Abstract
Onset of the adaptive immune response in mice infected with Mycobacterium tuberculosis is accompanied by slowing of bacterial replication and establishment of a chronic infection. Stabilization of bacterial numbers during the chronic phase of infection is dependent on the activity of the gamma interferon (IFN-γ)-inducible nitric oxide synthase (NOS2). Previously, we described a differential signature-tagged mutagenesis screen designed to identify M. tuberculosis “counterimmune” mechanisms and reported the isolation of three mutants in the H37Rv strain background containing transposon insertions in the rv0072, rv0405, and rv2958c genes. These mutants were impaired for replication and virulence in NOS2−/− mice but were growth-proficient and virulent in IFN-γ−/− mice, suggesting that the disrupted genes were required for bacterial resistance to an IFN-γ-dependent immune mechanism other than NOS2. Here, we report that the attenuation of these strains is attributable to an underlying transposon-independent deficiency in biosynthesis of phthiocerol dimycocerosate (PDIM), a cell wall lipid that is required for full virulence in mice. We performed whole-genome resequencing of a PDIM-deficient clone and identified a spontaneous point mutation in the putative polyketide synthase PpsD that results in a G44C amino acid substitution. We demonstrate by complementation with the wild-type ppsD gene and reversion of the ppsD gene to the wild-type sequence that the ppsD(G44C) point mutation is responsible for PDIM deficiency, virulence attenuation in NOS2−/− and wild-type C57BL/6 mice, and a growth advantage in vitro in liquid culture. We conclude that PDIM biosynthesis is required for M. tuberculosis resistance to an IFN-γ-mediated immune response that is independent of NOS2.
INTRODUCTION
Pathogenic mycobacteria possess a unique array of complex cell wall-associated lipids. The most abundant of these lipids, the phthiocerol dimycocerosates (PDIMs) (Fig. 1), are among the best characterized (23). PDIMs contain long-chain diols esterified by methyl-branched fatty acid chains. As early as 1974, it was recognized that a spontaneously arising PDIM-deficient variant of the laboratory strain H37Rv was attenuated in a guinea pig model of infection (11). Shortly thereafter, it was shown that the in vivo survival of an avirulent Mycobacterium tuberculosis strain was enhanced by coating the bacteria with cholesterol oleate and purified PDIM (16). A genetic link between PDIM production and virulence was not established until a quarter century later, when a large chromosomal locus was identified as playing an essential role in the biosynthesis and export of PDIM (3, 4, 6). Transposon insertions within the fadD26, fadD28, mmpL7, and drrC genes, and in the putative transcriptional promoter region upstream of the fadD26 gene, were identified in strains deficient in surface-localized PDIM. The fadD26 and fadD28 mutants apparently fail to synthesize PDIM, whereas the mmpL7 and drrC mutants produce PDIM but accumulate it intracellularly, thus implicating these genes in transmembrane transport of PDIM to the cell surface. Strains deficient in production or surface localization of PDIM are markedly attenuated for growth in the lungs of intravenously (3, 6) or intranasally (28) infected mice, and in gamma interferon (IFN-γ)-activated but not in nonactivated macrophages (28).
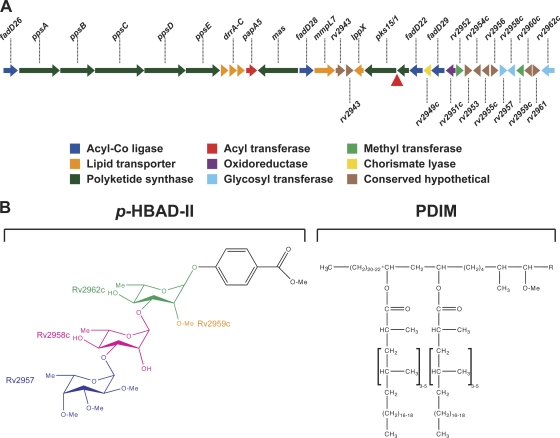
Fig. 1.
PDIM and p-HBAD biosynthesis in M. tuberculosis. (A) Genomic locus responsible for PDIM and p-HBAD-II biosynthesis. In M. tuberculosis H37Rv and other members of the Euro-American lineage, a frameshift mutation (red triangle) disrupts the pks15/1 open reading frame. Other M. tuberculosis lineages have an intact pks15/1 locus that encodes a functional polyketide synthase. (B) Structures of p-HBAD-II and PDIM. The polyketide synthase Pks15/1 adds malonyl coenzyme A (malonyl-CoA) units to p-hydroxybenzoic acid to generate p-hydroxyphenylalkanoic acid derivatives, which are precursors of PGL biosynthesis (5). M. tuberculosis 37Rv lacks Pks15/1 activity and produces tri-glycosylated p-hydroxybenzoic acid (p-HBAD-II) and PDIM instead. The Rv2962c, Rv2958c, and Rv2957 glycosyl transferases add rhamnose → rhamnose → fucose to p-hydroxyphenylalkanoic acid (25), and the Rv2959c methyltransferase O-methylates the C2 ring position of the proximal rhamnosyl residue (24).
Mutant strains that lack surface-localized PDIM display enhanced membrane permeability (3), but the precise role of PDIM in virulence of M. tuberculosis is unclear. The attenuated growth of PDIM-deficient M. tuberculosis in IFN-γ-activated macrophages is reversed by treatment of the infected macrophages with Nω-nitro-l-arginine methyl ester (l-NAME), a small-molecule inhibitor of the mammalian inducible nitric oxide synthase (NOS2), suggesting that PDIM might play a role in countering the impact of this important host antimicrobial mechanism (28). However, PDIM-deficient bacteria do not show increased sensitivity to reactive nitrogen species (RNS) in vitro (3), suggesting that M. tuberculosis might be sensitized to RNS during intracellular growth or that the impact of l-NAME on intracellular bacteria might be indirect, possibly via modulation of expression of host (9) or pathogen (29, 35) genes that are RNS responsive. A detailed comparative analysis of PDIM-proficient and PDIM-deficient M. tuberculosis strains in macrophages revealed that PDIM participates in receptor-dependent phagocytosis and inhibition of phagosome acidification (2). PDIM insertion in model membranes caused alterations in membrane fluidity, suggesting that physical changes to the host cell membrane caused by interaction with PDIM may influence the uptake and ultimate cellular destination of M. tuberculosis (2).
Whereas all M. tuberculosis clinical isolates apparently produce PDIM, only a subset of isolates produce the closely related phenolic glycolipids (PGLs). The PGLs comprise a PDIM lipid core that is terminated by a glycosylated aromatic nucleus (26). A 7-bp deletion in the pks15/1 polyketide synthase locus, resulting in a translational frameshift, is responsible for the lack of PGL production by the H37Rv laboratory strain and all other strains of the Euro-American M. tuberculosis lineage (5, 10). This frameshift abolishes production of the Pks15/1 polyketide synthase that is responsible for biosynthesis of the PGL precursor p-hydroxyphenylalkanoate from p-hydroxybenzoic acid (5). In the absence of Pks15/1 function, glycosylated p-hydroxybenzoic acid methyl esters (p-HBADs) accumulate and are released into the culture medium (5, 25). Three glycosyltransferases, encoded by the rv2962c, rv2958c, and rv2957 genes, are thought to add, successively, two rhamnosyl residues and one fucosyl residue to the p-hydroxybenzoic acid moiety (Fig. 1B) (25). M. tuberculosis H37Rv produces the triglycosylated p-HBAD (p-HBAD-II). Disruption of rv2962 abolishes p-HBAD production entirely; disruption of the rv2958c or rv2957 gene results in the production of monoglycosylated p-HBAD (p-HBAD-I) (25).
In M. tuberculosis strains that produce PGL, a role for PGL in immune modulation and virulence has been reported (26). Whether the p-HBAD moieties secreted by PGL-negative M. tuberculosis strains might also play a role in immune modulation and virulence is not known. Previously, we described a genetic screen that was designed to identify M. tuberculosis genes involved in countering IFN-γ-dependent host immune mechanisms other than NOS2 (12). Disruption of these “counterimmune” (cim) genes severely attenuated growth and virulence in NOS2−/− mice but had little or no impact on bacterial growth and virulence in IFN-γ−/− mice. One of the cim mutants identified in this study contained a transposon (Tn) insertion in the rv2958c gene, suggesting that secreted p-HBAD-II might, like full-length PGL, modulate the interaction of the bacterium and the host immune response. Here, we describe further studies to elucidate the role of the rv2962c, rv2958c, and rv2957 glycosyltransferases in M. tuberculosis virulence and counterimmunity. In contrast to our previous report, we find that these genes are dispensable for bacterial growth and survival in wild-type (C57BL/6) and NOS2−/− mice. We demonstrate that the phenotypes we had previously ascribed to disruption of the rv2958c gene are attributable, instead, to the spontaneous loss of PDIM production in the rv2958c::Tn mutant. Whole-genome resequencing of a spontaneous PDIM-deficient variant that arose during in vitro cultivation of M. tuberculosis H37Rv identified a single-nucleotide polymorphism (SNP) in the ppsD gene, which encodes a modular polyketide synthase putatively involved in PDIM biosynthesis (34). We demonstrate by complementation and reversion that the spontaneous point mutation in the ppsD gene is responsible for both the defect in PDIM production and attenuation of virulence in mice. We additionally show that the spontaneous PDIM deficient variant has an in vitro growth advantage that allows it to replace the PDIM-proficient parental strain during subculture. We suggest that spontaneous loss of PDIM production is likely to be a common phenomenon that calls for a reexamination of published genetic studies of M. tuberculosis in which complementation analysis was not done or was unsuccessful.
MATERIALS AND METHODS
Bacteriology.
M. tuberculosis strains from the McKinney lab were H37Rv (parental strain) and Tn5370 (Tn) mutagenized derivatives rv0072::Tn, rv0405::Tn, and rv2958c::Tn, described previously (12). M. tuberculosis strains from the Guilhot lab were H37Rv (parental strain), Δrv2957, Δrv2958c, Δrv2959c, and Δrv2962c, described previously (24, 25). Bacteria were cultured at 37°C in Middlebrook 7H9 broth (Difco) containing 10% albumin-dextrose-saline, 0.5% glycerol, and 0.05% Tween 80 or on Middlebook 7H10 agar (Difco) containing 10% oleic acid-albumin-dextrose-catalase (BD Biosciences) and 0.5% glycerol. Cycloheximide was added at 10 μg ml−1 to prevent fungal contamination, as needed. Kanamycin (15 μg ml−1), hygromycin (50 μg ml−1), and sucrose (2%) were added to the growth media, as needed. Frozen stocks were prepared by growing liquid cultures in 7H9 broth to mid-log phase (optical density at 600 nm [OD600] = 0.5) and freezing in aliquots at −80°C after the addition of glycerol (15% [vol/vol]).
Mouse infections.
Male and female C57BL/6, NOS2−/−, and IFN-γ−/− mice, 5 to 8 weeks of age, were purchased from Jackson Laboratories and housed in The Rockefeller University's Laboratory Animal Research Center or the EPFL Center of Phenogenomics under specific-pathogen-free conditions. Bacteria were grown to mid-log phase (OD600 = 0.5) in 7H9 broth, collected by centrifugation (2,500 × g, 15 min), resuspended in phosphate-buffered saline containing 0.05% Tween 80 (PBST), and centrifuged at a low speed (150 × g, 5 min) to remove clumps. The declumped supernatant was adjusted to an OD600 of 0.1 (∼108 CFU ml−1) and further diluted 2-fold before being loaded into the nebulizer. Mice were infected by the aerosol route with ∼100 CFU using a custom-built aerosol exposure chamber (Department of Mechanical Engineering, University of Wisconsin, Madison, WI) and an exposure time of 15 min, as described previously (37). Infected mice were euthanized by CO2 overdose. Bacterial CFU were enumerated by plating serially diluted lung homogenates on 7H10 agar and counting colonies after 3 to 4 weeks at 37°C. For survival experiments, infected mice were monitored twice daily, and animals that showed signs of illness (ruffled fur, immobility, hunched posture, labored breathing) were euthanized by CO2 overdose and scored as “died.” The animal protocols for these studies were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of The Rockefeller University and by the chief veterinarian of the Swiss Federal Institute of Technology Lausanne (EPFL), the Service de la Consommation et des Affaires Vétérinaires of the Canton of Vaud, and the Swiss Office Vétérinaire Fédéral.
p-HBAD-II glycosylation and PDIM production.
p-HBAD-II biosynthesis by wild-type and mutant strains of M. tuberculosis was analyzed by thin-layer chromatography (TLC) of extracted and purified glycolipids, as described previously (25). PDIM biosynthesis was analyzed by growing bacteria to mid-log phase and labeling 10 ml of culture for 24 to 48 h with 10 μCi of [1-14C]-propionate (specific activity of 54 Ci mol−1 [Amersham] or 55.9 Ci mol−1 [Campro Scientific]). Apolar lipids were extracted essentially as described previously (32). Labeled cells were collected by centrifugation (2,500 × g, 10 min), resuspended in 5 ml of 10:1 (vol/vol) methanol/0.3% NaCl, and 5 ml of petroleum ether was added. Samples were vortexed vigorously for 4 min and phase-separated by centrifugation (750 × g, 10 min). The upper layer was removed, and the extraction was repeated with an additional 5 ml of petroleum ether. Remaining bacteria in the combined petroleum ether fraction were killed by the addition of an equal volume of chloroform. Extracts were reduced to ∼10 ml by overnight evaporation and spotted (25 to 30 μl) on a silica gel 60 F254 TLC plate (5 by 10 cm; Merck). TLC plates were developed in petroleum ether/diethyl ether (9:1 [vol/vol]), air-dried, and visualized using a Typhoon PhosphorImager (Amersham Biosciences).
Whole-genome resequencing.
High-quality genomic DNA was prepared from PDIM-positive and PDIM-negative isolates of H37Rv by the cetyltrimethylammonium bromide (CTAB)-lysozyme method (17). Genomic DNA fragment sequencing libraries were prepared using a genomic DNA sample prep kit (Illumina) according to the manufacturer's instructions, with 5 μg of purified genomic DNA. Each genomic DNA fragment library was sequenced on one lane of an Illumina genome analyzer IIx sequencing chip using a single-read cluster generation kit v2 (Illumina) and a 36-cycle sequencing kit v2 (Illumina). Image analysis and base calling were done using the Illumina Pipeline software package, v1.32.
A total of 6.2 and 5.3 million reads 36 bases in length were obtained for the PDIM-positive and PDIM-negative H37Rv clones, respectively. These sequence reads were mapped to the reference M. tuberculosis H37Rv sequence using Maq v0.7.1 via ungapped alignments allowing up to two mismatches per read. This method mapped 95% and 89% of the PDIM-positive and PDIM-negative sequence reads, respectively, to the H37Rv genome. Maq was also used for SNP calling and filtering out low-quality SNPs using the SNP filter designed for single-end reads (18). SNPs identified in the ppsA and ppsD genes were further confirmed by PCR and sequencing using the primer pairs ppsAF/ppsAR and ppsDF/ppsDR (Table 1).
Table 1.
Oligonucleotide primersa
| Name | Sequence (5′ to 3′) |
|---|---|
| ppsAF | GCGAGGACCTGGTCGGTATC |
| ppsAR | GGCCTTGTTGAGGTTGGTC |
| ppsBF | GAACTCTGCCACGAGCTGG |
| ppsBR | GCACCGATGACGAGCTG |
| ppsDF | GTCTTAATTAAGGAAACCCTGGGACTCGAC |
| ppsDR | TAGGCGCGCCGCCAAGTGAATTGCCACCAG |
| ppsDF2 | CTGGAATTCTAAGAAGGAGATATACATATGACAAGTCTGGCGGAGC |
| ppsDR2 | CTGGTTAACTCGGGGATGCTCACAGGTC |
| ppsDR4 | AAGCTTGAGGGCGGATGTGAT |
| MVinsF | AGCGAGGACAACTTGAGC |
| ppsDF4 | CGATTCGCGCTCAGCTAGAC |
| pJGR | AAATGCCGATATCCTATTGGC |
| pJGF | GTGGACCTCGACGACCTC |
Restriction enzyme sites are underlined. The strong ribosome-binding site in primer ppsDF2 is indicated in italics.
Plasmid construction.
Oligonucleotide primers used for plasmid construction are listed in Table 1. For complementation of the ppsD(G44C) mutation, the full-length ppsD gene was amplified from PDIM-positive H37Rv genomic DNA using primers ppsDF2 and ppsDR2 that contain EcoRI and HpaI restriction sites, respectively. The ppsDF2 primer additionally harbors a strong ribosome-binding site immediately upstream of the translational start site of the ppsD gene. The resulting PCR products were cloned in pCR2.1-TOPO (Invitrogen) and sequenced. Clones were identified that contained wild-type ppsD sequence between a unique NcoI restriction site and the 3′ end of the ppsD gene and between a unique HindIII site and the unique NcoI site. The first 979 bp of the ppsD gene 5′ to the unique HindIII site were amplified by PCR from PDIM-positive H37Rv genomic DNA using primers ppsDF2 and ppsDR4, cloned in pCR2.1-TOPO, and sequenced. The ppsD 5′-HindIII fragment was digested out of the pCR2.1 cloning vector with EcoRI and HindIII and cloned downstream of the strong constitutive hsp60 promoter in pMV361, a vector that integrates at the attB site on the M. tuberculosis chromosome and contains a Kanr marker for selection (33). The resulting pMV361-ppsD 5′-HindIII construct was digested with HindIII and HpaI, and the HindIII-NcoI and NcoI-3′ fragments of the ppsD gene were ligated together in the plasmid. The resulting full-length ppsD complementation construct, pAT223, was confirmed by sequencing.
For allelic exchange of the ppsD(G44C) point mutation, the wild-type ppsD sequence approximately 450 bp up- and downstream of the point mutation was amplified from PDIM-positive H37Rv genomic DNA using primers ppsDF and ppsDR, which contain PacI and AscI restriction sites, respectively. The resulting PCR product was digested with PacI and AscI and ligated to PacI/AscI-digested pJG1100, a suicide vector that contains Kanr and Hygr markers for selection of recombinants and the sacB counterselectable marker that confers sucrose sensitivity, to generate pAT221. The construct was confirmed by sequencing.
Strain construction.
Oligonucleotide primers used in PCR confirmation of strains are listed in Table 1. For complementation of the ppsD(G44C) point mutation with pAT223, the plasmid was electroporated into the PDIM-negative H37Rv ppsD(G44C) strain, and Kanr colonies were selected. The presence of pAT223 was confirmed by PCR using primers MVinsF and ppsDR4. Reversion of the ppsD(G44C) point mutation to the wild-type sequence was accomplished by a two-step homologous recombination method. The pAT221 allelic exchange vector was electroporated into the PDIM-negative H37Rv ppsD(G44C) strain, and Kanr Hygr colonies were selected. Isolates containing the pAT221 vector integrated at the ppsD gene were identified by PCR with the primer pairs ppsDF4/pJGR and pJGF/ppsDR4. These isolates were grown in 7H9 medium without antibiotic selection to mid-log phase and plated on 7H10 agar containing 2% sucrose to select isolates that had undergone a second recombination. Excision of the plasmid in sucrose-resistant isolates was confirmed by PCR using primers ppsDF4/ppsDR4, and the resulting PCR product was sequenced to determine whether the wild-type ppsD sequence or the ppsD(G44C) mutant sequence was present.
Statistics.
Student's unpaired t test (one-tailed) was used to assess statistical significance of pairwise comparisons between groups of mice infected with PDIM-positive or PDIM-negative bacteria. The Mantel-Cox log-rank test was used for comparison of Kaplan-Meier plots of mouse survival. P values were calculated using GraphPad Prism 4.0 software (GraphPad Software, Inc.). P values of <0.05 were considered significant.
RESULTS
A strain of M. tuberculosis with a Tn insertion in the rv2958c gene is attenuated in wild-type, NOS2−/−, and IFN-γ−/− mice.
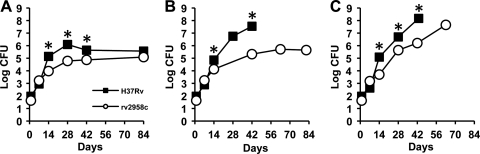
Previously, we reported the results of a pilot signature-tagged mutagenesis screen to identify M. tuberculosis genes involved in countering the impact of IFN-γ-dependent immune mechanisms other than NOS2 (12). This was accomplished by parallel screening of Tn-induced mutants in intravenously infected gene knockout mice to identify mutants that were attenuated for growth and virulence in NOS2−/− mice but unimpaired for growth and virulence in IFN-γ−/− mice. One of the mutants identified in this screen contained a Tn insertion in the rv2958c gene, encoding a putative rhamnosyl transferase involved in biosynthesis of p-HBAD-II (25). To confirm the phenotypes observed in the screen, we infected mice by the aerosol route with the H37Rv parental strain or with the rv2958c::Tn mutant derived from it. Consistent with the results of the screen, growth of the rv2958c::Tn mutant was attenuated in C57BL/6 (wild-type) mice (Fig. 2A) and in NOS2−/− mice (Fig. 2B). In contrast to our previous results, however, we found that growth of the rv2958c::Tn mutant was also somewhat attenuated in IFN-γ−/− mice (Fig. 2C). We do not know the reason for this discrepancy. A possible explanation is that screening and retesting of mutants in our previous report were done in mice infected by the intravenous route (12), whereas the experiments reported here were done in mice infected by the respiratory route, which is the natural route of infection.
Fig. 2.
Growth kinetics of M. tuberculosis strains H37Rv (wild type) and rv2958c::Tn in wild-type and immunodeficient mice. C57BL/6 (A), NOS2−/− (B), and IFN-γ−/− (C) mice were aerosol infected with M. tuberculosis strains H37Rv (squares) or rv2958c::Tn (circles). These strains were described previously (12). Groups of mice were sacrificed at the indicated time points, and bacterial CFU were enumerated by plating lung homogenates on 7H10 agar and scoring colonies after 3 to 4 weeks of incubation at 37°C. Symbols represent means (n = 4 or 5 mice per group per time point); error bars indicate standard errors. Asterisks indicate a statistically significant difference (P < 0.05) between the groups. Representative results of two independent experiments are shown.
Role of p-HBAD-II in M. tuberculosis growth in C57BL/6 and NOS2−/− mice.
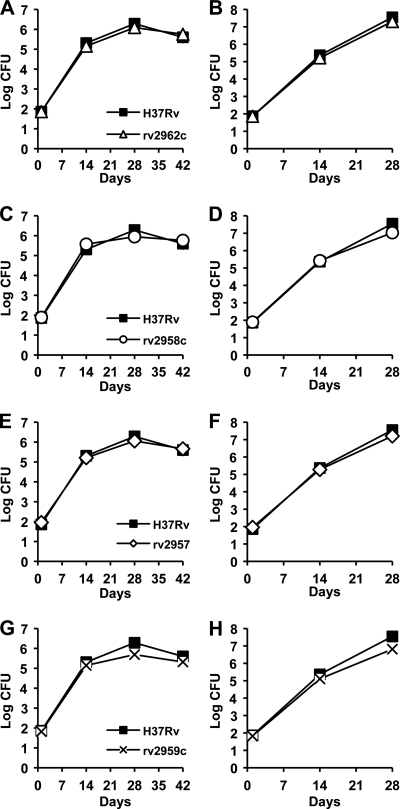
Concurrent with our report describing the results of our pilot screen to identify M. tuberculosis “counterimmune” (cim) mutants (12), another group reported the construction of deletion mutations in the M. tuberculosis rv2962c, rv2958c, and rv2957 genes and the roles of the putative glycosyl transferases encoded by these genes in the biosynthesis of p-HBAD-II (Fig. 1) (25). In a separate report, the same group also described the construction of a deletion mutation in the rv2959c gene, encoding a methyltransferase responsible for O-methylation of the first rhamnosyl residue linked to the p-hydroxybenzoic acid moiety of p-HBAD-II (Fig. 1) (24). In order to elucidate the contributions of these genes to M. tuberculosis virulence and counterimmunity, our groups jointly investigated the growth kinetics and virulence of the Δrv2962c (Fig. 3A and B), Δrv2958c (Fig. 3C and D), Δrv2957 (Fig. 3E and F), and Δrv2959c (Fig. 3G and H) mutants in aerosol-infected mice. We found that none of these mutants was attenuated for growth and persistence in aerosol-infected C57BL/6 (Fig. 3A, C, E, and G) or NOS2−/− (Fig. 3B, D, F, and H) mice. These results were in sharp contrast to the phenotype of the rv2958c::Tn mutant isolated in our screen, which was markedly attenuated in both C57BL/6 and NOS2−/− mice (Fig. 2) (12).
Fig. 3.
p-HBAD-II biosynthesis is not required for M. tuberculosis growth and survival in wild-type or immunodeficient mice. C57BL/6 (A, C, E, and G) and NOS2−/− (B, D, F, and H) mice were aerosol infected with M. tuberculosis strains H37Rv (A to H, squares), Δrv2962c (A and B, triangles), Δrv2958c (C and D, circles), Δrv2958c (E and F, diamonds), or Δrv2959c (G and H, crosses). These strains were described previously (24, 25). Groups of mice were sacrificed at the indicated time points, and bacterial CFU were enumerated by plating lung homogenates on 7H10 agar and scoring colonies after 3 to 4 weeks of incubation at 37°C. Symbols represent means (n = 4 or 5 mice per group per time point); error bars indicate standard errors. This experiment was performed once.
Loss of PDIM production in the M. tuberculosis rv2958c::Tn strain.
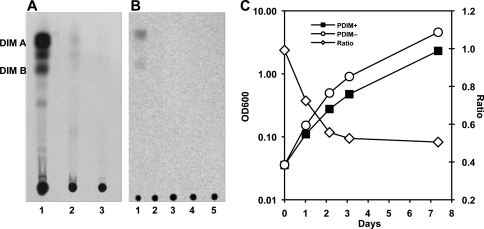
Analysis of bacterial cell wall lipid fractions by thin-layer chromatography revealed that the rv2958c::Tn mutant (12) was deficient in PDIM production (Fig. 4A, lane 3), compared to the H37Rv parental strain (Fig. 4A, lane 2). Loss of Rv2958c function per se was not responsible for loss of PDIM, because we found that the Δrv2958c mutant (25) produced wild-type levels of PDIM (not shown). Two additional mutants identified in our counterimmune screen (12), containing Tn insertions in the rv0072 (Fig. 4B, lane 4) and rv0405 (Fig. 4B, lane 5) genes, were also found to be PDIM deficient. We speculated that loss of PDIM production might have occurred during the isolation of the rv2958c::Tn, rv0072::Tn, and rv0405::Tn mutants, which involves clonal selection and expansion of bacteria arising from a single transposition event. We therefore derived eight independent cell lines from our parental H37Rv frozen stock, which had been used to generate the Tn-induced mutants that were screened in mice (12), by plating for single colonies on nonselective media and analyzing the clonal cell lines derived from randomly selected individual colonies for PDIM production. Of the eight clonal cell lines that we analyzed, four were PDIM deficient (not shown), suggesting that our parental H37Rv stock (Fig. 4A, lane 2; Fig. 4B, lane 1) comprised a mixture of PDIM-positive and PDIM-negative variants.
Fig. 4.
PDIM deficiency confers an in vitro growth advantage in M. tuberculosis H37Rv. (A and B) Thin-layer chromatographic analysis of PDIM biosynthesis. Bacteria were labeled with [14C]propionate, which preferentially labels PDIM (6), and cell wall lipids were extracted and separated by thin-layer chromatography. (A) M. tuberculosis strains. H37Rv, Guilhot lab (25) (lane 1); H37Rv, McKinney lab (12) (lane 2); rv2958c::Tn (12) (lane 3). (B) M. tuberculosis strains, McKinney lab (12). H37Rv (lane 1), H37Rv after subculture (lane 2), rv2958c::Tn (lane 3), rv0072::Tn (lane 4), rv0405::Tn (lane 5). (C) Independently derived subclones of PDIM-positive H37Rv (squares) and PDIM-negative H37Rv (circles) were grown in 7H9 broth with aeration at 37°C. Growth of the cultures was monitored by withdrawing aliquots and measuring the OD600 at the indicated time points (plotted on the primary y axis). The (PDIM-positive OD600)/(PDIM-negative OD600) ratios at each time point are plotted on the secondary y axis (diamonds). Results are representative of three independent experiments.
Consistent with this interpretation, we found that PDIM production was dramatically reduced in the McKinney lab's parental H37Rv strain (Fig. 4A, lane 2) compared to that in the Guilhot lab's H37Rv strain (Fig. 4A, lane 1). We also found that PDIM production by the McKinney lab's parental H37Rv strain was undetectable after only four rounds of in vitro subculture from frozen stocks (Fig. 4B, lane 2). These observations suggest that spontaneously arising PDIM deficiency might confer a growth advantage in vitro, thus promoting replacement of the parental PDIM-positive strain with PDIM-negative variants during in vitro passage. To test this idea, we compared the in vitro growth kinetics of independently derived PDIM-positive and PDIM-negative H37Rv clones (see above) and found that the PDIM-negative bacteria had a significant and reproducible growth advantage over the PDIM-positive bacteria (Fig. 4C). We found that the in vitro growth advantage of PDIM-negative variants was particularly pronounced during the early outgrowth of cultures inoculated from frozen stocks (data not shown), which might reflect differential recovery of PDIM-positive and PDIM-negative strains from the stress caused by freezing/thawing.
Growth kinetics of PDIM-negative H37Rv, rv2958c::Tn, rv0072::Tn, and rv0405::Tn strains in C57BL/6, NOS2−/−, and IFN-γ−/− mice.
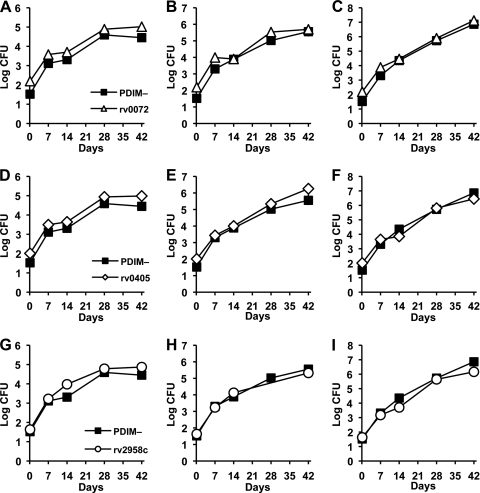
Previously, we reported that M. tuberculosis rv0072::Tn, rv0405::Tn, and rv2958c::Tn mutants were severely attenuated for growth and virulence in intravenously infected NOS2−/− mice but were only slightly attenuated in IFN-γ−/− mice (12). Our subsequent discovery that these strains were PDIM deficient (Fig. 4A), presumably due to selection of preexisting PDIM-negative variants in our parental H37Rv stock during Tn mutagenesis, suggested that the attenuation of these mutants in mice might be due, at least in part, to their PDIM deficiency. To test this idea, we compared the growth kinetics of these mutants with a PDIM-negative H37Rv clone in aerosol-infected mice (Fig. 5). We found that the rv0072::Tn (Fig. 5A to C), rv0405::Tn (Fig. 5D to F), and rv2958c::Tn (Fig. 5G to I) mutants grew with kinetics similar to those of the PDIM-negative H37Rv clone in C57BL/6 (Fig. 5A, D, and G), NOS2−/− (Fig. 5B, E, and H), and IFN-γ−/− (Fig. 5C, F, and I) mice. These observations suggest that the in vivo phenotypes we reported previously for these mutants (12) were probably due to the spontaneous loss of PDIM production in these strains, rather than disruption of the rv2958c, rv0072, or rv0405 genes per se.
Fig. 5.
Growth kinetics of PDIM-negative H37Rv and M. tuberculosis rv0072::Tn, rv0405::Tn, and rv2958c::Tn mutants in wild-type and immunodeficient mice. C57BL/6 (A, D, and G), NOS2−/− (B, E, and H), and IFN-γ−/− (C, F, and I) mice were aerosol infected with M. tuberculosis H37Rv PDIM-negative variant (A to I, squares), rv0072::Tn (A to C, triangles), rv0405::Tn (D to F, diamonds), or rv2958c::Tn (G to I, circles). The PDIM-negative variant of H37Rv is described herein; the Tn mutant strains were described previously (12). Groups of mice were sacrificed at the indicated time points, and bacterial CFU were enumerated by plating lung homogenates on 7H10 agar and scoring colonies after 3 to 4 weeks of incubation at 37°C. Symbols represent means (n = 4 or 5 mice per group per time point); error bars indicate standard errors. This experiment was performed once.
Growth kinetics of PDIM-positive and PDIM-negative clones of H37Rv in C57BL/6, NOS2−/−, and IFN-γ−/− mice.
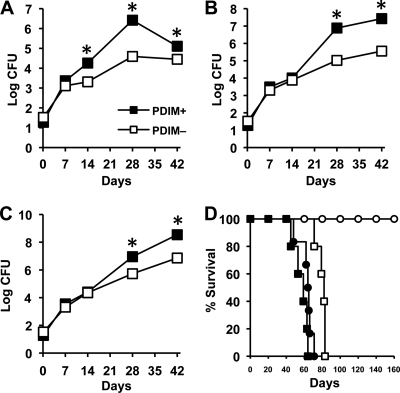
To further test the idea that PDIM deficiency might contribute to the in vivo attenuation of our rv2958c::Tn, rv0072::Tn, and rv0405::Tn mutants, we compared the growth kinetics of PDIM-positive and PDIM-negative clones of H37Rv in aerosol-infected mice. We found that, compared to the PDIM-positive H37Rv clone, the PDIM-negative H37Rv clone was markedly attenuated for growth in C57BL/6 (Fig. 6A), NOS2−/− (Fig. 6B), and IFN-γ−/− (Fig. 6C) mice. Virulence of the PDIM-negative H37Rv clone, measured in terms of survival time of infected mice, was also attenuated in NOS2−/− mice (P = 0.0005) and, to a lesser extent, in IFN-γ−/− mice (P = 0.002) (Fig. 6D). Median survival time (MST) of NOS2−/− mice was >200 days after infection with PDIM-negative H37Rv, compared to 64 days postinfection for NOS2−/− mice infected with PDIM-positive H37Rv (Fig. 6D). The MST of IFN-γ−/− mice was also longer for mice infected with PDIM-negative H37Rv (MST, 82 days) compared to mice infected with PDIM-positive H37Rv (MST, 59 days) (Fig. 6D). These data strongly suggest that the in vivo attenuation of the “counterimmune” mutants described in our previous report (12) can be attributed to the fortuitous loss of PDIM production in these strains.
Fig. 6.
Growth kinetics of PDIM-positive and PDIM-negative subclones of M. tuberculosis H37Rv in wild-type and immunodeficient mice. (A to C) C57BL/6 (A), NOS2−/− (B), and IFN-γ−/− (C) mice were aerosol-infected with PDIM-positive (filled squares) or PDIM-negative (open squares) subclones of M. tuberculosis H37Rv. Groups of mice were sacrificed at the indicated time points, and bacterial CFU were enumerated by plating lung homogenates on 7H10 agar and scoring colonies after 3 to 4 weeks of incubation at 37°C. Symbols represent means (n = 4 or 5 mice per group per time point); error bars indicate standard errors. Asterisks indicate statistically significant differences (P < 0.05) between the groups. (D) Survival of NOS2−/− (circles) and IFN-γ−/− (squares) mice (n = 5 or 6 per group) after aerosol infection with PDIM-positive (filled symbols) or PDIM-negative (open symbols) subclones of M. tuberculosis H37Rv. This experiment was performed once.
Whole-genome resequencing of PDIM-negative H37Rv identifies a point mutation, ppsD(G44C), responsible for PDIM deficiency.
Since our results demonstrated a correlation between spontaneous loss of the ability to produce PDIM and attenuation in the mouse model of infection, we sought to identify the mutation responsible for PDIM deficiency in a PDIM-negative clone. Analysis of the PDIM biosynthesis locus (Fig. 1) by Southern blotting did not identify any major insertions or genetic rearrangements (data not shown), suggesting that a point mutation might be the cause of PDIM deficiency. To identify putative point mutations that could contribute to PDIM deficiency, we performed whole-genome resequencing of PDIM-positive and PDIM-negative H37Rv clones using an Illumina genome analyzer platform. In comparison to the H37Rv reference sequence, our PDIM-positive H37Rv clone had 161 putative SNPs and 15 putative small sequence insertions or deletions (indels). The PDIM-negative H37Rv clone had 151 putative SNPs and 15 putative indels. These sequence alterations included 72 polymorphisms (57 SNPs and 15 indels) that were recently identified by whole-genome resequencing of six H37Rv isolates and that are likely to be errors within the H37Rv reference sequence (14). Analysis of the remaining sequence polymorphisms, excluding those in the highly repetitive GC-rich PE_PGRS coding regions, revealed 22 putative SNPs unique to the PDIM-negative H37Rv clone.
Among the SNPs present only in the PDIM-negative H37Rv isolate were two mutations in genes required for PDIM biosynthesis: a putative A to C at position 2544 in the ppsA gene and a G to T at position 130 in the ppsD gene. Only the G to T in the ppsD gene was predicted to be nonsynonymous, generating a glycine-to-cysteine transition at position 44 in the PpsD protein. PpsD is a modular polyketide synthase, and the G44C point mutation identified in our PDIM-negative clone is located within the putative β-ketoacyl acyl carrier protein synthase enzymatic domain (34). We reasoned that the G44C mutation might interfere with the activity of PpsD, since an additional cysteine residue could cause formation of aberrant disulfide bonds, thereby preventing proper protein folding or interfering with the function of the active site cysteine residue. We confirmed that the ppsD(G44C) point mutation was unique to the PDIM-negative clone by PCR and sequencing. We were not able to confirm the point mutation in the ppsA gene by sequencing of a PCR product, suggesting that there are likely to be some false positives among the SNPs identified by whole-genome resequencing.
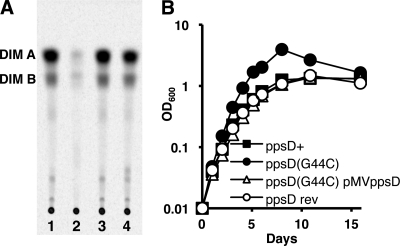
We attempted to restore PDIM production to the PDIM-negative H37Rv ppsD(G44C) mutant using two approaches. We complemented the ppsD(G44C) mutation in trans by providing a wild-type copy of the ppsD gene under the control of a strong constitutive promoter on the vector pMV361, which integrates in the M. tuberculosis chromosome at the unique attB site. We also reverted the ppsD(G44C) mutation in the PDIM-negative clone to the wild-type ppsD sequence by a two-step homologous recombination method. Both strains were tested for the ability to produce PDIM in vitro by thin-layer chromatography of extractable cell wall lipids. Production of PDIM was restored by either complementation or reversion (ppsD rev) of the ppsD(G44C) point mutation (Fig. 7A), confirming that this spontaneous mutation is responsible for the deficiency in PDIM production of our PDIM-negative H37Rv clone.
Fig. 7.
The ppsD(G44C) point mutation is responsible for PDIM deficiency and the in vitro growth advantage of the H37Rv PDIM-negative subclone. (A) Thin-layer chromatographic analysis of PDIM biosynthesis. Bacteria were labeled with [14C]propionate, which is preferentially incorporated into PDIM (6), and cell wall lipids were extracted and separated by thin-layer chromatography. M. tuberculosis H37Rv strains: ppsD+ (lane 1), ppsD(G44C) (lane 2), ppsD rev (lane 3), ppsD(G44C) pMV361-ppsD (lane 4). Results are representative of two independent experiments. (B) Bacteria were grown in 7H9 broth with aeration at 37°C. Growth was monitored by withdrawing aliquots and measuring the OD600 at the indicated time points. M. tuberculosis H37Rv strains: ppsD+ (filled squares), ppsD(G44C) (filled circles), ppsD rev (open circles), ppsD(G44C) pMV361-ppsD (open triangles). Results are representative of three independent experiments.
The ppsD(G44C) point mutation enhances growth of H37Rv in vitro.
Since the ppsD(G44C) spontaneous point mutation was responsible for PDIM deficiency, we sought to establish whether the in vitro and in vivo growth characteristics of our PDIM-negative H37Rv clone could also be attributed to this point mutation. We compared the in vitro growth kinetics of the PDIM-positive ppsD+ clone, the PDIM-negative ppsD(G44C) mutant, the ppsD(G44C) pMV-ppsD complemented mutant, and the strain in which the ppsD gene was reverted to the wild-type sequence (ppsD rev) in standard liquid culture conditions. The PDIM-negative ppsD(G44C) mutant consistently grew at a higher rate and to a higher cell density in liquid culture than the PDIM-positive ppsD+ clone (Fig. 7B). Either complementation or reversion of the ppsD(G44C) point mutation restored growth to a rate similar to the PDIM-positive ppsD+ clone (Fig. 7B). These data demonstrate that the ppsD(G44C) point mutation is responsible for the enhanced in vitro growth rate of the PDIM-negative H37Rv clone.
The ppsD(G44C) point mutation attenuates replication and virulence of H37Rv in mice.
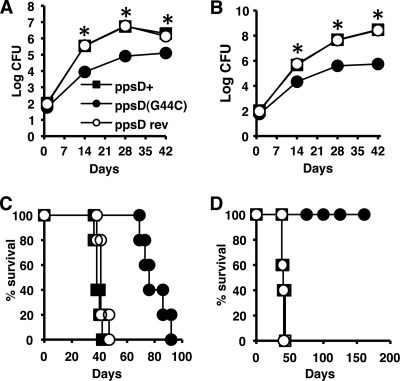
Next, we tested whether the spontaneous ppsD(G44C) mutation was also responsible for attenuation of the PDIM-negative H37Rv clone in the mouse model of infection. For these experiments, we focused on the strain in which ppsD was reverted to the wild-type sequence (ppsD rev), because the complemented strain exhibited a moderately reduced in vitro growth rate (Fig. 7B). The PDIM-positive ppsD+, PDIM-negative ppsD(G44C), and ppsD rev strains were introduced into the lungs of C57BL/6, NOS2−/−, and IFN-γ−/− mice by the aerosol route, and replication of the bacteria in the lungs and survival of the mice were monitored. Consistent with previous experiments, the PDIM-negative ppsD(G44C) mutant was attenuated for growth in the lungs of C57BL/6 (Fig. 8A) and NOS2−/− (Fig. 8B) mice. Virulence of the PDIM-negative ppsD(G44C) mutant, measured by survival time of infected mice, was likewise modestly attenuated in IFN-γ−/− mice (Fig. 8C) (P = 0.0018) and dramatically attenuated in NOS2−/− mice (Fig. 8D) (P = 0.0025). Reversion of the ppsD(G44C) spontaneous point mutation to the wild-type ppsD sequence reversed each of these phenotypes. The ppsD rev strain replicated in the lungs of C57BL/6 (Fig. 8A) and NOS2−/− (Fig. 8B) mice with kinetics identical to those of the PDIM-positive ppsD+ clone. In addition, reversion to the wild-type ppsD sequence restored virulence in IFN-γ−/− (Fig. 8C) and NOS2−/− (Fig. 8D) mice. Survival times of IFN-γ−/− and NOS2−/− mice infected with PDIM-positive ppsD+ H37Rv or the ppsD rev strain were not significantly different (for IFN-γ−/− mice, P = 0.159; for NOS2−/− mice, P = 0.398). These data confirm that a spontaneous point mutation, ppsD(G44C), was responsible for both loss of PDIM production and attenuation of the PDIM-negative H37Rv clone in mice.
Fig. 8.
Reversion of the ppsD(G44C) point mutation restores wild-type levels of growth and virulence in mice. (A to D) Mice were aerosol infected with ppsD+ (filled squares), ppsD(G44C) (filled circles), or ppsD rev (open circles) strains of M. tuberculosis H37Rv. (A and B) Bacterial growth in the lungs of C57BL/6 (A) and NOS2−/− (B) mice. Groups of mice were sacrificed at the indicated time points, and bacterial CFU were enumerated by plating lung homogenates on 7H10 and scoring colonies after 3 to 4 weeks of incubation at 37°C. Symbols represent means (n = 4 mice per group); error bars indicate standard errors. Asterisks indicate statistically significant differences (P < 0.05) in comparisons of ppsD(G44C) versus ppsD+ and ppsD(G44C) versus ppsD rev strains. This experiment was performed once. (C and D) Survival of IFN-γ−/− (C) and iNOS−/− (D) mice (n = 5 mice per group). This experiment was performed once.
DISCUSSION
Long-term persistence in the face of innate and adaptive immune responses is a hallmark of tuberculosis, yet little is known about the “counterimmune” mechanisms that promote the persistence of M. tuberculosis in immunocompetent hosts. We previously identified the glycosyltransferase encoded by the rv2958c gene as a putative factor required to counter the impact of IFN-γ-dependent immune mechanisms other than NOS2 (12). Here, we tested mutants harboring targeted deletions of genes that are required for biosynthesis of secreted p-HBAD, including the rv2958c gene, for their ability to replicate in the lungs of mice. Although none of the Δrv2957, Δrv2958c, Δrv2959c, or Δrv2962c mutants were attenuated for growth, it remains possible that secreted p-HBADs play a role in virulence modulation that is not reflected in the bacterial growth kinetics, as shown previously for PGL (26).
We attribute the discrepancy between the phenotypes of the rv2958c::Tn and Δrv2958c mutants in the mouse infection model to the spontaneous loss of PDIM production in our H37Rv parental strain and the rv2958c::Tn mutant derived from it. Our observations demonstrate a role for PDIM in countering the impact of an IFN-γ-dependent, NOS2-independent immune mechanism, in addition to the previously postulated role for PDIM in protecting the bacteria from the cidal activity of RNS (28). Consistent with this idea, we found that the survival time of IFN-γ−/− and NOS2−/− mice was not significantly different (P = 0.07) after infection with the PDIM-positive H37Rv clone. In contrast, following infection with the PDIM-negative H37Rv clone, NOS2−/− mice survived significantly longer than IFN-γ−/− mice (P < 0.001). Our results are also consistent with the results of a recent screen for mutants that specifically alter growth of M. tuberculosis in NOS2−/− mice, which identified two independent mutations in the drrA gene encoding a putative PDIM transporter (22). The immune mechanism responsible for this IFN-γ-dependent, NOS2-independent attenuation and the countermechanism by which PDIM confers protection are currently unknown.
We demonstrated that a point mutation, ppsD(G44C), in a PDIM-negative clone derived from our H37Rv parental strain was responsible for both a defect in PDIM production and attenuation in mice. To our knowledge, this is the first direct evidence that PpsD is required for PDIM biosynthesis and virulence. We consistently observed a low level of res- idual PDIM production by the ppsD(G44C) mutant. The ppsD(G44C) mutant was a clonal isolate from a single colony, suggesting that weak PDIM production is a property of this strain. It is possible that reversion of the G44C point mutation or compensatory mutations elsewhere in ppsD that restore PDIM synthesis occur spontaneously at some low frequency. It is unlikely that such strains would become a significant fraction of the population, however, since the ppsD(G44C) mutant grows at a higher rate. We therefore favor the possibility that the PpsDG44C mutant protein retains some residual activity that enables weak PDIM production. The low level of PDIM produced by the ppsD(G44C) mutant is apparently not sufficient, however, to support normal replication in the lungs of mice.
A correlation was previously observed between PDIM deficiency and a growth advantage in liquid culture (7). Similarly, our PDIM-negative ppsD(G44C) mutant had an enhanced in vitro growth rate and grew to a higher cell density than PDIM-positive clones. The in vitro growth advantage of such spontaneous PDIM-negative variants could explain why they are able to supplant the parental strain during repeated cycles of growth in vitro, for example, during mutant strain construction. Indeed PDIM-negative variants were isolated with significantly higher frequency in a culture that was serially passaged than in a nonpassaged control (7). It has also been suggested that spontaneous PDIM deficient variants might be selected by genetic manipulations involving electroporation or bacteriophage infections (11, 26). Since our results indicate that these spontaneous PDIM-deficient variants are attenuated for virulence, care should be taken to minimize the number of passages during genetic manipulation of M. tuberculosis and to ensure that strains used in animal infection studies are PDIM proficient.
Spontaneously arising PDIM-deficient variants have been described previously (1, 7, 8, 11, 15, 19–21) and are probably more common than has been reported or realized. Although spontaneous PDIM deficiency has most frequently been associated with the H37Rv strain, it has also recently been described for the M. tuberculosis Erdman strain and the clinical isolate HN878 (7, 20). Many of the attenuated M. tuberculosis mutants described in the scientific literature have not been complemented, and in other cases, complementation restored virulence only partially. Some of these mutants might have acquired unrecognized secondary mutations causing PDIM deficiency, which could be a factor contributing to their attenuation. Moreover, a number of genes have been implicated in PDIM synthesis, based on the PDIM-negative phenotypes of the corresponding mutants, in the absence of confirmatory complementation (for example, see references 13, 27, 28, 30, 31, and 36). Some of these mutant strains might contain unrecognized secondary mutations that are the true cause of their PDIM deficiency. This phenomenon might also explain the observation that Mycobacterium leprae produces PDIM despite lacking functional copies of certain polyketide biosynthesis genes that were reported to be essential for PDIM production in M. tuberculosis (discussed in reference 36).
Construction and characterization of random or targeted gene-disrupted mutants is a powerful technique to assign biological functions to biochemical pathways in mycobacteria. However, our findings underscore the idea that any functional assignment must be tentative in the absence of complementation analysis, even in cases where the mutation is not polar on the expression of neighboring genes.
ACKNOWLEDGMENTS
We thank Peter Giannakis and Laetitia Martin for expert technical assistance with animal experiments and Keith Harshman and Jérome Thomas at the University of Lausanne core facility for performing the whole-genome resequencing reactions.
This work was supported by a Robert D. Watkins Graduate Fellowship from the American Society for Microbiology (M.A.K.), the William Randolph Hearst Endowed Scholarship Fund (K.B.H.), NIH MSTP grant GM07739 (M.A.K. and K.B.H.) for the Tri-Institutional MD/PhD Program of Weill-Cornell Medical School, Rockefeller University, and the Sloan-Kettering Institute, an Irvington Institute Postdoctoral Fellowship of the Cancer Research Institute (A.D.T.), SystemsX, The Swiss Initiative in Systems Biology (S.U.), and National Institutes of Health grant AI046392 (J.D.M.).
Footnotes
Published ahead of print on 16 May 2011.
REFERENCES
- 1. Andreu N., Gibert I. 2008. Cell population heterogeneity in Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb.) 88:553–559 [DOI] [PubMed] [Google Scholar]
- 2. Astarie-Dequeker C., et al. 2009. Phthiocerol dimycocerosates of M. tuberculosis participate in macrophage invasion by inducing changes in the organization of plasma membrane lipids. PLoS Pathog. 5:e1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Camacho L. R., et al. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis: evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276:19845–19854 [DOI] [PubMed] [Google Scholar]
- 4. Camacho L. R., Ensergueix D., Perez E., Gicquel B., Guilhot C. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257–267 [DOI] [PubMed] [Google Scholar]
- 5. Constant P., et al. 2002. Role of the pks15/1 gene in the biosynthesis of phenolglycolipids in the Mycobacterium tuberculosis complex: evidence that all strains synthesize glycosylated p-hydroxybenzoic acid methyl esters and that strains devoid of phenolglycolipids harbor a frameshift mutation in the pks15/1 gene. J. Biol. Chem. 277:38148–38158 [DOI] [PubMed] [Google Scholar]
- 6. Cox J. S., Chen B., McNeil M., Jacobs W. R., Jr. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79–83 [DOI] [PubMed] [Google Scholar]
- 7. Domenech P., Reed M. B. 2009. Rapid and spontaneous loss of phthiocerol dimycocerosate (PDIM) from Mycobacterium tuberculosis grown in vitro: implications for virulence studies. Microbiol. 155:3532–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Domenech P., et al. 2004. The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J. Biol. Chem. 279:21257–21265 [DOI] [PubMed] [Google Scholar]
- 9. Ehrt S., et al. 2001. Reprogramming of the macrophage transcriptome in response to interferon-γ and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J. Exp. Med. 194:1123–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gagneux S., et al. 2006. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 103:2869–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goren M. B., Brokl O., Schaefer W. B. 1974. Lipids of putative relevance to virulence in Mycobacterium tuberculosis: phthiocerol dimycocerosate and the attenuation indicator lipid. Infect. Immun. 9:150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hisert K. B., et al. 2004. Identification of Mycobacterium tuberculosis counterimmune (cim) mutants in immunodeficient mice by differential screening. Infect. Immun. 72:5315–5321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hotter G. S., et al. 2005. Transposon mutagenesis of Mb0100 at the ppe1-nrp locus in Mycobacterium bovis disrupts phthiocerol dimycocerosate (PDIM) and glycosylphenol-PDIM biosynthesis, producing an avirulent strain with vaccine properties at least equal to those of M. bovis BCG. J. Bacteriol. 187:2267–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ioerger T. R., et al. 2010. Variation among genome sequences of H37Rv strains of Mycobacterium tuberculosis from multiple laboratories. J. Bacteriol. 192:3645–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kana B. D., et al. 2008. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol. Microbiol. 67:672–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kondo E., Kanai K. 1976. A suggested role of a host-parasite lipid complex in mycobacterial infection. Jpn. J. Med. Sci. Biol. 29:199–201 [DOI] [PubMed] [Google Scholar]
- 17. Larsen M. H., Biermann K., Tandberg S., Hsu T., Jacobs W. R. J. 2007. Genetic manipulation of Mycobacterium tuberculosis. Curr. Protoc. Microbiol. Chapter 10:Unit 10A.2 [DOI] [PubMed] [Google Scholar]
- 18. Li H., Ruan J., Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Manjunatha U. H., et al. 2006. Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci., U. S. A. 103:431–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marrero J., Rhee K. Y., Schnappinger D., Pethe K., Ehrt S. 2010. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc. Natl. Acad. Sci., U. S. A. 107:9819–9824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsunaga I., et al. 2004. Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells. J. Exp. Med. 200:1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murry J. P., Pandey A. K., Sassetti C. M., Rubin E. J. 2009. Phthiocerol dimycocerosate transport is required for resisting interferon-γ-independent immunity. J. Infect. Dis. 200:774–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Onwueme K. C., Vos C. J., Zurita J., Ferreras J. A., Quadri L. E. 2005. The dimycocerosate ester polyketide virulence factors of mycobacteria. Prog. Lipid Res. 44:259–302 [DOI] [PubMed] [Google Scholar]
- 24. Perez E., et al. 2004. Molecular dissection of the role of two methyltransferases in the biosynthesis of phenolglycolipids and phthiocerol dimycoserosate in the Mycobacterium tuberculosis complex. J. Biol. Chem. 279:42584–42592 [DOI] [PubMed] [Google Scholar]
- 25. Pérez E., et al. 2004. Characterization of three glycosyltransferases involved in the biosynthesis of the phenolic glycolipid antigens from the Mycobacterium tuberculosis complex. J. Biol. Chem. 279:42574–42583 [DOI] [PubMed] [Google Scholar]
- 26. Reed M. B., et al. 2004. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431:84–87 [DOI] [PubMed] [Google Scholar]
- 27. Rousseau C., et al. 2003. Virulence attenuation of two Mas-like polyketide synthase mutants of Mycobacterium tuberculosis. Microbiology 149:1837–1847 [DOI] [PubMed] [Google Scholar]
- 28. Rousseau C., et al. 2004. Production of phthiocerol dimycocerosates protects Mycobacterium tuberculosis from the cidal activity of reactive nitrogen intermediates produced by macrophages and modulates the early immune response to infection. Cell. Microbiol. 6:277–287 [DOI] [PubMed] [Google Scholar]
- 29. Schnappinger D., et al. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sirakova T. D., Dubey V. S., Cynamon M. H., Kolattukudy P. E. 2003. Attenuation of Mycobacterium tuberculosis by disruption of a mas-like gene or a chalcone synthase-like gene, which causes deficiency in dimycocerosyl phthiocerol synthesis. J. Bacteriol. 185:2999–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sirakova T. D., Dubey V. S., Kim H.-J., Cynamon M. H., Kolattukudy P. E. 2003. The largest open reading frame (pks12) in the Mycobacterium tuberculosis genome is involved in pathogenesis and dimycocerosyl phthiocerol synthesis. Infect. Immun. 71:3794–3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Slayden R. A., Barry C. E. I. 2001. Analysis of the lipids of Mycobacterium tuberculosis, p. 229–245 In Parish T., Stoker N. G. (ed.), Mycobacterium tuberculosis protocols. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 33. Stover C. K., et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460 [DOI] [PubMed] [Google Scholar]
- 34. Trivedi O. A., et al. 2005. Dissecting the mechanism and assembly of a complex virulence mycobacterial lipid. Mol. Cell 17:631–643 [DOI] [PubMed] [Google Scholar]
- 35. Voskuil M. I., et al. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Waddell S. J., et al. 2005. Inactivation of polyketide synthase and related genes results in the loss of complex lipids in Mycobacterium tuberculosis H37Rv. Lett. Appl. Microbiol. 40:201–206 [DOI] [PubMed] [Google Scholar]
- 37. Wiegeshaus E. H., McMurray D. N., Grover A. A., Harding G. E., Smith D. W. 1970. Host-parasite relationships in experimental airborne tuberculosis. III. Relevance of microbial enumeration to acquired resistance in guinea pigs. Am. Rev. Respir. Dis. 102:422–429 [DOI] [PubMed] [Google Scholar]