 0.4% NO3−, which acts as an alternative electron acceptor (59); this medium was also used for the aerobic cultures. As shown in Fig. 1A, A549 cells lost their viability upon treatment with aerobic culture supernatants as assessed by the MTT cell viability assay. After a 6-h treatment, the mean OD570 value derived from live cells had decreased to ∼13% of that of the control treatment. In contrast, A549 cells treated with anaerobic culture supernatants remained viable (Fig. 1A). The contrasting cytotoxic activities of these two culture supernatants were further confirmed by a trypan blue stain assay (Fig. 1B). We next examined whether the differential cytotoxic effects of the two supernatants were reflected by host cell morphological changes. As shown in Fig. 1C, A549 cells treated with aerobic culture supernatants completely lost their normal cellular morphology, while such changes in cell shape were not observed in cells treated with anaerobic culture supernatants.
Fig. 1.
Cytotoxic activity of P. aeruginosa culture supernatants toward A549 cells. (A) Relative viability of A549 human epithelial cells treated with cell-free culture supernatants (CS) of PAO1 grown in LB containing 0.4% NO3− either aerobically (21% O2) or anaerobically (0% O2). After a 6-h treatment, 30 μl of MTT reagent (5 mg/ml) was added to the cells and the plate was read at 570 nm. The relative viability is shown as a ratio of OD570 values from control medium treatment versus CS treatment. The values shown are the means ± SD from three independent experiments. *, P < 0.01 versus treatment with anaerobic CS. (B) Relative viability of A549 cells as assessed by trypan blue staining assay. Experimental conditions for A549 cell growth and treatment were identical to those described for panel A. Nonviable (i.e., blue-stained) cells were counted and divided by the total number of counted cells. *, P < 0.01 versus treatment with anaerobic CS. (C) Morphological changes of A549 cells in response to the treatment with LBN medium (LB medium containing 0.4% NO3−), aerobic PAO1 CS, or anaerobic PAO1 CS. A549 cells were treated for 6 h before photos were taken. The images were acquired using a Zeiss Axiovert 200 inverted microscope at a ×100 magnification.
Elastase secretion is reduced under anaerobic growth conditions.
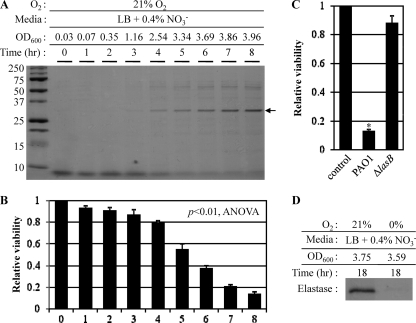
Elastase, a major virulence factor (3), is the most abundant protein secreted into the culture media during aerobic growth of P. aeruginosa (Fig. 2A, black arrow). We first monitored the time profile of elastase secretion during aerobic growth of PAO1. As growth time progressed, increased levels of elastase were detected in the culture supernatant (Fig. 2A). A band corresponding to elastase started to appear after 3 h, when the culture reached an OD600 of ∼1.16, and continued to increase steadily as the culture reached stationary phase. As expected, the elastase band was not observed in the culture supernatants of the ΔlasB mutant (see Fig. S1 in the supplemental material). To investigate the potential role of elastase in the observed cytotoxicity of the aerobic culture supernatants, we treated A549 human airway epithelial cells with the culture supernatants of PAO1 harvested at various time points. As shown in Fig. 2B, the viability of A549 cells decreased in proportion to the level of elastase present in the culture supernatants. The relative survival rate for each treatment was normalized using the value obtained from the control treatment, in which A549 cells were treated with the medium, LB plus 0.4% NO3−. As expected, the culture supernatant of a mutant with the elastase-encoding lasB gene interrupted by transposon insertion failed to kill A549 cells under the same experimental conditions (Fig. 2C), providing conclusive evidence that elastase was responsible for the cytotoxic activity against A549 cells.
Fig. 2.
Elastase is responsible for A549 cell death, and secretion of elastase is significantly repressed during anaerobic growth. (A) Time-dependent accumulation of elastase in the culture supernatant (CS) of aerobic PAO1 culture. Bacteria grown overnight in LB at 37°C were inoculated at 1:100 in LB + 0.4% NO3−, and growth was monitored by measuring OD600. Aliquots of the culture were harvested every hour, and protein contents present in each CS were analyzed by SDS-PAGE. The protein band that corresponds to the mature elastase is shown with an arrow. (B) Effect of increasing levels of elastase on A549 cell viability. The viability of A549 cells (1 × 104 cells) treated with the same set of CSs for 6 h was monitored using an MTT assay as described in Fig. 1. The differences in the mean values among the treatment groups are statistically different (P < 0.01, ANOVA). (C) A549 cell viability in response to the CS of a lasB-deficient mutant. The mutant was grown aerobically for 8 h in LB plus 0.4% (wt/vol) NO3−. Assay conditions were identical to those described for panel B. *, P < 0.01 versus treatment with the aerobic CS of ΔlasB mutant. (D) The level of elastase secreted into the culture medium during anaerobic growth. PAO1 was grown in LB plus 0.4% (wt/vol) NO3− inside an anaerobic chamber. The cell density (OD600) after an 18-h cultivation in a flask stirred with a magnetic bar to ensure homogeneous mixing was 3.59. The level of elastase was analyzed by SDS-PAGE.
We then examined whether the noncytotoxic nature of the anaerobic culture supernatant was due to a factor associated with the modulation of elastase secretion. Indeed, the levels of elastase present in anaerobic culture supernatants were significantly lower in our SDS-PAGE analysis than those present in the aerobic supernatants (Fig. 2D). Again, the final cell densities (OD600 of ∼3.75 versus ∼3.59) after 18 h of growth were similar between these two cultures, implying that the anaerobiosis-specific repression of elastase secretion was not due to retarded bacterial growth. Together, these results suggest that elastase secretion is highly suppressed during anaerobic growth and such repression is responsible for the loss of cytotoxicity toward A549 human airway epithelial cells.
Next, we examined whether the anaerobiosis-induced decrease in elastase secretion was also observed in other P. aeruginosa strains. As in the case of PAO1, PA14 and three nonmucoid pneumonia patient isolates produced sufficient levels of elastase during aerobic growth. In contrast, PAK, a highly piliated P. aeruginosa strain, and FRD1, a mucoid CF patient isolate, produced a negligible or significantly decreased level of elastase, respectively (see Fig. S2A in the supplemental material). During anaerobic growth, however, all tested strains produced very low levels of elastase, thereby further confirming our results in PAO1 (Fig. S2A). Again, relative cytotoxicity was directly proportional to the amount of elastase present in the culture supernatants (Fig. S2B).
Production, but not secretion, of elastase was decreased under anaerobic conditions.
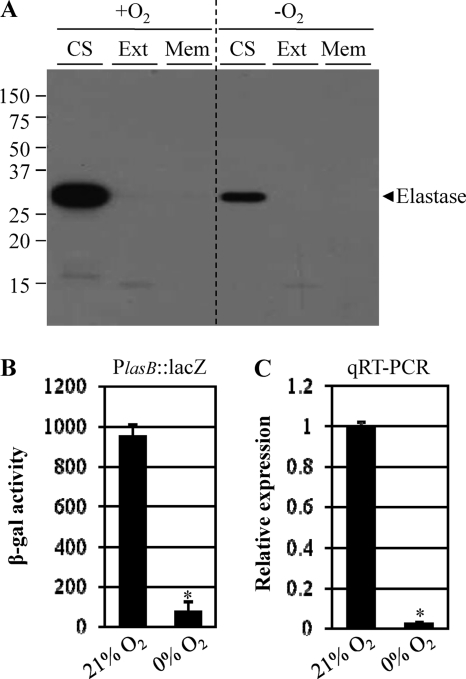
Elastase has been reported to be synthesized as an ∼53-kDa preproenzyme containing an ∼2.4-kDa signal sequence and to be translocated to the periplasmic space after synthesis. The resultant periplasmic protein is then further processed to generate the mature secretory ∼33-kDa elastase and ∼18-kDa propeptide, which acts as an elastase inhibitor by forming a complex with the processed elastase (22). To determine whether the decrease in elastase secretion during anaerobic growth was caused by decreased production or decreased levels of secretion, we analyzed levels of elastase in each of three different cellular fractions—the culture supernatant (CS), the cell extract (Ext), and the membrane fraction (Mem)—by Western blot analysis. As shown in Fig. 3A, elastase was detected only in the CS fractions and a significantly larger amount of elastase was present in the aerobic CS than in the anaerobic CS, further validating the results shown in Fig. 2D. Cell extracts and membrane fractions in both preparations did not contain either unprocessed or processed elastase. This suggests that the reduced elastase secretion observed during anaerobic growth was due not to incomplete posttranslational processes but to decreased production per se.
Fig. 3.
Anaerobiosis-induced suppression of elastase secretion is controlled at the transcriptional level. (A) Western blot analysis of elastase in each of three cellular fractions: the culture supernatant (CS), the cell extract (Ext), and the membrane fraction (Mem) of PAO1 grown aerobically (21% O2) versus anaerobically (0% O2). Because the level of elastase secretion is dependent on cell density, bacteria were grown to similar cell densities before harvest (OD600 of ∼3.0) under the two conditions. The arrowhead shown to the right indicates elastase. (B) β-Galactosidase activity of PAO1 harboring a chromosomal copy of a lasB promoter-lacZ fusion gene. Reporter cells were inoculated in LB plus 0.4% (wt/vol) NO3− and grown to an OD600 of ∼3.0 under both conditions. *, P < 0.01 versus β-galactosidase activity of PAO1 grown aerobically. (C) Quantitative RT-PCR analysis of the expression of the lasB gene encoding elastase in PAO1. qRT-PCR was conducted using cDNA synthesized from 2 μg total RNA extracted from PAO1 grown to an OD600 of ∼3.0 under both conditions. Transcript levels of the lasB gene were normalized to those of the rpoD gene transcript. *, P < 0.01 versus lasB transcript level in PAO1 grown aerobically.
To examine whether the differential elastase production is mirrored in lasB transcription, we compared the lasB transcript levels in PAO1 grown aerobically and that grown anaerobically. Since lasB gene expression occurs in a cell density-dependent manner, bacteria were grown to an OD600 of ∼3.0 under both conditions. Figure 3B and C shows β-galactosidase activity of PAO1 harboring a chromosomal copy of the lasB promoter-lacZ fusion and quantitative RT-PCR analysis of lasB mRNA, respectively. In these two independent assays, mRNA expression of the lasB gene was >12-fold and >40-fold decreased, respectively, in bacteria grown by anaerobic respiration, further corroborating the idea that anaerobiosis-induced suppression of elastase secretion is regulated at the transcriptional level.
Production of 3-oxo-C12-HSL and C4-HSL was suppressed under anaerobic growth conditions.
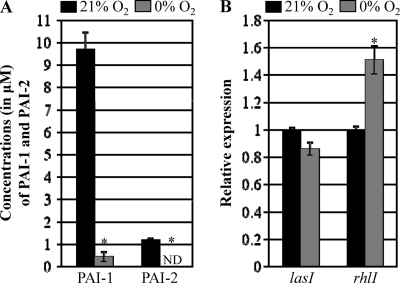
Although the LasI/R QS system plays a dominant role in the complex QS hierarchy in P. aeruginosa (34), three QS systems, namely, LasI/R, RhlI/R, and PQS, all participate in a complex signaling network to regulate lasB gene expression (29). Recently, it was reported that production of PQS is completely abrogated during anaerobic growth of P. aeruginosa (43, 51). Given the fact that synthesis of the PQS signal molecule is coordinately regulated by the LasI/R and RhlI/R components of the QS system (28), this finding suggests that these two QS systems might also be differentially modulated during anaerobic and aerobic growth. To gain a better understanding of the molecular basis of the anaerobiosis-induced suppression of lasB transcription, we measured the levels of 3-oxo-C12-HSL (PAI-1) and C4-HSL (PAI-2) in cell-free culture supernatants of PAO1 grown either aerobically or anaerobically. PAI-1 and PAI-2 were quantified using GC-MS and a CV026 reporter-based bioassay, respectively. In our GC-MS analysis using commercially purchased purified PAI-1 as a standard, we found that aerobic culture supernatant contained ∼9.72 μM PAI-1. In contrast, only ∼0.44 μM was detected in the cell-free supernatant of anaerobic cultures (Fig. 4A). We then measured the level of PAI-2 in cell-free culture supernatants of PAO1 using C. violaceum CV026, a reporter strain that produces violacein in response to exogenously added PAI-2 (27). The level of PAI-2 in aerobic cultures grown to an OD600 of ∼3.0 was determined to be 1.23 ± 0.05 μM, a value ∼7.9 times lower than PAI-1 (Fig. 4A). In contrast, the level of PAI-2 in the cell-free supernatant of an equally dense anaerobic culture was below the detection limits. Together, these results indicate that during anaerobic growth, production of PAI-1 and PAI-2 is highly suppressed, likely rendering P. aeruginosa QS incompetent under such conditions. We then examined whether the substantially decreased production of autoinducers is associated with altered expression of the lasI and rhlI genes. Figure 4B shows that lasI gene expression levels were similar in bacteria irrespective of the growth conditions, while the expression of rhlI was rather increased in PAO1 grown under anaerobic conditions. This suggests that the dramatically reduced levels of PAI-1 and PAI-2 observed under anaerobic growth conditions are not attributable to transcriptional regulation of the genes involved in their synthesis.
Fig. 4.
Quantification of PAI-1 and PAI-2 in the culture supernatants of PAO1 grown either aerobically or anaerobically. (A) Concentrations (μM) of two homoserine lactone-based autoinducers (PAI-1 and PAI-2) in aerobic (21% O2) and anaerobic (0% O2) CSs. Bacterial cells were grown to similar final densities (i.e., OD600 of ∼3.0) under the two conditions. Autoinducers were extracted using acidified ethyl acetate, and PAI-1 and PAI-2 were analyzed with GC-MS/MS and a CV026 reporter cell assay, respectively. Quantification was performed using CSs obtained from three independent cultures, and results are displayed as means ± SD. ND, not detected. *, P < 0.01 versus autoinducer level produced aerobically. (B) mRNA transcript levels of lasI and rhlI in PAO1 cells grown aerobically (black bars) and anaerobically (gray bars) as assessed by qRT-PCR analysis. qRT-PCR was conducted using cDNA synthesized from 2 μg total RNA extracted from PAO1 grown either aerobically (21% O2) or anaerobically (0% O2). Transcript levels of tested genes were normalized to those of the rpoD gene transcript. Three independent experiments were performed, and mean values ± SD are displayed in each bar. *, P < 0.01 versus rhlI transcript in PAO1 grown aerobically.
Transcription of most virulence-associated genes was suppressed during anaerobic growth.
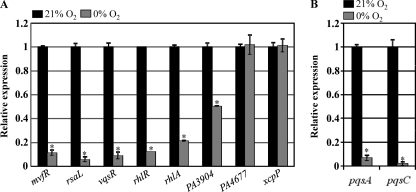
Next, we sought to elucidate the effect of suppressed synthesis of PAI-1 and PAI-2 on the expression of downstream virulence genes. We tested eight genes reported to be directly regulated by LasR (17). Figure 5A shows the relative expression of these selected genes as measured by quantitative RT-PCR analysis. Similar to the case of lasB, transcript levels of the mvfR, rsaL, vqsR, and rhlR genes, which encode major regulators in the P. aeruginosa QS system, were ∼10-fold lower in bacteria grown by anaerobic respiration compared to their aerobically grown counterparts. Transcript levels of rhlA, which encodes rhamnosyltransferase (34), and PA3904, a hypothetical gene with unknown function (45), were ∼20% and ∼50% of the transcript levels observed under aerobic growth, respectively. In contrast, mRNA levels of PA4677 (45) and xcpP (5) were similar in cells grown under either condition. These results suggest that anaerobiosis downregulated the expression of several, but not all, LasR-regulated genes.
Fig. 5.
Anaerobiosis-induced transcriptional modulation of genes involved in QS regulation. (A) Quantitative RT-PCR analysis of genes that were previously determined to be directly regulated by LasR. Assay conditions were identical to those described in Fig. 4B. Three independent experiments were performed, and mean values ± SD are displayed in each bar. *, P < 0.01 versus transcript levels in PAO1 grown aerobically. (B) Downstream effects of suppressed transcription of mvfR on the expression of pqsA and pqsC as assessed by qRT-PCR. *, P < 0.01 versus transcript levels in PAO1 grown aerobically.
Next, we examined the downstream effects of anaerobiosis-induced suppression of mvfR, a transcriptional regulator of the synthesis of the PQS signal molecule (14). To address this question, we analyzed transcript levels of two genes known to be regulated by MvfR, namely, pqsA and pqsC, which encode enzymes involved in PQS synthesis (56). As shown in Fig. 5B, expression levels of pqsA and pqsC during anaerobic growth were only ∼6.7% and ∼2% of those during aerobic growth, respectively. This result provides further basis for the significant suppression of PQS synthesis during anaerobic growth.
Addition of autoinducers restored lasB transcription during anaerobic growth.
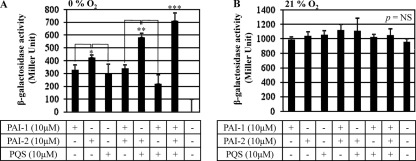
To further verify that anaerobiosis-induced abrogation of lasB transcription was due to an insufficient level of autoinducers, we examined whether lasB transcription was activated by the addition of exogenous autoinducers. Higher levels of lasB transcription, expressed as β-galactosidase activity, were observed after the addition of 10 μM PAI-2 (Fig. 6, first three bars) than after addition of PAI-1 or PQS, suggesting that PAI-2 plays a more important role in inducing lasB transcription under anaerobic conditions than the other two autoinducers. When pairs of autoinducers were added, the pair of PAI-2 and PQS was more effective at activating lasB transcription than the other two pairs (i.e., PAI-1 + PQS and PAI-1 + PAI-2), which failed to induce additive β-galactosidase activity (Fig. 6, fourth to sixth bars from left). The highest level of lasB transcription was achieved when all three autoinducers were added: ∼74% of the level observed for aerobic growth (Fig. 3B). Again, no significant lasB transcription occurred in the control treatment (bacteria treated with the same concentration of MeOH used to dissolve the signal molecules) (Fig. 6, first bar from right). When the same experiment was repeated under aerobic conditions, during which autoinducers are normally produced, no significant changes in β-galactosidase activity were observed in response to the added extraneous autoinducers (Fig. 6B). This result further proves that the anaerobiosis-induced suppression of lasB transcription is due to a lack of autoinducers and also suggests that PAI-1, PAI-2, and PQS are all required for maximal lasB transcription.
Fig. 6.
Exogenously supplemented autoinducers restore lasB transcription during anaerobic growth. PAI-1, PAI-2, and PQS, either alone or in combination, were added to the anaerobic (A) or aerobic (B) culture of the PlasB::lacZ reporter strain at a final concentration of 10 μM. β-Galactosidase activity was assessed using cells grown for 18 h under both conditions. Three independent experiments were performed, and mean values ± SD are displayed in each bar. *, P < 0.01 versus the other two treatments with PAI-1 (first bar) or PQS (third bar); **, P < 0.01 versus the other two treatments with PAI-1/PAI-2 (fourth bar) or PAI-1/PQS (sixth bar); ***, P < 0.01 versus all other treatments. Methanol was used as a vehicle control (last bar). P = NS (not significant) in all comparisons (ANOVA).
DISCUSSION
LasR, the most upstream QS regulon in the P. aeruginosa QS hierarchy, regulates the expression of more than 300 virulence-associated genes (17). However, recent genetic studies using diverse P. aeruginosa clinical isolates reported that adaptive mutations in the lasR gene occur spontaneously in the course of chronic airway infection in CF (9, 12, 19, 42). Phenotype changes conferred on P. aeruginosa by these mutations include (i) facilitated growth on amino acids present in relatively large quantities in CF airways (1, 12), (ii) an efficient shift to an anaerobic mode of growth using nitrate over oxygen (20), and (iii) elevated antibiotic resistance (12, 20). Therefore, frequent identification of lasR mutants suggests that P. aeruginosa may acquire the mutation to increase its survival fitness in a harsh host environment at the expense of its ability to regulate QS-mediated virulence properties. Furthermore, this notion also supports the idea that QS machinery may be dispensable once chronic infection is successfully established in the patient airway.
Our study was initiated by the observation that cell-free supernatants obtained from anaerobic cultures of PAO1 failed to kill A549 airway epithelial cells, while aerobic culture supernatants were cytotoxic. Because expression of virulence traits is dependent on QS, this result led us to further characterize QS-controlled virulence regulation of P. aeruginosa under anaerobic growth conditions. Our subsequent analysis demonstrated that (i) the observed cytotoxicity is mediated by the secretion of elastase (Fig. 2) and (ii) the anaerobiosis-induced loss of cytotoxicity is due to the suppressed level of lasB gene transcription (Fig. 3), which was later found to be mediated by the substantially reduced autoinducer synthesis (Fig. 4) and subsequent decrease in transcription of QS regulators (Fig. 5).
The data presented in Fig. 4A indicate that the concentrations of PAI-1 and PAI-2 in the culture medium during aerobic growth were 9.72 μM and 1.23 μM, respectively, yielding a value of PAI-2/(PAI-1 + PAI-2) of ∼0.11, consistent with the previous findings of Singh and colleagues (46). In contrast, our data suggested that PAO1 produced significantly suppressed levels of PAI-1 and nondetectable PAI-2 during anaerobic growth, respectively. Together with previous reports revealing that the production of PQS is also highly inhibited during anaerobic growth (43, 51), these findings suggest that (i) the three major autoinducers mediating P. aeruginosa QS are either not produced or produced at significantly lower levels during anaerobic growth and (ii) QS may therefore not be functional in PAO1 growing under anaerobic conditions. It is of particular interest that the levels of PAI-1 and PAI-2 detected in sputum samples isolated from CF patients colonized with up to 108 CFU/ml of P. aeruginosa were significantly lower than those produced in laboratory aerobic cultures (46). Likewise, Erikson and colleagues also reported that autoinducers detected in an independent set of CF sputa were present at very low levels, with concentrations of PAI-2 lower than those of PAI-1 (15). Because anaerobiosis suppresses the production of autoinducers, as was demonstrated in this study, these findings further corroborate that anaerobic respiration is likely a major mode of bacterial growth in the CF mucus (59).
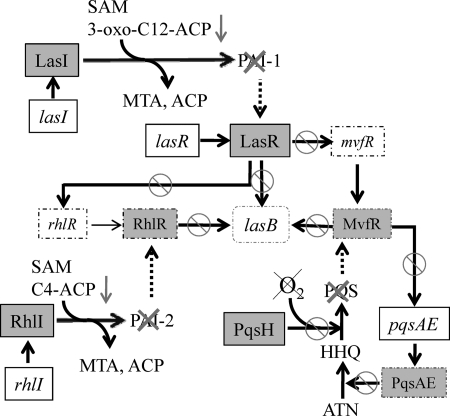
Schertzer and colleagues recently elucidated the molecular basis behind the abrogated synthesis of PQS during growth without oxygen (43). Because oxygen and NADH are required as cofactors for the enzymatic synthesis of PQS from its precursor, 2-heptyl-4-quinolone (HHQ), a lack of oxygen prevents the enzymatic conversion of HHQ to PQS from taking place. Thus, the suppressed synthesis of PQS during anaerobic growth is due not to the anaerobiosis-induced altered expression of genes, whose products are involved in PQS synthesis, but to the absence of molecular oxygen that is physically required for terminal hydroxylation of HHQ (Fig. 7). It appears, however, that molecular oxygen is not directly involved in the synthesis of HSL-based autoinducers. Synthesis of 3-oxo-C12-HSL and C4-HSL by their cognate autoinducer synthase (i.e., LasI and RhlI) requires S-adenosylmethionine (SAM) and 3-oxo-C12-acyl carrier protein or the N-butyryl acyl carrier protein, respectively (18, 38). Our qRT-PCR analysis results shown in Fig. 4B demonstrated that transcript levels of lasI and rhlI were either similar or increased during anaerobic growth compared with those under aerobic growth, suggesting that the anaerobiosis-specific inhibition of autoinducer synthesis is not likely caused by downregulation of associated genes. Interestingly, two independent genome-wide microarray analyses revealed that gene expression of QS-regulated acyl carrier proteins (PA0999, PA1869, PA3333, and PA3334) was invariably decreased during anaerobic growth (16, 52). Although more analysis at the protein level is necessary to allow more robust conclusions, it is likely that suppressed synthesis of PAI-1 and PAI-2 may be due to a limitation of acyl carrier proteins (Fig. 7, downward arrows).
Fig. 7.
A simplified model of P. aeruginosa QS regulation under anaerobic conditions. During anaerobic growth, production of three major QS signal molecules, PAI-1, PAI-2, and PQS, is highly suppressed, rendering P. aeruginosa incapable of QS. Gene names are shown in italic, while protein names are shown in roman inside gray squares. Downregulated gene transcripts and proteins are shown in squares with dashed lines. Anaerobiosis-induced suppression of lasB transcription (shown in the center) is mediated by inactivation of PAI-1- (top), PAI-2- (left), and PQS-mediated (right) QS. Abbreviations and symbols: SAM, S-adenosylmethionine; ACP, acyl carrier protein; MTA, 5′-methylthioadenosine; HHQ, 2-heptyl-4-quinolone; ANT, anthranilate; arrow, activation or production; arrow with circle and slash, suppressed contribution. The lack of oxygen is denoted by a cross. Dashed arrows indicate suppressed association of QS signals with their cognate regulator proteins.
Our autoinducer “add-back” experiments demonstrated that lasB transcription was restored to intermediate levels by the addition of each of three autoinducer molecules (Fig. 6). These results indicated that (i) virtually none of the autoinducers were produced to sufficient levels during anaerobic growth and (ii) each autoinducer has a distinct role in inducing lasB expression. Consistent with this notion, lasB transcription was restored to its highest level in the presence of all three autoinducers, suggesting that maximal activation of lasB gene transcription is due to the combined effects of the three autoinducers. It is also noteworthy that PAO1 growing anaerobically responded better to PAI-2 either alone or with PQS to activate lasB transcription (Fig. 6), suggesting that during anaerobic growth, PAI-2-mediated QS may play a more important role than QS induced by the action of other signaling molecules. It was recently reported that PAI-2 production, albeit delayed, still occurs in a lasR mutant strain and that the RhlI/R QS system can override, at least in part, the effects of lasR mutations (13). In addition, comprehensive chronological genetic analysis using a large number of CF isolates revealed that isolates that lost the ability to produce PAI-1 appeared earlier than strains that were unable to produce both PAI-1 and PAI-2 (2). This finding indicates that bacteria may lose the LasI/R QS system more readily than the RhlI/R system during the course of chronic airway infection. Together, these findings suggest that P. aeruginosa may have evolved a mechanism by which it can express virulence factors in response to PAI-2 when the LasI/R system is not available.
QS has been studied extensively as an obvious target to alleviate bacterial virulence (37). Such approaches have been considered to be advantageous because targeting QS may not impose selective pressure for the development of resistance as we have witnessed with antibiotics. Our results, however, indicate that QS per se is not actively occurring in P. aeruginosa growing in an anaerobic environment. Given the fact that local regions with reduced oxygen tension exist in the CF mucus airway, these results clearly suggest that we need to change the way we understand P. aeruginosa pathogenic mechanisms and thus deal with P. aeruginosa infections. We anticipate that the data provided in this study will prompt further investigations with the ultimate goal of eradicating this persistent colonizer from anaerobic mucus layers.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare and Family Affairs, Republic of Korea, A090951 (to S.S.Y.), and by a grant from the National Research Foundation of Korea (NRF) funded by the Korea Government (MEST), no. 2009-0087951 (to S.S.Y.). This work was also supported by grants from the National Research Foundation of Korea (NRF) funded by the Korean Government, no. 2010-0015901 and no. 2010-0015901 (to J.-H.L.).
We are thankful to Kyung-Hee Choi (Wonkwang University, South Korea) for providing the pUC18T mini-Tn7T-Gm plasmid DNA. We are also indebted to Efrat Kessler (Tel Aviv University, Israel) for kindly providing us with the antielastase antibody.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 9 May 2011.
REFERENCES
- 1. Barth A. L., Pitt T. L. 1996. The high amino-acid content of sputum from cystic fibrosis patients promotes growth of auxotrophic Pseudomonas aeruginosa. J. Med. Microbiol. 45:110–119 [DOI] [PubMed] [Google Scholar]
- 2. Bjarnsholt T., et al. 2010. Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS One 5:e10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blackwood L. L., Stone R. M., Iglewski B. H., Pennington J. E. 1983. Evaluation of Pseudomonas aeruginosa exotoxin A and elastase as virulence factors in acute lung infection. Infect. Immun. 39:198–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Byrd M. S., Pang B., Mishra M., Swords W. E., Wozniak D. J. 2010. The Pseudomonas aeruginosa exopolysaccharide Psl facilitates surface adherence and NF-κB activation in A549 cells. mBio 1(3):e00140–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chapon-Herve V., et al. 1997. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol. Microbiol. 24:1169–1178 [DOI] [PubMed] [Google Scholar]
- 6. Choi K. H., DeShazer D., Schweizer H. P. 2006. Mini-Tn7 insertion in bacteria with multiple glmS-linked attTn7 sites: example Burkholderia mallei ATCC 23344. Nat. Protoc. 1:162–169 [DOI] [PubMed] [Google Scholar]
- 7. Choi K. H., Schweizer H. P. 2006. Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1:153–161 [DOI] [PubMed] [Google Scholar]
- 8. Chun C. K., Ozer E. A., Welsh M. J., Zabner J., Greenberg E. P. 2004. Inactivation of a Pseudomonas aeruginosa quorum-sensing signal by human airway epithelia. Proc. Natl. Acad. Sci. U. S. A. 101:3587–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ciofu O., Mandsberg L. F., Bjarnsholt T., Wassermann T., Hoiby N. 2010. Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants. Microbiology 156:1108–1119 [DOI] [PubMed] [Google Scholar]
- 10. Clatworthy A. E., et al. 2009. Pseudomonas aeruginosa infection of zebrafish involves both host and pathogen determinants. Infect. Immun. 77:1293–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crosa J. H. 1989. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol. Rev. 53:517–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. D'Argenio D. A., et al. 2007. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 64:512–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dekimpe V., Deziel E. 2009. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 155:712–723 [DOI] [PubMed] [Google Scholar]
- 14. Deziel E., et al. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. U. S. A. 101:1339–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Erickson D. L., et al. 2002. Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infect. Immun. 70:1783–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Filiatrault M. J., et al. 2005. Effect of anaerobiosis and nitrate on gene expression in Pseudomonas aeruginosa. Infect. Immun. 73:3764–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gilbert K. B., Kim T. H., Gupta R., Greenberg E. P., Schuster M. 2009. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol. Microbiol. 73:1072–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gould T. A., Schweizer H. P., Churchill M. E. 2004. Structure of the Pseudomonas aeruginosa acyl-homoserinelactone synthase LasI. Mol. Microbiol. 53:1135–1146 [DOI] [PubMed] [Google Scholar]
- 19. Hoffman L. R., et al. 2009. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J. Cyst. Fibros. 8:66–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoffman L. R., et al. 2010. Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathog. 6:e1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones K. L., et al. 2000. Elevation of nitrotyrosine and nitrate concentrations in cystic fibrosis sputum. Pediatr. Pulmonol. 30:79–85 [DOI] [PubMed] [Google Scholar]
- 22. Kessler E., Safrin M. 1994. The propeptide of Pseudomonas aeruginosa elastase acts an elastase inhibitor. J. Biol. Chem. 269:22726–22731 [PubMed] [Google Scholar]
- 23. Kownatzki R., Tummler B., Doring G. 1987. Rhamnolipid of Pseudomonas aeruginosa in sputum of cystic fibrosis patients. Lancet i:1026–1027 [DOI] [PubMed] [Google Scholar]
- 24. Lau G. W., Hassett D. J., Ran H., Kong F. 2004. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 10:599–606 [DOI] [PubMed] [Google Scholar]
- 25. Lee D. G., et al. 2006. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 7:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsui H., et al. 1998. Evidence for periciliary liquid layer depletion, not abnormal ion composition, in the pathogenesis of cystic fibrosis airways disease. Cell 95:1005–1015 [DOI] [PubMed] [Google Scholar]
- 27. McClean K. H., et al. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703–3711 [DOI] [PubMed] [Google Scholar]
- 28. McGrath S., Wade D. S., Pesci E. C. 2004. Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol. Lett. 230:27–34 [DOI] [PubMed] [Google Scholar]
- 29. McKnight S. L., Iglewski B. H., Pesci E. C. 2000. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182:2702–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McRipley R. J., Sbarra A. J. 1967. Role of the phagocyte in host-parasite interactions. XI. Relationship between stimulated oxidative metabolism and hydrogen peroxide formation, and intracellular killing. J. Bacteriol. 94:1417–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ojoo J. C., Mulrennan S. A., Kastelik J. A., Morice A. H., Redington A. E. 2005. Exhaled breath condensate pH and exhaled nitric oxide in allergic asthma and in cystic fibrosis. Thorax 60:22–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O'May C. Y., Reid D. W., Kirov S. M. 2006. Anaerobic culture conditions favor biofilm-like phenotypes in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. FEMS Immunol. Med. Microbiol. 48:373–380 [DOI] [PubMed] [Google Scholar]
- 33. Papaioannou E., et al. 2009. Quorum-quenching acylase reduces the virulence of Pseudomonas aeruginosa in a Caenorhabditis elegans infection model. Antimicrob. Agents Chemother. 53:4891–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pearson J. P., Pesci E. C., Iglewski B. H. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pesci E. C., Iglewski B. H. 1997. The chain of command in Pseudomonas quorum sensing. Trends Microbiol. 5:132–134 [DOI] [PubMed] [Google Scholar]
- 36. Pesci E. C., et al. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96:11229–11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rasmussen T. B., Givskov M. 2006. Quorum sensing inhibitors: a bargain of effects. Microbiology 152:895–904 [DOI] [PubMed] [Google Scholar]
- 38. Raychaudhuri A., Jerga A., Tipton P. A. 2005. Chemical mechanism and substrate specificity of RhlI, an acylhomoserine lactone synthase from Pseudomonas aeruginosa. Biochemistry 44:2974–2981 [DOI] [PubMed] [Google Scholar]
- 39. Sabra W., Kim E. J., Zeng A. P. 2002. Physiological responses of Pseudomonas aeruginosa PAO1 to oxidative stress in controlled microaerobic and aerobic cultures. Microbiology 148:3195–3202 [DOI] [PubMed] [Google Scholar]
- 40. Sanderson K., Wescombe L., Kirov S. M., Champion A., Reid D. W. 2008. Bacterial cyanogenesis occurs in the cystic fibrosis lung. Eur. Respir. J. 32:329–333 [DOI] [PubMed] [Google Scholar]
- 41. Sawa T., et al. 1998. In vitro cellular toxicity predicts Pseudomonas aeruginosa virulence in lung infections. Infect. Immun. 66:3242–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schaber J. A., et al. 2004. Analysis of quorum sensing-deficient clinical isolates of Pseudomonas aeruginosa. J. Med. Microbiol. 53:841–853 [DOI] [PubMed] [Google Scholar]
- 43. Schertzer J. W., Brown S. A., Whiteley M. 2010. Oxygen levels rapidly modulate Pseudomonas aeruginosa social behaviours via substrate limitation of PqsH. Mol. Microbiol. 77:1527–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schuster M., Greenberg E. P. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296:73–81 [DOI] [PubMed] [Google Scholar]
- 45. Schuster M., Urbanowski M. L., Greenberg E. P. 2004. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. U. S. A. 101:15833–15839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singh P. K., et al. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762–764 [DOI] [PubMed] [Google Scholar]
- 47. Strom M. S., Nunn D., Lory S. 1991. Multiple roles of the pilus biogenesis protein pilD: involvement of pilD in excretion of enzymes from Pseudomonas aeruginosa. J. Bacteriol. 173:1175–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tang H. B., et al. 1996. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect. Immun. 64:37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tollefson A. E., Ryerse J. S., Scaria A., Hermiston T. W., Wold W. S. 1996. The E3-11.6-kDa adenovirus death protein (ADP) is required for efficient cell death: characterization of cells infected with adp mutants. Virology 220:152–162 [DOI] [PubMed] [Google Scholar]
- 50. Tommassen J., Filloux A., Bally M., Murgier M., Lazdunski A. 1992. Protein secretion in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 9:73–90 [DOI] [PubMed] [Google Scholar]
- 51. Toyofuku M., et al. 2008. Influence of the Pseudomonas quinolone signal on denitrification in Pseudomonas aeruginosa. J. Bacteriol. 190:7947–7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wagner V. E., Bushnell D., Passador L., Brooks A. I., Iglewski B. H. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wick M. J., Hamood A. N., Iglewski B. H. 1990. Analysis of the structure-function relationship of Pseudomonas aeruginosa exotoxin A. Mol. Microbiol. 4:527–535 [DOI] [PubMed] [Google Scholar]
- 54. Worlitzsch D., et al. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109:317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xiao G., et al. 2006. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol. Microbiol. 62:1689–1699 [DOI] [PubMed] [Google Scholar]
- 56. Xiao G., He J., Rahme L. G. 2006. Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology 152:1679–1686 [DOI] [PubMed] [Google Scholar]
- 57. Yoon M. Y., Lee K. M., Jeong S. H., Kim J., Yoon S. S. 2010. Heterogeneous virulence potential and high antibiotic resistance of Pseudomonas aeruginosa strains isolated from Korean pneumonia patients. J. Microbiol. 48:518–525 [DOI] [PubMed] [Google Scholar]
- 58. Yoon M. Y., Lee K. M., Park Y., Yoon S. S. 2011. Contribution of cell elongation to the biofilm formation of Pseudomonas aeruginosa during anaerobic respiration. PLoS One. 6:e16105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yoon S. S., et al. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3:593–603 [DOI] [PubMed] [Google Scholar]
- 60. Yoon S. S., et al. 2007. Two-pronged survival strategy for the major cystic fibrosis pathogen, Pseudomonas aeruginosa, lacking the capacity to degrade nitric oxide during anaerobic respiration. EMBO J. 26:3662–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yoon S. S., Mekalanos J. J. 2006. 2,3-Butanediol synthesis and the emergence of the Vibrio cholerae El Tor biotype. Infect. Immun. 74:6547–6556 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.